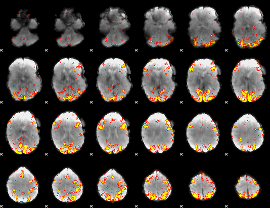
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
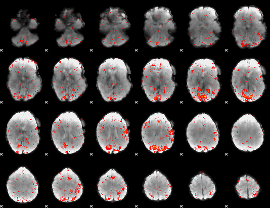
GLM result group-wise
 |
GLM binary result thresholded by 1
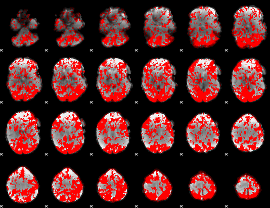 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
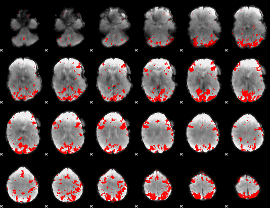 |
GLM Z-score result thresholded by 1
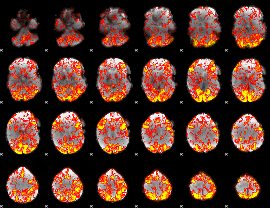 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
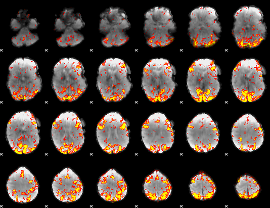 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:379; Jaccard:0.10913
 |
Component:379; Jaccard:0.13268
 |
Component:379; Jaccard:0.16237
 |
Component:186; Jaccard:0.19807
 |
Component:186; Jaccard:0.24366
 |
Component:186; Jaccard:0.29324
 |
Component:186; Jaccard:0.34253
 |
Correlation:0.073203
  |
Correlation:0.073203
  |
Correlation:0.073203
  |
Correlation:0.13369
  |
Correlation:0.13369
  |
Correlation:0.13369
  |
Correlation:0.13369
  |
Component:186; Jaccard:0.10419
 |
Component:186; Jaccard:0.12818
 |
Component:186; Jaccard:0.1593
 |
Component:379; Jaccard:0.19627
 |
Component:379; Jaccard:0.23102
 |
Component:379; Jaccard:0.26297
 |
Component:379; Jaccard:0.28746
 |
Correlation:0.13369
  |
Correlation:0.13369
  |
Correlation:0.13369
  |
Correlation:0.073203
  |
Correlation:0.073203
  |
Correlation:0.073203
  |
Correlation:0.073203
  |
Component:267; Jaccard:0.099358
 |
Component:267; Jaccard:0.11691
 |
Component:267; Jaccard:0.13689
 |
Component:267; Jaccard:0.15997
 |
Component:267; Jaccard:0.18171
 |
Component:267; Jaccard:0.20045
 |
Component:267; Jaccard:0.21463
 |
Correlation:0.029019
  |
Correlation:0.029019
  |
Correlation:0.029019
  |
Correlation:0.029019
  |
Correlation:0.029019
  |
Correlation:0.029019
  |
Correlation:0.029019
  |
Component:185; Jaccard:0.09434
 |
Component:317; Jaccard:0.10956
 |
Component:317; Jaccard:0.12755
 |
Component:290; Jaccard:0.1492
 |
Component:290; Jaccard:0.17186
 |
Component:290; Jaccard:0.19474
 |
Component:290; Jaccard:0.21279
 |
Correlation:-0.019072
  |
Correlation:0.23973
  |
Correlation:0.23973
  |
Correlation:0.15867
  |
Correlation:0.15867
  |
Correlation:0.15867
  |
Correlation:0.15867
  |
Component:317; Jaccard:0.093579
 |
Component:290; Jaccard:0.10601
 |
Component:290; Jaccard:0.12655
 |
Component:317; Jaccard:0.14778
 |
Component:317; Jaccard:0.16713
 |
Component:317; Jaccard:0.18449
 |
Component:317; Jaccard:0.19565
 |
Correlation:0.23973
  |
Correlation:0.15867
  |
Correlation:0.15867
  |
Correlation:0.23973
  |
Correlation:0.23973
  |
Correlation:0.23973
  |
Correlation:0.23973
  |
Component:342; Jaccard:0.090907
 |
Component:185; Jaccard:0.10582
 |
Component:149; Jaccard:0.12171
 |
Component:149; Jaccard:0.14153
 |
Component:149; Jaccard:0.16228
 |
Component:149; Jaccard:0.18045
 |
Component:149; Jaccard:0.19369
 |
Correlation:0.23519
  |
Correlation:-0.019072
  |
Correlation:0.12719
  |
Correlation:0.12719
  |
Correlation:0.12719
  |
Correlation:0.12719
  |
Correlation:0.12719
  |
Component:311; Jaccard:0.09014
 |
Component:342; Jaccard:0.10461
 |
Component:342; Jaccard:0.11953
 |
Component:342; Jaccard:0.13369
 |
Component:342; Jaccard:0.14595
 |
Component:342; Jaccard:0.15382
 |
Component:127; Jaccard:0.16093
 |
Correlation:0.12114
  |
Correlation:0.23519
  |
Correlation:0.23519
  |
Correlation:0.23519
  |
Correlation:0.23519
  |
Correlation:0.23519
  |
Correlation:0.08962
  |
Component:290; Jaccard:0.088791
 |
Component:149; Jaccard:0.10262
 |
Component:185; Jaccard:0.11654
 |
Component:185; Jaccard:0.12682
 |
Component:185; Jaccard:0.13526
 |
Component:127; Jaccard:0.14863
 |
Component:342; Jaccard:0.15867
 |
Correlation:0.15867
  |
Correlation:0.12719
  |
Correlation:-0.019072
  |
Correlation:-0.019072
  |
Correlation:-0.019072
  |
Correlation:0.08962
  |
Correlation:0.23519
  |
Component:153; Jaccard:0.08835
 |
Component:311; Jaccard:0.099825
 |
Component:311; Jaccard:0.10925
 |
Component:311; Jaccard:0.11841
 |
Component:127; Jaccard:0.13343
 |
Component:207; Jaccard:0.14697
 |
Component:207; Jaccard:0.15834
 |
Correlation:-0.083391
  |
Correlation:0.12114
  |
Correlation:0.12114
  |
Correlation:0.12114
  |
Correlation:0.08962
  |
Correlation:0.030771
  |
Correlation:0.030771
  |
Component:149; Jaccard:0.086384
 |
Component:153; Jaccard:0.098537
 |
Component:153; Jaccard:0.10837
 |
Component:127; Jaccard:0.11763
 |
Component:207; Jaccard:0.13291
 |
Component:185; Jaccard:0.14062
 |
Component:352; Jaccard:0.14717
 |
Correlation:0.12719
  |
Correlation:-0.083391
  |
Correlation:-0.083391
  |
Correlation:0.08962
  |
Correlation:0.030771
  |
Correlation:-0.019072
  |
Correlation:0.25211
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:379; Jaccard:0.11259
 |
Component:379; Jaccard:0.1341
 |
Component:379; Jaccard:0.15946
 |
Component:186; Jaccard:0.19211
 |
Component:186; Jaccard:0.22977
 |
Component:186; Jaccard:0.26864
 |
Component:186; Jaccard:0.30701
 |
Correlation:0.34692
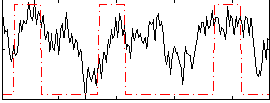 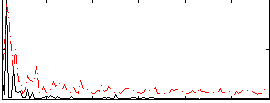 |
Correlation:0.34692
  |
Correlation:0.34692
  |
Correlation:0.2314
  |
Correlation:0.2314
  |
Correlation:0.2314
  |
Correlation:0.2314
  |
Component:186; Jaccard:0.10796
 |
Component:186; Jaccard:0.13045
 |
Component:186; Jaccard:0.15872
 |
Component:379; Jaccard:0.18816
 |
Component:379; Jaccard:0.21805
 |
Component:379; Jaccard:0.24635
 |
Component:379; Jaccard:0.26913
 |
Correlation:0.2314
  |
Correlation:0.2314
  |
Correlation:0.2314
  |
Correlation:0.34692
  |
Correlation:0.34692
  |
Correlation:0.34692
  |
Correlation:0.34692
  |
Component:267; Jaccard:0.10584
 |
Component:267; Jaccard:0.12379
 |
Component:267; Jaccard:0.14418
 |
Component:267; Jaccard:0.16567
 |
Component:290; Jaccard:0.19256
 |
Component:290; Jaccard:0.21917
 |
Component:290; Jaccard:0.24391
 |
Correlation:0.23001
  |
Correlation:0.23001
  |
Correlation:0.23001
  |
Correlation:0.23001
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Component:079; Jaccard:0.10082
 |
Component:290; Jaccard:0.11474
 |
Component:290; Jaccard:0.13801
 |
Component:290; Jaccard:0.16466
 |
Component:267; Jaccard:0.18616
 |
Component:267; Jaccard:0.2039
 |
Component:267; Jaccard:0.22048
 |
Correlation:0.1201
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.23001
  |
Correlation:0.23001
  |
Correlation:0.23001
  |
Component:311; Jaccard:0.10068
 |
Component:311; Jaccard:0.11361
 |
Component:311; Jaccard:0.12662
 |
Component:311; Jaccard:0.13894
 |
Component:317; Jaccard:0.15427
 |
Component:317; Jaccard:0.16699
 |
Component:207; Jaccard:0.17837
 |
Correlation:0.1952
 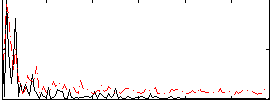 |
Correlation:0.1952
  |
Correlation:0.1952
  |
Correlation:0.1952
  |
Correlation:0.36105
  |
Correlation:0.36105
  |
Correlation:0.39362
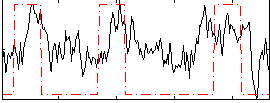  |
Component:185; Jaccard:0.097038
 |
Component:079; Jaccard:0.11199
 |
Component:079; Jaccard:0.12353
 |
Component:317; Jaccard:0.13839
 |
Component:127; Jaccard:0.14879
 |
Component:127; Jaccard:0.16526
 |
Component:127; Jaccard:0.17766
 |
Correlation:0.26718
  |
Correlation:0.1201
  |
Correlation:0.1201
  |
Correlation:0.36105
  |
Correlation:0.27308
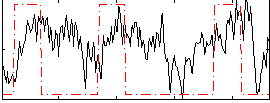 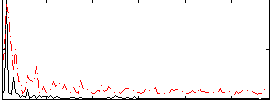 |
Correlation:0.27308
  |
Correlation:0.27308
  |
Component:290; Jaccard:0.095425
 |
Component:317; Jaccard:0.10762
 |
Component:317; Jaccard:0.12265
 |
Component:079; Jaccard:0.13341
 |
Component:311; Jaccard:0.14755
 |
Component:207; Jaccard:0.16368
 |
Component:317; Jaccard:0.17552
 |
Correlation:0.30943
  |
Correlation:0.36105
  |
Correlation:0.36105
  |
Correlation:0.1201
  |
Correlation:0.1952
  |
Correlation:0.39362
  |
Correlation:0.36105
  |
Component:317; Jaccard:0.094431
 |
Component:185; Jaccard:0.10708
 |
Component:185; Jaccard:0.11753
 |
Component:127; Jaccard:0.13082
 |
Component:207; Jaccard:0.14731
 |
Component:149; Jaccard:0.15883
 |
Component:149; Jaccard:0.16961
 |
Correlation:0.36105
  |
Correlation:0.26718
  |
Correlation:0.26718
  |
Correlation:0.27308
  |
Correlation:0.39362
  |
Correlation:0.24246
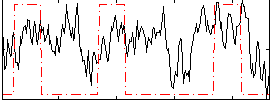  |
Correlation:0.24246
  |
Component:153; Jaccard:0.090731
 |
Component:342; Jaccard:0.10099
 |
Component:149; Jaccard:0.1148
 |
Component:149; Jaccard:0.13055
 |
Component:149; Jaccard:0.1451
 |
Component:311; Jaccard:0.15497
 |
Component:311; Jaccard:0.15735
 |
Correlation:0.18497
  |
Correlation:0.20819
 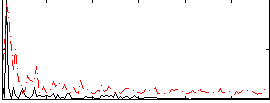 |
Correlation:0.24246
  |
Correlation:0.24246
  |
Correlation:0.24246
  |
Correlation:0.1952
  |
Correlation:0.1952
  |
Component:342; Jaccard:0.089811
 |
Component:149; Jaccard:0.099576
 |
Component:127; Jaccard:0.1131
 |
Component:207; Jaccard:0.12889
 |
Component:079; Jaccard:0.1417
 |
Component:079; Jaccard:0.14688
 |
Component:079; Jaccard:0.14899
 |
Correlation:0.20819
  |
Correlation:0.24246
  |
Correlation:0.27308
  |
Correlation:0.39362
  |
Correlation:0.1201
  |
Correlation:0.1201
  |
Correlation:0.1201
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
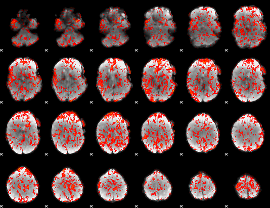 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
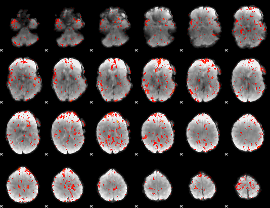 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
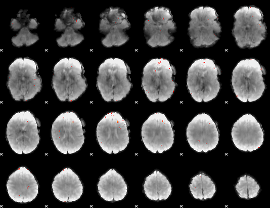 |
Component:298; Jaccard:0.097261
 |
Component:298; Jaccard:0.10499
 |
Component:298; Jaccard:0.099432
 |
Component:039; Jaccard:0.090205
 |
Component:039; Jaccard:0.065408
 |
Component:039; Jaccard:0.035789
 |
Component:039; Jaccard:0.012766
 |
Correlation:0.11651
  |
Correlation:0.11651
  |
Correlation:0.11651
  |
Correlation:0.067775
 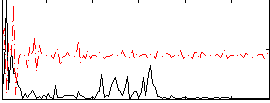 |
Correlation:0.067775
  |
Correlation:0.067775
  |
Correlation:0.067775
  |
Component:275; Jaccard:0.07718
 |
Component:039; Jaccard:0.090072
 |
Component:039; Jaccard:0.099003
 |
Component:298; Jaccard:0.077715
 |
Component:031; Jaccard:0.046277
 |
Component:144; Jaccard:0.024669
 |
Component:144; Jaccard:0.012764
 |
Correlation:0.23847
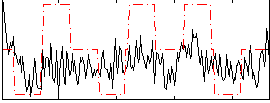  |
Correlation:0.067775
  |
Correlation:0.067775
  |
Correlation:0.11651
  |
Correlation:0.18577
  |
Correlation:0.1663
  |
Correlation:0.1663
  |
Component:039; Jaccard:0.074208
 |
Component:031; Jaccard:0.082496
 |
Component:031; Jaccard:0.083514
 |
Component:031; Jaccard:0.069395
 |
Component:298; Jaccard:0.044031
 |
Component:031; Jaccard:0.024017
 |
Component:275; Jaccard:0.010122
 |
Correlation:0.067775
  |
Correlation:0.18577
  |
Correlation:0.18577
  |
Correlation:0.18577
  |
Correlation:0.11651
  |
Correlation:0.18577
  |
Correlation:0.23847
  |
Component:375; Jaccard:0.072784
 |
Component:275; Jaccard:0.082264
 |
Component:275; Jaccard:0.081403
 |
Component:375; Jaccard:0.06481
 |
Component:275; Jaccard:0.04288
 |
Component:275; Jaccard:0.023672
 |
Component:031; Jaccard:0.009601
 |
Correlation:0.20397
  |
Correlation:0.23847
  |
Correlation:0.23847
  |
Correlation:0.20397
  |
Correlation:0.23847
  |
Correlation:0.23847
  |
Correlation:0.18577
  |
Component:031; Jaccard:0.072444
 |
Component:375; Jaccard:0.081377
 |
Component:375; Jaccard:0.079081
 |
Component:275; Jaccard:0.064203
 |
Component:314; Jaccard:0.042518
 |
Component:314; Jaccard:0.022115
 |
Component:314; Jaccard:0.009476
 |
Correlation:0.18577
  |
Correlation:0.20397
  |
Correlation:0.20397
  |
Correlation:0.23847
  |
Correlation:0.0093997
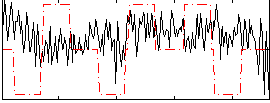  |
Correlation:0.0093997
  |
Correlation:0.0093997
  |
Component:274; Jaccard:0.066919
 |
Component:236; Jaccard:0.071264
 |
Component:314; Jaccard:0.071011
 |
Component:314; Jaccard:0.061428
 |
Component:144; Jaccard:0.041624
 |
Component:298; Jaccard:0.021785
 |
Component:298; Jaccard:0.009259
 |
Correlation:0.20981
 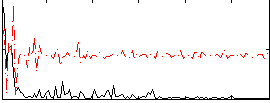 |
Correlation:0.1334
 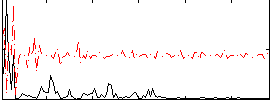 |
Correlation:0.0093997
  |
Correlation:0.0093997
  |
Correlation:0.1663
  |
Correlation:0.11651
  |
Correlation:0.11651
  |
Component:236; Jaccard:0.066871
 |
Component:274; Jaccard:0.070483
 |
Component:236; Jaccard:0.06939
 |
Component:144; Jaccard:0.059862
 |
Component:375; Jaccard:0.037606
 |
Component:265; Jaccard:0.020807
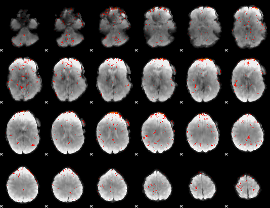 |
Component:274; Jaccard:0.008816
 |
Correlation:0.1334
  |
Correlation:0.20981
  |
Correlation:0.1334
  |
Correlation:0.1663
  |
Correlation:0.20397
  |
Correlation:-0.04239
 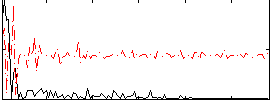 |
Correlation:0.20981
  |
Component:346; Jaccard:0.066854
 |
Component:291; Jaccard:0.068578
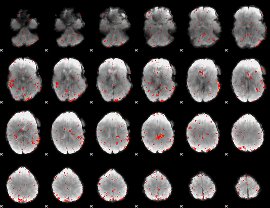 |
Component:274; Jaccard:0.069255
 |
Component:236; Jaccard:0.058297
 |
Component:274; Jaccard:0.036716
 |
Component:274; Jaccard:0.01966
 |
Component:259; Jaccard:0.008452
 |
Correlation:0.13974
  |
Correlation:0.27989
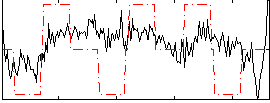  |
Correlation:0.20981
  |
Correlation:0.1334
  |
Correlation:0.20981
  |
Correlation:0.20981
  |
Correlation:-0.11089
  |
Component:291; Jaccard:0.065356
 |
Component:346; Jaccard:0.068156
 |
Component:144; Jaccard:0.068072
 |
Component:274; Jaccard:0.05746
 |
Component:236; Jaccard:0.035681
 |
Component:236; Jaccard:0.018299
 |
Component:265; Jaccard:0.008277
 |
Correlation:0.27989
  |
Correlation:0.13974
  |
Correlation:0.1663
  |
Correlation:0.20981
  |
Correlation:0.1334
  |
Correlation:0.1334
  |
Correlation:-0.04239
  |
Component:105; Jaccard:0.064153
 |
Component:140; Jaccard:0.067002
 |
Component:082; Jaccard:0.065336
 |
Component:265; Jaccard:0.052826
 |
Component:265; Jaccard:0.03567
 |
Component:375; Jaccard:0.017015
 |
Component:236; Jaccard:0.007738
 |
Correlation:0.15691
  |
Correlation:0.18153
  |
Correlation:0.1568
  |
Correlation:-0.04239
  |
Correlation:-0.04239
  |
Correlation:0.20397
  |
Correlation:0.1334
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:079; Jaccard:0.14256
 |
Component:079; Jaccard:0.16593
 |
Component:079; Jaccard:0.16927
 |
Component:079; Jaccard:0.15014
 |
Component:290; Jaccard:0.11682
 |
Component:290; Jaccard:0.083604
 |
Component:290; Jaccard:0.053959
 |
Correlation:0.063261
  |
Correlation:0.063261
  |
Correlation:0.063261
  |
Correlation:0.063261
  |
Correlation:0.089355
  |
Correlation:0.089355
  |
Correlation:0.089355
  |
Component:290; Jaccard:0.11267
 |
Component:290; Jaccard:0.13657
 |
Component:290; Jaccard:0.15
 |
Component:290; Jaccard:0.14493
 |
Component:079; Jaccard:0.11275
 |
Component:079; Jaccard:0.073289
 |
Component:079; Jaccard:0.045255
 |
Correlation:0.089355
  |
Correlation:0.089355
  |
Correlation:0.089355
  |
Correlation:0.089355
  |
Correlation:0.063261
  |
Correlation:0.063261
  |
Correlation:0.063261
  |
Component:384; Jaccard:0.10334
 |
Component:384; Jaccard:0.11527
 |
Component:048; Jaccard:0.11964
 |
Component:048; Jaccard:0.11153
 |
Component:048; Jaccard:0.08584
 |
Component:048; Jaccard:0.058622
 |
Component:127; Jaccard:0.037773
 |
Correlation:0.3648
  |
Correlation:0.3648
  |
Correlation:0.040287
  |
Correlation:0.040287
  |
Correlation:0.040287
  |
Correlation:0.040287
  |
Correlation:0.10874
  |
Component:311; Jaccard:0.098104
 |
Component:048; Jaccard:0.10863
 |
Component:127; Jaccard:0.11117
 |
Component:127; Jaccard:0.1019
 |
Component:127; Jaccard:0.080125
 |
Component:127; Jaccard:0.055919
 |
Component:048; Jaccard:0.037743
 |
Correlation:0.043896
  |
Correlation:0.040287
  |
Correlation:0.10874
  |
Correlation:0.10874
  |
Correlation:0.10874
  |
Correlation:0.10874
  |
Correlation:0.040287
  |
Component:267; Jaccard:0.095404
 |
Component:267; Jaccard:0.1058
 |
Component:384; Jaccard:0.11098
 |
Component:207; Jaccard:0.093034
 |
Component:207; Jaccard:0.065913
 |
Component:023; Jaccard:0.049103
 |
Component:023; Jaccard:0.031283
 |
Correlation:0.11913
  |
Correlation:0.11913
  |
Correlation:0.3648
  |
Correlation:0.21506
  |
Correlation:0.21506
  |
Correlation:0.1231
  |
Correlation:0.1231
  |
Component:207; Jaccard:0.092971
 |
Component:207; Jaccard:0.10497
 |
Component:207; Jaccard:0.10609
 |
Component:384; Jaccard:0.092176
 |
Component:023; Jaccard:0.065108
 |
Component:207; Jaccard:0.045125
 |
Component:207; Jaccard:0.02755
 |
Correlation:0.21506
  |
Correlation:0.21506
  |
Correlation:0.21506
  |
Correlation:0.3648
  |
Correlation:0.1231
  |
Correlation:0.21506
  |
Correlation:0.21506
  |
Component:048; Jaccard:0.089335
 |
Component:127; Jaccard:0.10392
 |
Component:267; Jaccard:0.10485
 |
Component:267; Jaccard:0.089886
 |
Component:267; Jaccard:0.063838
 |
Component:267; Jaccard:0.041167
 |
Component:186; Jaccard:0.025223
 |
Correlation:0.040287
  |
Correlation:0.10874
  |
Correlation:0.11913
  |
Correlation:0.11913
  |
Correlation:0.11913
  |
Correlation:0.11913
  |
Correlation:0.057914
  |
Component:127; Jaccard:0.088385
 |
Component:311; Jaccard:0.10232
 |
Component:311; Jaccard:0.094919
 |
Component:311; Jaccard:0.076279
 |
Component:384; Jaccard:0.061915
 |
Component:186; Jaccard:0.041032
 |
Component:267; Jaccard:0.024829
 |
Correlation:0.10874
  |
Correlation:0.043896
  |
Correlation:0.043896
  |
Correlation:0.043896
  |
Correlation:0.3648
  |
Correlation:0.057914
  |
Correlation:0.11913
  |
Component:366; Jaccard:0.083618
 |
Component:366; Jaccard:0.086714
 |
Component:186; Jaccard:0.081933
 |
Component:186; Jaccard:0.072464
 |
Component:186; Jaccard:0.056256
 |
Component:387; Jaccard:0.03629
 |
Component:311; Jaccard:0.021858
 |
Correlation:0.085104
  |
Correlation:0.085104
  |
Correlation:0.057914
  |
Correlation:0.057914
  |
Correlation:0.057914
  |
Correlation:-0.082149
  |
Correlation:0.043896
  |
Component:186; Jaccard:0.079705
 |
Component:387; Jaccard:0.083916
 |
Component:366; Jaccard:0.080141
 |
Component:023; Jaccard:0.072266
 |
Component:311; Jaccard:0.054636
 |
Component:311; Jaccard:0.034834
 |
Component:387; Jaccard:0.021825
 |
Correlation:0.057914
  |
Correlation:-0.082149
  |
Correlation:0.085104
  |
Correlation:0.1231
  |
Correlation:0.043896
  |
Correlation:0.043896
  |
Correlation:-0.082149
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
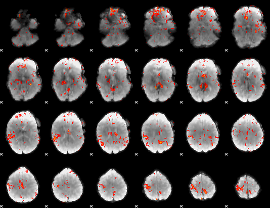 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:188; Jaccard:0.099779
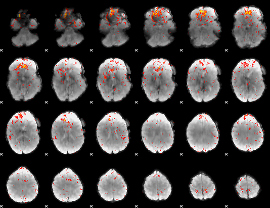 |
Component:188; Jaccard:0.11508
 |
Component:188; Jaccard:0.12692
 |
Component:188; Jaccard:0.12653
 |
Component:188; Jaccard:0.11444
 |
Component:188; Jaccard:0.091268
 |
Component:188; Jaccard:0.062087
 |
Correlation:0.31444
  |
Correlation:0.31444
  |
Correlation:0.31444
  |
Correlation:0.31444
  |
Correlation:0.31444
  |
Correlation:0.31444
  |
Correlation:0.31444
  |
Component:373; Jaccard:0.098726
 |
Component:373; Jaccard:0.10795
 |
Component:328; Jaccard:0.11723
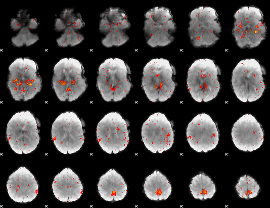 |
Component:328; Jaccard:0.1237
 |
Component:328; Jaccard:0.11258
 |
Component:328; Jaccard:0.08669
 |
Component:328; Jaccard:0.056158
 |
Correlation:0.3177
 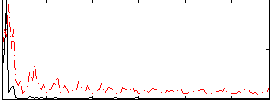 |
Correlation:0.3177
  |
Correlation:0.14639
  |
Correlation:0.14639
  |
Correlation:0.14639
  |
Correlation:0.14639
  |
Correlation:0.14639
  |
Component:002; Jaccard:0.088705
 |
Component:328; Jaccard:0.10365
 |
Component:373; Jaccard:0.11305
 |
Component:373; Jaccard:0.10737
 |
Component:373; Jaccard:0.093827
 |
Component:373; Jaccard:0.073145
 |
Component:373; Jaccard:0.05019
 |
Correlation:0.22621
  |
Correlation:0.14639
  |
Correlation:0.3177
  |
Correlation:0.3177
  |
Correlation:0.3177
  |
Correlation:0.3177
  |
Correlation:0.3177
  |
Component:328; Jaccard:0.088247
 |
Component:002; Jaccard:0.10074
 |
Component:002; Jaccard:0.10715
 |
Component:002; Jaccard:0.10566
 |
Component:002; Jaccard:0.093047
 |
Component:002; Jaccard:0.069711
 |
Component:294; Jaccard:0.042779
 |
Correlation:0.14639
  |
Correlation:0.22621
  |
Correlation:0.22621
  |
Correlation:0.22621
  |
Correlation:0.22621
  |
Correlation:0.22621
  |
Correlation:-0.025671
  |
Component:292; Jaccard:0.087912
 |
Component:260; Jaccard:0.095977
 |
Component:260; Jaccard:0.10147
 |
Component:260; Jaccard:0.099871
 |
Component:260; Jaccard:0.086781
 |
Component:294; Jaccard:0.068475
 |
Component:260; Jaccard:0.042734
 |
Correlation:0.17651
  |
Correlation:0.11389
  |
Correlation:0.11389
  |
Correlation:0.11389
  |
Correlation:0.11389
  |
Correlation:-0.025671
  |
Correlation:0.11389
  |
Component:260; Jaccard:0.087004
 |
Component:292; Jaccard:0.089904
 |
Component:294; Jaccard:0.093024
 |
Component:294; Jaccard:0.094425
 |
Component:294; Jaccard:0.084305
 |
Component:260; Jaccard:0.063893
 |
Component:002; Jaccard:0.041115
 |
Correlation:0.11389
  |
Correlation:0.17651
  |
Correlation:-0.025671
  |
Correlation:-0.025671
  |
Correlation:-0.025671
  |
Correlation:0.11389
  |
Correlation:0.22621
  |
Component:295; Jaccard:0.084702
 |
Component:295; Jaccard:0.086824
 |
Component:293; Jaccard:0.087034
 |
Component:273; Jaccard:0.081442
 |
Component:273; Jaccard:0.071522
 |
Component:273; Jaccard:0.053558
 |
Component:057; Jaccard:0.034034
 |
Correlation:-0.0088191
  |
Correlation:-0.0088191
  |
Correlation:0.0044612
  |
Correlation:0.067528
  |
Correlation:0.067528
  |
Correlation:0.067528
  |
Correlation:0.089744
  |
Component:298; Jaccard:0.08235
 |
Component:294; Jaccard:0.08616
 |
Component:292; Jaccard:0.08574
 |
Component:293; Jaccard:0.080253
 |
Component:222; Jaccard:0.065305
 |
Component:222; Jaccard:0.050663
 |
Component:222; Jaccard:0.032467
 |
Correlation:0.013348
  |
Correlation:-0.025671
  |
Correlation:0.17651
  |
Correlation:0.0044612
  |
Correlation:0.010564
  |
Correlation:0.010564
  |
Correlation:0.010564
  |
Component:293; Jaccard:0.081533
 |
Component:293; Jaccard:0.085864
 |
Component:295; Jaccard:0.083712
 |
Component:292; Jaccard:0.077296
 |
Component:293; Jaccard:0.064094
 |
Component:057; Jaccard:0.050416
 |
Component:273; Jaccard:0.031507
 |
Correlation:0.0044612
  |
Correlation:0.0044612
  |
Correlation:-0.0088191
  |
Correlation:0.17651
  |
Correlation:0.0044612
  |
Correlation:0.089744
  |
Correlation:0.067528
  |
Component:393; Jaccard:0.078786
 |
Component:069; Jaccard:0.081491
 |
Component:273; Jaccard:0.081931
 |
Component:069; Jaccard:0.074425
 |
Component:057; Jaccard:0.063312
 |
Component:292; Jaccard:0.04742
 |
Component:292; Jaccard:0.029445
 |
Correlation:0.08407
  |
Correlation:0.1033
  |
Correlation:0.067528
  |
Correlation:0.1033
  |
Correlation:0.089744
  |
Correlation:0.17651
  |
Correlation:0.17651
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
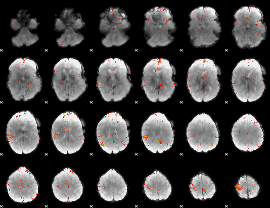 |
Component:298; Jaccard:0.11269
 |
Component:298; Jaccard:0.12464
 |
Component:298; Jaccard:0.13799
 |
Component:298; Jaccard:0.14264
 |
Component:298; Jaccard:0.13506
 |
Component:298; Jaccard:0.11753
 |
Component:375; Jaccard:0.10158
 |
Correlation:0.20992
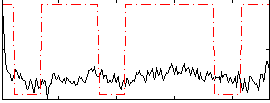  |
Correlation:0.20992
  |
Correlation:0.20992
  |
Correlation:0.20992
  |
Correlation:0.20992
  |
Correlation:0.20992
  |
Correlation:0.18349
 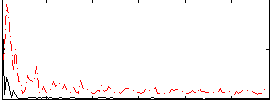 |
Component:373; Jaccard:0.091073
 |
Component:373; Jaccard:0.1006
 |
Component:373; Jaccard:0.10913
 |
Component:375; Jaccard:0.11723
 |
Component:375; Jaccard:0.12236
 |
Component:375; Jaccard:0.11702
 |
Component:298; Jaccard:0.092131
 |
Correlation:-0.19563
 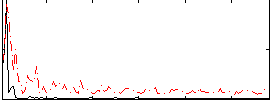 |
Correlation:-0.19563
  |
Correlation:-0.19563
  |
Correlation:0.18349
  |
Correlation:0.18349
  |
Correlation:0.18349
  |
Correlation:0.20992
  |
Component:236; Jaccard:0.087882
 |
Component:236; Jaccard:0.094742
 |
Component:375; Jaccard:0.10741
 |
Component:373; Jaccard:0.11216
 |
Component:260; Jaccard:0.10882
 |
Component:260; Jaccard:0.10028
 |
Component:260; Jaccard:0.087636
 |
Correlation:0.19945
  |
Correlation:0.19945
  |
Correlation:0.18349
  |
Correlation:-0.19563
  |
Correlation:0.067988
  |
Correlation:0.067988
  |
Correlation:0.067988
  |
Component:082; Jaccard:0.08252
 |
Component:082; Jaccard:0.094265
 |
Component:082; Jaccard:0.10347
 |
Component:082; Jaccard:0.10848
 |
Component:082; Jaccard:0.1087
 |
Component:082; Jaccard:0.099285
 |
Component:082; Jaccard:0.083421
 |
Correlation:0.24597
  |
Correlation:0.24597
  |
Correlation:0.24597
  |
Correlation:0.24597
  |
Correlation:0.24597
  |
Correlation:0.24597
  |
Correlation:0.24597
  |
Component:275; Jaccard:0.082509
 |
Component:375; Jaccard:0.09355
 |
Component:260; Jaccard:0.09948
 |
Component:260; Jaccard:0.10602
 |
Component:373; Jaccard:0.10711
 |
Component:373; Jaccard:0.093852
 |
Component:373; Jaccard:0.076638
 |
Correlation:0.2398
  |
Correlation:0.18349
  |
Correlation:0.067988
  |
Correlation:0.067988
  |
Correlation:-0.19563
  |
Correlation:-0.19563
  |
Correlation:-0.19563
  |
Component:292; Jaccard:0.081501
 |
Component:260; Jaccard:0.089701
 |
Component:236; Jaccard:0.099068
 |
Component:236; Jaccard:0.098963
 |
Component:294; Jaccard:0.098957
 |
Component:294; Jaccard:0.090119
 |
Component:294; Jaccard:0.076583
 |
Correlation:-0.070117
  |
Correlation:0.067988
  |
Correlation:0.19945
  |
Correlation:0.19945
  |
Correlation:0.11328
  |
Correlation:0.11328
  |
Correlation:0.11328
  |
Component:375; Jaccard:0.080653
 |
Component:275; Jaccard:0.087843
 |
Component:294; Jaccard:0.090454
 |
Component:294; Jaccard:0.097565
 |
Component:236; Jaccard:0.091658
 |
Component:236; Jaccard:0.076758
 |
Component:140; Jaccard:0.065975
 |
Correlation:0.18349
  |
Correlation:0.2398
  |
Correlation:0.11328
  |
Correlation:0.11328
  |
Correlation:0.19945
  |
Correlation:0.19945
  |
Correlation:0.22159
  |
Component:260; Jaccard:0.080413
 |
Component:292; Jaccard:0.08546
 |
Component:275; Jaccard:0.089856
 |
Component:275; Jaccard:0.089567
 |
Component:069; Jaccard:0.085644
 |
Component:069; Jaccard:0.07387
 |
Component:093; Jaccard:0.060989
 |
Correlation:0.067988
  |
Correlation:-0.070117
  |
Correlation:0.2398
  |
Correlation:0.2398
  |
Correlation:-0.0052363
  |
Correlation:-0.0052363
  |
Correlation:0.19724
  |
Component:295; Jaccard:0.079733
 |
Component:295; Jaccard:0.085117
 |
Component:295; Jaccard:0.088306
 |
Component:069; Jaccard:0.087667
 |
Component:093; Jaccard:0.084024
 |
Component:140; Jaccard:0.073777
 |
Component:069; Jaccard:0.060745
 |
Correlation:0.059384
  |
Correlation:0.059384
  |
Correlation:0.059384
  |
Correlation:-0.0052363
  |
Correlation:0.19724
  |
Correlation:0.22159
  |
Correlation:-0.0052363
  |
Component:069; Jaccard:0.079243
 |
Component:069; Jaccard:0.084556
 |
Component:292; Jaccard:0.087386
 |
Component:295; Jaccard:0.087362
 |
Component:275; Jaccard:0.081096
 |
Component:093; Jaccard:0.073615
 |
Component:236; Jaccard:0.05791
 |
Correlation:-0.0052363
  |
Correlation:-0.0052363
  |
Correlation:-0.070117
  |
Correlation:0.059384
  |
Correlation:0.2398
  |
Correlation:0.19724
  |
Correlation:0.19945
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































