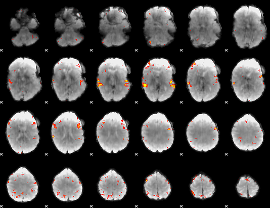
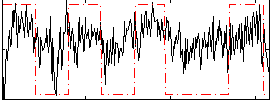
Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
GLM result group-wise
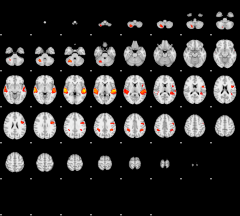 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
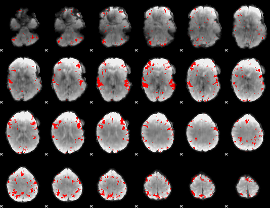 |
GLM binary result thresholded by 3.5
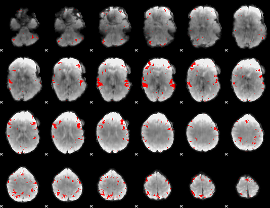 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
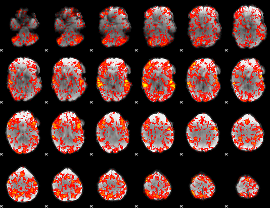 |
GLM Z-score result thresholded by 1.5
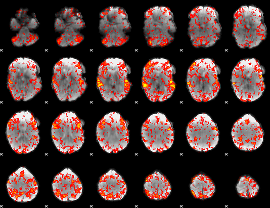 |
GLM Z-score result thresholded by 2
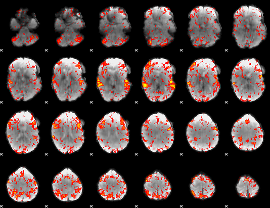 |
GLM Z-score result thresholded by 2.5
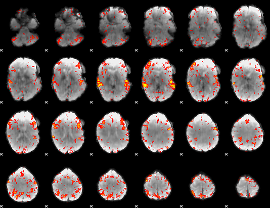 |
GLM Z-score result thresholded by 3
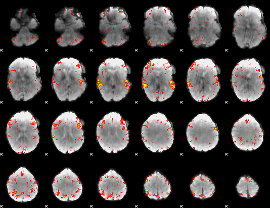 |
GLM Z-score result thresholded by 3.5
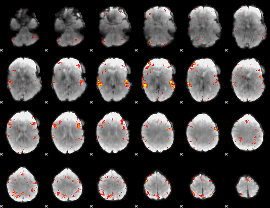 |
GLM Z-score result thresholded by 4
 |
Component:301; Jaccard:0.13236
 |
Component:301; Jaccard:0.15678
 |
Component:301; Jaccard:0.17552
 |
Component:301; Jaccard:0.17917
 |
Component:301; Jaccard:0.16003
 |
Component:301; Jaccard:0.12588
 |
Component:046; Jaccard:0.096225
 |
Correlation:0.33477
  |
Correlation:0.33477
  |
Correlation:0.33477
  |
Correlation:0.33477
  |
Correlation:0.33477
  |
Correlation:0.33477
  |
Correlation:-0.2814
  |
Component:197; Jaccard:0.12041
 |
Component:046; Jaccard:0.14071
 |
Component:141; Jaccard:0.16113
 |
Component:141; Jaccard:0.17153
 |
Component:141; Jaccard:0.15524
 |
Component:046; Jaccard:0.12428
 |
Component:301; Jaccard:0.088906
 |
Correlation:0.43424
 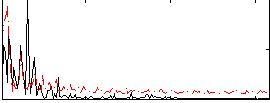 |
Correlation:-0.2814
  |
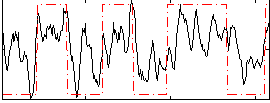
Correlation:0.60613
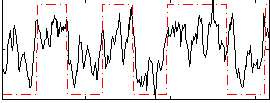  |
Correlation:0.60613
  |
Correlation:0.60613
  |
Correlation:-0.2814
  |
Correlation:0.33477
  |
Component:046; Jaccard:0.11952
 |
Component:299; Jaccard:0.14056
 |
Component:299; Jaccard:0.16093
 |
Component:299; Jaccard:0.16769
 |
Component:299; Jaccard:0.15273
 |
Component:299; Jaccard:0.12124
 |
Component:141; Jaccard:0.080382
 |
Correlation:-0.2814
  |
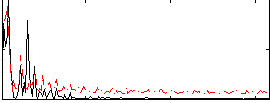
Correlation:0.56707
  |
Correlation:0.56707
  |
Correlation:0.56707
  |
Correlation:0.56707
  |
Correlation:0.56707
  |
Correlation:0.60613
  |
Component:299; Jaccard:0.11758
 |
Component:141; Jaccard:0.13777
 |
Component:046; Jaccard:0.15718
 |
Component:046; Jaccard:0.16067
 |
Component:046; Jaccard:0.14819
 |
Component:141; Jaccard:0.12101
 |
Component:299; Jaccard:0.079192
 |
Correlation:0.56707
  |
Correlation:0.60613
  |
Correlation:-0.2814
  |
Correlation:-0.2814
  |
Correlation:-0.2814
  |
Correlation:0.60613
  |
Correlation:0.56707
  |
Component:141; Jaccard:0.11335
 |
Component:197; Jaccard:0.1358
 |
Component:197; Jaccard:0.14131
 |
Component:205; Jaccard:0.14565
 |
Component:205; Jaccard:0.13187
 |
Component:205; Jaccard:0.10448
 |
Component:205; Jaccard:0.066277
 |
Correlation:0.60613
  |
Correlation:0.43424
  |
Correlation:0.43424
  |
Correlation:0.602
  |
Correlation:0.602
  |
Correlation:0.602
  |
Correlation:0.602
  |
Component:349; Jaccard:0.10546
 |
Component:205; Jaccard:0.12083
 |
Component:205; Jaccard:0.14018
 |
Component:144; Jaccard:0.13545
 |
Component:144; Jaccard:0.12107
 |
Component:144; Jaccard:0.092329
 |
Component:144; Jaccard:0.061967
 |
Correlation:0.47316
 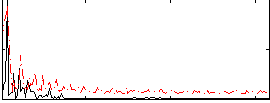 |
Correlation:0.602
  |
Correlation:0.602
  |
Correlation:0.51898
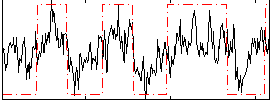 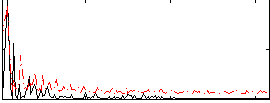 |
Correlation:0.51898
  |
Correlation:0.51898
  |
Correlation:0.51898
  |
Component:118; Jaccard:0.10417
 |
Component:349; Jaccard:0.1205
 |
Component:144; Jaccard:0.13196
 |
Component:197; Jaccard:0.13368
 |
Component:197; Jaccard:0.11077
 |
Component:118; Jaccard:0.084687
 |
Component:118; Jaccard:0.061896
 |
Correlation:0.12505
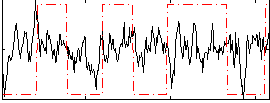  |
Correlation:0.47316
  |
Correlation:0.51898
  |
Correlation:0.43424
  |
Correlation:0.43424
  |
Correlation:0.12505
  |
Correlation:0.12505
  |
Component:344; Jaccard:0.10278
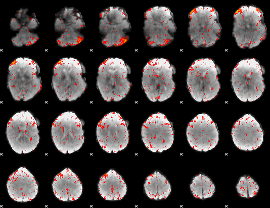 |
Component:144; Jaccard:0.11731
 |
Component:349; Jaccard:0.12962
 |
Component:349; Jaccard:0.12654
 |
Component:349; Jaccard:0.10557
 |
Component:197; Jaccard:0.079281
 |
Component:197; Jaccard:0.052203
 |
Correlation:0.3178
  |
Correlation:0.51898
  |
Correlation:0.47316
  |
Correlation:0.47316
  |
Correlation:0.47316
  |
Correlation:0.43424
  |
Correlation:0.43424
  |
Component:205; Jaccard:0.10057
 |
Component:118; Jaccard:0.11472
 |
Component:118; Jaccard:0.12108
 |
Component:118; Jaccard:0.11831
 |
Component:118; Jaccard:0.10423
 |
Component:349; Jaccard:0.076776
 |
Component:349; Jaccard:0.047352
 |
Correlation:0.602
  |
Correlation:0.12505
  |
Correlation:0.12505
  |
Correlation:0.12505
  |
Correlation:0.12505
  |
Correlation:0.47316
  |
Correlation:0.47316
  |
Component:144; Jaccard:0.098271
 |
Component:344; Jaccard:0.11035
 |
Component:344; Jaccard:0.11238
 |
Component:397; Jaccard:0.10613
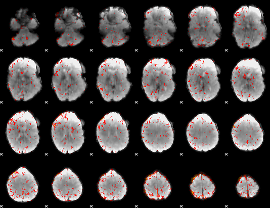 |
Component:397; Jaccard:0.089637
 |
Component:397; Jaccard:0.067199
 |
Component:397; Jaccard:0.047074
 |
Correlation:0.51898
  |
Correlation:0.3178
  |
Correlation:0.3178
  |
Correlation:0.31733
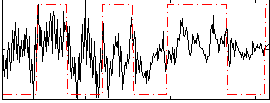 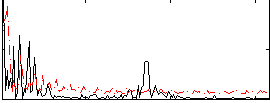 |
Correlation:0.31733
  |
Correlation:0.31733
  |
Correlation:0.31733
  |
Cope: 02:STORY BACK
|
GLM result group-wise
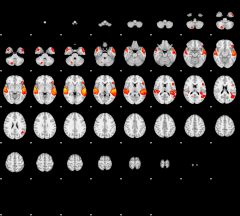 |
GLM binary result thresholded by 1
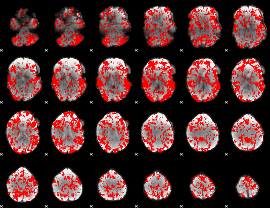 |
GLM binary result thresholded by 1.5
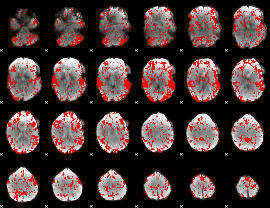 |
GLM binary result thresholded by 2
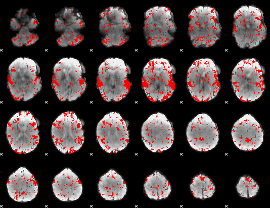 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
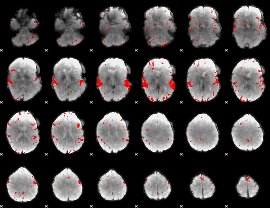 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
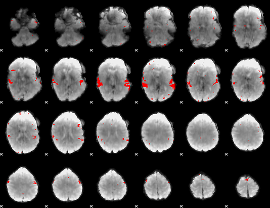 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
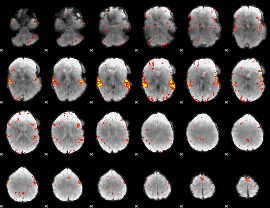 |
GLM Z-score result thresholded by 3.5
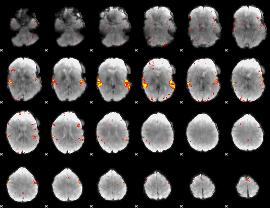 |
GLM Z-score result thresholded by 4
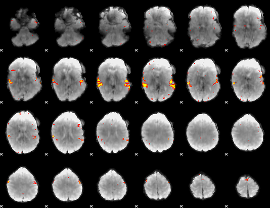 |
Component:046; Jaccard:0.14792
 |
Component:046; Jaccard:0.19795
 |
Component:046; Jaccard:0.25319
 |
Component:046; Jaccard:0.28465
 |
Component:046; Jaccard:0.26445
 |
Component:046; Jaccard:0.21284
 |
Component:046; Jaccard:0.16346
 |
Correlation:0.22124
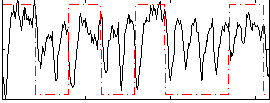  |
Correlation:0.22124
  |
Correlation:0.22124
  |
Correlation:0.22124
  |
Correlation:0.22124
  |
Correlation:0.22124
  |
Correlation:0.22124
  |
Component:301; Jaccard:0.12028
 |
Component:301; Jaccard:0.13892
 |
Component:301; Jaccard:0.14385
 |
Component:385; Jaccard:0.12966
 |
Component:385; Jaccard:0.11512
 |
Component:385; Jaccard:0.08866
 |
Component:385; Jaccard:0.069455
 |
Correlation:-0.42389
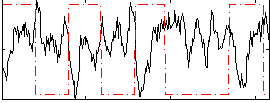 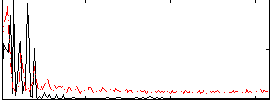 |
Correlation:-0.42389
  |
Correlation:-0.42389
  |
Correlation:0.33831
 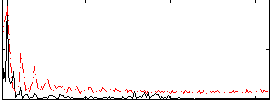 |
Correlation:0.33831
  |
Correlation:0.33831
  |
Correlation:0.33831
  |
Component:336; Jaccard:0.11426
 |
Component:336; Jaccard:0.12932
 |
Component:336; Jaccard:0.13489
 |
Component:301; Jaccard:0.12958
 |
Component:376; Jaccard:0.1017
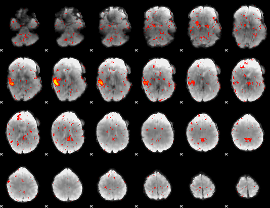 |
Component:376; Jaccard:0.083411
 |
Component:376; Jaccard:0.065128
 |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:-0.42389
  |
Correlation:0.32908
 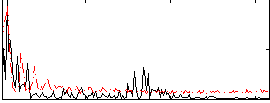 |
Correlation:0.32908
  |
Correlation:0.32908
  |
Component:118; Jaccard:0.1015
 |
Component:118; Jaccard:0.11463
 |
Component:385; Jaccard:0.1278
 |
Component:336; Jaccard:0.12209
 |
Component:301; Jaccard:0.10049
 |
Component:301; Jaccard:0.069452
 |
Component:118; Jaccard:0.050691
 |
Correlation:-0.18823
 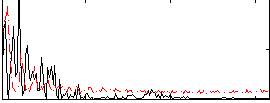 |
Correlation:-0.18823
  |
Correlation:0.33831
  |
Correlation:0.25002
  |
Correlation:-0.42389
  |
Correlation:-0.42389
  |
Correlation:-0.18823
  |
Component:385; Jaccard:0.09111
 |
Component:385; Jaccard:0.11047
 |
Component:118; Jaccard:0.11979
 |
Component:118; Jaccard:0.11333
 |
Component:336; Jaccard:0.097871
 |
Component:118; Jaccard:0.069292
 |
Component:301; Jaccard:0.047408
 |
Correlation:0.33831
  |
Correlation:0.33831
  |
Correlation:-0.18823
  |
Correlation:-0.18823
  |
Correlation:0.25002
  |
Correlation:-0.18823
  |
Correlation:-0.42389
  |
Component:356; Jaccard:0.087677
 |
Component:074; Jaccard:0.095805
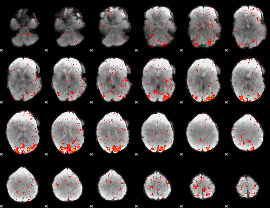 |
Component:376; Jaccard:0.10361
 |
Component:376; Jaccard:0.11052
 |
Component:118; Jaccard:0.09199
 |
Component:336; Jaccard:0.067728
 |
Component:336; Jaccard:0.045657
 |
Correlation:0.35467
  |
Correlation:0.11238
  |
Correlation:0.32908
  |
Correlation:0.32908
  |
Correlation:-0.18823
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Component:074; Jaccard:0.086534
 |
Component:356; Jaccard:0.095255
 |
Component:074; Jaccard:0.098072
 |
Component:356; Jaccard:0.089522
 |
Component:356; Jaccard:0.070174
 |
Component:279; Jaccard:0.051993
 |
Component:279; Jaccard:0.039128
 |
Correlation:0.11238
  |
Correlation:0.35467
  |
Correlation:0.11238
  |
Correlation:0.35467
  |
Correlation:0.35467
  |
Correlation:0.40044
  |
Correlation:0.40044
  |
Component:344; Jaccard:0.086191
 |
Component:376; Jaccard:0.091858
 |
Component:356; Jaccard:0.095545
 |
Component:074; Jaccard:0.085243
 |
Component:279; Jaccard:0.067368
 |
Component:356; Jaccard:0.051595
 |
Component:356; Jaccard:0.038378
 |
Correlation:-0.35649
  |
Correlation:0.32908
  |
Correlation:0.35467
  |
Correlation:0.11238
  |
Correlation:0.40044
  |
Correlation:0.35467
  |
Correlation:0.35467
  |
Component:275; Jaccard:0.084334
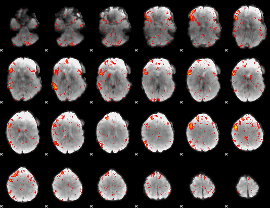 |
Component:279; Jaccard:0.087746
 |
Component:279; Jaccard:0.090249
 |
Component:279; Jaccard:0.083618
 |
Component:074; Jaccard:0.063509
 |
Component:074; Jaccard:0.040954
 |
Component:027; Jaccard:0.027982
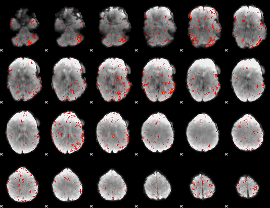 |
Correlation:-0.047329
  |
Correlation:0.40044
  |
Correlation:0.40044
  |
Correlation:0.40044
  |
Correlation:0.11238
  |
Correlation:0.11238
  |
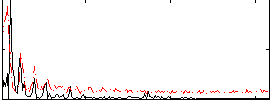
Correlation:-0.021774
  |
Component:386; Jaccard:0.082769
 |
Component:344; Jaccard:0.086327
 |
Component:342; Jaccard:0.085446
 |
Component:342; Jaccard:0.073292
 |
Component:342; Jaccard:0.055771
 |
Component:382; Jaccard:0.037027
 |
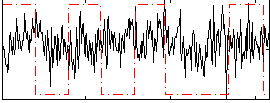
Component:006; Jaccard:0.027937
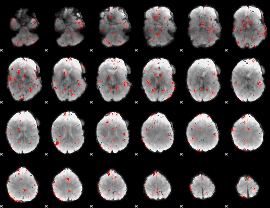 |
Correlation:-0.24585
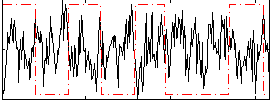 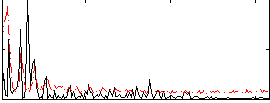 |
Correlation:-0.35649
  |
Correlation:0.025069
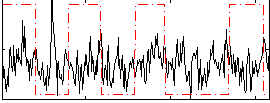 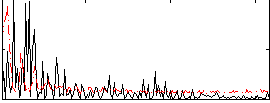 |
Correlation:0.025069
  |
Correlation:0.025069
  |
Correlation:-0.019014
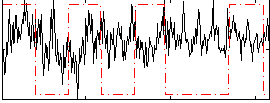 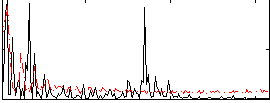 |
Correlation:0.3317
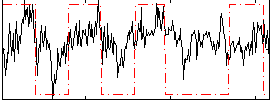 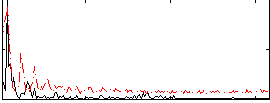 |
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
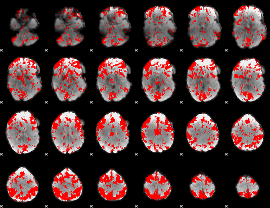 |
GLM binary result thresholded by 2
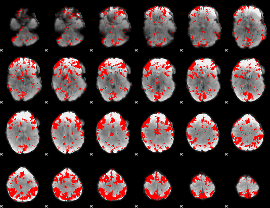 |
GLM binary result thresholded by 2.5
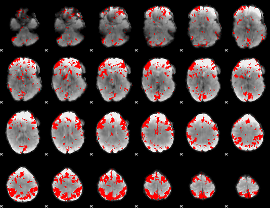 |
GLM binary result thresholded by 3
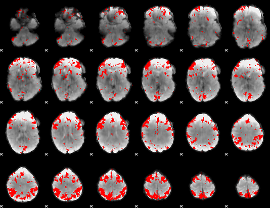 |
GLM binary result thresholded by 3.5
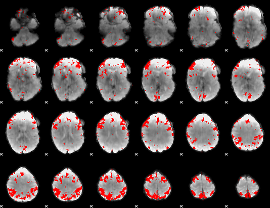 |
GLM binary result thresholded by 4
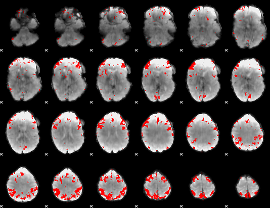 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
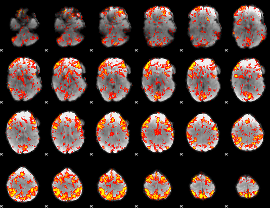 |
GLM Z-score result thresholded by 2
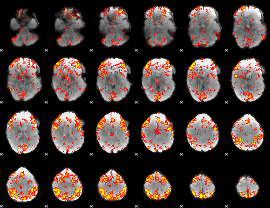 |
GLM Z-score result thresholded by 2.5
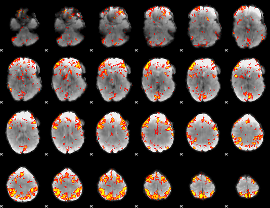 |
GLM Z-score result thresholded by 3
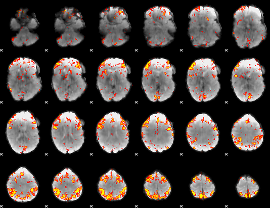 |
GLM Z-score result thresholded by 3.5
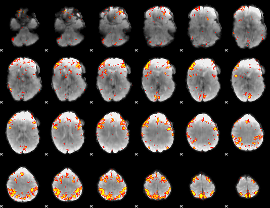 |
GLM Z-score result thresholded by 4
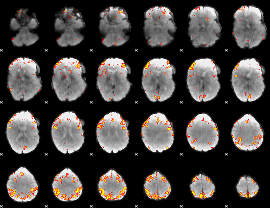 |
Component:299; Jaccard:0.16168
 |
Component:299; Jaccard:0.19644
 |
Component:299; Jaccard:0.23388
 |
Component:205; Jaccard:0.27487
 |
Component:205; Jaccard:0.31233
 |
Component:205; Jaccard:0.33449
 |
Component:205; Jaccard:0.34118
 |
Correlation:0.5945
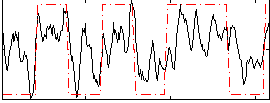 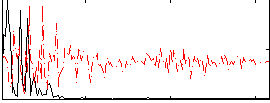 |
Correlation:0.5945
  |
Correlation:0.5945
  |
Correlation:0.6133
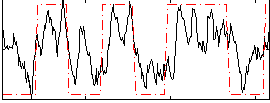 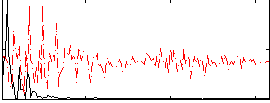 |
Correlation:0.6133
  |
Correlation:0.6133
  |
Correlation:0.6133
  |
Component:205; Jaccard:0.14815
 |
Component:205; Jaccard:0.18683
 |
Component:205; Jaccard:0.23089
 |
Component:299; Jaccard:0.26758
 |
Component:299; Jaccard:0.29406
 |
Component:299; Jaccard:0.30382
 |
Component:299; Jaccard:0.30057
 |
Correlation:0.6133
  |
Correlation:0.6133
  |
Correlation:0.6133
  |
Correlation:0.5945
  |
Correlation:0.5945
  |
Correlation:0.5945
  |
Correlation:0.5945
  |
Component:349; Jaccard:0.13945
 |
Component:349; Jaccard:0.16088
 |
Component:349; Jaccard:0.18127
 |
Component:349; Jaccard:0.19456
 |
Component:141; Jaccard:0.20626
 |
Component:141; Jaccard:0.20757
 |
Component:141; Jaccard:0.19825
 |
Correlation:0.48796
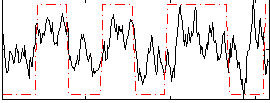  |
Correlation:0.48796
  |
Correlation:0.48796
  |
Correlation:0.48796
  |
Correlation:0.65749
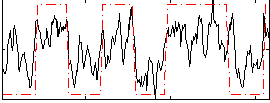 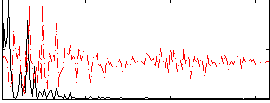 |
Correlation:0.65749
  |
Correlation:0.65749
  |
Component:197; Jaccard:0.13762
 |
Component:141; Jaccard:0.15307
 |
Component:141; Jaccard:0.17534
 |
Component:141; Jaccard:0.19398
 |
Component:349; Jaccard:0.2006
 |
Component:349; Jaccard:0.1988
 |
Component:207; Jaccard:0.1887
 |
Correlation:0.47247
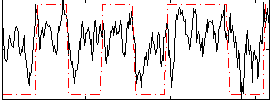 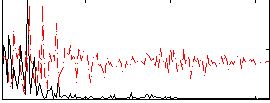 |
Correlation:0.65749
  |
Correlation:0.65749
  |
Correlation:0.65749
  |
Correlation:0.48796
  |
Correlation:0.48796
  |
Correlation:0.58427
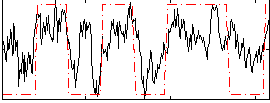 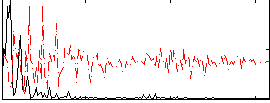 |
Component:141; Jaccard:0.13063
 |
Component:197; Jaccard:0.1508
 |
Component:207; Jaccard:0.16901
 |
Component:207; Jaccard:0.18418
 |
Component:207; Jaccard:0.19232
 |
Component:207; Jaccard:0.19446
 |
Component:349; Jaccard:0.18729
 |
Correlation:0.65749
  |
Correlation:0.47247
  |
Correlation:0.58427
  |
Correlation:0.58427
  |
Correlation:0.58427
  |
Correlation:0.58427
  |
Correlation:0.48796
  |
Component:207; Jaccard:0.12918
 |
Component:207; Jaccard:0.15013
 |
Component:197; Jaccard:0.15922
 |
Component:144; Jaccard:0.17056
 |
Component:144; Jaccard:0.17626
 |
Component:144; Jaccard:0.17427
 |
Component:144; Jaccard:0.16627
 |
Correlation:0.58427
  |
Correlation:0.58427
  |
Correlation:0.47247
  |
Correlation:0.54245
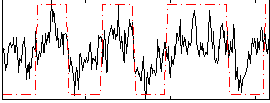 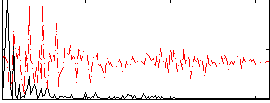 |
Correlation:0.54245
  |
Correlation:0.54245
  |
Correlation:0.54245
  |
Component:292; Jaccard:0.12371
 |
Component:144; Jaccard:0.14013
 |
Component:144; Jaccard:0.15803
 |
Component:197; Jaccard:0.16138
 |
Component:197; Jaccard:0.15872
 |
Component:292; Jaccard:0.15222
 |
Component:292; Jaccard:0.14163
 |
Correlation:0.44562
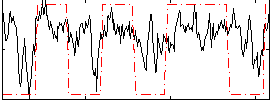 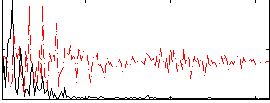 |
Correlation:0.54245
  |
Correlation:0.54245
  |
Correlation:0.47247
  |
Correlation:0.47247
  |
Correlation:0.44562
  |
Correlation:0.44562
  |
Component:144; Jaccard:0.12072
 |
Component:292; Jaccard:0.13846
 |
Component:292; Jaccard:0.15023
 |
Component:292; Jaccard:0.15758
 |
Component:292; Jaccard:0.15744
 |
Component:197; Jaccard:0.1487
 |
Component:197; Jaccard:0.13559
 |
Correlation:0.54245
  |
Correlation:0.44562
  |
Correlation:0.44562
  |
Correlation:0.44562
  |
Correlation:0.44562
  |
Correlation:0.47247
  |
Correlation:0.47247
  |
Component:351; Jaccard:0.11214
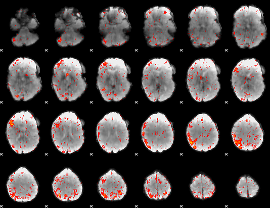 |
Component:352; Jaccard:0.12408
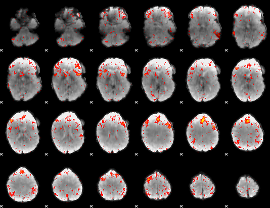 |
Component:352; Jaccard:0.13375
 |
Component:352; Jaccard:0.13848
 |
Component:352; Jaccard:0.13899
 |
Component:352; Jaccard:0.13468
 |
Component:352; Jaccard:0.12633
 |
Correlation:0.48699
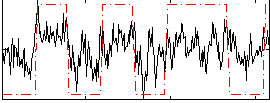 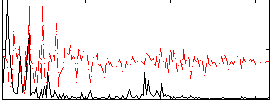 |
Correlation:0.43311
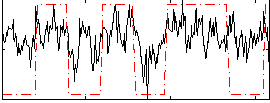  |
Correlation:0.43311
  |
Correlation:0.43311
  |
Correlation:0.43311
  |
Correlation:0.43311
  |
Correlation:0.43311
  |
Component:352; Jaccard:0.11175
 |
Component:351; Jaccard:0.12247
 |
Component:351; Jaccard:0.13153
 |
Component:351; Jaccard:0.13562
 |
Component:351; Jaccard:0.13558
 |
Component:313; Jaccard:0.12907
 |
Component:313; Jaccard:0.12238
 |
Correlation:0.43311
  |
Correlation:0.48699
  |
Correlation:0.48699
  |
Correlation:0.48699
  |
Correlation:0.48699
  |
Correlation:0.42494
  |
Correlation:0.42494
  |
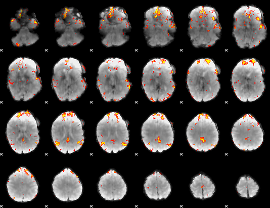
Cope: 04:STORY-MATH BACK
|
GLM result group-wise
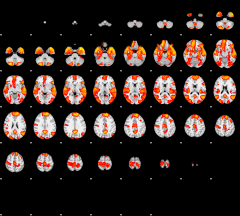 |
GLM binary result thresholded by 1
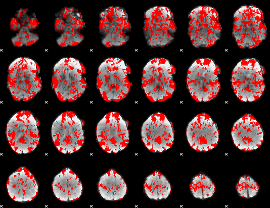 |
GLM binary result thresholded by 1.5
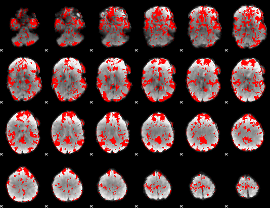 |
GLM binary result thresholded by 2
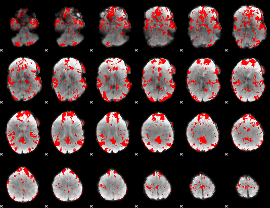 |
GLM binary result thresholded by 2.5
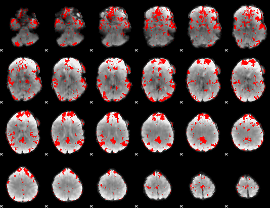 |
GLM binary result thresholded by 3
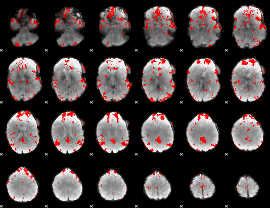 |
GLM binary result thresholded by 3.5
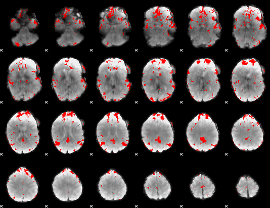 |
GLM binary result thresholded by 4
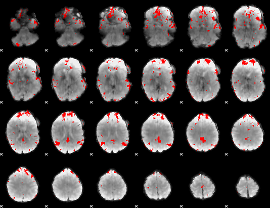 |
GLM Z-score result thresholded by 1
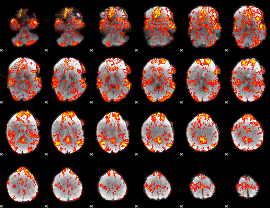 |
GLM Z-score result thresholded by 1.5
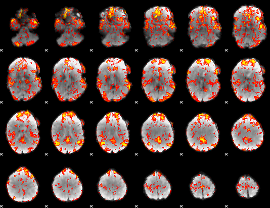 |
GLM Z-score result thresholded by 2
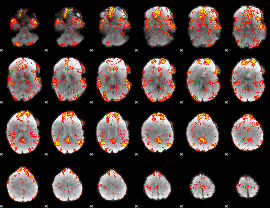 |
GLM Z-score result thresholded by 2.5
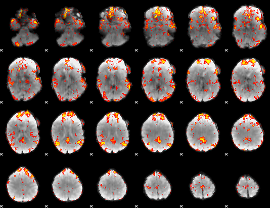 |
GLM Z-score result thresholded by 3
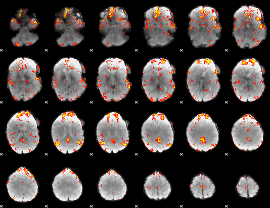 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:279; Jaccard:0.14582
 |
Component:279; Jaccard:0.1772
 |
Component:279; Jaccard:0.212
 |
Component:279; Jaccard:0.24493
 |
Component:279; Jaccard:0.27183
 |
Component:279; Jaccard:0.2822
 |
Component:279; Jaccard:0.27958
 |
Correlation:0.43227
  |
Correlation:0.43227
  |
Correlation:0.43227
  |
Correlation:0.43227
  |
Correlation:0.43227
  |
Correlation:0.43227
  |
Correlation:0.43227
  |
Component:307; Jaccard:0.12071
 |
Component:307; Jaccard:0.1407
 |
Component:307; Jaccard:0.16048
 |
Component:307; Jaccard:0.17519
 |
Component:307; Jaccard:0.18317
 |
Component:229; Jaccard:0.18212
 |
Component:229; Jaccard:0.18078
 |
Correlation:0.55685
  |
Correlation:0.55685
  |
Correlation:0.55685
  |
Correlation:0.55685
  |
Correlation:0.55685
  |
Correlation:0.35013
  |
Correlation:0.35013
  |
Component:336; Jaccard:0.11683
 |
Component:229; Jaccard:0.12733
 |
Component:229; Jaccard:0.14861
 |
Component:229; Jaccard:0.16471
 |
Component:229; Jaccard:0.17739
 |
Component:307; Jaccard:0.17982
 |
Component:307; Jaccard:0.16912
 |
Correlation:0.27484
 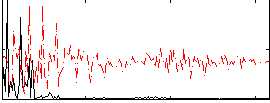 |
Correlation:0.35013
  |
Correlation:0.35013
  |
Correlation:0.35013
  |
Correlation:0.35013
  |
Correlation:0.55685
  |
Correlation:0.55685
  |
Component:394; Jaccard:0.11366
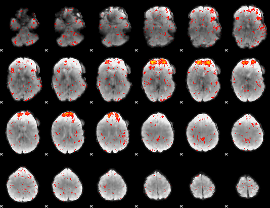 |
Component:336; Jaccard:0.12722
 |
Component:394; Jaccard:0.13799
 |
Component:394; Jaccard:0.1452
 |
Component:385; Jaccard:0.15113
 |
Component:385; Jaccard:0.14887
 |
Component:385; Jaccard:0.14081
 |
Correlation:0.45616
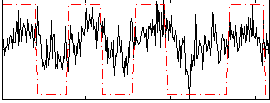 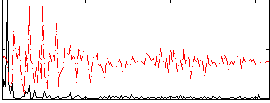 |
Correlation:0.27484
  |
Correlation:0.45616
  |
Correlation:0.45616
  |
Correlation:0.36215
  |
Correlation:0.36215
  |
Correlation:0.36215
  |
Component:229; Jaccard:0.10874
 |
Component:394; Jaccard:0.1271
 |
Component:336; Jaccard:0.13422
 |
Component:385; Jaccard:0.14501
 |
Component:394; Jaccard:0.14792
 |
Component:394; Jaccard:0.14296
 |
Component:394; Jaccard:0.13171
 |
Correlation:0.35013
  |
Correlation:0.45616
  |
Correlation:0.27484
  |
Correlation:0.36215
  |
Correlation:0.45616
  |
Correlation:0.45616
  |
Correlation:0.45616
  |
Component:356; Jaccard:0.10864
 |
Component:356; Jaccard:0.12085
 |
Component:385; Jaccard:0.13389
 |
Component:356; Jaccard:0.13892
 |
Component:356; Jaccard:0.14238
 |
Component:356; Jaccard:0.13888
 |
Component:356; Jaccard:0.12813
 |
Correlation:0.36315
  |
Correlation:0.36315
  |
Correlation:0.36215
  |
Correlation:0.36315
  |
Correlation:0.36315
  |
Correlation:0.36315
  |
Correlation:0.36315
  |
Component:385; Jaccard:0.10446
 |
Component:385; Jaccard:0.11919
 |
Component:356; Jaccard:0.13179
 |
Component:336; Jaccard:0.13373
 |
Component:019; Jaccard:0.13035
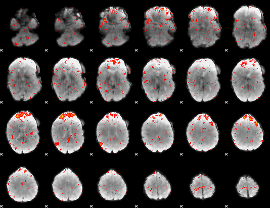 |
Component:129; Jaccard:0.12892
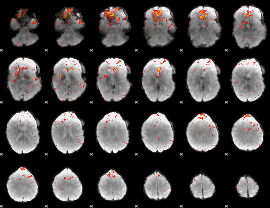 |
Component:129; Jaccard:0.1277
 |
Correlation:0.36215
  |
Correlation:0.36215
  |
Correlation:0.36315
  |
Correlation:0.27484
  |
Correlation:0.501
  |
Correlation:0.28021
  |
Correlation:0.28021
  |
Component:019; Jaccard:0.10315
 |
Component:019; Jaccard:0.1148
 |
Component:019; Jaccard:0.12473
 |
Component:019; Jaccard:0.13136
 |
Component:336; Jaccard:0.12774
 |
Component:019; Jaccard:0.12309
 |
Component:019; Jaccard:0.11352
 |
Correlation:0.501
  |
Correlation:0.501
  |
Correlation:0.501
  |
Correlation:0.501
  |
Correlation:0.27484
  |
Correlation:0.501
  |
Correlation:0.501
  |
Component:277; Jaccard:0.098892
 |
Component:277; Jaccard:0.10338
 |
Component:248; Jaccard:0.11029
 |
Component:129; Jaccard:0.11469
 |
Component:129; Jaccard:0.12509
 |
Component:336; Jaccard:0.11737
 |
Component:248; Jaccard:0.10085
 |
Correlation:0.013404
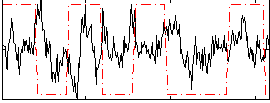 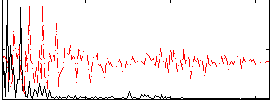 |
Correlation:0.013404
  |
Correlation:0.17801
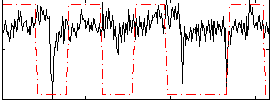 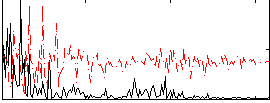 |
Correlation:0.28021
  |
Correlation:0.28021
  |
Correlation:0.27484
  |
Correlation:0.17801
  |
Component:161; Jaccard:0.094171
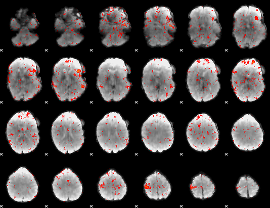 |
Component:248; Jaccard:0.10227
 |
Component:277; Jaccard:0.10503
 |
Component:248; Jaccard:0.11388
 |
Component:248; Jaccard:0.11515
 |
Component:248; Jaccard:0.11183
 |
Component:336; Jaccard:0.09922
 |
Correlation:0.29951
  |
Correlation:0.17801
  |
Correlation:0.013404
  |
Correlation:0.17801
  |
Correlation:0.17801
  |
Correlation:0.17801
  |
Correlation:0.27484
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
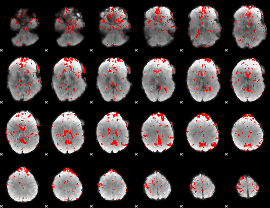 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
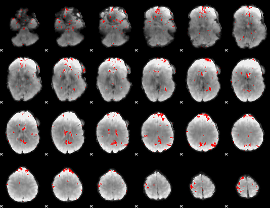 |
GLM binary result thresholded by 3
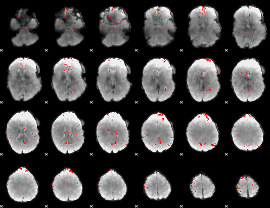 |
GLM binary result thresholded by 3.5
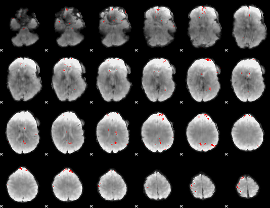 |
GLM binary result thresholded by 4
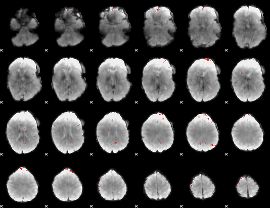 |
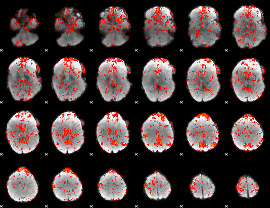
GLM Z-score result thresholded by 1
 |
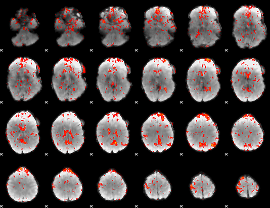
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
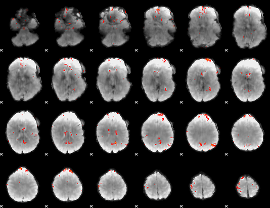
GLM Z-score result thresholded by 3
 |
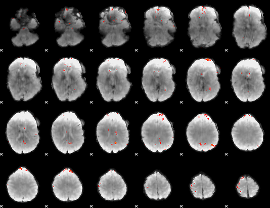
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
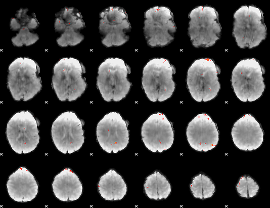 |
Component:156; Jaccard:0.13645
 |
Component:156; Jaccard:0.14195
 |
Component:156; Jaccard:0.13141
 |
Component:307; Jaccard:0.10523
 |
Component:307; Jaccard:0.07577
 |
Component:307; Jaccard:0.047656
 |
Component:307; Jaccard:0.025662
 |
Correlation:-0.033619
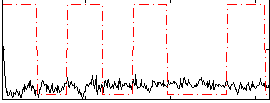 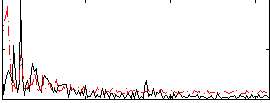 |
Correlation:-0.033619
  |
Correlation:-0.033619
  |
Correlation:0.55384
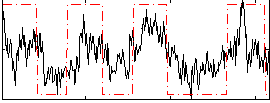  |
Correlation:0.55384
  |
Correlation:0.55384
  |
Correlation:0.55384
  |
Component:134; Jaccard:0.11924
 |
Component:307; Jaccard:0.12529
 |
Component:307; Jaccard:0.12401
 |
Component:156; Jaccard:0.10409
 |
Component:156; Jaccard:0.067969
 |
Component:156; Jaccard:0.036065
 |
Component:098; Jaccard:0.016746
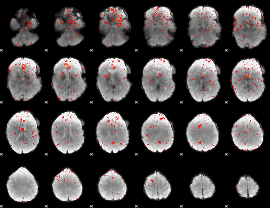 |
Correlation:-0.028045
  |
Correlation:0.55384
  |
Correlation:0.55384
  |
Correlation:-0.033619
  |
Correlation:-0.033619
  |
Correlation:-0.033619
  |
Correlation:0.19541
  |
Component:307; Jaccard:0.11462
 |
Component:134; Jaccard:0.11675
 |
Component:098; Jaccard:0.11303
 |
Component:098; Jaccard:0.091842
 |
Component:098; Jaccard:0.061828
 |
Component:098; Jaccard:0.033744
 |
Component:156; Jaccard:0.016573
 |
Correlation:0.55384
  |
Correlation:-0.028045
  |
Correlation:0.19541
  |
Correlation:0.19541
  |
Correlation:0.19541
  |
Correlation:0.19541
  |
Correlation:-0.033619
  |
Component:102; Jaccard:0.11287
 |
Component:098; Jaccard:0.1167
 |
Component:134; Jaccard:0.10341
 |
Component:102; Jaccard:0.076896
 |
Component:102; Jaccard:0.052046
 |
Component:102; Jaccard:0.027825
 |
Component:134; Jaccard:0.013491
 |
Correlation:0.13252
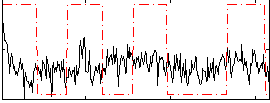 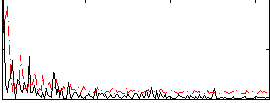 |
Correlation:0.19541
  |
Correlation:-0.028045
  |
Correlation:0.13252
  |
Correlation:0.13252
  |
Correlation:0.13252
  |
Correlation:-0.028045
  |
Component:098; Jaccard:0.10485
 |
Component:102; Jaccard:0.11197
 |
Component:102; Jaccard:0.1002
 |
Component:134; Jaccard:0.07681
 |
Component:134; Jaccard:0.048737
 |
Component:134; Jaccard:0.026981
 |
Component:331; Jaccard:0.013299
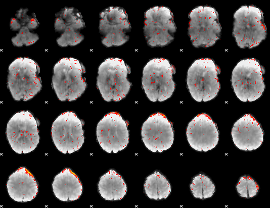 |
Correlation:0.19541
  |
Correlation:0.13252
  |
Correlation:0.13252
  |
Correlation:-0.028045
  |
Correlation:-0.028045
  |
Correlation:-0.028045
  |
Correlation:0.16118
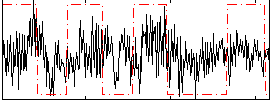  |
Component:176; Jaccard:0.088935
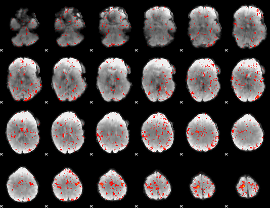 |
Component:279; Jaccard:0.082444
 |
Component:279; Jaccard:0.073969
 |
Component:279; Jaccard:0.057564
 |
Component:279; Jaccard:0.038418
 |
Component:129; Jaccard:0.023089
 |
Component:102; Jaccard:0.013078
 |
Correlation:0.093862
  |
Correlation:0.44555
  |
Correlation:0.44555
  |
Correlation:0.44555
  |
Correlation:0.44555
  |
Correlation:0.2669
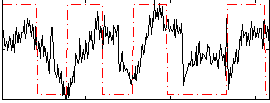 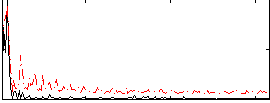 |
Correlation:0.13252
  |
Component:279; Jaccard:0.082283
 |
Component:176; Jaccard:0.082424
 |
Component:373; Jaccard:0.071439
 |
Component:373; Jaccard:0.057072
 |
Component:373; Jaccard:0.03802
 |
Component:331; Jaccard:0.023021
 |
Component:248; Jaccard:0.011459
 |
Correlation:0.44555
  |
Correlation:0.093862
  |
Correlation:0.37409
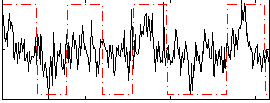 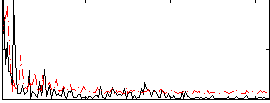 |
Correlation:0.37409
  |
Correlation:0.37409
  |
Correlation:0.16118
  |
Correlation:0.17944
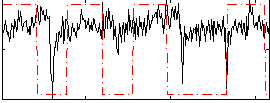 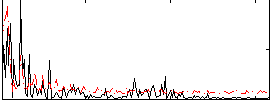 |
Component:248; Jaccard:0.078906
 |
Component:248; Jaccard:0.078975
 |
Component:248; Jaccard:0.070335
 |
Component:248; Jaccard:0.055326
 |
Component:129; Jaccard:0.03769
 |
Component:279; Jaccard:0.022022
 |
Component:279; Jaccard:0.010819
 |
Correlation:0.17944
  |
Correlation:0.17944
  |
Correlation:0.17944
  |
Correlation:0.17944
  |
Correlation:0.2669
  |
Correlation:0.44555
  |
Correlation:0.44555
  |
Component:019; Jaccard:0.078825
 |
Component:229; Jaccard:0.077774
 |
Component:176; Jaccard:0.069989
 |
Component:229; Jaccard:0.054209
 |
Component:229; Jaccard:0.036904
 |
Component:032; Jaccard:0.02152
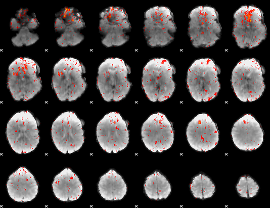 |
Component:373; Jaccard:0.010806
 |
Correlation:0.48803
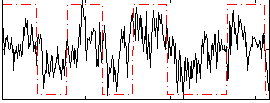 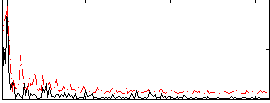 |
Correlation:0.32509
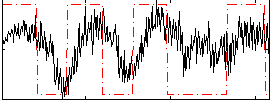 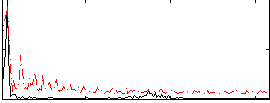 |
Correlation:0.093862
  |
Correlation:0.32509
  |
Correlation:0.32509
  |
Correlation:0.16017
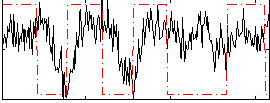  |
Correlation:0.37409
  |
Component:229; Jaccard:0.078556
 |
Component:373; Jaccard:0.077616
 |
Component:129; Jaccard:0.068202
 |
Component:129; Jaccard:0.054127
 |
Component:331; Jaccard:0.036826
 |
Component:248; Jaccard:0.021407
 |
Component:032; Jaccard:0.010696
 |
Correlation:0.32509
  |
Correlation:0.37409
  |
Correlation:0.2669
  |
Correlation:0.2669
  |
Correlation:0.16118
  |
Correlation:0.17944
  |
Correlation:0.16017
  |
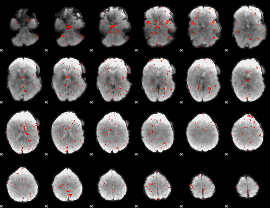
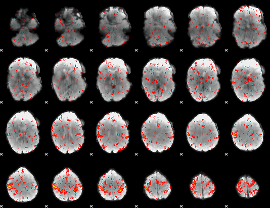
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
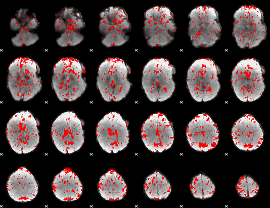 |
GLM binary result thresholded by 1.5
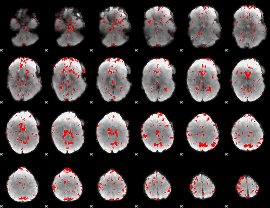 |
GLM binary result thresholded by 2
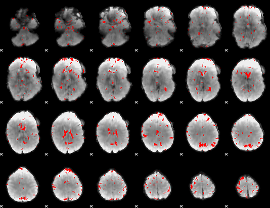 |
GLM binary result thresholded by 2.5
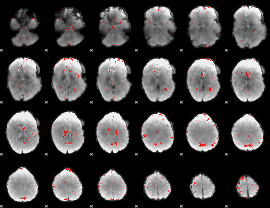 |
GLM binary result thresholded by 3
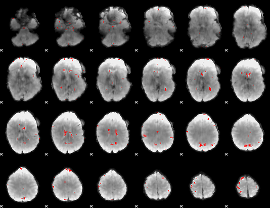 |
GLM binary result thresholded by 3.5
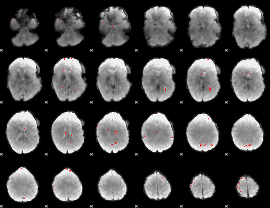 |
GLM binary result thresholded by 4
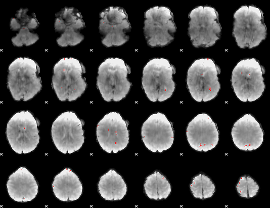 |
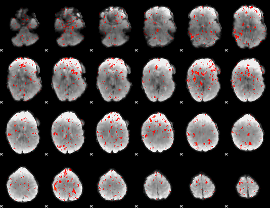
GLM Z-score result thresholded by 1
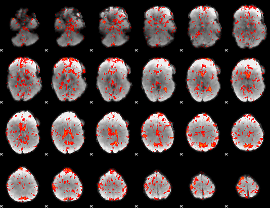 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
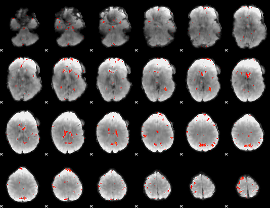 |
GLM Z-score result thresholded by 3
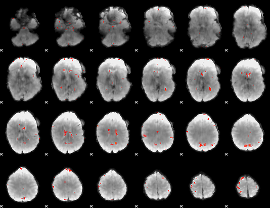 |
GLM Z-score result thresholded by 3.5
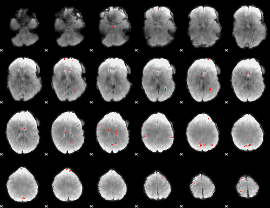 |
GLM Z-score result thresholded by 4
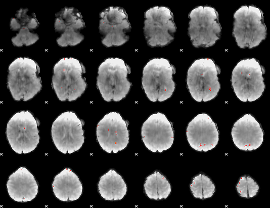 |
Component:156; Jaccard:0.18188
 |
Component:156; Jaccard:0.19468
 |
Component:156; Jaccard:0.1774
 |
Component:156; Jaccard:0.13039
 |
Component:156; Jaccard:0.077766
 |
Component:156; Jaccard:0.041915
 |
Component:156; Jaccard:0.018266
 |
Correlation:0.042113
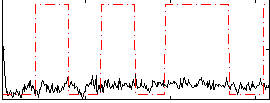 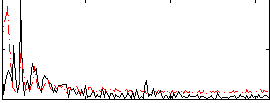 |
Correlation:0.042113
  |
Correlation:0.042113
  |
Correlation:0.042113
  |
Correlation:0.042113
  |
Correlation:0.042113
  |
Correlation:0.042113
  |
Component:134; Jaccard:0.13027
 |
Component:134; Jaccard:0.12501
 |
Component:134; Jaccard:0.098818
 |
Component:134; Jaccard:0.06518
 |
Component:134; Jaccard:0.035685
 |
Component:102; Jaccard:0.017653
 |
Component:134; Jaccard:0.008098
 |
Correlation:0.040315
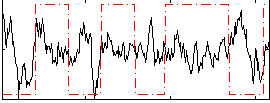  |
Correlation:0.040315
  |
Correlation:0.040315
  |
Correlation:0.040315
  |
Correlation:0.040315
  |
Correlation:-0.13851
  |
Correlation:0.040315
  |
Component:102; Jaccard:0.11605
 |
Component:102; Jaccard:0.11038
 |
Component:102; Jaccard:0.091718
 |
Component:102; Jaccard:0.063841
 |
Component:102; Jaccard:0.035147
 |
Component:134; Jaccard:0.01755
 |
Component:165; Jaccard:0.007926
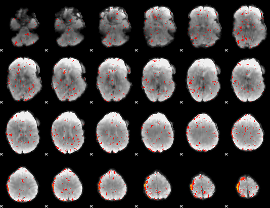 |
Correlation:-0.13851
  |
Correlation:-0.13851
  |
Correlation:-0.13851
  |
Correlation:-0.13851
  |
Correlation:-0.13851
  |
Correlation:0.040315
  |
Correlation:-0.058524
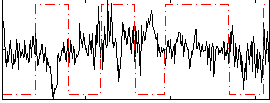 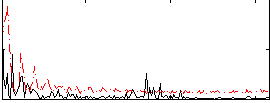 |
Component:057; Jaccard:0.091983
 |
Component:057; Jaccard:0.088611
 |
Component:057; Jaccard:0.074796
 |
Component:057; Jaccard:0.050438
 |
Component:109; Jaccard:0.031443
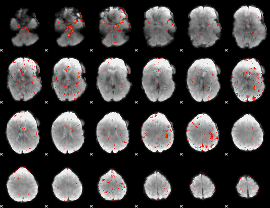 |
Component:165; Jaccard:0.016562
 |
Component:102; Jaccard:0.007741
 |
Correlation:0.27422
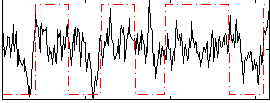  |
Correlation:0.27422
  |
Correlation:0.27422
  |
Correlation:0.27422
  |
Correlation:-0.07125
  |
Correlation:-0.058524
  |
Correlation:-0.13851
  |
Component:292; Jaccard:0.084695
 |
Component:292; Jaccard:0.08093
 |
Component:109; Jaccard:0.066743
 |
Component:109; Jaccard:0.049402
 |
Component:057; Jaccard:0.029546
 |
Component:109; Jaccard:0.015855
 |
Component:183; Jaccard:0.007044
 |
Correlation:0.44598
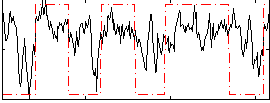 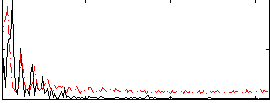 |
Correlation:0.44598
  |
Correlation:-0.07125
  |
Correlation:-0.07125
  |
Correlation:0.27422
  |
Correlation:-0.07125
  |
Correlation:-0.033618
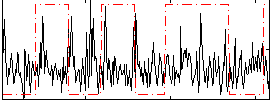 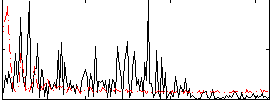 |
Component:387; Jaccard:0.0837
 |
Component:109; Jaccard:0.075074
 |
Component:292; Jaccard:0.06094
 |
Component:047; Jaccard:0.040369
 |
Component:165; Jaccard:0.027424
 |
Component:183; Jaccard:0.015583
 |
Component:057; Jaccard:0.006665
 |
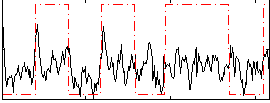
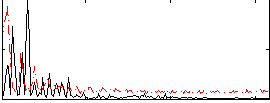
Correlation:0.29518
  |
Correlation:-0.07125
  |
Correlation:0.44598
  |
Correlation:0.17557
  |
Correlation:-0.058524
  |
Correlation:-0.033618
  |
Correlation:0.27422
  |
Component:176; Jaccard:0.079356
 |
Component:047; Jaccard:0.073683
 |
Component:047; Jaccard:0.05986
 |
Component:183; Jaccard:0.040331
 |
Component:183; Jaccard:0.027173
 |
Component:057; Jaccard:0.014311
 |
Component:176; Jaccard:0.006157
 |
Correlation:-0.098183
  |
Correlation:0.17557
  |
Correlation:0.17557
  |
Correlation:-0.033618
  |
Correlation:-0.033618
  |
Correlation:0.27422
  |
Correlation:-0.098183
  |
Component:047; Jaccard:0.078787
 |
Component:387; Jaccard:0.071623
 |
Component:176; Jaccard:0.057103
 |
Component:292; Jaccard:0.040191
 |
Component:292; Jaccard:0.022334
 |
Component:176; Jaccard:0.013261
 |
Component:109; Jaccard:0.006088
 |
Correlation:0.17557
  |
Correlation:0.29518
  |
Correlation:-0.098183
  |
Correlation:0.44598
  |
Correlation:0.44598
  |
Correlation:-0.098183
  |
Correlation:-0.07125
  |
Component:080; Jaccard:0.075557
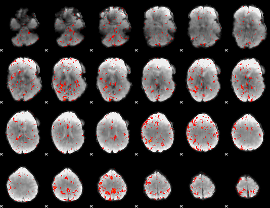 |
Component:176; Jaccard:0.071273
 |
Component:387; Jaccard:0.054368
 |
Component:165; Jaccard:0.03986
 |
Component:124; Jaccard:0.022008
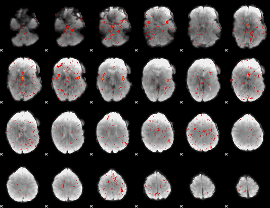 |
Component:080; Jaccard:0.011838
 |
Component:047; Jaccard:0.005862
 |
Correlation:0.18648
 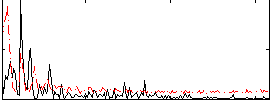 |
Correlation:-0.098183
  |
Correlation:0.29518
  |
Correlation:-0.058524
  |
Correlation:0.10063
 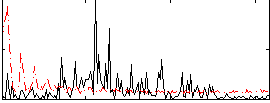 |
Correlation:0.18648
  |
Correlation:0.17557
  |
Component:231; Jaccard:0.075242
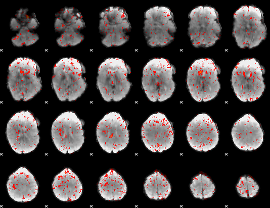 |
Component:080; Jaccard:0.068891
 |
Component:165; Jaccard:0.054088
 |
Component:176; Jaccard:0.039317
 |
Component:176; Jaccard:0.021956
 |
Component:047; Jaccard:0.011779
 |
Component:080; Jaccard:0.005769
 |
Correlation:0.17442
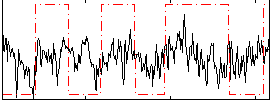 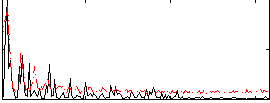 |
Correlation:0.18648
  |
Correlation:-0.058524
  |
Correlation:-0.098183
  |
Correlation:-0.098183
  |
Correlation:0.17557
  |
Correlation:0.18648
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































