Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
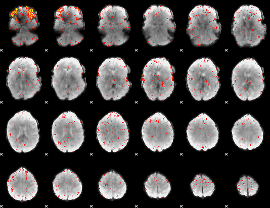
GLM result group-wise
 |
GLM binary result thresholded by 1
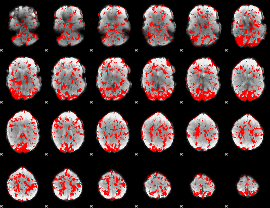 |
GLM binary result thresholded by 1.5
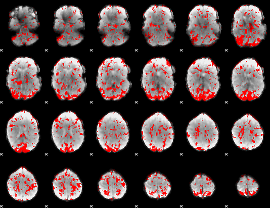 |
GLM binary result thresholded by 2
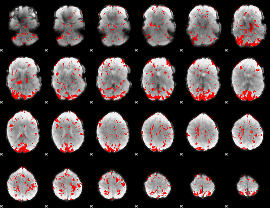 |
GLM binary result thresholded by 2.5
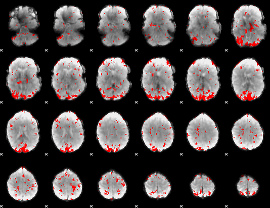 |
GLM binary result thresholded by 3
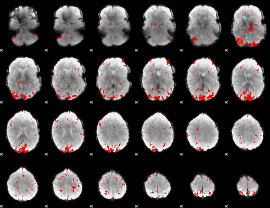 |
GLM binary result thresholded by 3.5
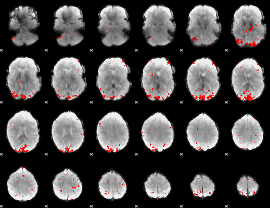 |
GLM binary result thresholded by 4
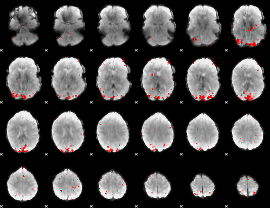 |
GLM Z-score result thresholded by 1
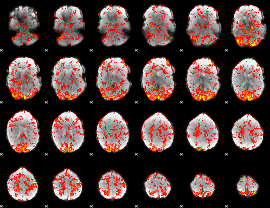 |
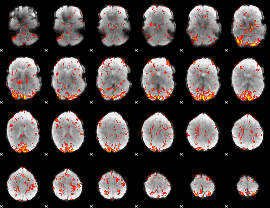
GLM Z-score result thresholded by 1.5
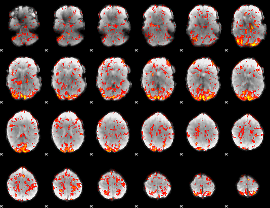 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
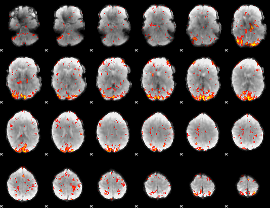 |
GLM Z-score result thresholded by 3
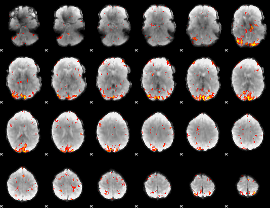 |
GLM Z-score result thresholded by 3.5
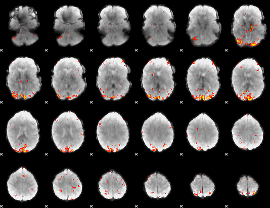 |
GLM Z-score result thresholded by 4
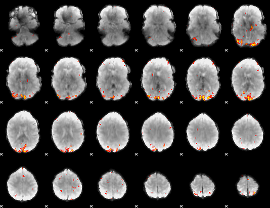 |
Component:285; Jaccard:0.1578
 |
Component:285; Jaccard:0.19375
 |
Component:285; Jaccard:0.22787
 |
Component:285; Jaccard:0.25184
 |
Component:285; Jaccard:0.25583
 |
Component:285; Jaccard:0.23332
 |
Component:285; Jaccard:0.19277
 |
Correlation:0.20943
  |
Correlation:0.20943
  |
Correlation:0.20943
  |
Correlation:0.20943
  |
Correlation:0.20943
  |
Correlation:0.20943
  |
Correlation:0.20943
  |
Component:194; Jaccard:0.11534
 |
Component:194; Jaccard:0.13379
 |
Component:194; Jaccard:0.15081
 |
Component:194; Jaccard:0.16083
 |
Component:194; Jaccard:0.158
 |
Component:194; Jaccard:0.14355
 |
Component:194; Jaccard:0.12214
 |
Correlation:0.21285
  |
Correlation:0.21285
  |
Correlation:0.21285
  |
Correlation:0.21285
  |
Correlation:0.21285
  |
Correlation:0.21285
  |
Correlation:0.21285
  |
Component:315; Jaccard:0.11114
 |
Component:315; Jaccard:0.12234
 |
Component:315; Jaccard:0.1304
 |
Component:315; Jaccard:0.13064
 |
Component:179; Jaccard:0.13243
 |
Component:179; Jaccard:0.12803
 |
Component:179; Jaccard:0.1149
 |
Correlation:0.090866
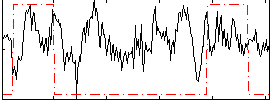 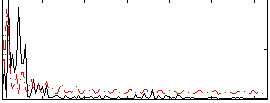 |
Correlation:0.090866
  |
Correlation:0.090866
  |
Correlation:0.090866
  |
Correlation:0.288
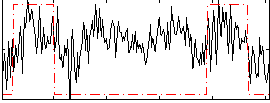 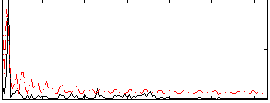 |
Correlation:0.288
  |
Correlation:0.288
  |
Component:307; Jaccard:0.099917
 |
Component:307; Jaccard:0.10949
 |
Component:307; Jaccard:0.11379
 |
Component:179; Jaccard:0.12442
 |
Component:315; Jaccard:0.11969
 |
Component:315; Jaccard:0.1037
 |
Component:259; Jaccard:0.092742
 |
Correlation:0.075183
  |
Correlation:0.075183
  |
Correlation:0.075183
  |
Correlation:0.288
  |
Correlation:0.090866
  |
Correlation:0.090866
  |
Correlation:0.066407
  |
Component:239; Jaccard:0.093025
 |
Component:016; Jaccard:0.095982
 |
Component:179; Jaccard:0.10946
 |
Component:307; Jaccard:0.11358
 |
Component:259; Jaccard:0.10402
 |
Component:259; Jaccard:0.10135
 |
Component:315; Jaccard:0.086762
 |
Correlation:0.049096
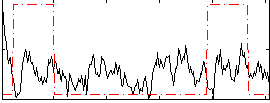  |
Correlation:0.14176
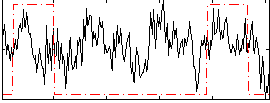  |
Correlation:0.288
  |
Correlation:0.075183
  |
Correlation:0.066407
  |
Correlation:0.066407
  |
Correlation:0.090866
  |
Component:016; Jaccard:0.088063
 |
Component:179; Jaccard:0.09332
 |
Component:016; Jaccard:0.10089
 |
Component:259; Jaccard:0.10244
 |
Component:307; Jaccard:0.10314
 |
Component:307; Jaccard:0.086563
 |
Component:255; Jaccard:0.077732
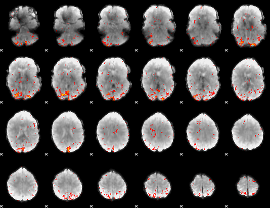 |
Correlation:0.14176
  |
Correlation:0.288
  |
Correlation:0.14176
  |
Correlation:0.066407
  |
Correlation:0.075183
  |
Correlation:0.075183
  |
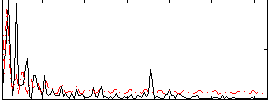
Correlation:0.017491
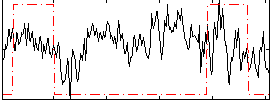 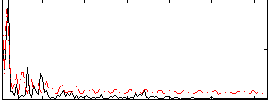 |
Component:138; Jaccard:0.086916
 |
Component:239; Jaccard:0.09147
 |
Component:259; Jaccard:0.094758
 |
Component:016; Jaccard:0.10135
 |
Component:016; Jaccard:0.093588
 |
Component:255; Jaccard:0.085772
 |
Component:307; Jaccard:0.071899
 |
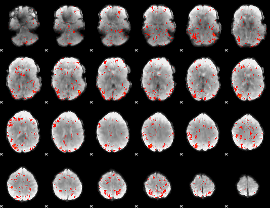
Correlation:-0.10821
  |
Correlation:0.049096
  |
Correlation:0.066407
  |
Correlation:0.14176
  |
Correlation:0.14176
  |
Correlation:0.017491
  |
Correlation:0.075183
  |
Component:032; Jaccard:0.086559
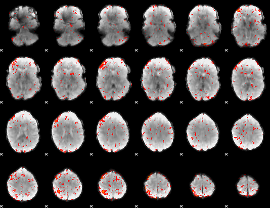 |
Component:138; Jaccard:0.091199
 |
Component:032; Jaccard:0.0933
 |
Component:032; Jaccard:0.090894
 |
Component:255; Jaccard:0.088796
 |
Component:016; Jaccard:0.081536
 |
Component:016; Jaccard:0.065894
 |
Correlation:0.055541
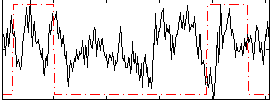  |
Correlation:-0.10821
  |
Correlation:0.055541
  |
Correlation:0.055541
  |
Correlation:0.017491
  |
Correlation:0.14176
  |
Correlation:0.14176
  |
Component:288; Jaccard:0.086148
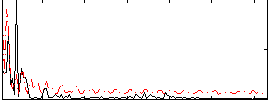
 |
Component:032; Jaccard:0.09055
 |
Component:138; Jaccard:0.091902
 |
Component:006; Jaccard:0.089749
 |
Component:006; Jaccard:0.084204
 |
Component:032; Jaccard:0.071391
 |
Component:032; Jaccard:0.056427
 |
Correlation:0.072872
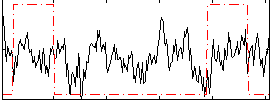  |
Correlation:0.055541
  |
Correlation:-0.10821
  |
Correlation:0.18197
  |
Correlation:0.18197
  |
Correlation:0.055541
  |
Correlation:0.055541
  |
Component:006; Jaccard:0.07981
 |
Component:288; Jaccard:0.089211
 |
Component:006; Jaccard:0.090052
 |
Component:255; Jaccard:0.086382
 |
Component:032; Jaccard:0.08332
 |
Component:006; Jaccard:0.069272
 |
Component:338; Jaccard:0.056251
 |
Correlation:0.18197
  |
Correlation:0.072872
  |
Correlation:0.18197
  |
Correlation:0.017491
  |
Correlation:0.055541
  |
Correlation:0.18197
  |
Correlation:0.11469
  |
Cope: 02:REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
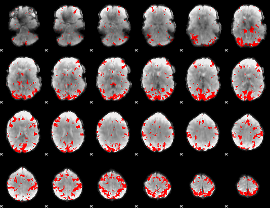 |
GLM binary result thresholded by 3.5
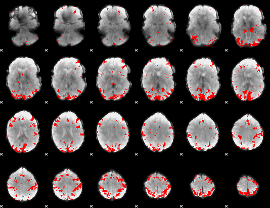 |
GLM binary result thresholded by 4
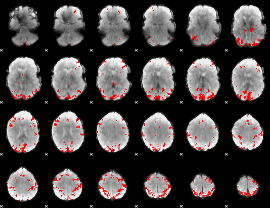 |
GLM Z-score result thresholded by 1
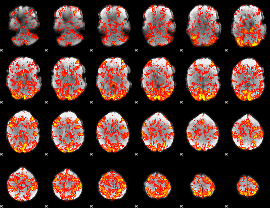 |
GLM Z-score result thresholded by 1.5
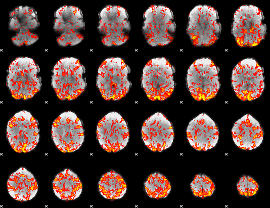 |
GLM Z-score result thresholded by 2
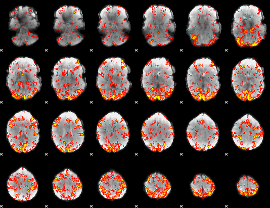 |
GLM Z-score result thresholded by 2.5
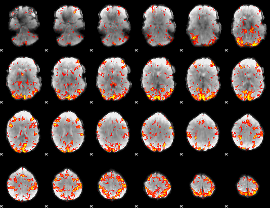 |
GLM Z-score result thresholded by 3
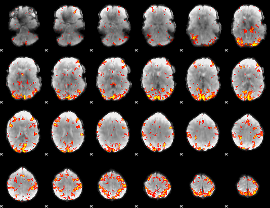 |
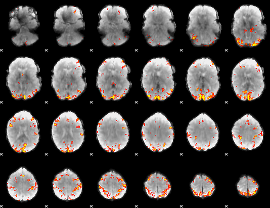
GLM Z-score result thresholded by 3.5
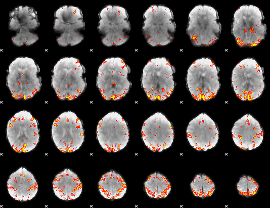 |
GLM Z-score result thresholded by 4
 |
Component:285; Jaccard:0.12898
 |
Component:285; Jaccard:0.16355
 |
Component:285; Jaccard:0.20272
 |
Component:285; Jaccard:0.24309
 |
Component:285; Jaccard:0.27645
 |
Component:285; Jaccard:0.29369
 |
Component:285; Jaccard:0.29375
 |
Correlation:0.36567
  |
Correlation:0.36567
  |
Correlation:0.36567
  |
Correlation:0.36567
  |
Correlation:0.36567
  |
Correlation:0.36567
  |
Correlation:0.36567
  |
Component:239; Jaccard:0.11785
 |
Component:189; Jaccard:0.13626
 |
Component:189; Jaccard:0.15861
 |
Component:189; Jaccard:0.17386
 |
Component:189; Jaccard:0.18395
 |
Component:189; Jaccard:0.18155
 |
Component:189; Jaccard:0.17128
 |
Correlation:0.037866
  |
Correlation:0.19258
  |
Correlation:0.19258
  |
Correlation:0.19258
  |
Correlation:0.19258
  |
Correlation:0.19258
  |
Correlation:0.19258
  |
Component:189; Jaccard:0.11453
 |
Component:239; Jaccard:0.13176
 |
Component:138; Jaccard:0.14663
 |
Component:138; Jaccard:0.15839
 |
Component:006; Jaccard:0.16347
 |
Component:006; Jaccard:0.16532
 |
Component:006; Jaccard:0.15793
 |
Correlation:0.19258
  |
Correlation:0.037866
  |
Correlation:0.25053
 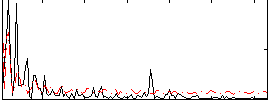 |
Correlation:0.25053
  |
Correlation:0.10624
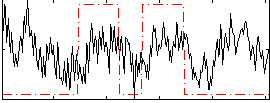 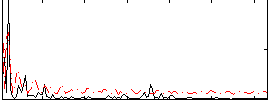 |
Correlation:0.10624
  |
Correlation:0.10624
  |
Component:138; Jaccard:0.11197
 |
Component:138; Jaccard:0.13062
 |
Component:239; Jaccard:0.14403
 |
Component:006; Jaccard:0.15499
 |
Component:138; Jaccard:0.16096
 |
Component:016; Jaccard:0.15749
 |
Component:016; Jaccard:0.14826
 |
Correlation:0.25053
  |
Correlation:0.25053
  |
Correlation:0.037866
  |
Correlation:0.10624
  |
Correlation:0.25053
  |
Correlation:0.27149
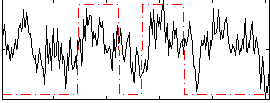  |
Correlation:0.27149
  |
Component:182; Jaccard:0.10885
 |
Component:182; Jaccard:0.12195
 |
Component:006; Jaccard:0.13752
 |
Component:239; Jaccard:0.1523
 |
Component:016; Jaccard:0.15693
 |
Component:307; Jaccard:0.15523
 |
Component:307; Jaccard:0.14703
 |
Correlation:0.11562
  |
Correlation:0.11562
  |
Correlation:0.10624
  |
Correlation:0.037866
  |
Correlation:0.27149
  |
Correlation:0.26315
  |
Correlation:0.26315
  |
Component:202; Jaccard:0.099027
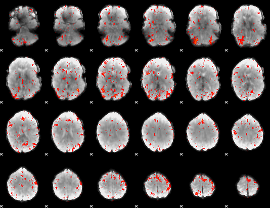 |
Component:006; Jaccard:0.11801
 |
Component:182; Jaccard:0.13397
 |
Component:016; Jaccard:0.14759
 |
Component:307; Jaccard:0.15482
 |
Component:138; Jaccard:0.15507
 |
Component:138; Jaccard:0.14457
 |
Correlation:0.2336
  |
Correlation:0.10624
  |
Correlation:0.11562
  |
Correlation:0.27149
  |
Correlation:0.26315
  |
Correlation:0.25053
  |
Correlation:0.25053
  |
Component:006; Jaccard:0.098467
 |
Component:016; Jaccard:0.11618
 |
Component:016; Jaccard:0.13391
 |
Component:307; Jaccard:0.14622
 |
Component:239; Jaccard:0.15382
 |
Component:239; Jaccard:0.14488
 |
Component:194; Jaccard:0.13728
 |
Correlation:0.10624
  |
Correlation:0.27149
  |
Correlation:0.27149
  |
Correlation:0.26315
  |
Correlation:0.037866
  |
Correlation:0.037866
  |
Correlation:0.20441
  |
Component:016; Jaccard:0.098462
 |
Component:307; Jaccard:0.11488
 |
Component:307; Jaccard:0.13188
 |
Component:182; Jaccard:0.14237
 |
Component:182; Jaccard:0.14335
 |
Component:194; Jaccard:0.14297
 |
Component:239; Jaccard:0.13006
 |
Correlation:0.27149
  |
Correlation:0.26315
  |
Correlation:0.26315
  |
Correlation:0.11562
  |
Correlation:0.11562
  |
Correlation:0.20441
  |
Correlation:0.037866
  |
Component:307; Jaccard:0.09743
 |
Component:202; Jaccard:0.11168
 |
Component:194; Jaccard:0.12375
 |
Component:194; Jaccard:0.13629
 |
Component:194; Jaccard:0.14163
 |
Component:182; Jaccard:0.13894
 |
Component:182; Jaccard:0.12601
 |
Correlation:0.26315
  |
Correlation:0.2336
  |
Correlation:0.20441
  |
Correlation:0.20441
  |
Correlation:0.20441
  |
Correlation:0.11562
  |
Correlation:0.11562
  |
Component:194; Jaccard:0.094911
 |
Component:194; Jaccard:0.10945
 |
Component:202; Jaccard:0.12272
 |
Component:288; Jaccard:0.12988
 |
Component:288; Jaccard:0.13401
 |
Component:288; Jaccard:0.13242
 |
Component:288; Jaccard:0.12386
 |
Correlation:0.20441
  |
Correlation:0.20441
  |
Correlation:0.2336
  |
Correlation:0.097327
  |
Correlation:0.097327
  |
Correlation:0.097327
  |
Correlation:0.097327
  |
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
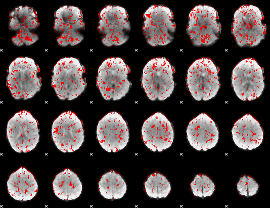 |
GLM binary result thresholded by 1.5
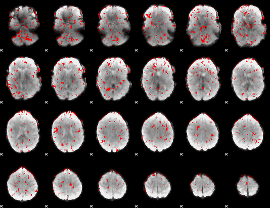 |
GLM binary result thresholded by 2
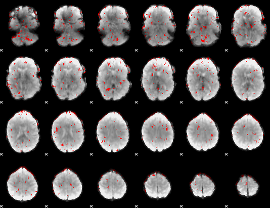 |
GLM binary result thresholded by 2.5
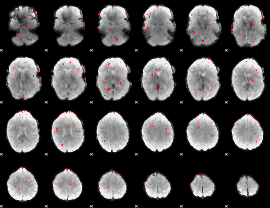 |
GLM binary result thresholded by 3
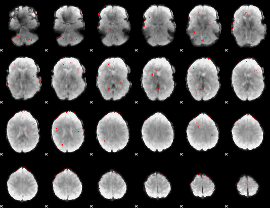 |
GLM binary result thresholded by 3.5
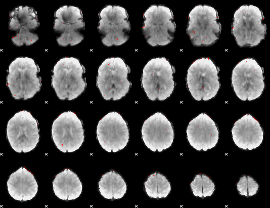 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
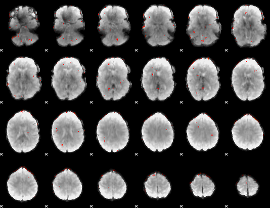 |
GLM Z-score result thresholded by 3.5
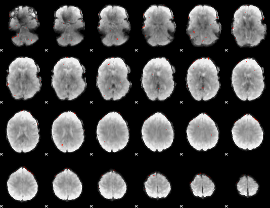 |
GLM Z-score result thresholded by 4
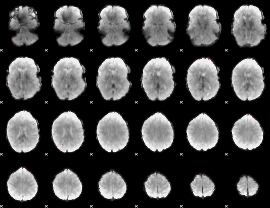 |
Component:267; Jaccard:0.14064
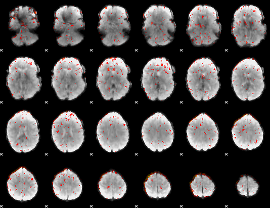 |
Component:267; Jaccard:0.15823
 |
Component:267; Jaccard:0.15879
 |
Component:267; Jaccard:0.13923
 |
Component:174; Jaccard:0.0975
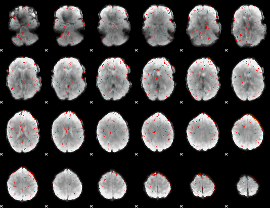 |
Component:174; Jaccard:0.06088
 |
Component:174; Jaccard:0.035377
 |
Correlation:0.47214
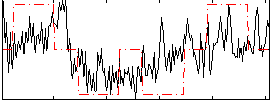  |
Correlation:0.47214
  |
Correlation:0.47214
  |
Correlation:0.47214
  |
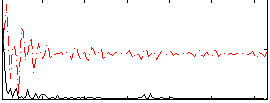
Correlation:0.48144
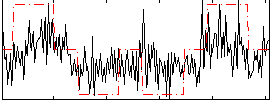 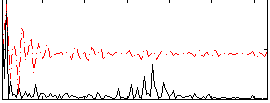 |
Correlation:0.48144
  |
Correlation:0.48144
  |
Component:210; Jaccard:0.11553
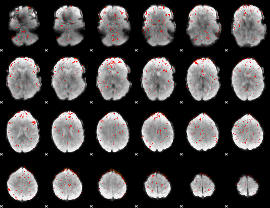 |
Component:174; Jaccard:0.12915
 |
Component:174; Jaccard:0.13626
 |
Component:174; Jaccard:0.12918
 |
Component:267; Jaccard:0.096657
 |
Component:267; Jaccard:0.056378
 |
Component:267; Jaccard:0.027534
 |
Correlation:0.39409
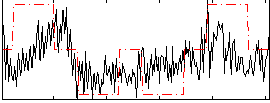 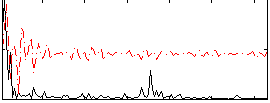 |
Correlation:0.48144
  |
Correlation:0.48144
  |
Correlation:0.48144
  |
Correlation:0.47214
  |
Correlation:0.47214
  |
Correlation:0.47214
  |
Component:174; Jaccard:0.1123
 |
Component:210; Jaccard:0.12814
 |
Component:210; Jaccard:0.12733
 |
Component:210; Jaccard:0.11348
 |
Component:210; Jaccard:0.082211
 |
Component:210; Jaccard:0.049591
 |
Component:210; Jaccard:0.025727
 |
Correlation:0.48144
  |
Correlation:0.39409
  |
Correlation:0.39409
  |
Correlation:0.39409
  |
Correlation:0.39409
  |
Correlation:0.39409
  |
Correlation:0.39409
  |
Component:319; Jaccard:0.1001
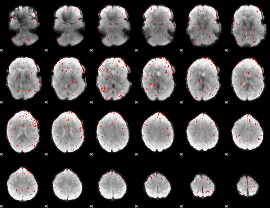 |
Component:319; Jaccard:0.11061
 |
Component:319; Jaccard:0.10969
 |
Component:093; Jaccard:0.096442
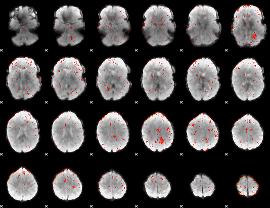 |
Component:093; Jaccard:0.068845
 |
Component:093; Jaccard:0.041429
 |
Component:093; Jaccard:0.020229
 |
Correlation:0.4672
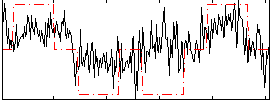 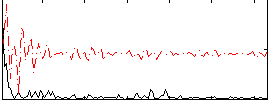 |
Correlation:0.4672
  |
Correlation:0.4672
  |
Correlation:0.38434
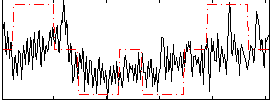  |
Correlation:0.38434
  |
Correlation:0.38434
  |
Correlation:0.38434
  |
Component:093; Jaccard:0.099563
 |
Component:093; Jaccard:0.11011
 |
Component:093; Jaccard:0.10944
 |
Component:319; Jaccard:0.09412
 |
Component:319; Jaccard:0.06618
 |
Component:319; Jaccard:0.039045
 |
Component:372; Jaccard:0.020184
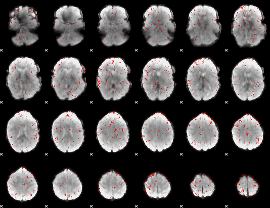 |
Correlation:0.38434
  |
Correlation:0.38434
  |
Correlation:0.38434
  |
Correlation:0.4672
  |
Correlation:0.4672
  |
Correlation:0.4672
  |
Correlation:0.32366
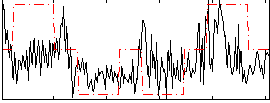 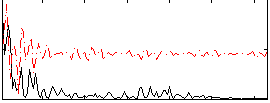 |
Component:372; Jaccard:0.096256
 |
Component:372; Jaccard:0.10459
 |
Component:372; Jaccard:0.10351
 |
Component:372; Jaccard:0.088296
 |
Component:372; Jaccard:0.061223
 |
Component:372; Jaccard:0.037741
 |
Component:319; Jaccard:0.019044
 |
Correlation:0.32366
  |
Correlation:0.32366
  |
Correlation:0.32366
  |
Correlation:0.32366
  |
Correlation:0.32366
  |
Correlation:0.32366
  |
Correlation:0.4672
  |
Component:001; Jaccard:0.091269
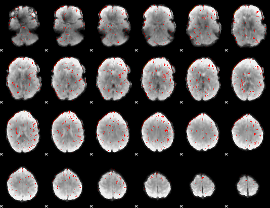 |
Component:001; Jaccard:0.094564
 |
Component:001; Jaccard:0.088628
 |
Component:001; Jaccard:0.073918
 |
Component:001; Jaccard:0.050322
 |
Component:177; Jaccard:0.032307
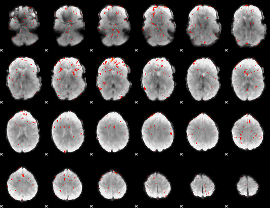 |
Component:177; Jaccard:0.017321
 |
Correlation:0.35383
  |
Correlation:0.35383
  |
Correlation:0.35383
  |
Correlation:0.35383
  |
Correlation:0.35383
  |
Correlation:0.33798
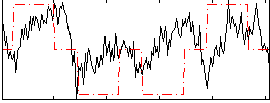 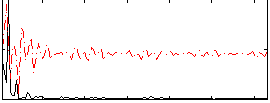 |
Correlation:0.33798
  |
Component:206; Jaccard:0.074292
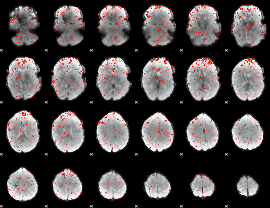 |
Component:274; Jaccard:0.078241
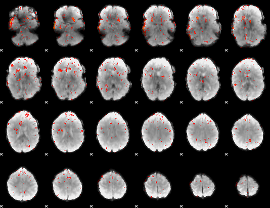 |
Component:274; Jaccard:0.075386
 |
Component:274; Jaccard:0.064704
 |
Component:177; Jaccard:0.047681
 |
Component:309; Jaccard:0.030098
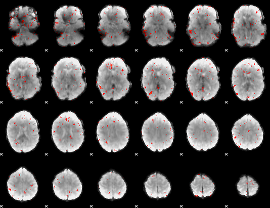 |
Component:309; Jaccard:0.015579
 |
Correlation:0.19762
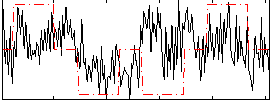 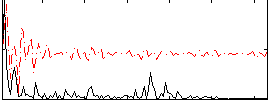 |
Correlation:0.53718
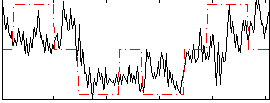  |
Correlation:0.53718
  |
Correlation:0.53718
  |
Correlation:0.33798
  |
Correlation:0.40317
  |
Correlation:0.40317
  |
Component:274; Jaccard:0.074106
 |
Component:309; Jaccard:0.075376
 |
Component:309; Jaccard:0.072076
 |
Component:309; Jaccard:0.060658
 |
Component:274; Jaccard:0.047472
 |
Component:274; Jaccard:0.02987
 |
Component:274; Jaccard:0.014974
 |
Correlation:0.53718
  |
Correlation:0.40317
  |
Correlation:0.40317
  |
Correlation:0.40317
  |
Correlation:0.53718
  |
Correlation:0.53718
  |
Correlation:0.53718
  |
Component:396; Jaccard:0.072839
 |
Component:396; Jaccard:0.072215
 |
Component:177; Jaccard:0.068739
 |
Component:177; Jaccard:0.060656
 |
Component:309; Jaccard:0.045131
 |
Component:001; Jaccard:0.029215
 |
Component:001; Jaccard:0.01429
 |
Correlation:0.28955
  |
Correlation:0.28955
  |
Correlation:0.33798
  |
Correlation:0.33798
  |
Correlation:0.40317
  |
Correlation:0.35383
  |
Correlation:0.35383
  |
Cope: 04:neg_PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
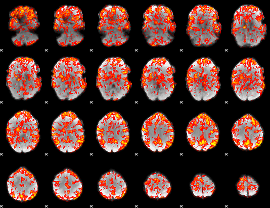 |
GLM Z-score result thresholded by 1.5
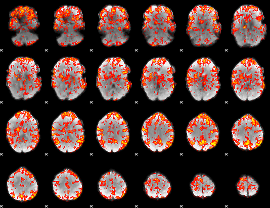 |
GLM Z-score result thresholded by 2
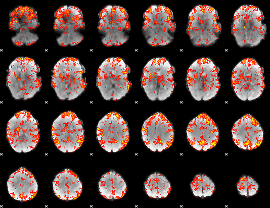 |
GLM Z-score result thresholded by 2.5
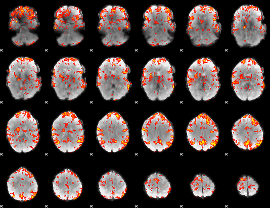 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
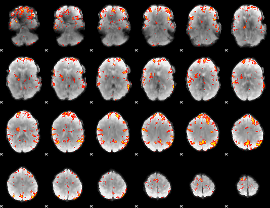 |
GLM Z-score result thresholded by 4
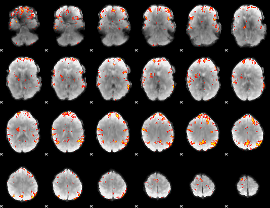 |
Component:322; Jaccard:0.12407
 |
Component:322; Jaccard:0.14968
 |
Component:322; Jaccard:0.1796
 |
Component:322; Jaccard:0.20883
 |
Component:322; Jaccard:0.23062
 |
Component:322; Jaccard:0.23735
 |
Component:322; Jaccard:0.22681
 |
Correlation:0.35355
  |
Correlation:0.35355
  |
Correlation:0.35355
  |
Correlation:0.35355
  |
Correlation:0.35355
  |
Correlation:0.35355
  |
Correlation:0.35355
  |
Component:377; Jaccard:0.10937
 |
Component:306; Jaccard:0.12525
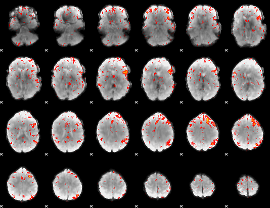 |
Component:306; Jaccard:0.14304
 |
Component:306; Jaccard:0.15828
 |
Component:306; Jaccard:0.16924
 |
Component:306; Jaccard:0.17098
 |
Component:306; Jaccard:0.16177
 |
Correlation:0.29843
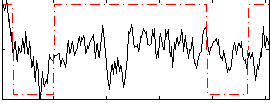 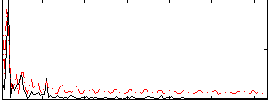 |
Correlation:0.069208
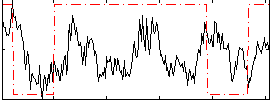  |
Correlation:0.069208
  |
Correlation:0.069208
  |
Correlation:0.069208
  |
Correlation:0.069208
  |
Correlation:0.069208
  |
Component:326; Jaccard:0.10769
 |
Component:326; Jaccard:0.1203
 |
Component:326; Jaccard:0.13116
 |
Component:308; Jaccard:0.14507
 |
Component:308; Jaccard:0.15582
 |
Component:308; Jaccard:0.15948
 |
Component:280; Jaccard:0.15113
 |
Correlation:0.019711
  |
Correlation:0.019711
  |
Correlation:0.019711
  |
Correlation:0.30367
  |
Correlation:0.30367
  |
Correlation:0.30367
  |
Correlation:0.13477
  |
Component:306; Jaccard:0.10749
 |
Component:377; Jaccard:0.11972
 |
Component:308; Jaccard:0.12924
 |
Component:280; Jaccard:0.14034
 |
Component:283; Jaccard:0.15009
 |
Component:280; Jaccard:0.15414
 |
Component:308; Jaccard:0.14999
 |
Correlation:0.069208
  |
Correlation:0.29843
  |
Correlation:0.30367
  |
Correlation:0.13477
  |
Correlation:0.20134
  |
Correlation:0.13477
  |
Correlation:0.30367
  |
Component:218; Jaccard:0.10033
 |
Component:218; Jaccard:0.11482
 |
Component:377; Jaccard:0.12799
 |
Component:283; Jaccard:0.13972
 |
Component:280; Jaccard:0.14981
 |
Component:283; Jaccard:0.15413
 |
Component:283; Jaccard:0.14609
 |
Correlation:0.13903
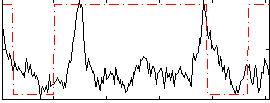 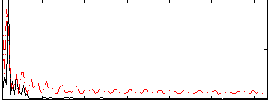 |
Correlation:0.13903
  |
Correlation:0.29843
  |
Correlation:0.20134
  |
Correlation:0.13477
  |
Correlation:0.20134
  |
Correlation:0.20134
  |
Component:236; Jaccard:0.095447
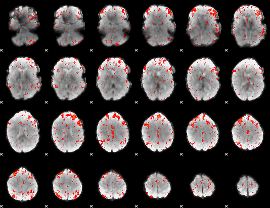 |
Component:308; Jaccard:0.112
 |
Component:218; Jaccard:0.12745
 |
Component:326; Jaccard:0.13842
 |
Component:218; Jaccard:0.14248
 |
Component:218; Jaccard:0.13904
 |
Component:167; Jaccard:0.12964
 |
Correlation:0.10692
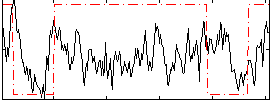  |
Correlation:0.30367
  |
Correlation:0.13903
  |
Correlation:0.019711
  |
Correlation:0.13903
  |
Correlation:0.13903
  |
Correlation:0.062892
  |
Component:280; Jaccard:0.095399
 |
Component:280; Jaccard:0.11014
 |
Component:280; Jaccard:0.12589
 |
Component:218; Jaccard:0.13761
 |
Component:326; Jaccard:0.14035
 |
Component:167; Jaccard:0.13713
 |
Component:218; Jaccard:0.12668
 |
Correlation:0.13477
  |
Correlation:0.13477
  |
Correlation:0.13477
  |
Correlation:0.13903
  |
Correlation:0.019711
  |
Correlation:0.062892
  |
Correlation:0.13903
  |
Component:308; Jaccard:0.094933
 |
Component:283; Jaccard:0.10854
 |
Component:283; Jaccard:0.12474
 |
Component:377; Jaccard:0.13231
 |
Component:167; Jaccard:0.13689
 |
Component:326; Jaccard:0.13619
 |
Component:326; Jaccard:0.1254
 |
Correlation:0.30367
  |
Correlation:0.20134
  |
Correlation:0.20134
  |
Correlation:0.29843
  |
Correlation:0.062892
  |
Correlation:0.019711
  |
Correlation:0.019711
  |
Component:283; Jaccard:0.094601
 |
Component:167; Jaccard:0.10763
 |
Component:167; Jaccard:0.11952
 |
Component:167; Jaccard:0.13056
 |
Component:377; Jaccard:0.12661
 |
Component:230; Jaccard:0.12329
 |
Component:268; Jaccard:0.1131
 |
Correlation:0.20134
  |
Correlation:0.062892
  |
Correlation:0.062892
  |
Correlation:0.062892
  |
Correlation:0.29843
  |
Correlation:0.059159
  |
Correlation:-0.11248
  |
Component:167; Jaccard:0.094567
 |
Component:383; Jaccard:0.10112
 |
Component:230; Jaccard:0.10974
 |
Component:230; Jaccard:0.1189
 |
Component:230; Jaccard:0.12399
 |
Component:268; Jaccard:0.11758
 |
Component:230; Jaccard:0.11138
 |
Correlation:0.062892
  |
Correlation:0.080881
  |
Correlation:0.059159
  |
Correlation:0.059159
  |
Correlation:0.059159
  |
Correlation:-0.11248
  |
Correlation:0.059159
  |
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
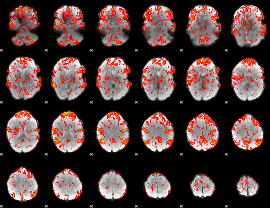 |
GLM Z-score result thresholded by 1.5
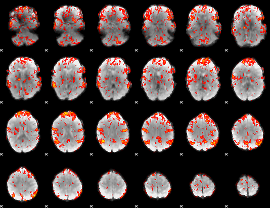 |
GLM Z-score result thresholded by 2
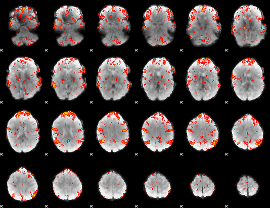 |
GLM Z-score result thresholded by 2.5
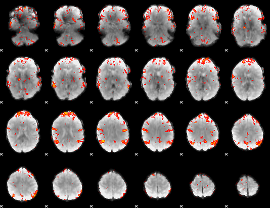 |
GLM Z-score result thresholded by 3
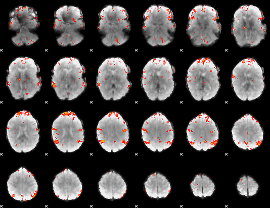 |
GLM Z-score result thresholded by 3.5
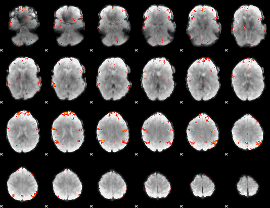 |
GLM Z-score result thresholded by 4
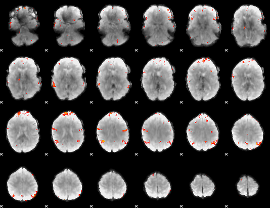 |
Component:167; Jaccard:0.13241
 |
Component:167; Jaccard:0.15483
 |
Component:167; Jaccard:0.17301
 |
Component:167; Jaccard:0.17731
 |
Component:167; Jaccard:0.16431
 |
Component:167; Jaccard:0.13433
 |
Component:167; Jaccard:0.097375
 |
Correlation:0.052551
  |
Correlation:0.052551
  |
Correlation:0.052551
  |
Correlation:0.052551
  |
Correlation:0.052551
  |
Correlation:0.052551
  |
Correlation:0.052551
  |
Component:205; Jaccard:0.11956
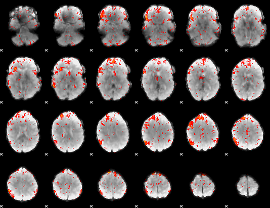 |
Component:205; Jaccard:0.12954
 |
Component:230; Jaccard:0.13836
 |
Component:268; Jaccard:0.14562
 |
Component:268; Jaccard:0.14394
 |
Component:268; Jaccard:0.12521
 |
Component:268; Jaccard:0.09402
 |
Correlation:0.10136
  |
Correlation:0.10136
  |
Correlation:0.042074
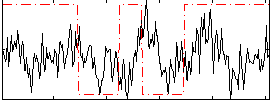 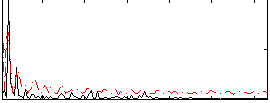 |
Correlation:0.13607
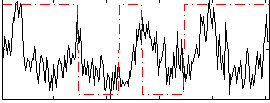  |
Correlation:0.13607
  |
Correlation:0.13607
  |
Correlation:0.13607
  |
Component:218; Jaccard:0.1128
 |
Component:230; Jaccard:0.12778
 |
Component:268; Jaccard:0.13457
 |
Component:230; Jaccard:0.14024
 |
Component:230; Jaccard:0.13003
 |
Component:230; Jaccard:0.10561
 |
Component:230; Jaccard:0.076202
 |
Correlation:-0.0019061
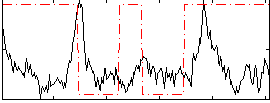 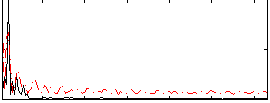 |
Correlation:0.042074
  |
Correlation:0.13607
  |
Correlation:0.042074
  |
Correlation:0.042074
  |
Correlation:0.042074
  |
Correlation:0.042074
  |
Component:230; Jaccard:0.11194
 |
Component:218; Jaccard:0.12487
 |
Component:205; Jaccard:0.13317
 |
Component:308; Jaccard:0.13322
 |
Component:308; Jaccard:0.119
 |
Component:218; Jaccard:0.096603
 |
Component:218; Jaccard:0.070356
 |
Correlation:0.042074
  |
Correlation:-0.0019061
  |
Correlation:0.10136
  |
Correlation:-0.0023097
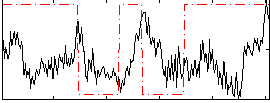 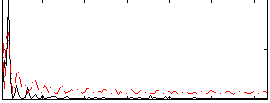 |
Correlation:-0.0023097
  |
Correlation:-0.0019061
  |
Correlation:-0.0019061
  |
Component:308; Jaccard:0.10768
 |
Component:308; Jaccard:0.12212
 |
Component:218; Jaccard:0.13283
 |
Component:218; Jaccard:0.13066
 |
Component:218; Jaccard:0.11834
 |
Component:308; Jaccard:0.095574
 |
Component:308; Jaccard:0.066464
 |
Correlation:-0.0023097
  |
Correlation:-0.0023097
  |
Correlation:-0.0019061
  |
Correlation:-0.0019061
  |
Correlation:-0.0019061
  |
Correlation:-0.0023097
  |
Correlation:-0.0023097
  |
Component:322; Jaccard:0.10673
 |
Component:268; Jaccard:0.11735
 |
Component:308; Jaccard:0.13204
 |
Component:205; Jaccard:0.12608
 |
Component:205; Jaccard:0.10515
 |
Component:283; Jaccard:0.084545
 |
Component:283; Jaccard:0.059118
 |
Correlation:0.1073
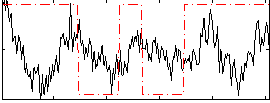  |
Correlation:0.13607
  |
Correlation:-0.0023097
  |
Correlation:0.10136
  |
Correlation:0.10136
  |
Correlation:-0.0023511
  |
Correlation:-0.0023511
  |
Component:268; Jaccard:0.10095
 |
Component:322; Jaccard:0.11576
 |
Component:322; Jaccard:0.11935
 |
Component:283; Jaccard:0.11656
 |
Component:283; Jaccard:0.10383
 |
Component:205; Jaccard:0.079594
 |
Component:280; Jaccard:0.054869
 |
Correlation:0.13607
  |
Correlation:0.1073
  |
Correlation:0.1073
  |
Correlation:-0.0023511
  |
Correlation:-0.0023511
  |
Correlation:0.10136
  |
Correlation:-0.055551
  |
Component:326; Jaccard:0.10043
 |
Component:283; Jaccard:0.10811
 |
Component:283; Jaccard:0.11628
 |
Component:322; Jaccard:0.11486
 |
Component:322; Jaccard:0.099297
 |
Component:322; Jaccard:0.078184
 |
Component:326; Jaccard:0.054099
 |
Correlation:-0.055497
  |
Correlation:-0.0023511
  |
Correlation:-0.0023511
  |
Correlation:0.1073
  |
Correlation:0.1073
  |
Correlation:0.1073
  |
Correlation:-0.055497
  |
Component:283; Jaccard:0.097696
 |
Component:326; Jaccard:0.10799
 |
Component:326; Jaccard:0.11174
 |
Component:326; Jaccard:0.10686
 |
Component:326; Jaccard:0.094726
 |
Component:326; Jaccard:0.076629
 |
Component:322; Jaccard:0.053529
 |
Correlation:-0.0023511
  |
Correlation:-0.055497
  |
Correlation:-0.055497
  |
Correlation:-0.055497
  |
Correlation:-0.055497
  |
Correlation:-0.055497
  |
Correlation:0.1073
  |
Component:198; Jaccard:0.095831
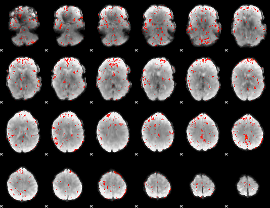 |
Component:198; Jaccard:0.10176
 |
Component:198; Jaccard:0.10073
 |
Component:306; Jaccard:0.096511
 |
Component:280; Jaccard:0.087688
 |
Component:280; Jaccard:0.074614
 |
Component:306; Jaccard:0.052382
 |
Correlation:0.22557
  |
Correlation:0.22557
  |
Correlation:0.22557
  |
Correlation:0.0099454
  |
Correlation:-0.055551
  |
Correlation:-0.055551
  |
Correlation:0.0099454
  |
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
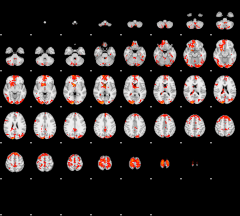 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
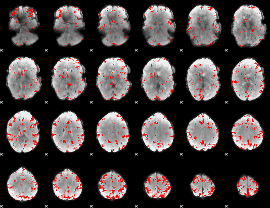 |
GLM binary result thresholded by 3.5
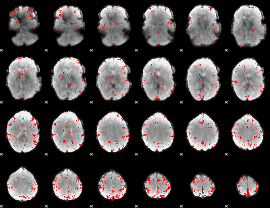 |
GLM binary result thresholded by 4
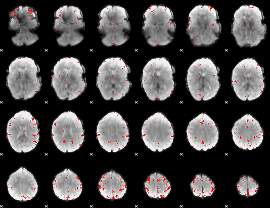 |
GLM Z-score result thresholded by 1
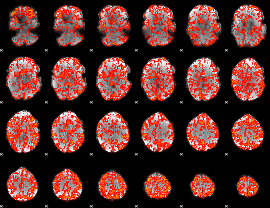 |
GLM Z-score result thresholded by 1.5
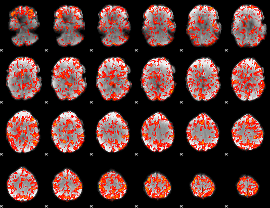 |
GLM Z-score result thresholded by 2
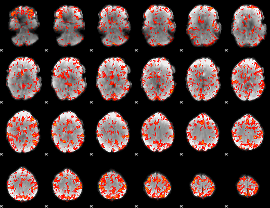 |
GLM Z-score result thresholded by 2.5
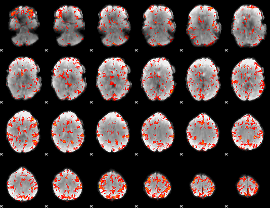 |
GLM Z-score result thresholded by 3
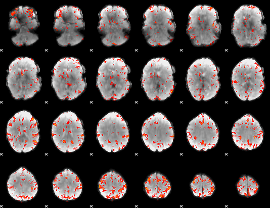 |
GLM Z-score result thresholded by 3.5
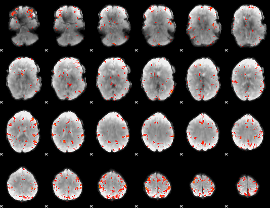 |
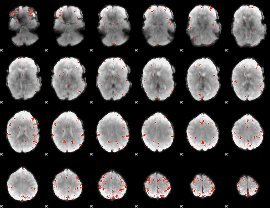
GLM Z-score result thresholded by 4
 |
Component:377; Jaccard:0.096195
 |
Component:189; Jaccard:0.10942
 |
Component:189; Jaccard:0.1288
 |
Component:189; Jaccard:0.14714
 |
Component:189; Jaccard:0.15344
 |
Component:189; Jaccard:0.14259
 |
Component:189; Jaccard:0.10911
 |
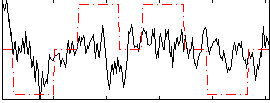
Correlation:0.27031
  |
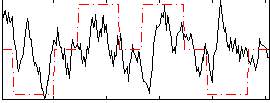
Correlation:0.19296
  |
Correlation:0.19296
  |
Correlation:0.19296
  |
Correlation:0.19296
  |
Correlation:0.19296
  |
Correlation:0.19296
  |
Component:189; Jaccard:0.09215
 |
Component:377; Jaccard:0.10749
 |
Component:377; Jaccard:0.11582
 |
Component:383; Jaccard:0.12287
 |
Component:383; Jaccard:0.12109
 |
Component:383; Jaccard:0.10846
 |
Component:383; Jaccard:0.086675
 |
Correlation:0.19296
  |
Correlation:0.27031
  |
Correlation:0.27031
  |
Correlation:0.14554
  |
Correlation:0.14554
  |
Correlation:0.14554
  |
Correlation:0.14554
  |
Component:383; Jaccard:0.090347
 |
Component:383; Jaccard:0.1032
 |
Component:383; Jaccard:0.11573
 |
Component:297; Jaccard:0.12057
 |
Component:297; Jaccard:0.11948
 |
Component:297; Jaccard:0.10546
 |
Component:076; Jaccard:0.083501
 |
Correlation:0.14554
  |
Correlation:0.14554
  |
Correlation:0.14554
  |
Correlation:0.26503
  |
Correlation:0.26503
  |
Correlation:0.26503
  |
Correlation:0.45316
  |
Component:182; Jaccard:0.090061
 |
Component:297; Jaccard:0.10098
 |
Component:297; Jaccard:0.11328
 |
Component:377; Jaccard:0.1172
 |
Component:076; Jaccard:0.11256
 |
Component:076; Jaccard:0.10219
 |
Component:297; Jaccard:0.078662
 |
Correlation:0.066787
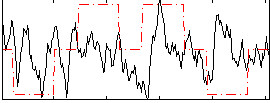  |
Correlation:0.26503
  |
Correlation:0.26503
  |
Correlation:0.27031
  |
Correlation:0.45316
  |
Correlation:0.45316
  |
Correlation:0.26503
  |
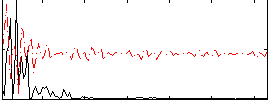
Component:173; Jaccard:0.08843
 |
Component:182; Jaccard:0.10052
 |
Component:182; Jaccard:0.10736
 |
Component:182; Jaccard:0.10945
 |
Component:377; Jaccard:0.10674
 |
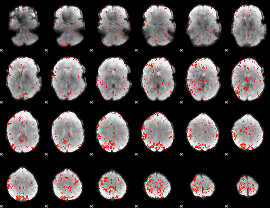
Component:012; Jaccard:0.091994
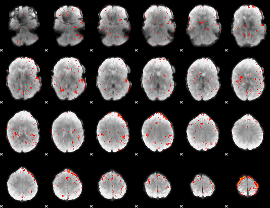 |
Component:012; Jaccard:0.076375
 |
Correlation:0.20525
  |
Correlation:0.066787
  |
Correlation:0.066787
  |
Correlation:0.066787
  |
Correlation:0.27031
  |
Correlation:0.24288
  |
Correlation:0.24288
  |
Component:297; Jaccard:0.087516
 |
Component:173; Jaccard:0.095772
 |
Component:173; Jaccard:0.10132
 |
Component:076; Jaccard:0.10662
 |
Component:182; Jaccard:0.10065
 |
Component:006; Jaccard:0.087895
 |
Component:006; Jaccard:0.068202
 |
Correlation:0.26503
  |
Correlation:0.20525
  |
Correlation:0.20525
  |
Correlation:0.45316
  |
Correlation:0.066787
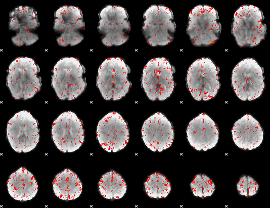
  |
Correlation:-0.044537
  |
Correlation:-0.044537
  |
Component:252; Jaccard:0.08406
 |
Component:241; Jaccard:0.090396
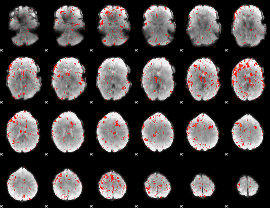 |
Component:241; Jaccard:0.097307
 |
Component:173; Jaccard:0.10153
 |
Component:151; Jaccard:0.096145
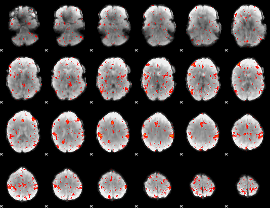 |
Component:377; Jaccard:0.085053
 |
Component:137; Jaccard:0.064192
 |
Correlation:0.15092
  |
Correlation:0.15972
  |
Correlation:0.15972
  |
Correlation:0.20525
  |
Correlation:-0.01768
  |
Correlation:0.27031
  |
Correlation:0.43335
  |
Component:322; Jaccard:0.082907
 |
Component:252; Jaccard:0.090251
 |
Component:151; Jaccard:0.095891
 |
Component:151; Jaccard:0.099562
 |
Component:006; Jaccard:0.095599
 |
Component:151; Jaccard:0.084133
 |
Component:037; Jaccard:0.063131
 |
Correlation:0.14482
  |
Correlation:0.15092
  |
Correlation:-0.01768
  |
Correlation:-0.01768
  |
Correlation:-0.044537
  |
Correlation:-0.01768
  |
Correlation:0.38713
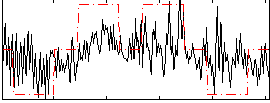 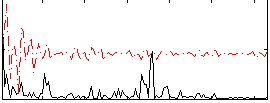 |
Component:241; Jaccard:0.08205
 |
Component:322; Jaccard:0.089944
 |
Component:076; Jaccard:0.095373
 |
Component:241; Jaccard:0.097908
 |
Component:012; Jaccard:0.09428
 |
Component:182; Jaccard:0.083576
 |
Component:376; Jaccard:0.062492
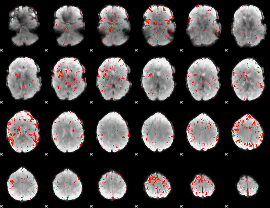 |
Correlation:0.15972
  |
Correlation:0.14482
  |
Correlation:0.45316
  |
Correlation:0.15972
  |
Correlation:0.24288
  |
Correlation:0.066787
  |
Correlation:0.043794
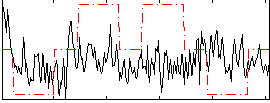  |
Component:239; Jaccard:0.081409
 |
Component:151; Jaccard:0.089006
 |
Component:322; Jaccard:0.094785
 |
Component:376; Jaccard:0.096512
 |
Component:376; Jaccard:0.092469
 |
Component:037; Jaccard:0.081865
 |
Component:151; Jaccard:0.061847
 |
Correlation:-0.0066045
  |
Correlation:-0.01768
  |
Correlation:0.14482
  |
Correlation:0.043794
  |
Correlation:0.043794
  |
Correlation:0.38713
  |
Correlation:-0.01768
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































