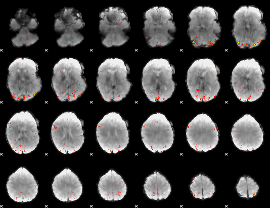
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:389; Jaccard:0.13311
 |
Component:389; Jaccard:0.17302
 |
Component:389; Jaccard:0.21434
 |
Component:389; Jaccard:0.24039
 |
Component:389; Jaccard:0.23554
 |
Component:389; Jaccard:0.19938
 |
Component:389; Jaccard:0.14728
 |
Correlation:0.27747
  |
Correlation:0.27747
  |
Correlation:0.27747
  |
Correlation:0.27747
  |
Correlation:0.27747
  |
Correlation:0.27747
  |
Correlation:0.27747
  |
Component:344; Jaccard:0.11561
 |
Component:344; Jaccard:0.13691
 |
Component:344; Jaccard:0.15458
 |
Component:344; Jaccard:0.15764
 |
Component:344; Jaccard:0.13618
 |
Component:393; Jaccard:0.10938
 |
Component:393; Jaccard:0.079179
 |
Correlation:-0.11499
  |
Correlation:-0.11499
  |
Correlation:-0.11499
  |
Correlation:-0.11499
  |
Correlation:-0.11499
  |
Correlation:-0.0016493
  |
Correlation:-0.0016493
  |
Component:166; Jaccard:0.10388
 |
Component:393; Jaccard:0.11731
 |
Component:393; Jaccard:0.13294
 |
Component:393; Jaccard:0.14204
 |
Component:393; Jaccard:0.13598
 |
Component:344; Jaccard:0.10028
 |
Component:344; Jaccard:0.065593
 |
Correlation:-0.20711
  |
Correlation:-0.0016493
  |
Correlation:-0.0016493
  |
Correlation:-0.0016493
  |
Correlation:-0.0016493
  |
Correlation:-0.11499
  |
Correlation:-0.11499
  |
Component:393; Jaccard:0.10009
 |
Component:166; Jaccard:0.11027
 |
Component:149; Jaccard:0.11902
 |
Component:149; Jaccard:0.12482
 |
Component:149; Jaccard:0.11236
 |
Component:042; Jaccard:0.085335
 |
Component:042; Jaccard:0.06261
 |
Correlation:-0.0016493
  |
Correlation:-0.20711
  |
Correlation:0.029448
  |
Correlation:0.029448
  |
Correlation:0.029448
  |
Correlation:0.54172
  |
Correlation:0.54172
  |
Component:238; Jaccard:0.097687
 |
Component:238; Jaccard:0.10837
 |
Component:238; Jaccard:0.11521
 |
Component:238; Jaccard:0.10921
 |
Component:042; Jaccard:0.099456
 |
Component:149; Jaccard:0.084858
 |
Component:149; Jaccard:0.055939
 |
Correlation:0.021129
  |
Correlation:0.021129
  |
Correlation:0.021129
  |
Correlation:0.021129
  |
Correlation:0.54172
  |
Correlation:0.029448
  |
Correlation:0.029448
  |
Component:242; Jaccard:0.092057
 |
Component:149; Jaccard:0.10112
 |
Component:166; Jaccard:0.11143
 |
Component:151; Jaccard:0.10874
 |
Component:151; Jaccard:0.098076
 |
Component:151; Jaccard:0.075981
 |
Component:151; Jaccard:0.051783
 |
Correlation:-0.080178
  |
Correlation:0.029448
  |
Correlation:-0.20711
  |
Correlation:-0.12844
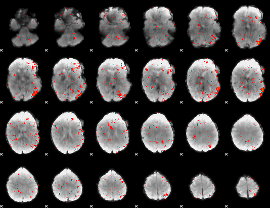
  |
Correlation:-0.12844
  |
Correlation:-0.12844
  |
Correlation:-0.12844
  |
Component:384; Jaccard:0.091087
 |
Component:384; Jaccard:0.096886
 |
Component:151; Jaccard:0.10646
 |
Component:042; Jaccard:0.1037
 |
Component:238; Jaccard:0.093541
 |
Component:238; Jaccard:0.069247
 |
Component:238; Jaccard:0.046599
 |
Correlation:-0.069225
  |
Correlation:-0.069225
  |
Correlation:-0.12844
  |
Correlation:0.54172
  |
Correlation:0.021129
  |
Correlation:0.021129
  |
Correlation:0.021129
  |
Component:056; Jaccard:0.08822
 |
Component:151; Jaccard:0.096345
 |
Component:042; Jaccard:0.099117
 |
Component:166; Jaccard:0.10244
 |
Component:166; Jaccard:0.079102
 |
Component:365; Jaccard:0.056129
 |
Component:365; Jaccard:0.042418
 |
Correlation:0.077593
  |
Correlation:-0.12844
  |
Correlation:0.54172
  |
Correlation:-0.20711
  |
Correlation:-0.20711
  |
Correlation:0.345
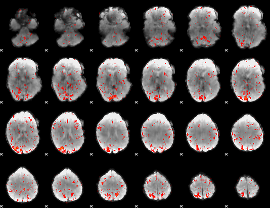
  |
Correlation:0.345
  |
Component:163; Jaccard:0.086571
 |
Component:056; Jaccard:0.095701
 |
Component:056; Jaccard:0.097695
 |
Component:163; Jaccard:0.09124
 |
Component:163; Jaccard:0.075752
 |
Component:111; Jaccard:0.053627
 |
Component:273; Jaccard:0.036194
 |
Correlation:0.081975
  |
Correlation:0.077593
  |
Correlation:0.077593
  |
Correlation:0.081975
  |
Correlation:0.081975
  |
Correlation:-0.13696
  |
Correlation:-0.020456
  |
Component:172; Jaccard:0.084335
 |
Component:242; Jaccard:0.095542
 |
Component:384; Jaccard:0.097564
 |
Component:056; Jaccard:0.090277
 |
Component:111; Jaccard:0.072847
 |
Component:163; Jaccard:0.053233
 |
Component:111; Jaccard:0.033744
 |
Correlation:0.017618
  |
Correlation:-0.080178
  |
Correlation:-0.069225
  |
Correlation:0.077593
  |
Correlation:-0.13696
  |
Correlation:0.081975
  |
Correlation:-0.13696
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
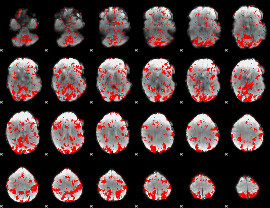 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
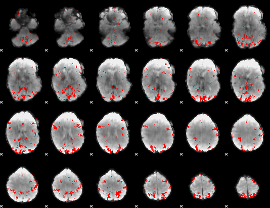 |
GLM binary result thresholded by 3.5
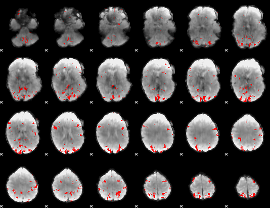 |
GLM binary result thresholded by 4
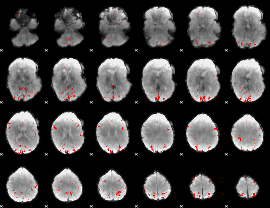 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
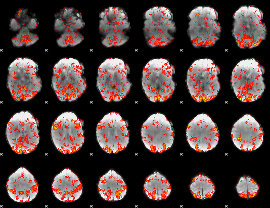 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
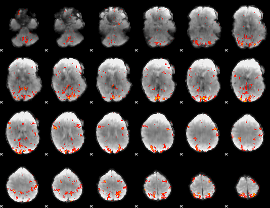 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:344; Jaccard:0.13333
 |
Component:344; Jaccard:0.16896
 |
Component:344; Jaccard:0.20387
 |
Component:344; Jaccard:0.23133
 |
Component:344; Jaccard:0.23493
 |
Component:344; Jaccard:0.20781
 |
Component:344; Jaccard:0.16365
 |
Correlation:0.3354
  |
Correlation:0.3354
  |
Correlation:0.3354
  |
Correlation:0.3354
  |
Correlation:0.3354
  |
Correlation:0.3354
  |
Correlation:0.3354
  |
Component:166; Jaccard:0.11557
 |
Component:389; Jaccard:0.13625
 |
Component:389; Jaccard:0.15814
 |
Component:389; Jaccard:0.1716
 |
Component:151; Jaccard:0.17591
 |
Component:151; Jaccard:0.16607
 |
Component:151; Jaccard:0.14271
 |
Correlation:0.29223
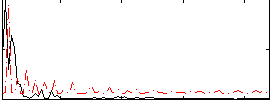
  |
Correlation:-0.093293
  |
Correlation:-0.093293
  |
Correlation:-0.093293
  |
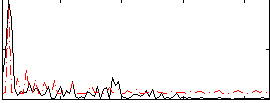
Correlation:0.31106
  |
Correlation:0.31106
  |
Correlation:0.31106
  |
Component:389; Jaccard:0.11367
 |
Component:166; Jaccard:0.12922
 |
Component:151; Jaccard:0.14918
 |
Component:151; Jaccard:0.16746
 |
Component:389; Jaccard:0.16452
 |
Component:389; Jaccard:0.14888
 |
Component:389; Jaccard:0.11572
 |
Correlation:-0.093293
  |
Correlation:0.29223
  |
Correlation:0.31106
  |
Correlation:0.31106
  |
Correlation:-0.093293
  |
Correlation:-0.093293
  |
Correlation:-0.093293
  |
Component:242; Jaccard:0.1044
 |
Component:151; Jaccard:0.12515
 |
Component:166; Jaccard:0.14016
 |
Component:393; Jaccard:0.14474
 |
Component:393; Jaccard:0.1386
 |
Component:393; Jaccard:0.12799
 |
Component:393; Jaccard:0.10207
 |
Correlation:0.025102
  |
Correlation:0.31106
  |
Correlation:0.29223
  |
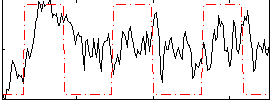
Correlation:0.196
  |
Correlation:0.196
  |
Correlation:0.196
  |
Correlation:0.196
  |
Component:111; Jaccard:0.10311
 |
Component:393; Jaccard:0.11972
 |
Component:393; Jaccard:0.13638
 |
Component:166; Jaccard:0.14095
 |
Component:166; Jaccard:0.12859
 |
Component:111; Jaccard:0.1088
 |
Component:111; Jaccard:0.083674
 |
Correlation:0.31195
  |
Correlation:0.196
  |
Correlation:0.196
  |
Correlation:0.29223
  |
Correlation:0.29223
  |
Correlation:0.31195
  |
Correlation:0.31195
  |
Component:393; Jaccard:0.10236
 |
Component:111; Jaccard:0.1165
 |
Component:111; Jaccard:0.12728
 |
Component:111; Jaccard:0.13048
 |
Component:111; Jaccard:0.1215
 |
Component:149; Jaccard:0.10755
 |
Component:149; Jaccard:0.082963
 |
Correlation:0.196
  |
Correlation:0.31195
  |
Correlation:0.31195
  |
Correlation:0.31195
  |
Correlation:0.31195
  |
Correlation:0.05548
  |
Correlation:0.05548
  |
Component:384; Jaccard:0.10083
 |
Component:242; Jaccard:0.11207
 |
Component:238; Jaccard:0.12096
 |
Component:238; Jaccard:0.12372
 |
Component:149; Jaccard:0.11912
 |
Component:166; Jaccard:0.10347
 |
Component:166; Jaccard:0.076704
 |
Correlation:0.15309
  |
Correlation:0.025102
  |
Correlation:0.1643
  |
Correlation:0.1643
  |
Correlation:0.05548
  |
Correlation:0.29223
  |
Correlation:0.29223
  |
Component:151; Jaccard:0.10079
 |
Component:238; Jaccard:0.11197
 |
Component:384; Jaccard:0.11611
 |
Component:149; Jaccard:0.11966
 |
Component:238; Jaccard:0.11591
 |
Component:238; Jaccard:0.097897
 |
Component:238; Jaccard:0.076103
 |
Correlation:0.31106
  |
Correlation:0.1643
  |
Correlation:0.15309
  |
Correlation:0.05548
  |
Correlation:0.1643
  |
Correlation:0.1643
  |
Correlation:0.1643
  |
Component:238; Jaccard:0.098499
 |
Component:384; Jaccard:0.10948
 |
Component:242; Jaccard:0.11476
 |
Component:384; Jaccard:0.11289
 |
Component:163; Jaccard:0.10294
 |
Component:163; Jaccard:0.08589
 |
Component:163; Jaccard:0.064172
 |
Correlation:0.1643
  |
Correlation:0.15309
  |
Correlation:0.025102
  |
Correlation:0.15309
  |
Correlation:0.0186
  |
Correlation:0.0186
  |
Correlation:0.0186
  |
Component:163; Jaccard:0.091969
 |
Component:163; Jaccard:0.10075
 |
Component:149; Jaccard:0.10906
 |
Component:163; Jaccard:0.11138
 |
Component:384; Jaccard:0.10014
 |
Component:273; Jaccard:0.083238
 |
Component:241; Jaccard:0.064097
 |
Correlation:0.0186
  |
Correlation:0.0186
  |
Correlation:0.05548
  |
Correlation:0.0186
  |
Correlation:0.15309
  |
Correlation:0.19711
 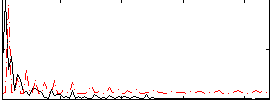 |
Correlation:0.48843
  |
Cope: 03:FACES-SHAPES BACK
|
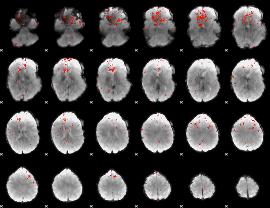
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
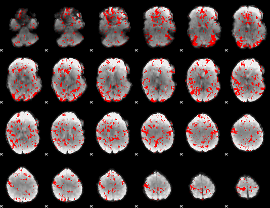 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
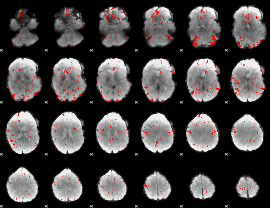 |
GLM binary result thresholded by 3
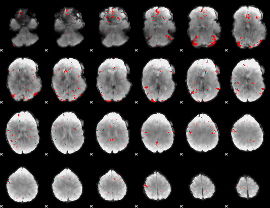 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
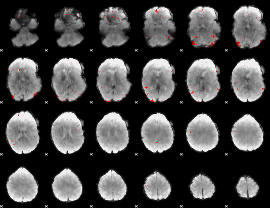 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
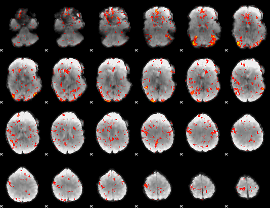 |
GLM Z-score result thresholded by 2.5
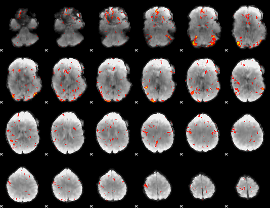 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:042; Jaccard:0.14448
 |
Component:042; Jaccard:0.20596
 |
Component:042; Jaccard:0.2667
 |
Component:042; Jaccard:0.29147
 |
Component:042; Jaccard:0.25585
 |
Component:042; Jaccard:0.19324
 |
Component:042; Jaccard:0.13134
 |
Correlation:0.52931
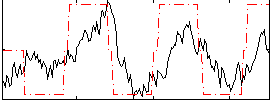 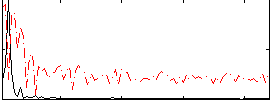 |
Correlation:0.52931
  |
Correlation:0.52931
  |
Correlation:0.52931
  |
Correlation:0.52931
  |
Correlation:0.52931
  |
Correlation:0.52931
  |
Component:363; Jaccard:0.091797
 |
Component:363; Jaccard:0.097131
 |
Component:184; Jaccard:0.10643
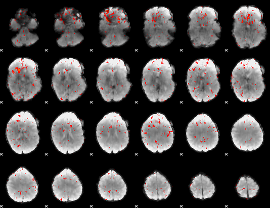 |
Component:184; Jaccard:0.098463
 |
Component:184; Jaccard:0.071506
 |
Component:184; Jaccard:0.045904
 |
Component:389; Jaccard:0.031951
 |
Correlation:0.28787
  |
Correlation:0.28787
  |
Correlation:0.28091
  |
Correlation:0.28091
  |
Correlation:0.28091
  |
Correlation:0.28091
  |
Correlation:0.20111
  |
Component:389; Jaccard:0.084198
 |
Component:184; Jaccard:0.096349
 |
Component:363; Jaccard:0.095475
 |
Component:389; Jaccard:0.081473
 |
Component:389; Jaccard:0.061517
 |
Component:389; Jaccard:0.044459
 |
Component:365; Jaccard:0.030248
 |
Correlation:0.20111
  |
Correlation:0.28091
  |
Correlation:0.28787
  |
Correlation:0.20111
  |
Correlation:0.20111
  |
Correlation:0.20111
  |
Correlation:0.32315
  |
Component:126; Jaccard:0.082132
 |
Component:389; Jaccard:0.092632
 |
Component:389; Jaccard:0.093067
 |
Component:234; Jaccard:0.078369
 |
Component:234; Jaccard:0.059122
 |
Component:365; Jaccard:0.041141
 |
Component:234; Jaccard:0.025637
 |
Correlation:0.18406
  |
Correlation:0.20111
  |
Correlation:0.20111
  |
Correlation:0.30613
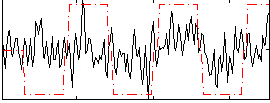  |
Correlation:0.30613
  |
Correlation:0.32315
  |
Correlation:0.30613
  |
Component:068; Jaccard:0.081575
 |
Component:068; Jaccard:0.087533
 |
Component:234; Jaccard:0.086529
 |
Component:363; Jaccard:0.077545
 |
Component:363; Jaccard:0.05523
 |
Component:363; Jaccard:0.038269
 |
Component:184; Jaccard:0.025538
 |
Correlation:0.37845
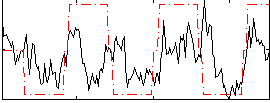 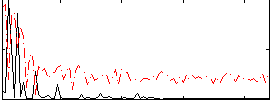 |
Correlation:0.37845
  |
Correlation:0.30613
  |
Correlation:0.28787
  |
Correlation:0.28787
  |
Correlation:0.28787
  |
Correlation:0.28091
  |
Component:184; Jaccard:0.080321
 |
Component:234; Jaccard:0.080821
 |
Component:068; Jaccard:0.083127
 |
Component:068; Jaccard:0.066321
 |
Component:365; Jaccard:0.052091
 |
Component:234; Jaccard:0.036897
 |
Component:363; Jaccard:0.024434
 |
Correlation:0.28091
  |
Correlation:0.30613
  |
Correlation:0.37845
  |
Correlation:0.37845
  |
Correlation:0.32315
  |
Correlation:0.30613
  |
Correlation:0.28787
  |
Component:101; Jaccard:0.074045
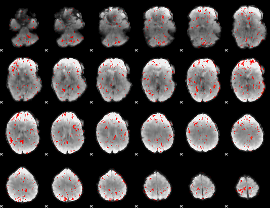 |
Component:126; Jaccard:0.078938
 |
Component:019; Jaccard:0.068109
 |
Component:204; Jaccard:0.062801
 |
Component:204; Jaccard:0.051424
 |
Component:204; Jaccard:0.036621
 |
Component:204; Jaccard:0.022253
 |
Correlation:0.23731
  |
Correlation:0.18406
  |
Correlation:0.20972
  |
Correlation:0.038304
  |
Correlation:0.038304
  |
Correlation:0.038304
  |
Correlation:0.038304
  |
Component:019; Jaccard:0.073013
 |
Component:019; Jaccard:0.07373
 |
Component:044; Jaccard:0.066984
 |
Component:365; Jaccard:0.061013
 |
Component:059; Jaccard:0.051358
 |
Component:059; Jaccard:0.034441
 |
Component:147; Jaccard:0.019387
 |
Correlation:0.20972
  |
Correlation:0.20972
  |
Correlation:0.2287
  |
Correlation:0.32315
  |
Correlation:0.24299
  |
Correlation:0.24299
  |
Correlation:0.11317
  |
Component:382; Jaccard:0.070418
 |
Component:101; Jaccard:0.07329
 |
Component:204; Jaccard:0.066391
 |
Component:059; Jaccard:0.060179
 |
Component:068; Jaccard:0.046785
 |
Component:068; Jaccard:0.031673
 |
Component:059; Jaccard:0.019079
 |
Correlation:0.1383
  |
Correlation:0.23731
  |
Correlation:0.038304
  |
Correlation:0.24299
  |
Correlation:0.37845
  |
Correlation:0.37845
  |
Correlation:0.24299
  |
Component:339; Jaccard:0.069964
 |
Component:382; Jaccard:0.07071
 |
Component:126; Jaccard:0.065953
 |
Component:147; Jaccard:0.05747
 |
Component:147; Jaccard:0.043584
 |
Component:147; Jaccard:0.029912
 |
Component:068; Jaccard:0.018522
 |
Correlation:0.026594
  |
Correlation:0.1383
  |
Correlation:0.18406
  |
Correlation:0.11317
  |
Correlation:0.11317
  |
Correlation:0.11317
  |
Correlation:0.37845
  |
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
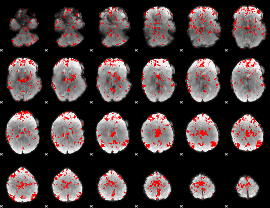 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
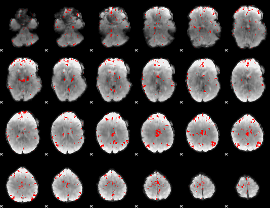 |
GLM binary result thresholded by 3.5
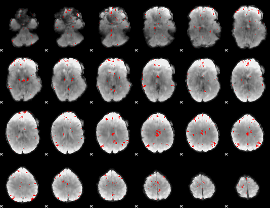 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
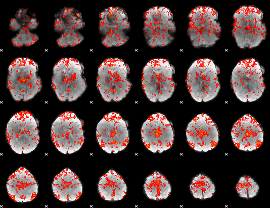 |
GLM Z-score result thresholded by 2
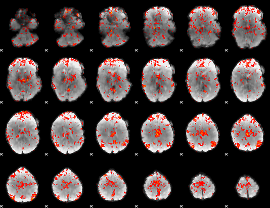 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
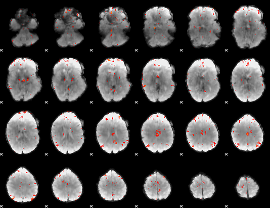 |
GLM Z-score result thresholded by 4
 |
Component:015; Jaccard:0.099332
 |
Component:015; Jaccard:0.12494
 |
Component:015; Jaccard:0.14887
 |
Component:015; Jaccard:0.16142
 |
Component:015; Jaccard:0.1492
 |
Component:015; Jaccard:0.108
 |
Component:015; Jaccard:0.069083
 |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.11231
  |
Component:377; Jaccard:0.095275
 |
Component:377; Jaccard:0.10932
 |
Component:145; Jaccard:0.12212
 |
Component:145; Jaccard:0.13575
 |
Component:145; Jaccard:0.13205
 |
Component:145; Jaccard:0.10343
 |
Component:145; Jaccard:0.068751
 |
Correlation:0.22727
  |
Correlation:0.22727
  |
Correlation:0.15531
  |
Correlation:0.15531
  |
Correlation:0.15531
  |
Correlation:0.15531
  |
Correlation:0.15531
  |
Component:040; Jaccard:0.085193
 |
Component:145; Jaccard:0.10091
 |
Component:377; Jaccard:0.11788
 |
Component:377; Jaccard:0.11623
 |
Component:377; Jaccard:0.10047
 |
Component:377; Jaccard:0.074988
 |
Component:377; Jaccard:0.050315
 |
Correlation:0.29316
  |
Correlation:0.15531
  |
Correlation:0.22727
  |
Correlation:0.22727
  |
Correlation:0.22727
  |
Correlation:0.22727
  |
Correlation:0.22727
  |
Component:078; Jaccard:0.082829
 |
Component:040; Jaccard:0.099075
 |
Component:040; Jaccard:0.10793
 |
Component:040; Jaccard:0.10806
 |
Component:244; Jaccard:0.090596
 |
Component:244; Jaccard:0.065723
 |
Component:244; Jaccard:0.039822
 |
Correlation:0.26406
  |
Correlation:0.29316
  |
Correlation:0.29316
  |
Correlation:0.29316
  |
Correlation:0.12679
 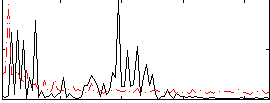 |
Correlation:0.12679
  |
Correlation:0.12679
  |
Component:145; Jaccard:0.081627
 |
Component:078; Jaccard:0.094752
 |
Component:078; Jaccard:0.10443
 |
Component:078; Jaccard:0.10276
 |
Component:040; Jaccard:0.090289
 |
Component:078; Jaccard:0.06205
 |
Component:040; Jaccard:0.036095
 |
Correlation:0.15531
  |
Correlation:0.26406
  |
Correlation:0.26406
  |
Correlation:0.26406
  |
Correlation:0.29316
  |
Correlation:0.26406
  |
Correlation:0.29316
  |
Component:070; Jaccard:0.081507
 |
Component:070; Jaccard:0.089518
 |
Component:244; Jaccard:0.096999
 |
Component:244; Jaccard:0.10077
 |
Component:078; Jaccard:0.087075
 |
Component:040; Jaccard:0.061619
 |
Component:158; Jaccard:0.035218
 |
Correlation:0.21555
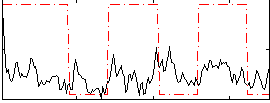  |
Correlation:0.21555
  |
Correlation:0.12679
  |
Correlation:0.12679
  |
Correlation:0.26406
  |
Correlation:0.29316
  |
Correlation:-0.049039
  |
Component:216; Jaccard:0.080667
 |
Component:244; Jaccard:0.086752
 |
Component:070; Jaccard:0.093039
 |
Component:070; Jaccard:0.086947
 |
Component:070; Jaccard:0.072042
 |
Component:158; Jaccard:0.053383
 |
Component:078; Jaccard:0.03517
 |
Correlation:0.021486
  |
Correlation:0.12679
  |
Correlation:0.21555
  |
Correlation:0.21555
  |
Correlation:0.21555
  |
Correlation:-0.049039
  |
Correlation:0.26406
  |
Component:313; Jaccard:0.080183
 |
Component:297; Jaccard:0.084634
 |
Component:297; Jaccard:0.087721
 |
Component:320; Jaccard:0.083543
 |
Component:320; Jaccard:0.071179
 |
Component:320; Jaccard:0.051628
 |
Component:320; Jaccard:0.031718
 |
Correlation:0.14062
  |
Correlation:0.22588
  |
Correlation:0.22588
  |
Correlation:0.24389
  |
Correlation:0.24389
  |
Correlation:0.24389
  |
Correlation:0.24389
  |
Component:297; Jaccard:0.07743
 |
Component:313; Jaccard:0.084592
 |
Component:313; Jaccard:0.085748
 |
Component:129; Jaccard:0.081761
 |
Component:158; Jaccard:0.070323
 |
Component:070; Jaccard:0.051318
 |
Component:070; Jaccard:0.030997
 |
Correlation:0.22588
  |
Correlation:0.14062
  |
Correlation:0.14062
  |
Correlation:0.19423
 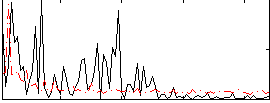 |
Correlation:-0.049039
  |
Correlation:0.21555
  |
Correlation:0.21555
  |
Component:065; Jaccard:0.075594
 |
Component:216; Jaccard:0.080982
 |
Component:129; Jaccard:0.084489
 |
Component:297; Jaccard:0.080762
 |
Component:297; Jaccard:0.063338
 |
Component:351; Jaccard:0.046306
 |
Component:204; Jaccard:0.02957
 |
Correlation:0.12077
  |
Correlation:0.021486
  |
Correlation:0.19423
  |
Correlation:0.22588
  |
Correlation:0.22588
  |
Correlation:0.052351
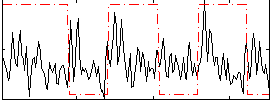  |
Correlation:0.060002
 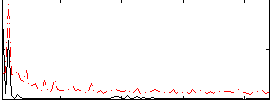 |
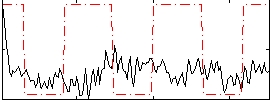
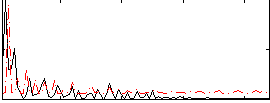
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
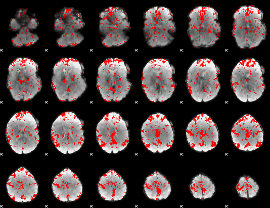 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
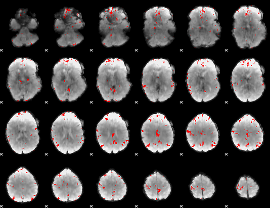 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
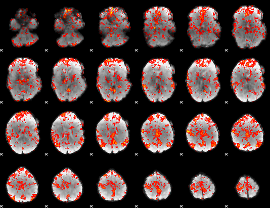 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:068; Jaccard:0.10046
 |
Component:068; Jaccard:0.121
 |
Component:015; Jaccard:0.14181
 |
Component:068; Jaccard:0.15128
 |
Component:145; Jaccard:0.14535
 |
Component:145; Jaccard:0.12476
 |
Component:145; Jaccard:0.093116
 |
Correlation:0.39241
  |
Correlation:0.39241
  |
Correlation:0.093277
  |
Correlation:0.39241
  |
Correlation:0.10267
  |
Correlation:0.10267
  |
Correlation:0.10267
  |
Component:015; Jaccard:0.098107
 |
Component:015; Jaccard:0.12005
 |
Component:068; Jaccard:0.13946
 |
Component:015; Jaccard:0.15044
 |
Component:015; Jaccard:0.14444
 |
Component:068; Jaccard:0.11628
 |
Component:015; Jaccard:0.076244
 |
Correlation:0.093277
  |
Correlation:0.093277
  |
Correlation:0.39241
  |
Correlation:0.093277
  |
Correlation:0.093277
  |
Correlation:0.39241
  |
Correlation:0.093277
  |
Component:377; Jaccard:0.090455
 |
Component:145; Jaccard:0.10338
 |
Component:145; Jaccard:0.12596
 |
Component:145; Jaccard:0.1445
 |
Component:068; Jaccard:0.14258
 |
Component:015; Jaccard:0.11386
 |
Component:068; Jaccard:0.075314
 |
Correlation:-0.057248
  |
Correlation:0.10267
  |
Correlation:0.10267
  |
Correlation:0.10267
  |
Correlation:0.39241
  |
Correlation:0.093277
  |
Correlation:0.39241
  |
Component:216; Jaccard:0.088148
 |
Component:377; Jaccard:0.10281
 |
Component:377; Jaccard:0.11183
 |
Component:377; Jaccard:0.11515
 |
Component:377; Jaccard:0.1068
 |
Component:204; Jaccard:0.090332
 |
Component:204; Jaccard:0.072543
 |
Correlation:0.011541
  |
Correlation:-0.057248
  |
Correlation:-0.057248
  |
Correlation:-0.057248
  |
Correlation:-0.057248
  |
Correlation:0.13062
  |
Correlation:0.13062
  |
Component:070; Jaccard:0.084497
 |
Component:070; Jaccard:0.094774
 |
Component:070; Jaccard:0.1019
 |
Component:204; Jaccard:0.10437
 |
Component:204; Jaccard:0.10495
 |
Component:377; Jaccard:0.085371
 |
Component:377; Jaccard:0.06049
 |
Correlation:-0.049197
  |
Correlation:-0.049197
  |
Correlation:-0.049197
  |
Correlation:0.13062
  |
Correlation:0.13062
  |
Correlation:-0.057248
  |
Correlation:-0.057248
  |
Component:145; Jaccard:0.083384
 |
Component:216; Jaccard:0.09369
 |
Component:204; Jaccard:0.094435
 |
Component:070; Jaccard:0.099912
 |
Component:070; Jaccard:0.088035
 |
Component:349; Jaccard:0.070588
 |
Component:349; Jaccard:0.052878
 |
Correlation:0.10267
  |
Correlation:0.011541
  |
Correlation:0.13062
  |
Correlation:-0.049197
  |
Correlation:-0.049197
  |
Correlation:0.16116
  |
Correlation:0.16116
  |
Component:126; Jaccard:0.079321
 |
Component:039; Jaccard:0.088226
 |
Component:039; Jaccard:0.094094
 |
Component:039; Jaccard:0.093062
 |
Component:349; Jaccard:0.082472
 |
Component:070; Jaccard:0.067267
 |
Component:351; Jaccard:0.047662
 |
Correlation:0.23823
  |
Correlation:0.11518
  |
Correlation:0.11518
  |
Correlation:0.11518
  |
Correlation:0.16116
  |
Correlation:-0.049197
  |
Correlation:-0.011782
 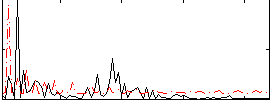 |
Component:039; Jaccard:0.079196
 |
Component:078; Jaccard:0.084818
 |
Component:216; Jaccard:0.090835
 |
Component:078; Jaccard:0.089051
 |
Component:039; Jaccard:0.081
 |
Component:099; Jaccard:0.064773
 |
Component:070; Jaccard:0.045797
 |
Correlation:0.11518
  |
Correlation:0.065301
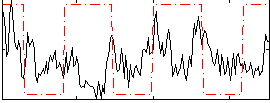 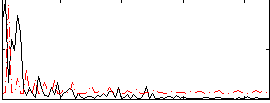 |
Correlation:0.011541
  |
Correlation:0.065301
  |
Correlation:0.11518
  |
Correlation:0.014669
 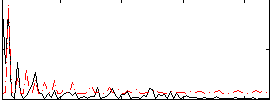 |
Correlation:-0.049197
  |
Component:313; Jaccard:0.078633
 |
Component:313; Jaccard:0.084797
 |
Component:078; Jaccard:0.089286
 |
Component:104; Jaccard:0.086734
 |
Component:099; Jaccard:0.078802
 |
Component:351; Jaccard:0.064231
 |
Component:099; Jaccard:0.044923
 |
Correlation:0.074842
  |
Correlation:0.074842
  |
Correlation:0.065301
  |
Correlation:0.17394
  |
Correlation:0.014669
  |
Correlation:-0.011782
  |
Correlation:0.014669
  |
Component:078; Jaccard:0.075917
 |
Component:126; Jaccard:0.081796
 |
Component:313; Jaccard:0.086624
 |
Component:349; Jaccard:0.085943
 |
Component:078; Jaccard:0.077525
 |
Component:039; Jaccard:0.061946
 |
Component:158; Jaccard:0.0423
 |
Correlation:0.065301
  |
Correlation:0.23823
  |
Correlation:0.074842
  |
Correlation:0.16116
  |
Correlation:0.065301
  |
Correlation:0.11518
  |
Correlation:0.1681
 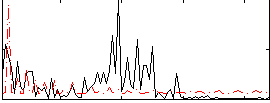 |
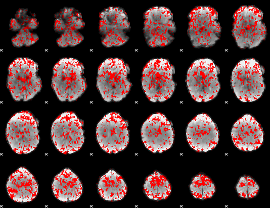
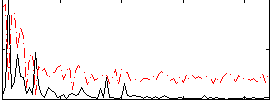
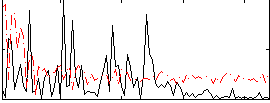
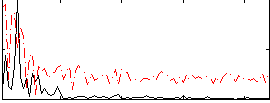
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
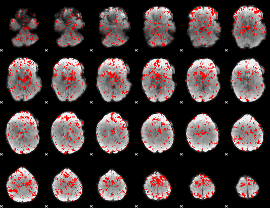 |
GLM binary result thresholded by 2
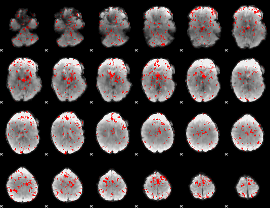 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
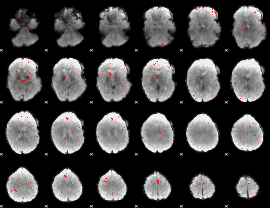 |
GLM binary result thresholded by 3.5
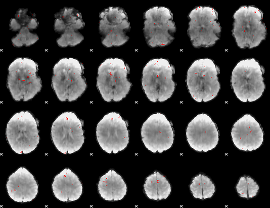 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
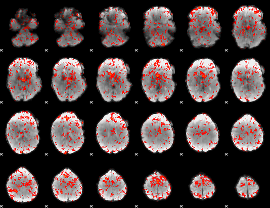 |
GLM Z-score result thresholded by 2
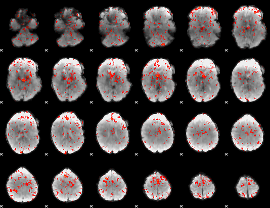 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:324; Jaccard:0.098852
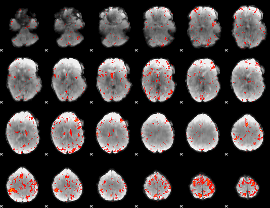 |
Component:324; Jaccard:0.099828
 |
Component:324; Jaccard:0.086205
 |
Component:171; Jaccard:0.070595
 |
Component:171; Jaccard:0.043215
 |
Component:171; Jaccard:0.016342
 |
Component:157; Jaccard:0.005827
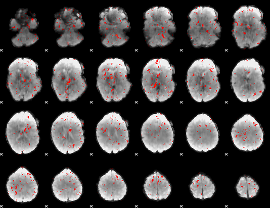 |
Correlation:0.40732
  |
Correlation:0.40732
  |
Correlation:0.40732
  |
Correlation:0.28261
  |
Correlation:0.28261
  |
Correlation:0.28261
  |
Correlation:0.16158
  |
Component:224; Jaccard:0.087241
 |
Component:224; Jaccard:0.090553
 |
Component:171; Jaccard:0.083961
 |
Component:040; Jaccard:0.061402
 |
Component:040; Jaccard:0.036291
 |
Component:167; Jaccard:0.016289
 |
Component:040; Jaccard:0.005772
 |
Correlation:0.30328
  |
Correlation:0.30328
  |
Correlation:0.28261
  |
Correlation:0.31974
  |
Correlation:0.31974
  |
Correlation:0.25353
  |
Correlation:0.31974
  |
Component:352; Jaccard:0.080094
 |
Component:352; Jaccard:0.083554
 |
Component:224; Jaccard:0.08383
 |
Component:224; Jaccard:0.061387
 |
Component:167; Jaccard:0.034614
 |
Component:040; Jaccard:0.015864
 |
Component:224; Jaccard:0.005244
 |
Correlation:0.35132
  |
Correlation:0.35132
  |
Correlation:0.30328
  |
Correlation:0.30328
  |
Correlation:0.25353
  |
Correlation:0.31974
  |
Correlation:0.30328
  |
Component:057; Jaccard:0.07639
 |
Component:057; Jaccard:0.080843
 |
Component:040; Jaccard:0.07747
 |
Component:057; Jaccard:0.057177
 |
Component:224; Jaccard:0.034163
 |
Component:057; Jaccard:0.015287
 |
Component:308; Jaccard:0.005127
 |
Correlation:0.39195
  |
Correlation:0.39195
  |
Correlation:0.31974
  |
Correlation:0.39195
  |
Correlation:0.30328
  |
Correlation:0.39195
  |
Correlation:0.21888
  |
Component:191; Jaccard:0.071507
 |
Component:171; Jaccard:0.078477
 |
Component:352; Jaccard:0.076757
 |
Component:324; Jaccard:0.056771
 |
Component:057; Jaccard:0.033832
 |
Component:224; Jaccard:0.015143
 |
Component:171; Jaccard:0.005079
 |
Correlation:0.17469
  |
Correlation:0.28261
  |
Correlation:0.35132
  |
Correlation:0.40732
  |
Correlation:0.39195
  |
Correlation:0.30328
  |
Correlation:0.28261
  |
Component:277; Jaccard:0.071182
 |
Component:040; Jaccard:0.078124
 |
Component:057; Jaccard:0.07656
 |
Component:352; Jaccard:0.055498
 |
Component:157; Jaccard:0.031636
 |
Component:360; Jaccard:0.013801
 |
Component:167; Jaccard:0.004844
 |
Correlation:0.15288
  |
Correlation:0.31974
  |
Correlation:0.39195
  |
Correlation:0.35132
  |
Correlation:0.16158
  |
Correlation:0.26454
  |
Correlation:0.25353
  |
Component:040; Jaccard:0.070262
 |
Component:191; Jaccard:0.076599
 |
Component:191; Jaccard:0.072353
 |
Component:167; Jaccard:0.054832
 |
Component:352; Jaccard:0.031107
 |
Component:324; Jaccard:0.013679
 |
Component:324; Jaccard:0.004719
 |
Correlation:0.31974
  |
Correlation:0.17469
  |
Correlation:0.17469
  |
Correlation:0.25353
  |
Correlation:0.35132
  |
Correlation:0.40732
  |
Correlation:0.40732
  |
Component:360; Jaccard:0.070055
 |
Component:360; Jaccard:0.075022
 |
Component:360; Jaccard:0.069401
 |
Component:191; Jaccard:0.054044
 |
Component:131; Jaccard:0.030293
 |
Component:394; Jaccard:0.013479
 |
Component:360; Jaccard:0.004648
 |
Correlation:0.26454
  |
Correlation:0.26454
  |
Correlation:0.26454
  |
Correlation:0.17469
  |
Correlation:0.25387
  |
Correlation:0.25008
  |
Correlation:0.26454
  |
Component:082; Jaccard:0.069774
 |
Component:272; Jaccard:0.072612
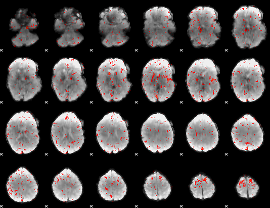 |
Component:272; Jaccard:0.068167
 |
Component:157; Jaccard:0.052575
 |
Component:244; Jaccard:0.030123
 |
Component:157; Jaccard:0.013206
 |
Component:394; Jaccard:0.004334
 |
Correlation:-0.040033
  |
Correlation:0.26001
  |
Correlation:0.26001
  |
Correlation:0.16158
  |
Correlation:0.10764
  |
Correlation:0.16158
  |
Correlation:0.25008
  |
Component:023; Jaccard:0.068205
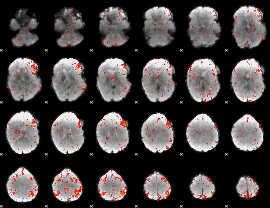 |
Component:082; Jaccard:0.07016
 |
Component:131; Jaccard:0.064679
 |
Component:272; Jaccard:0.051465
 |
Component:360; Jaccard:0.02971
 |
Component:352; Jaccard:0.01277
 |
Component:191; Jaccard:0.004328
 |
Correlation:0.15778
  |
Correlation:-0.040033
  |
Correlation:0.25387
  |
Correlation:0.26001
  |
Correlation:0.26454
  |
Correlation:0.35132
  |
Correlation:0.17469
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































