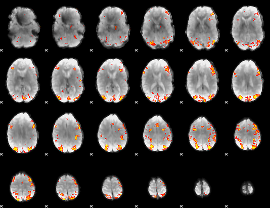
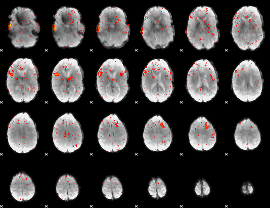
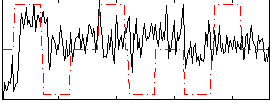
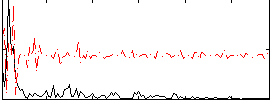
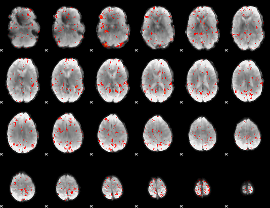
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
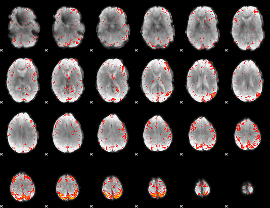
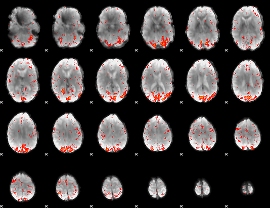
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
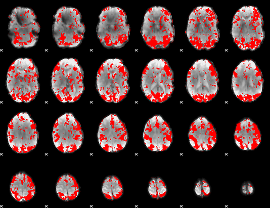 |
GLM binary result thresholded by 2
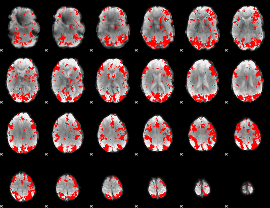 |
GLM binary result thresholded by 2.5
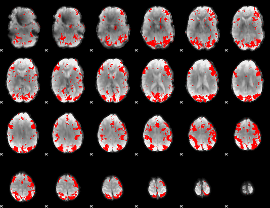 |
GLM binary result thresholded by 3
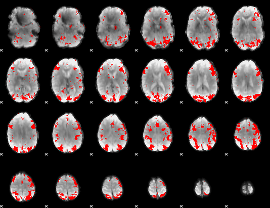 |
GLM binary result thresholded by 3.5
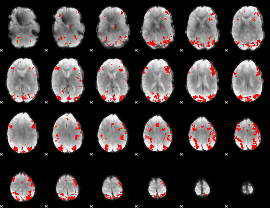 |
GLM binary result thresholded by 4
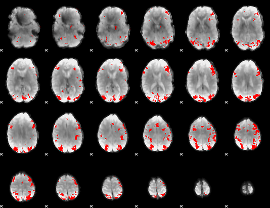 |
GLM Z-score result thresholded by 1
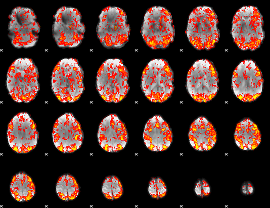 |
GLM Z-score result thresholded by 1.5
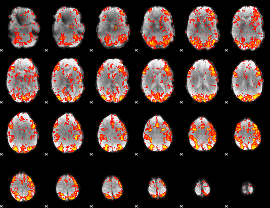 |
GLM Z-score result thresholded by 2
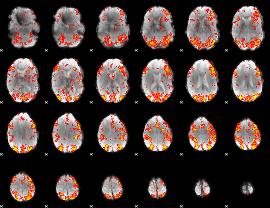 |
GLM Z-score result thresholded by 2.5
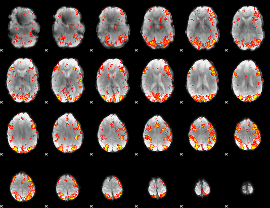 |
GLM Z-score result thresholded by 3
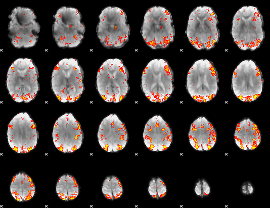 |
GLM Z-score result thresholded by 3.5
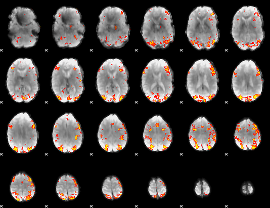 |
GLM Z-score result thresholded by 4
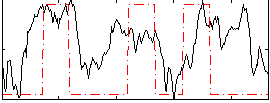
 |
Component:279; Jaccard:0.14991
 |
Component:279; Jaccard:0.20042
 |
Component:279; Jaccard:0.26006
 |
Component:279; Jaccard:0.31636
 |
Component:279; Jaccard:0.36405
 |
Component:279; Jaccard:0.38821
 |
Component:279; Jaccard:0.38713
 |
Correlation:0.34828
 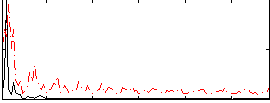 |
Correlation:0.34828
  |
Correlation:0.34828
  |
Correlation:0.34828
  |
Correlation:0.34828
  |
Correlation:0.34828
  |
Correlation:0.34828
  |
Component:105; Jaccard:0.13689
 |
Component:105; Jaccard:0.16304
 |
Component:105; Jaccard:0.18426
 |
Component:105; Jaccard:0.19381
 |
Component:105; Jaccard:0.19226
 |
Component:105; Jaccard:0.18015
 |
Component:105; Jaccard:0.16095
 |
Correlation:0.15665
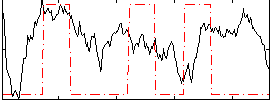 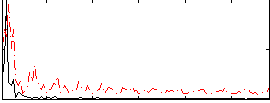 |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
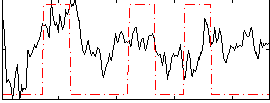
Component:169; Jaccard:0.13431
 |
Component:169; Jaccard:0.15516
 |
Component:169; Jaccard:0.17211
 |
Component:169; Jaccard:0.17878
 |
Component:169; Jaccard:0.17607
 |
Component:057; Jaccard:0.16787
 |
Component:057; Jaccard:0.15706
 |
Correlation:0.27407
 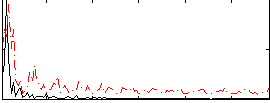 |
Correlation:0.27407
  |
Correlation:0.27407
  |
Correlation:0.27407
  |
Correlation:0.27407
  |
Correlation:0.20087
 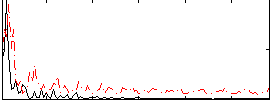 |
Correlation:0.20087
  |
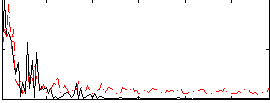
Component:378; Jaccard:0.11749
 |
Component:378; Jaccard:0.13919
 |
Component:378; Jaccard:0.15862
 |
Component:378; Jaccard:0.16857
 |
Component:057; Jaccard:0.17086
 |
Component:169; Jaccard:0.16468
 |
Component:169; Jaccard:0.14616
 |
Correlation:0.23049
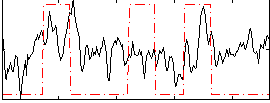  |
Correlation:0.23049
  |
Correlation:0.23049
  |
Correlation:0.23049
  |
Correlation:0.20087
  |
Correlation:0.27407
  |
Correlation:0.27407
  |
Component:301; Jaccard:0.11383
 |
Component:301; Jaccard:0.13486
 |
Component:057; Jaccard:0.15197
 |
Component:057; Jaccard:0.16514
 |
Component:378; Jaccard:0.16899
 |
Component:378; Jaccard:0.1601
 |
Component:322; Jaccard:0.14606
 |
Correlation:0.24214
  |
Correlation:0.24214
  |
Correlation:0.20087
  |
Correlation:0.20087
  |
Correlation:0.23049
  |
Correlation:0.23049
  |
Correlation:0.22714
  |
Component:061; Jaccard:0.11244
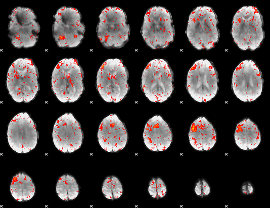 |
Component:057; Jaccard:0.13345
 |
Component:301; Jaccard:0.15024
 |
Component:301; Jaccard:0.15741
 |
Component:355; Jaccard:0.15848
 |
Component:355; Jaccard:0.15455
 |
Component:378; Jaccard:0.14562
 |
Correlation:0.21821
 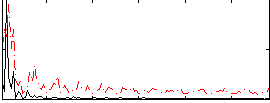 |
Correlation:0.20087
  |
Correlation:0.24214
  |
Correlation:0.24214
  |
Correlation:0.11312
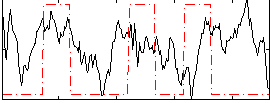 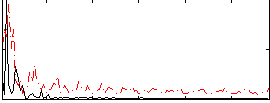 |
Correlation:0.11312
  |
Correlation:0.23049
  |
Component:057; Jaccard:0.11101
 |
Component:355; Jaccard:0.12802
 |
Component:355; Jaccard:0.14562
 |
Component:355; Jaccard:0.15686
 |
Component:301; Jaccard:0.15391
 |
Component:322; Jaccard:0.15202
 |
Component:355; Jaccard:0.14314
 |
Correlation:0.20087
  |
Correlation:0.11312
  |
Correlation:0.11312
  |
Correlation:0.11312
  |
Correlation:0.24214
  |
Correlation:0.22714
  |
Correlation:0.11312
  |
Component:355; Jaccard:0.10581
 |
Component:061; Jaccard:0.12408
 |
Component:322; Jaccard:0.13144
 |
Component:322; Jaccard:0.14372
 |
Component:322; Jaccard:0.15102
 |
Component:301; Jaccard:0.14396
 |
Component:301; Jaccard:0.12784
 |
Correlation:0.11312
  |
Correlation:0.21821
  |
Correlation:0.22714
  |
Correlation:0.22714
  |
Correlation:0.22714
  |
Correlation:0.24214
  |
Correlation:0.24214
  |
Component:280; Jaccard:0.10244
 |
Component:280; Jaccard:0.1202
 |
Component:280; Jaccard:0.13099
 |
Component:280; Jaccard:0.13642
 |
Component:280; Jaccard:0.13673
 |
Component:036; Jaccard:0.12872
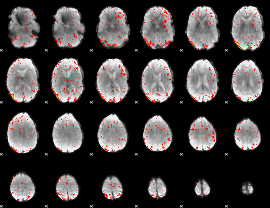 |
Component:036; Jaccard:0.12281
 |
Correlation:0.28787
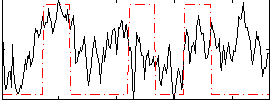 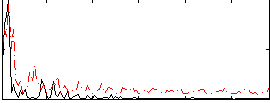 |
Correlation:0.28787
  |
Correlation:0.28787
  |
Correlation:0.28787
  |
Correlation:0.28787
  |
Correlation:0.01676
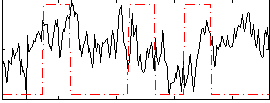 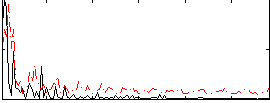 |
Correlation:0.01676
  |
Component:386; Jaccard:0.097879
 |
Component:294; Jaccard:0.11262
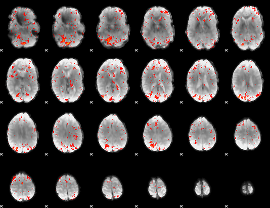 |
Component:061; Jaccard:0.13063
 |
Component:294; Jaccard:0.12832
 |
Component:036; Jaccard:0.13108
 |
Component:280; Jaccard:0.12846
 |
Component:280; Jaccard:0.11681
 |
Correlation:0.13999
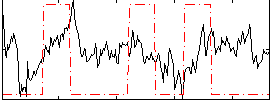 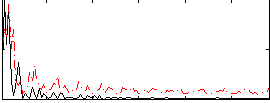 |
Correlation:0.26041
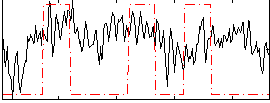 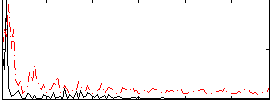 |
Correlation:0.21821
  |
Correlation:0.26041
  |
Correlation:0.01676
  |
Correlation:0.28787
  |
Correlation:0.28787
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
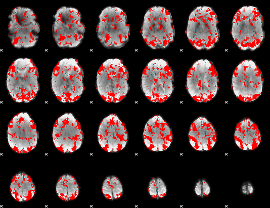 |
GLM binary result thresholded by 1.5
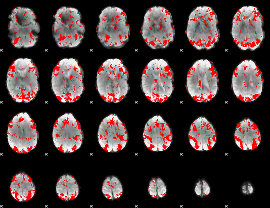 |
GLM binary result thresholded by 2
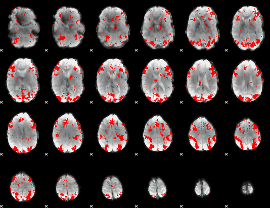 |
GLM binary result thresholded by 2.5
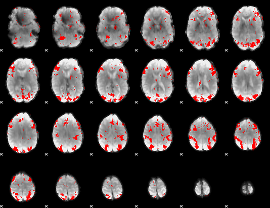 |
GLM binary result thresholded by 3
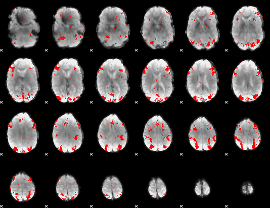 |
GLM binary result thresholded by 3.5
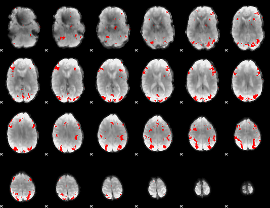 |
GLM binary result thresholded by 4
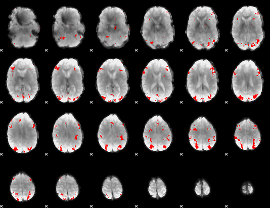 |
GLM Z-score result thresholded by 1
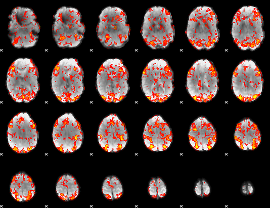 |
GLM Z-score result thresholded by 1.5
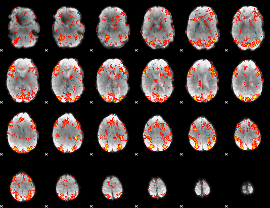 |
GLM Z-score result thresholded by 2
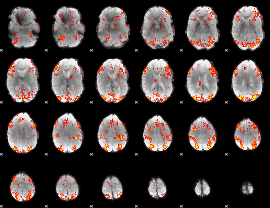 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
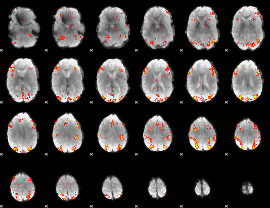 |
GLM Z-score result thresholded by 3.5
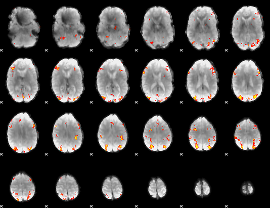
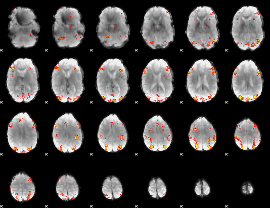 |
GLM Z-score result thresholded by 4
 |
Component:279; Jaccard:0.19342
 |
Component:279; Jaccard:0.24413
 |
Component:279; Jaccard:0.29412
 |
Component:279; Jaccard:0.33576
 |
Component:279; Jaccard:0.34852
 |
Component:279; Jaccard:0.32662
 |
Component:279; Jaccard:0.28154
 |
Correlation:0.074125
  |
Correlation:0.074125
  |
Correlation:0.074125
  |
Correlation:0.074125
  |
Correlation:0.074125
  |
Correlation:0.074125
  |
Correlation:0.074125
  |
Component:105; Jaccard:0.1508
 |
Component:105; Jaccard:0.17053
 |
Component:105; Jaccard:0.17863
 |
Component:105; Jaccard:0.17754
 |
Component:105; Jaccard:0.16516
 |
Component:355; Jaccard:0.14673
 |
Component:355; Jaccard:0.12586
 |
Correlation:-0.047327
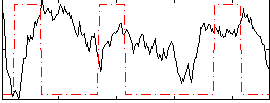 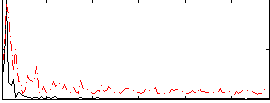 |
Correlation:-0.047327
  |
Correlation:-0.047327
  |
Correlation:-0.047327
  |
Correlation:-0.047327
  |
Correlation:-0.087466
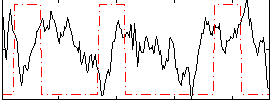 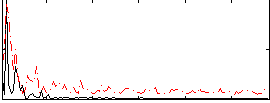 |
Correlation:-0.087466
  |
Component:355; Jaccard:0.12933
 |
Component:355; Jaccard:0.14914
 |
Component:355; Jaccard:0.16525
 |
Component:355; Jaccard:0.17142
 |
Component:355; Jaccard:0.16497
 |
Component:105; Jaccard:0.14187
 |
Component:322; Jaccard:0.1208
 |
Correlation:-0.087466
  |
Correlation:-0.087466
  |
Correlation:-0.087466
  |
Correlation:-0.087466
  |
Correlation:-0.087466
  |
Correlation:-0.047327
  |
Correlation:0.11963
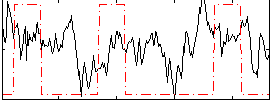 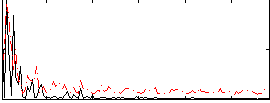 |
Component:057; Jaccard:0.12204
 |
Component:057; Jaccard:0.1382
 |
Component:057; Jaccard:0.15133
 |
Component:057; Jaccard:0.15837
 |
Component:057; Jaccard:0.15218
 |
Component:322; Jaccard:0.13766
 |
Component:057; Jaccard:0.11679
 |
Correlation:-0.050171
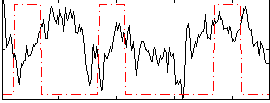 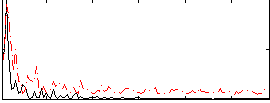 |
Correlation:-0.050171
  |
Correlation:-0.050171
  |
Correlation:-0.050171
  |
Correlation:-0.050171
  |
Correlation:0.11963
  |
Correlation:-0.050171
  |
Component:301; Jaccard:0.10713
 |
Component:322; Jaccard:0.12442
 |
Component:322; Jaccard:0.13993
 |
Component:322; Jaccard:0.14997
 |
Component:322; Jaccard:0.14837
 |
Component:057; Jaccard:0.13547
 |
Component:105; Jaccard:0.10986
 |
Correlation:-0.049676
  |
Correlation:0.11963
  |
Correlation:0.11963
  |
Correlation:0.11963
  |
Correlation:0.11963
  |
Correlation:-0.050171
  |
Correlation:-0.047327
  |
Component:322; Jaccard:0.10536
 |
Component:301; Jaccard:0.11645
 |
Component:326; Jaccard:0.12551
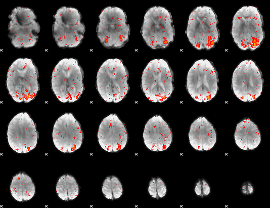 |
Component:326; Jaccard:0.13151
 |
Component:326; Jaccard:0.12847
 |
Component:326; Jaccard:0.119
 |
Component:326; Jaccard:0.099238
 |
Correlation:0.11963
  |
Correlation:-0.049676
  |
Correlation:0.22798
  |
Correlation:0.22798
  |
Correlation:0.22798
  |
Correlation:0.22798
  |
Correlation:0.22798
  |
Component:061; Jaccard:0.10203
 |
Component:326; Jaccard:0.11412
 |
Component:301; Jaccard:0.12034
 |
Component:301; Jaccard:0.11684
 |
Component:085; Jaccard:0.10973
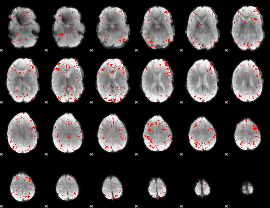 |
Component:085; Jaccard:0.097991
 |
Component:036; Jaccard:0.08505
 |
Correlation:-0.15884
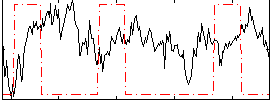 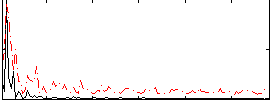 |
Correlation:0.22798
  |
Correlation:-0.049676
  |
Correlation:-0.049676
  |
Correlation:-0.034647
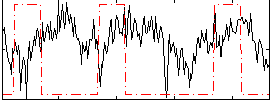 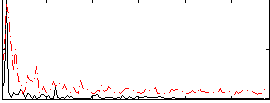 |
Correlation:-0.034647
  |
Correlation:0.055363
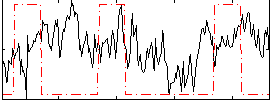 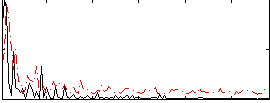 |
Component:326; Jaccard:0.098555
 |
Component:280; Jaccard:0.10571
 |
Component:085; Jaccard:0.11283
 |
Component:085; Jaccard:0.11371
 |
Component:301; Jaccard:0.10693
 |
Component:036; Jaccard:0.095925
 |
Component:085; Jaccard:0.083733
 |
Correlation:0.22798
  |
Correlation:-0.073133
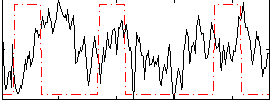  |
Correlation:-0.034647
  |
Correlation:-0.034647
  |
Correlation:-0.049676
  |
Correlation:0.055363
  |
Correlation:-0.034647
  |
Component:235; Jaccard:0.098531
 |
Component:061; Jaccard:0.10531
 |
Component:280; Jaccard:0.11067
 |
Component:280; Jaccard:0.10887
 |
Component:036; Jaccard:0.10449
 |
Component:301; Jaccard:0.089965
 |
Component:301; Jaccard:0.07135
 |
Correlation:0.19576
  |
Correlation:-0.15884
  |
Correlation:-0.073133
  |
Correlation:-0.073133
  |
Correlation:0.055363
  |
Correlation:-0.049676
  |
Correlation:-0.049676
  |
Component:169; Jaccard:0.097892
 |
Component:085; Jaccard:0.10419
 |
Component:036; Jaccard:0.10765
 |
Component:036; Jaccard:0.10787
 |
Component:280; Jaccard:0.1
 |
Component:280; Jaccard:0.087818
 |
Component:280; Jaccard:0.07116
 |
Correlation:-0.25192
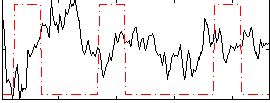 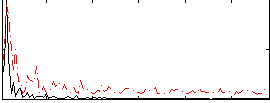 |
Correlation:-0.034647
  |
Correlation:0.055363
  |
Correlation:0.055363
  |
Correlation:-0.073133
  |
Correlation:-0.073133
  |
Correlation:-0.073133
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
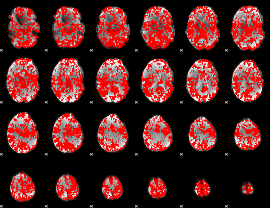 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
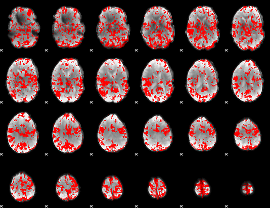 |
GLM binary result thresholded by 2.5
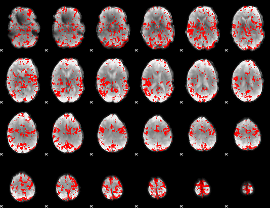 |
GLM binary result thresholded by 3
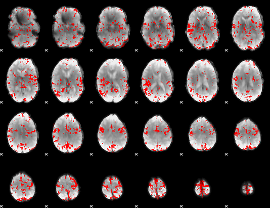 |
GLM binary result thresholded by 3.5
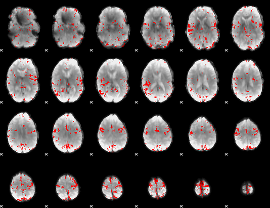 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
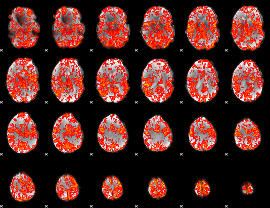 |
GLM Z-score result thresholded by 1.5
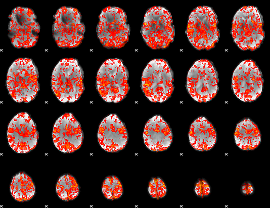 |
GLM Z-score result thresholded by 2
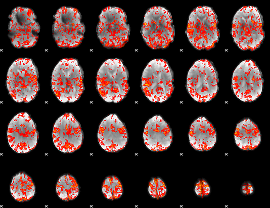 |
GLM Z-score result thresholded by 2.5
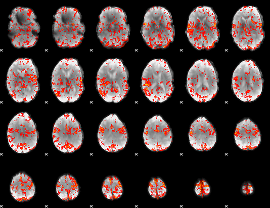 |
GLM Z-score result thresholded by 3
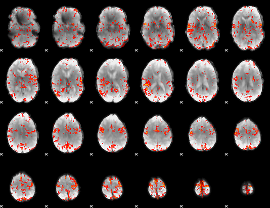 |
GLM Z-score result thresholded by 3.5
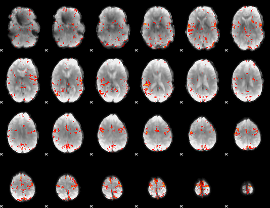 |
GLM Z-score result thresholded by 4
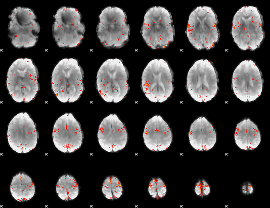 |
Component:246; Jaccard:0.090685
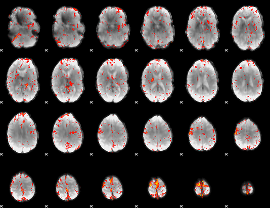 |
Component:246; Jaccard:0.10298
 |
Component:246; Jaccard:0.11617
 |
Component:246; Jaccard:0.12808
 |
Component:246; Jaccard:0.13475
 |
Component:246; Jaccard:0.12758
 |
Component:246; Jaccard:0.098181
 |
Correlation:0.26672
 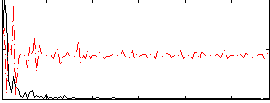 |
Correlation:0.26672
  |
Correlation:0.26672
  |
Correlation:0.26672
  |
Correlation:0.26672
  |
Correlation:0.26672
  |
Correlation:0.26672
  |
Component:080; Jaccard:0.087995
 |
Component:080; Jaccard:0.10096
 |
Component:080; Jaccard:0.11465
 |
Component:080; Jaccard:0.12577
 |
Component:080; Jaccard:0.12923
 |
Component:143; Jaccard:0.1222
 |
Component:143; Jaccard:0.0978
 |
Correlation:0.39062
 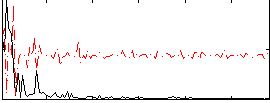 |
Correlation:0.39062
  |
Correlation:0.39062
  |
Correlation:0.39062
  |
Correlation:0.39062
  |
Correlation:0.20753
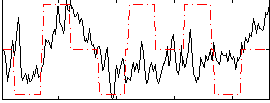 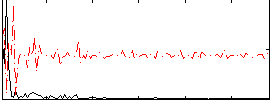 |
Correlation:0.20753
  |
Component:169; Jaccard:0.08223
 |
Component:143; Jaccard:0.09404
 |
Component:143; Jaccard:0.1084
 |
Component:143; Jaccard:0.12189
 |
Component:143; Jaccard:0.1284
 |
Component:080; Jaccard:0.12029
 |
Component:080; Jaccard:0.09615
 |
Correlation:0.31175
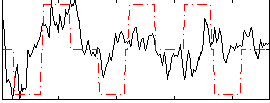  |
Correlation:0.20753
  |
Correlation:0.20753
  |
Correlation:0.20753
  |
Correlation:0.20753
  |
Correlation:0.39062
  |
Correlation:0.39062
  |
Component:143; Jaccard:0.081438
 |
Component:380; Jaccard:0.089808
 |
Component:380; Jaccard:0.10008
 |
Component:380; Jaccard:0.10834
 |
Component:383; Jaccard:0.11433
 |
Component:383; Jaccard:0.11043
 |
Component:383; Jaccard:0.09063
 |
Correlation:0.20753
  |
Correlation:0.22825
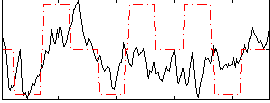 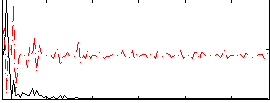 |
Correlation:0.22825
  |
Correlation:0.22825
  |
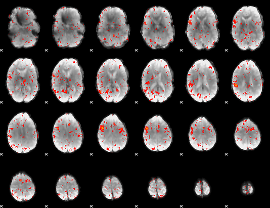
Correlation:0.2516
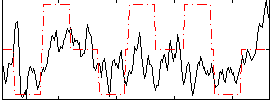  |
Correlation:0.2516
  |
Correlation:0.2516
  |
Component:380; Jaccard:0.079258
 |
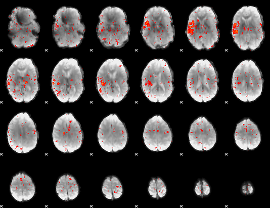
Component:191; Jaccard:0.085876
 |
Component:383; Jaccard:0.096929
 |
Component:191; Jaccard:0.10809
 |
Component:191; Jaccard:0.11054
 |
Component:191; Jaccard:0.099559
 |
Component:041; Jaccard:0.082612
 |
Correlation:0.22825
  |
Correlation:0.37147
 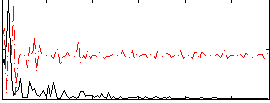 |
Correlation:0.2516
  |
Correlation:0.37147
  |
Correlation:0.37147
  |
Correlation:0.37147
  |
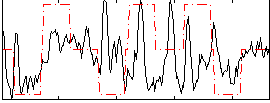
Correlation:0.3096
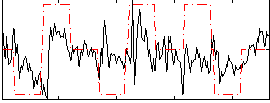  |
Component:191; Jaccard:0.076047
 |
Component:169; Jaccard:0.085702
 |
Component:191; Jaccard:0.096663
 |
Component:383; Jaccard:0.10767
 |
Component:380; Jaccard:0.1082
 |
Component:041; Jaccard:0.096188
 |
Component:288; Jaccard:0.075905
 |
Correlation:0.37147
  |
Correlation:0.31175
  |
Correlation:0.37147
  |
Correlation:0.2516
  |
Correlation:0.22825
  |
Correlation:0.3096
  |
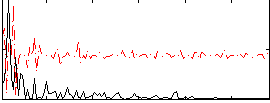
Correlation:0.1357
 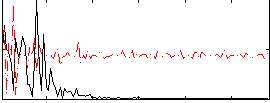 |
Component:383; Jaccard:0.075155
 |
Component:383; Jaccard:0.085156
 |
Component:288; Jaccard:0.091646
 |
Component:015; Jaccard:0.10227
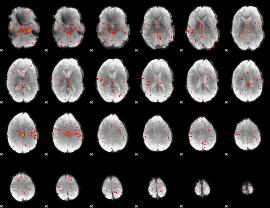 |
Component:288; Jaccard:0.10115
 |
Component:380; Jaccard:0.09533
 |
Component:191; Jaccard:0.074866
 |
Correlation:0.2516
  |
Correlation:0.2516
  |
Correlation:0.1357
  |
Correlation:0.17585
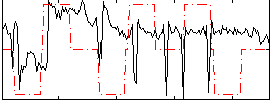 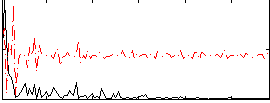 |
Correlation:0.1357
  |
Correlation:0.22825
  |
Correlation:0.37147
  |
Component:378; Jaccard:0.074038
 |
Component:288; Jaccard:0.081799
 |
Component:015; Jaccard:0.091034
 |
Component:288; Jaccard:0.09968
 |
Component:015; Jaccard:0.098691
 |
Component:288; Jaccard:0.09426
 |
Component:380; Jaccard:0.068769
 |
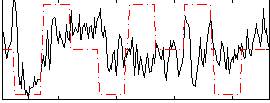
Correlation:0.24003
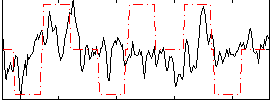  |
Correlation:0.1357
  |
Correlation:0.17585
  |
Correlation:0.1357
  |
Correlation:0.17585
  |
Correlation:0.1357
  |
Correlation:0.22825
  |
Component:288; Jaccard:0.072731
 |
Component:386; Jaccard:0.080366
 |
Component:053; Jaccard:0.087984
 |
Component:053; Jaccard:0.095044
 |
Component:053; Jaccard:0.098001
 |
Component:053; Jaccard:0.086376
 |
Component:053; Jaccard:0.066182
 |
Correlation:0.1357
  |
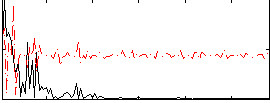
Correlation:0.19741
 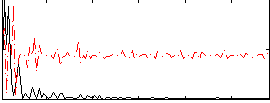 |
Correlation:0.18859
 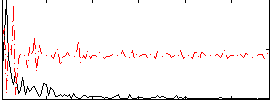 |
Correlation:0.18859
  |
Correlation:0.18859
  |
Correlation:0.18859
  |
Correlation:0.18859
  |
Component:386; Jaccard:0.072521
 |
Component:378; Jaccard:0.079813
 |
Component:198; Jaccard:0.086772
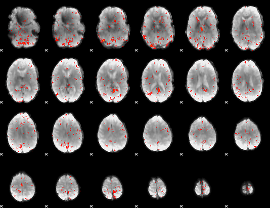 |
Component:198; Jaccard:0.093757
 |
Component:041; Jaccard:0.096648
 |
Component:361; Jaccard:0.085011
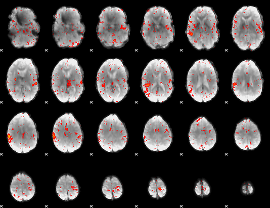 |
Component:361; Jaccard:0.063033
 |
Correlation:0.19741
  |
Correlation:0.24003
  |
Correlation:0.27003
 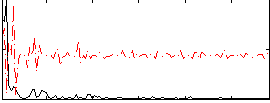 |
Correlation:0.27003
  |
Correlation:0.3096
  |
Correlation:0.31085
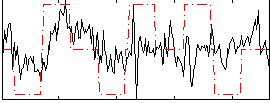 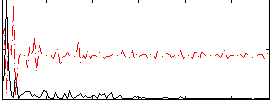 |
Correlation:0.31085
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
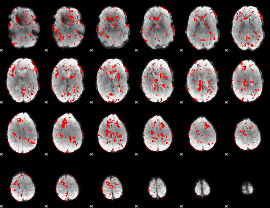 |
GLM binary result thresholded by 1.5
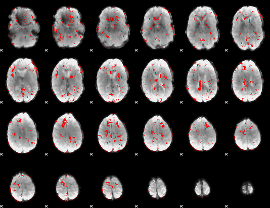 |
GLM binary result thresholded by 2
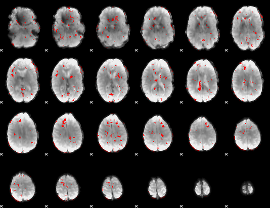 |
GLM binary result thresholded by 2.5
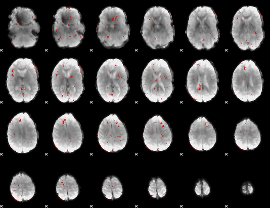 |
GLM binary result thresholded by 3
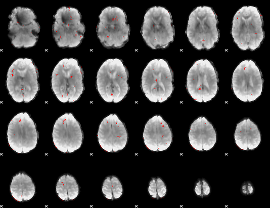 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
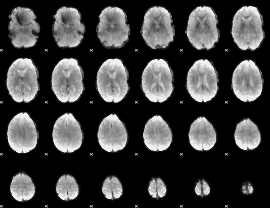 |
GLM Z-score result thresholded by 1
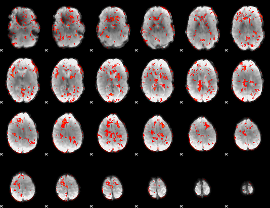 |
GLM Z-score result thresholded by 1.5
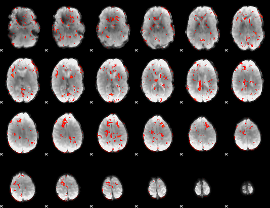 |
GLM Z-score result thresholded by 2
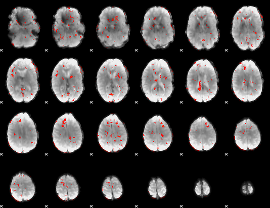 |
GLM Z-score result thresholded by 2.5
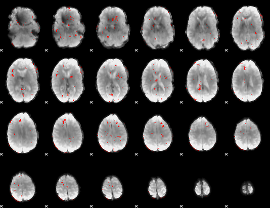 |
GLM Z-score result thresholded by 3
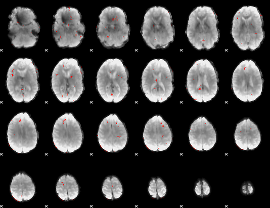 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
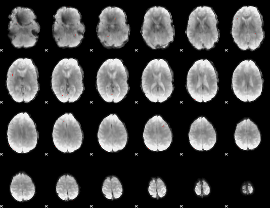 |
Component:078; Jaccard:0.096539
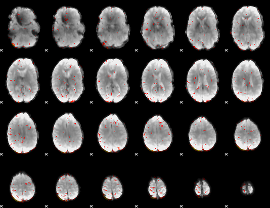 |
Component:078; Jaccard:0.10601
 |
Component:078; Jaccard:0.099713
 |
Component:078; Jaccard:0.07759
 |
Component:025; Jaccard:0.048666
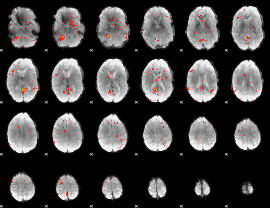 |
Component:025; Jaccard:0.021302
 |
Component:025; Jaccard:0.007374
 |
Correlation:0.24565
 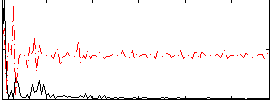 |
Correlation:0.24565
  |
Correlation:0.24565
  |
Correlation:0.24565
  |
Correlation:0.20407
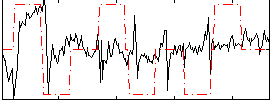  |
Correlation:0.20407
  |
Correlation:0.20407
  |
Component:025; Jaccard:0.08291
 |
Component:025; Jaccard:0.089881
 |
Component:025; Jaccard:0.089145
 |
Component:025; Jaccard:0.073431
 |
Component:078; Jaccard:0.04532
 |
Component:078; Jaccard:0.018916
 |
Component:351; Jaccard:0.004569
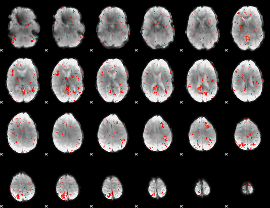 |
Correlation:0.20407
  |
Correlation:0.20407
  |
Correlation:0.20407
  |
Correlation:0.20407
  |
Correlation:0.24565
  |
Correlation:0.24565
  |
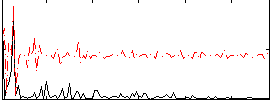
Correlation:0.21998
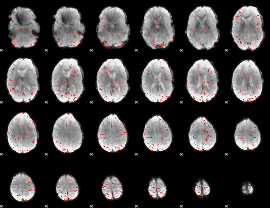
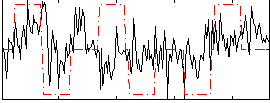  |
Component:342; Jaccard:0.079152
 |
Component:342; Jaccard:0.076132
 |
Component:050; Jaccard:0.065336
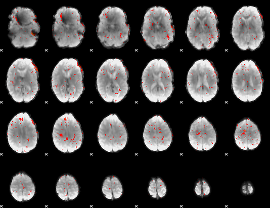 |
Component:050; Jaccard:0.051447
 |
Component:348; Jaccard:0.029579
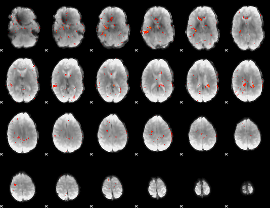 |
Component:351; Jaccard:0.013922
 |
Component:185; Jaccard:0.004432
 |
Correlation:0.044998
  |
Correlation:0.044998
  |
Correlation:0.031676
 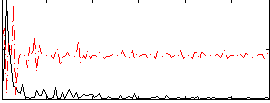 |
Correlation:0.031676
  |
Correlation:0.11036
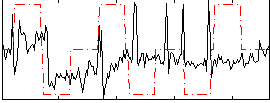 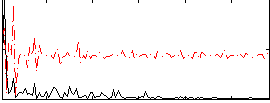 |
Correlation:0.21998
  |
Correlation:0.059115
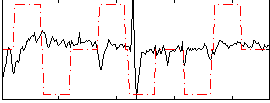 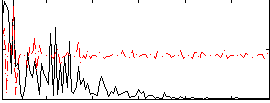 |
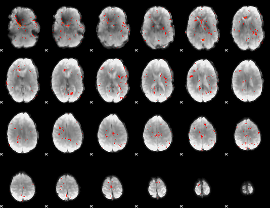
Component:235; Jaccard:0.069509
 |
Component:050; Jaccard:0.070594
 |
Component:342; Jaccard:0.060274
 |
Component:348; Jaccard:0.047085
 |
Component:351; Jaccard:0.029179
 |
Component:244; Jaccard:0.013749
 |
Component:244; Jaccard:0.004234
 |
Correlation:0.12227
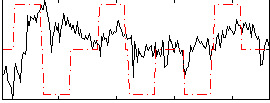 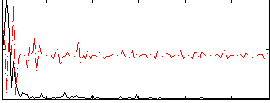 |
Correlation:0.031676
  |
Correlation:0.044998
  |
Correlation:0.11036
  |
Correlation:0.21998
  |
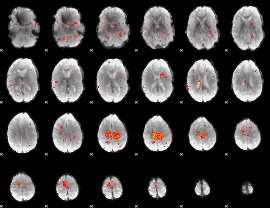
Correlation:0.1369
  |
Correlation:0.1369
  |
Component:363; Jaccard:0.069322
 |
Component:363; Jaccard:0.067922
 |
Component:348; Jaccard:0.058704
 |
Component:351; Jaccard:0.044993
 |
Component:050; Jaccard:0.02797
 |
Component:340; Jaccard:0.01276
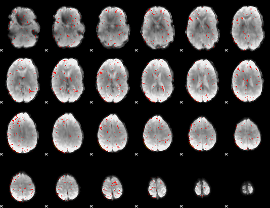 |
Component:278; Jaccard:0.004073
 |
Correlation:0.084064
  |
Correlation:0.084064
  |
Correlation:0.11036
  |
Correlation:0.21998
  |
Correlation:0.031676
  |
Correlation:0.22592
  |
Correlation:0.21484
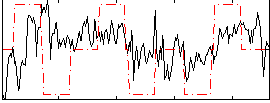 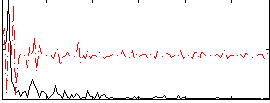 |
Component:185; Jaccard:0.067961
 |
Component:027; Jaccard:0.066447
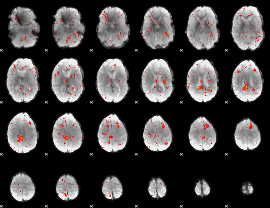 |
Component:351; Jaccard:0.057302
 |
Component:340; Jaccard:0.041084
 |
Component:244; Jaccard:0.026882
 |
Component:278; Jaccard:0.012498
 |
Component:243; Jaccard:0.003832
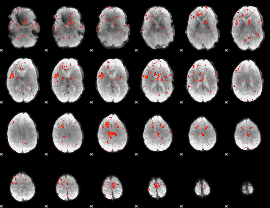 |
Correlation:0.059115
  |
Correlation:0.10446
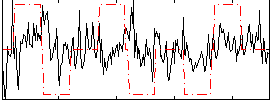 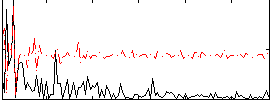 |
Correlation:0.21998
  |
Correlation:0.22592
  |
Correlation:0.1369
  |
Correlation:0.21484
  |
Correlation:-0.011239
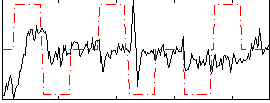 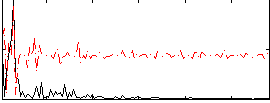 |
Component:027; Jaccard:0.067388
 |
Component:351; Jaccard:0.065326
 |
Component:363; Jaccard:0.057015
 |
Component:342; Jaccard:0.039172
 |
Component:340; Jaccard:0.025141
 |
Component:050; Jaccard:0.01159
 |
Component:078; Jaccard:0.003676
 |
Correlation:0.10446
  |
Correlation:0.21998
  |
Correlation:0.084064
  |
Correlation:0.044998
  |
Correlation:0.22592
  |
Correlation:0.031676
  |
Correlation:0.24565
  |
Component:050; Jaccard:0.066375
 |
Component:348; Jaccard:0.06502
 |
Component:027; Jaccard:0.054312
 |
Component:398; Jaccard:0.039065
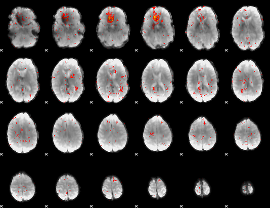 |
Component:278; Jaccard:0.023363
 |
Component:348; Jaccard:0.011441
 |
Component:027; Jaccard:0.003578
 |
Correlation:0.031676
  |
Correlation:0.11036
  |
Correlation:0.10446
  |
Correlation:0.31973
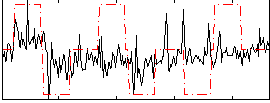 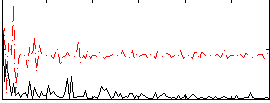 |
Correlation:0.21484
  |
Correlation:0.11036
  |
Correlation:0.10446
  |
Component:351; Jaccard:0.065337
 |
Component:185; Jaccard:0.064683
 |
Component:185; Jaccard:0.053712
 |
Component:244; Jaccard:0.038396
 |
Component:398; Jaccard:0.021879
 |
Component:185; Jaccard:0.010176
 |
Component:340; Jaccard:0.003389
 |
Correlation:0.21998
  |
Correlation:0.059115
  |
Correlation:0.059115
  |
Correlation:0.1369
  |
Correlation:0.31973
  |
Correlation:0.059115
  |
Correlation:0.22592
  |
Component:318; Jaccard:0.065018
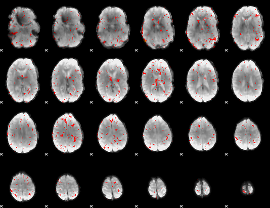 |
Component:284; Jaccard:0.061963
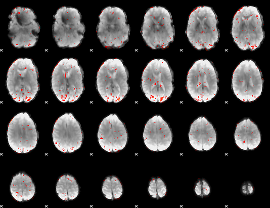 |
Component:340; Jaccard:0.053355
 |
Component:027; Jaccard:0.037698
 |
Component:185; Jaccard:0.021337
 |
Component:027; Jaccard:0.010159
 |
Component:170; Jaccard:0.003037
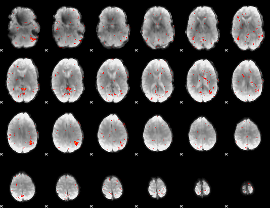 |
Correlation:0.24583
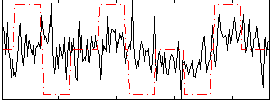 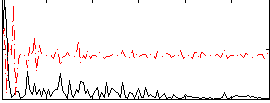 |
Correlation:0.13223
  |
Correlation:0.22592
  |
Correlation:0.10446
  |
Correlation:0.059115
  |
Correlation:0.10446
  |
Correlation:0.16829
  |
Cope: 05:neg_MATCH BACK
|
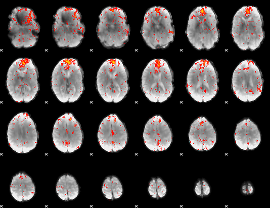
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
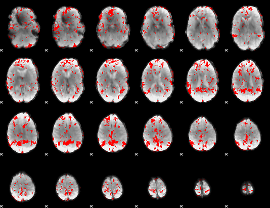 |
GLM binary result thresholded by 2
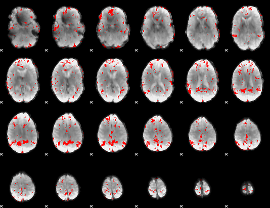 |
GLM binary result thresholded by 2.5
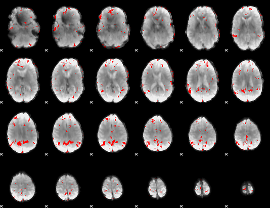 |
GLM binary result thresholded by 3
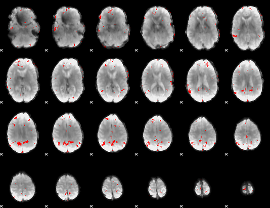 |
GLM binary result thresholded by 3.5
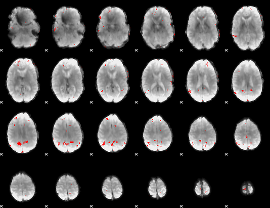 |
GLM binary result thresholded by 4
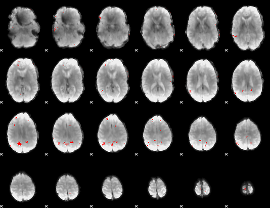 |
GLM Z-score result thresholded by 1
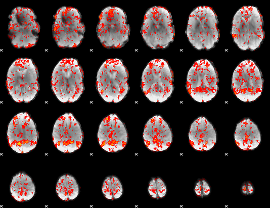 |
GLM Z-score result thresholded by 1.5
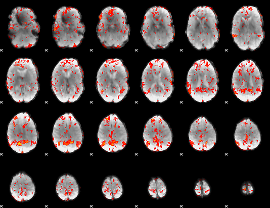 |
GLM Z-score result thresholded by 2
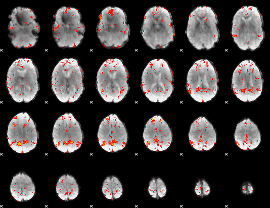 |
GLM Z-score result thresholded by 2.5
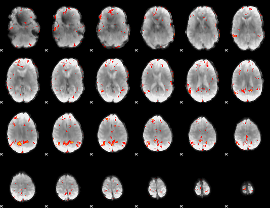 |
GLM Z-score result thresholded by 3
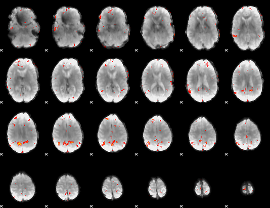 |
GLM Z-score result thresholded by 3.5
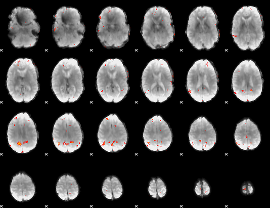 |
GLM Z-score result thresholded by 4
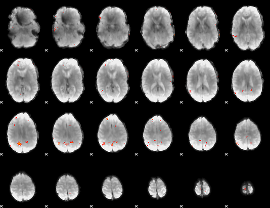 |
Component:172; Jaccard:0.12099
 |
Component:172; Jaccard:0.14485
 |
Component:172; Jaccard:0.15643
 |
Component:172; Jaccard:0.14441
 |
Component:172; Jaccard:0.11335
 |
Component:172; Jaccard:0.076868
 |
Component:172; Jaccard:0.048377
 |
Correlation:-0.039769
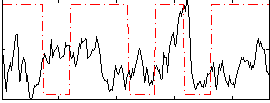 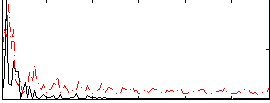 |
Correlation:-0.039769
  |
Correlation:-0.039769
  |
Correlation:-0.039769
  |
Correlation:-0.039769
  |
Correlation:-0.039769
  |
Correlation:-0.039769
  |
Component:390; Jaccard:0.11769
 |
Component:390; Jaccard:0.13084
 |
Component:390; Jaccard:0.12944
 |
Component:390; Jaccard:0.11196
 |
Component:100; Jaccard:0.085947
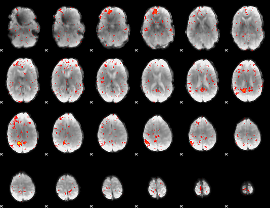 |
Component:100; Jaccard:0.062469
 |
Component:100; Jaccard:0.039152
 |
Correlation:0.010289
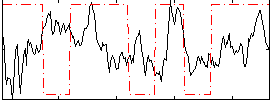 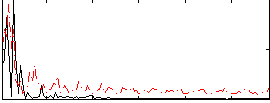 |
Correlation:0.010289
  |
Correlation:0.010289
  |
Correlation:0.010289
  |
Correlation:0.23506
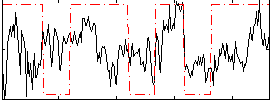 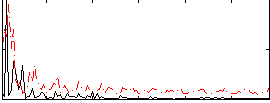 |
Correlation:0.23506
  |
Correlation:0.23506
  |
Component:059; Jaccard:0.10091
 |
Component:100; Jaccard:0.10629
 |
Component:100; Jaccard:0.11411
 |
Component:100; Jaccard:0.10752
 |
Component:390; Jaccard:0.081828
 |
Component:390; Jaccard:0.053943
 |
Component:390; Jaccard:0.034418
 |
Correlation:0.059224
  |
Correlation:0.23506
  |
Correlation:0.23506
  |
Correlation:0.23506
  |
Correlation:0.010289
  |
Correlation:0.010289
  |
Correlation:0.010289
  |
Component:100; Jaccard:0.092024
 |
Component:059; Jaccard:0.10549
 |
Component:059; Jaccard:0.098144
 |
Component:059; Jaccard:0.080537
 |
Component:059; Jaccard:0.053763
 |
Component:035; Jaccard:0.033671
 |
Component:079; Jaccard:0.021587
 |
Correlation:0.23506
  |
Correlation:0.059224
  |
Correlation:0.059224
  |
Correlation:0.059224
  |
Correlation:0.059224
  |
Correlation:0.12268
 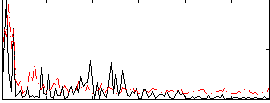 |
Correlation:0.11656
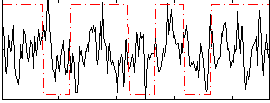 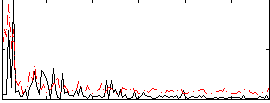 |
Component:283; Jaccard:0.088837
 |
Component:283; Jaccard:0.089715
 |
Component:283; Jaccard:0.079552
 |
Component:366; Jaccard:0.062306
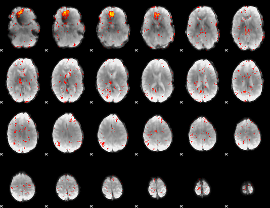 |
Component:035; Jaccard:0.051438
 |
Component:192; Jaccard:0.031685
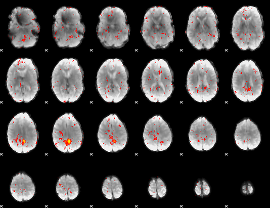 |
Component:192; Jaccard:0.019815
 |
Correlation:-0.11936
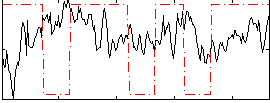  |
Correlation:-0.11936
  |
Correlation:-0.11936
  |
Correlation:0.1912
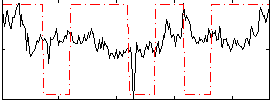  |
Correlation:0.12268
  |
Correlation:0.001416
  |
Correlation:0.001416
  |
Component:327; Jaccard:0.085169
 |
Component:327; Jaccard:0.08515
 |
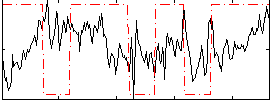
Component:162; Jaccard:0.075712
 |
Component:192; Jaccard:0.061807
 |
Component:192; Jaccard:0.046046
 |
Component:079; Jaccard:0.031291
 |
Component:366; Jaccard:0.019023
 |
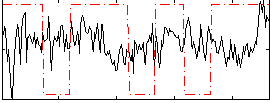
Correlation:0.044628
  |
Correlation:0.044628
  |
Correlation:0.0090049
 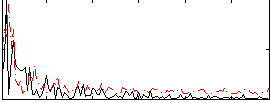 |
Correlation:0.001416
  |
Correlation:0.001416
  |
Correlation:0.11656
  |
Correlation:0.1912
  |
Component:162; Jaccard:0.080168
 |
Component:162; Jaccard:0.081678
 |
Component:327; Jaccard:0.075413
 |
Component:162; Jaccard:0.061423
 |
Component:382; Jaccard:0.045879
 |
Component:382; Jaccard:0.029547
 |
Component:382; Jaccard:0.017625
 |
Correlation:0.0090049
  |
Correlation:0.0090049
  |
Correlation:0.044628
  |
Correlation:0.0090049
  |
Correlation:-0.16805
  |
Correlation:-0.16805
  |
Correlation:-0.16805
  |
Component:232; Jaccard:0.078022
 |
Component:366; Jaccard:0.078727
 |
Component:366; Jaccard:0.07365
 |
Component:327; Jaccard:0.059208
 |
Component:079; Jaccard:0.044656
 |
Component:059; Jaccard:0.028988
 |
Component:035; Jaccard:0.017587
 |
Correlation:-0.027244
  |
Correlation:0.1912
  |
Correlation:0.1912
  |
Correlation:0.044628
  |
Correlation:0.11656
  |
Correlation:0.059224
  |
Correlation:0.12268
  |
Component:366; Jaccard:0.076156
 |
Component:232; Jaccard:0.07783
 |
Component:192; Jaccard:0.071532
 |
Component:035; Jaccard:0.059076
 |
Component:162; Jaccard:0.04458
 |
Component:366; Jaccard:0.028843
 |
Component:182; Jaccard:0.016715
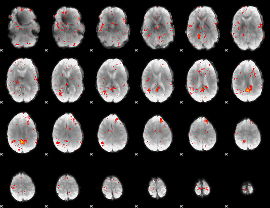 |
Correlation:0.1912
  |
Correlation:-0.027244
  |
Correlation:0.001416
  |
Correlation:0.12268
  |
Correlation:0.0090049
  |
Correlation:0.1912
  |
Correlation:0.0041217
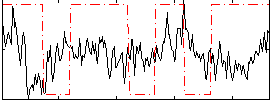 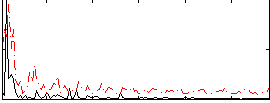 |
Component:346; Jaccard:0.07418
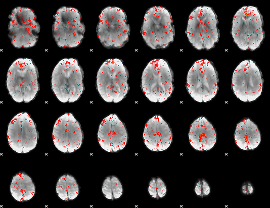 |
Component:383; Jaccard:0.075461
 |
Component:399; Jaccard:0.070355
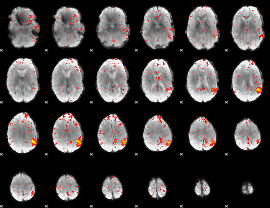 |
Component:283; Jaccard:0.058395
 |
Component:366; Jaccard:0.043152
 |
Component:162; Jaccard:0.026199
 |
Component:059; Jaccard:0.01558
 |
Correlation:-0.0069778
  |
Correlation:-0.039712
  |
Correlation:0.020292
  |
Correlation:-0.11936
  |
Correlation:0.1912
  |
Correlation:0.0090049
  |
Correlation:0.059224
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
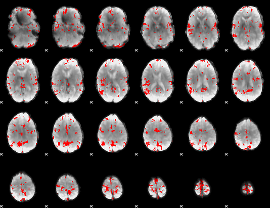 |
GLM binary result thresholded by 4
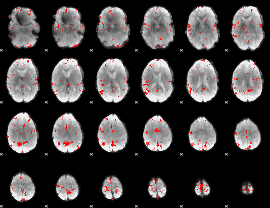 |
GLM Z-score result thresholded by 1
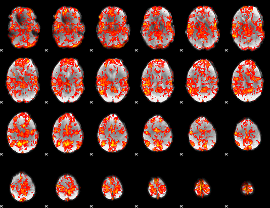 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
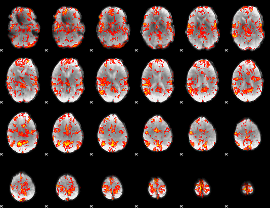 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:143; Jaccard:0.09669
 |
Component:143; Jaccard:0.11198
 |
Component:143; Jaccard:0.1294
 |
Component:143; Jaccard:0.14268
 |
Component:143; Jaccard:0.15114
 |
Component:172; Jaccard:0.15611
 |
Component:172; Jaccard:0.14984
 |
Correlation:0.34429
  |
Correlation:0.34429
  |
Correlation:0.34429
  |
Correlation:0.34429
  |
Correlation:0.34429
  |
Correlation:0.41899
  |
Correlation:0.41899
  |
Component:059; Jaccard:0.093414
 |
Component:383; Jaccard:0.1071
 |
Component:383; Jaccard:0.12297
 |
Component:383; Jaccard:0.13779
 |
Component:383; Jaccard:0.14999
 |
Component:383; Jaccard:0.15495
 |
Component:383; Jaccard:0.14545
 |
Correlation:0.36694
  |
Correlation:0.38479
  |
Correlation:0.38479
  |
Correlation:0.38479
  |
Correlation:0.38479
  |
Correlation:0.38479
  |
Correlation:0.38479
  |
Component:383; Jaccard:0.092538
 |
Component:059; Jaccard:0.10475
 |
Component:172; Jaccard:0.11732
 |
Component:172; Jaccard:0.13497
 |
Component:172; Jaccard:0.14924
 |
Component:143; Jaccard:0.15316
 |
Component:143; Jaccard:0.13528
 |
Correlation:0.38479
  |
Correlation:0.36694
  |
Correlation:0.41899
  |
Correlation:0.41899
  |
Correlation:0.41899
  |
Correlation:0.34429
  |
Correlation:0.34429
  |
Component:080; Jaccard:0.087803
 |
Component:172; Jaccard:0.10074
 |
Component:059; Jaccard:0.11669
 |
Component:059; Jaccard:0.12653
 |
Component:059; Jaccard:0.12934
 |
Component:297; Jaccard:0.12397
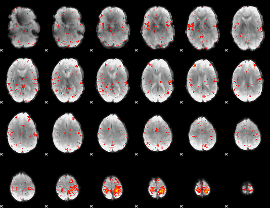 |
Component:297; Jaccard:0.10955
 |
Correlation:0.44374
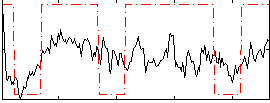  |
Correlation:0.41899
  |
Correlation:0.36694
  |
Correlation:0.36694
  |
Correlation:0.36694
  |
Correlation:0.39418
 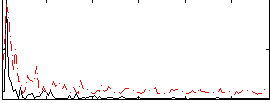 |
Correlation:0.39418
  |
Component:172; Jaccard:0.085824
 |
Component:297; Jaccard:0.096008
 |
Component:053; Jaccard:0.10778
 |
Component:297; Jaccard:0.11867
 |
Component:297; Jaccard:0.1257
 |
Component:059; Jaccard:0.12127
 |
Component:390; Jaccard:0.10336
 |
Correlation:0.41899
  |
Correlation:0.39418
  |
Correlation:0.33778
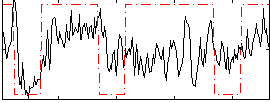  |
Correlation:0.39418
  |
Correlation:0.39418
  |
Correlation:0.36694
  |
Correlation:0.24366
  |
Component:246; Jaccard:0.085119
 |
Component:080; Jaccard:0.095896
 |
Component:297; Jaccard:0.10772
 |
Component:053; Jaccard:0.11762
 |
Component:053; Jaccard:0.12044
 |
Component:053; Jaccard:0.11504
 |
Component:059; Jaccard:0.10285
 |
Correlation:0.27374
 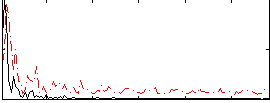 |
Correlation:0.44374
  |
Correlation:0.39418
  |
Correlation:0.33778
  |
Correlation:0.33778
  |
Correlation:0.33778
  |
Correlation:0.36694
  |
Component:297; Jaccard:0.084241
 |
Component:053; Jaccard:0.094368
 |
Component:080; Jaccard:0.10346
 |
Component:080; Jaccard:0.11003
 |
Component:390; Jaccard:0.11366
 |
Component:390; Jaccard:0.11338
 |
Component:053; Jaccard:0.10168
 |
Correlation:0.39418
  |
Correlation:0.33778
  |
Correlation:0.44374
  |
Correlation:0.44374
  |
Correlation:0.24366
  |
Correlation:0.24366
  |
Correlation:0.33778
  |
Component:380; Jaccard:0.083676
 |
Component:390; Jaccard:0.093038
 |
Component:390; Jaccard:0.10297
 |
Component:390; Jaccard:0.10928
 |
Component:080; Jaccard:0.1113
 |
Component:080; Jaccard:0.10606
 |
Component:182; Jaccard:0.096854
 |
Correlation:0.37041
 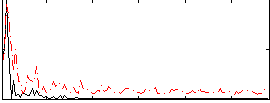 |
Correlation:0.24366
  |
Correlation:0.24366
  |
Correlation:0.24366
  |
Correlation:0.44374
  |
Correlation:0.44374
  |
Correlation:0.31389
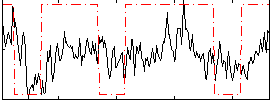 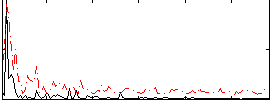 |
Component:390; Jaccard:0.083446
 |
Component:246; Jaccard:0.091491
 |
Component:288; Jaccard:0.099918
 |
Component:288; Jaccard:0.10679
 |
Component:288; Jaccard:0.10853
 |
Component:182; Jaccard:0.10359
 |
Component:100; Jaccard:0.093322
 |
Correlation:0.24366
  |
Correlation:0.27374
  |
Correlation:0.15982
  |
Correlation:0.15982
  |
Correlation:0.15982
  |
Correlation:0.31389
  |
Correlation:0.29232
  |
Component:053; Jaccard:0.081933
 |
Component:380; Jaccard:0.091405
 |
Component:380; Jaccard:0.098162
 |
Component:380; Jaccard:0.10252
 |
Component:380; Jaccard:0.10206
 |
Component:288; Jaccard:0.10073
 |
Component:080; Jaccard:0.090255
 |
Correlation:0.33778
  |
Correlation:0.37041
  |
Correlation:0.37041
  |
Correlation:0.37041
  |
Correlation:0.37041
  |
Correlation:0.15982
  |
Correlation:0.44374
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































