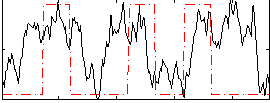
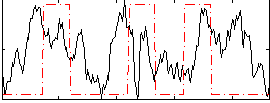
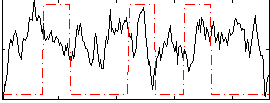
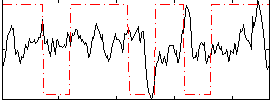
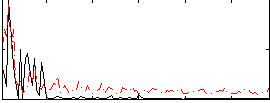
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
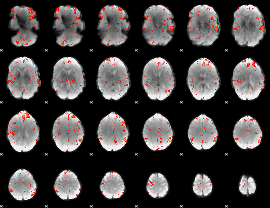
GLM result group-wise
 |
GLM binary result thresholded by 1
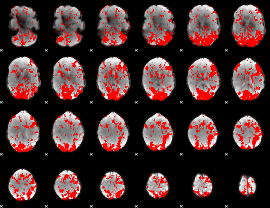 |
GLM binary result thresholded by 1.5
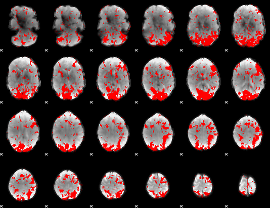 |
GLM binary result thresholded by 2
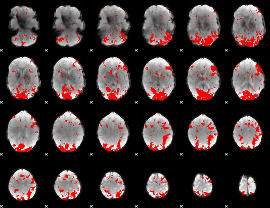 |
GLM binary result thresholded by 2.5
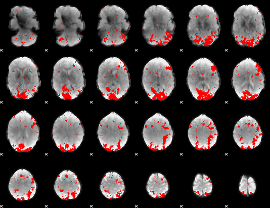 |
GLM binary result thresholded by 3
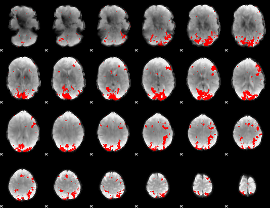 |
GLM binary result thresholded by 3.5
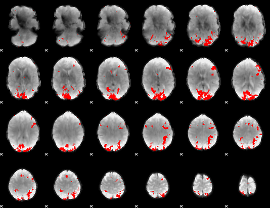 |
GLM binary result thresholded by 4
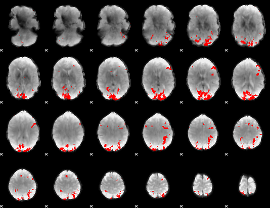 |
GLM Z-score result thresholded by 1
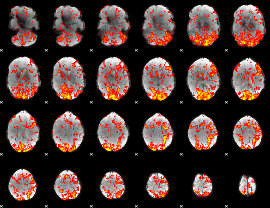 |
GLM Z-score result thresholded by 1.5
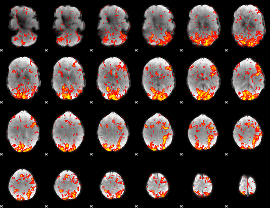 |
GLM Z-score result thresholded by 2
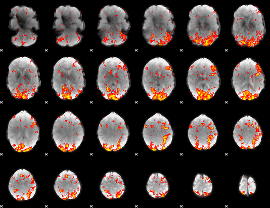 |
GLM Z-score result thresholded by 2.5
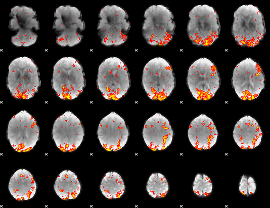 |
GLM Z-score result thresholded by 3
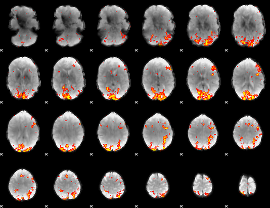 |
GLM Z-score result thresholded by 3.5
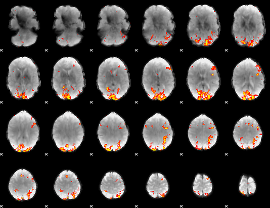 |
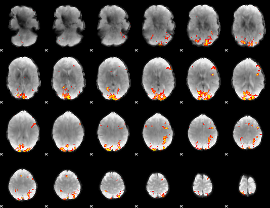
GLM Z-score result thresholded by 4
 |
Component:029; Jaccard:0.16934
 |
Component:029; Jaccard:0.21612
 |
Component:029; Jaccard:0.26513
 |
Component:029; Jaccard:0.30291
 |
Component:029; Jaccard:0.32469
 |
Component:029; Jaccard:0.31346
 |
Component:388; Jaccard:0.29827
 |
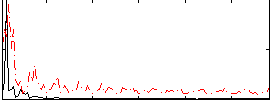
Correlation:0.10846
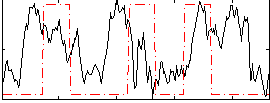 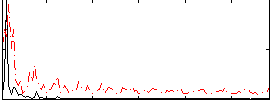 |
Correlation:0.10846
  |
Correlation:0.10846
  |
Correlation:0.10846
  |
Correlation:0.10846
  |
Correlation:0.10846
  |
Correlation:0.27904
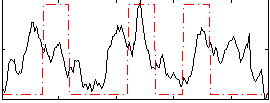 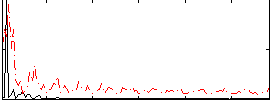 |
Component:331; Jaccard:0.15644
 |
Component:266; Jaccard:0.19134
 |
Component:388; Jaccard:0.23536
 |
Component:388; Jaccard:0.28038
 |
Component:388; Jaccard:0.30712
 |
Component:388; Jaccard:0.31213
 |
Component:029; Jaccard:0.28279
 |
Correlation:0.090534
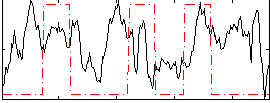 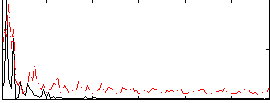 |
Correlation:0.24221
  |
Correlation:0.27904
  |
Correlation:0.27904
  |
Correlation:0.27904
  |
Correlation:0.27904
  |
Correlation:0.10846
  |
Component:266; Jaccard:0.14964
 |
Component:331; Jaccard:0.1887
 |
Component:266; Jaccard:0.23343
 |
Component:266; Jaccard:0.26553
 |
Component:266; Jaccard:0.27362
 |
Component:056; Jaccard:0.26588
 |
Component:056; Jaccard:0.2516
 |
Correlation:0.24221
  |
Correlation:0.090534
  |
Correlation:0.24221
  |
Correlation:0.24221
  |
Correlation:0.24221
  |
Correlation:0.13551
 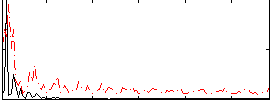 |
Correlation:0.13551
  |
Component:056; Jaccard:0.14855
 |
Component:056; Jaccard:0.18687
 |
Component:056; Jaccard:0.22696
 |
Component:056; Jaccard:0.25695
 |
Component:056; Jaccard:0.27104
 |
Component:266; Jaccard:0.25511
 |
Component:266; Jaccard:0.22716
 |
Correlation:0.13551
  |
Correlation:0.13551
  |
Correlation:0.13551
  |
Correlation:0.13551
  |
Correlation:0.13551
  |
Correlation:0.24221
  |
Correlation:0.24221
  |
Component:070; Jaccard:0.14348
 |
Component:388; Jaccard:0.18516
 |
Component:331; Jaccard:0.21481
 |
Component:331; Jaccard:0.22916
 |
Component:331; Jaccard:0.22688
 |
Component:331; Jaccard:0.21459
 |
Component:331; Jaccard:0.19111
 |
Correlation:0.18895
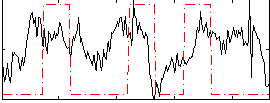 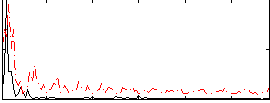 |
Correlation:0.27904
  |
Correlation:0.090534
  |
Correlation:0.090534
  |
Correlation:0.090534
  |
Correlation:0.090534
  |
Correlation:0.090534
  |
Component:388; Jaccard:0.14157
 |
Component:070; Jaccard:0.17001
 |
Component:070; Jaccard:0.19117
 |
Component:070; Jaccard:0.19804
 |
Component:070; Jaccard:0.19137
 |
Component:070; Jaccard:0.17644
 |
Component:070; Jaccard:0.15475
 |
Correlation:0.27904
  |
Correlation:0.18895
  |
Correlation:0.18895
  |
Correlation:0.18895
  |
Correlation:0.18895
  |
Correlation:0.18895
  |
Correlation:0.18895
  |
Component:282; Jaccard:0.12817
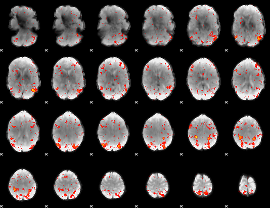 |
Component:142; Jaccard:0.14561
 |
Component:142; Jaccard:0.16949
 |
Component:142; Jaccard:0.18603
 |
Component:142; Jaccard:0.18978
 |
Component:142; Jaccard:0.17557
 |
Component:142; Jaccard:0.15231
 |
Correlation:0.22008
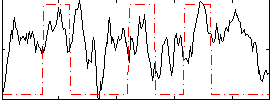  |
Correlation:0.19937
  |
Correlation:0.19937
  |
Correlation:0.19937
  |
Correlation:0.19937
  |
Correlation:0.19937
  |
Correlation:0.19937
  |
Component:376; Jaccard:0.12395
 |
Component:282; Jaccard:0.14463
 |
Component:376; Jaccard:0.15391
 |
Component:376; Jaccard:0.15752
 |
Component:376; Jaccard:0.15098
 |
Component:376; Jaccard:0.13563
 |
Component:376; Jaccard:0.11757
 |
Correlation:0.25555
  |
Correlation:0.22008
  |
Correlation:0.25555
  |
Correlation:0.25555
  |
Correlation:0.25555
  |
Correlation:0.25555
  |
Correlation:0.25555
  |
Component:379; Jaccard:0.12305
 |
Component:376; Jaccard:0.14117
 |
Component:379; Jaccard:0.1506
 |
Component:379; Jaccard:0.15103
 |
Component:379; Jaccard:0.14148
 |
Component:379; Jaccard:0.12488
 |
Component:190; Jaccard:0.10529
 |
Correlation:0.26116
  |
Correlation:0.25555
  |
Correlation:0.26116
  |
Correlation:0.26116
  |
Correlation:0.26116
  |
Correlation:0.26116
  |
Correlation:-0.0019876
 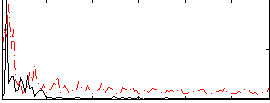 |
Component:347; Jaccard:0.12137
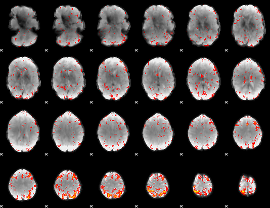 |
Component:379; Jaccard:0.14067
 |
Component:282; Jaccard:0.14998
 |
Component:282; Jaccard:0.14575
 |
Component:190; Jaccard:0.13256
 |
Component:190; Jaccard:0.1206
 |
Component:379; Jaccard:0.10339
 |
Correlation:0.16885
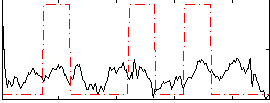  |
Correlation:0.26116
  |
Correlation:0.22008
  |
Correlation:0.22008
  |
Correlation:-0.0019876
  |
Correlation:-0.0019876
  |
Correlation:0.26116
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
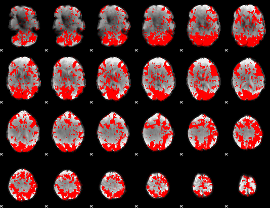 |
GLM binary result thresholded by 1.5
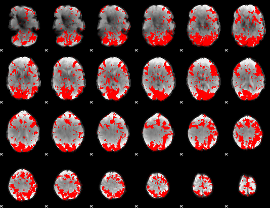 |
GLM binary result thresholded by 2
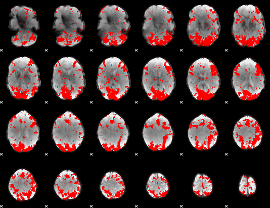 |
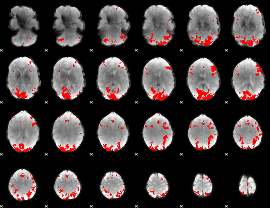
GLM binary result thresholded by 2.5
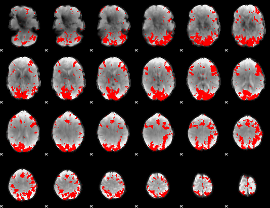 |
GLM binary result thresholded by 3
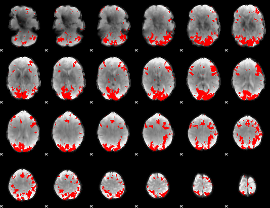 |
GLM binary result thresholded by 3.5
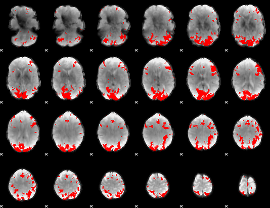 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
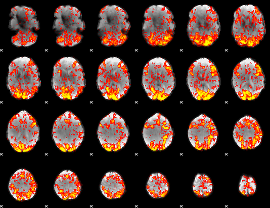 |
GLM Z-score result thresholded by 1.5
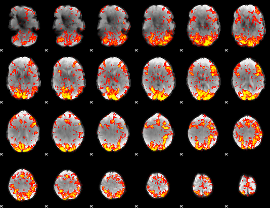 |
GLM Z-score result thresholded by 2
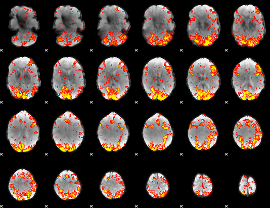 |
GLM Z-score result thresholded by 2.5
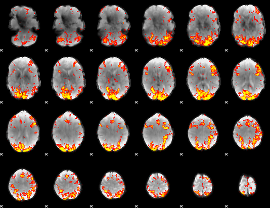 |
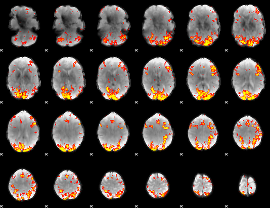
GLM Z-score result thresholded by 3
 |
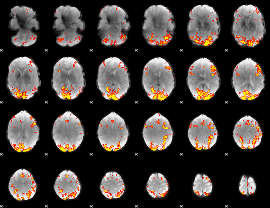
GLM Z-score result thresholded by 3.5
 |
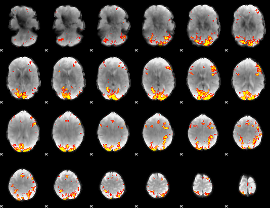
GLM Z-score result thresholded by 4
 |
Component:029; Jaccard:0.14363
 |
Component:029; Jaccard:0.181
 |
Component:029; Jaccard:0.22494
 |
Component:029; Jaccard:0.27065
 |
Component:029; Jaccard:0.31443
 |
Component:029; Jaccard:0.34721
 |
Component:029; Jaccard:0.36646
 |
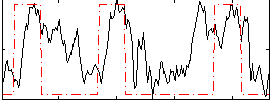
Correlation:0.41662
 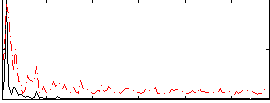 |
Correlation:0.41662
  |
Correlation:0.41662
  |
Correlation:0.41662
  |
Correlation:0.41662
  |
Correlation:0.41662
  |
Correlation:0.41662
  |
Component:331; Jaccard:0.14008
 |
Component:331; Jaccard:0.16991
 |
Component:331; Jaccard:0.19861
 |
Component:266; Jaccard:0.22567
 |
Component:266; Jaccard:0.25783
 |
Component:266; Jaccard:0.27906
 |
Component:388; Jaccard:0.30089
 |
Correlation:0.33604
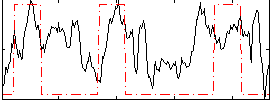 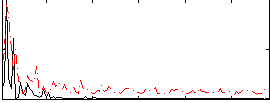 |
Correlation:0.33604
  |
Correlation:0.33604
  |
Correlation:0.26268
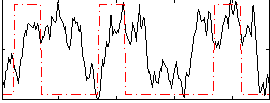 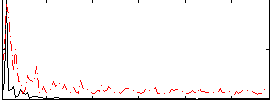 |
Correlation:0.26268
  |
Correlation:0.26268
  |
Correlation:0.29096
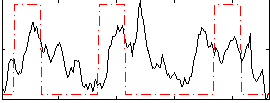 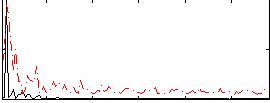 |
Component:070; Jaccard:0.13661
 |
Component:070; Jaccard:0.16285
 |
Component:266; Jaccard:0.19037
 |
Component:331; Jaccard:0.22312
 |
Component:388; Jaccard:0.24528
 |
Component:388; Jaccard:0.27713
 |
Component:266; Jaccard:0.28325
 |
Correlation:0.22593
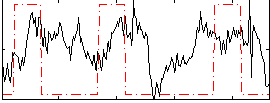 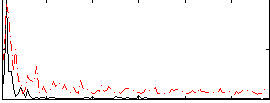 |
Correlation:0.22593
  |
Correlation:0.26268
  |
Correlation:0.33604
  |
Correlation:0.29096
  |
Correlation:0.29096
  |
Correlation:0.26268
  |
Component:144; Jaccard:0.1301
 |
Component:266; Jaccard:0.15433
 |
Component:070; Jaccard:0.18863
 |
Component:388; Jaccard:0.21221
 |
Component:331; Jaccard:0.23948
 |
Component:331; Jaccard:0.24562
 |
Component:056; Jaccard:0.25041
 |
Correlation:0.38322
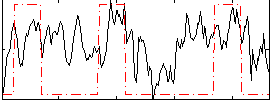 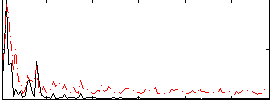 |
Correlation:0.26268
  |
Correlation:0.22593
  |
Correlation:0.29096
  |
Correlation:0.33604
  |
Correlation:0.33604
  |
Correlation:0.27113
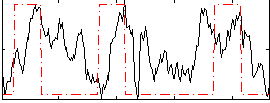 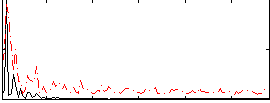 |
Component:056; Jaccard:0.12371
 |
Component:056; Jaccard:0.15063
 |
Component:056; Jaccard:0.17786
 |
Component:070; Jaccard:0.20789
 |
Component:056; Jaccard:0.22342
 |
Component:056; Jaccard:0.24178
 |
Component:331; Jaccard:0.24209
 |
Correlation:0.27113
  |
Correlation:0.27113
  |
Correlation:0.27113
  |
Correlation:0.22593
  |
Correlation:0.27113
  |
Correlation:0.27113
  |
Correlation:0.33604
  |
Component:266; Jaccard:0.12276
 |
Component:144; Jaccard:0.14691
 |
Component:388; Jaccard:0.17722
 |
Component:056; Jaccard:0.20285
 |
Component:070; Jaccard:0.22062
 |
Component:070; Jaccard:0.22165
 |
Component:070; Jaccard:0.21344
 |
Correlation:0.26268
  |
Correlation:0.38322
  |
Correlation:0.29096
  |
Correlation:0.27113
  |
Correlation:0.22593
  |
Correlation:0.22593
  |
Correlation:0.22593
  |
Component:379; Jaccard:0.12271
 |
Component:379; Jaccard:0.14633
 |
Component:379; Jaccard:0.16834
 |
Component:379; Jaccard:0.18438
 |
Component:379; Jaccard:0.19381
 |
Component:142; Jaccard:0.19662
 |
Component:142; Jaccard:0.19963
 |
Correlation:0.29187
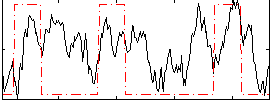 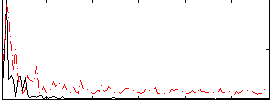 |
Correlation:0.29187
  |
Correlation:0.29187
  |
Correlation:0.29187
  |
Correlation:0.29187
  |
Correlation:0.34107
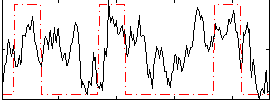  |
Correlation:0.34107
  |
Component:284; Jaccard:0.12216
 |
Component:388; Jaccard:0.14457
 |
Component:144; Jaccard:0.16331
 |
Component:144; Jaccard:0.17285
 |
Component:142; Jaccard:0.18809
 |
Component:379; Jaccard:0.19239
 |
Component:379; Jaccard:0.18238
 |
Correlation:0.24096
  |
Correlation:0.29096
  |
Correlation:0.38322
  |
Correlation:0.38322
  |
Correlation:0.34107
  |
Correlation:0.29187
  |
Correlation:0.29187
  |
Component:290; Jaccard:0.12093
 |
Component:284; Jaccard:0.14135
 |
Component:284; Jaccard:0.15687
 |
Component:142; Jaccard:0.17186
 |
Component:144; Jaccard:0.1773
 |
Component:190; Jaccard:0.1742
 |
Component:190; Jaccard:0.17182
 |
Correlation:0.053358
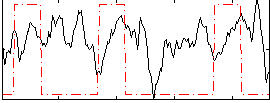 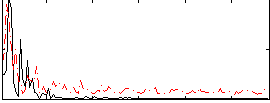 |
Correlation:0.24096
  |
Correlation:0.24096
  |
Correlation:0.34107
  |
Correlation:0.38322
  |
Correlation:0.30343
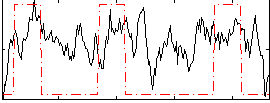 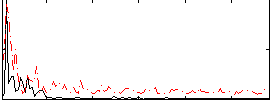 |
Correlation:0.30343
  |
Component:190; Jaccard:0.11801
 |
Component:190; Jaccard:0.13435
 |
Component:142; Jaccard:0.15174
 |
Component:284; Jaccard:0.16703
 |
Component:190; Jaccard:0.17185
 |
Component:144; Jaccard:0.17336
 |
Component:144; Jaccard:0.16166
 |
Correlation:0.30343
  |
Correlation:0.30343
  |
Correlation:0.34107
  |
Correlation:0.24096
  |
Correlation:0.30343
  |
Correlation:0.38322
  |
Correlation:0.38322
  |
Cope: 03:MATCH-REL BACK
|
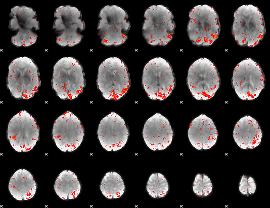
GLM result group-wise
 |
GLM binary result thresholded by 1
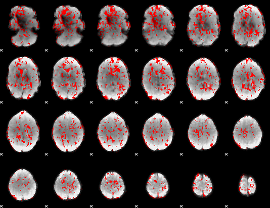 |
GLM binary result thresholded by 1.5
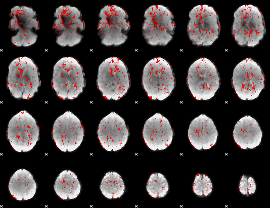 |
GLM binary result thresholded by 2
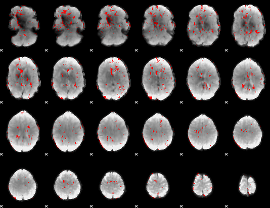 |
GLM binary result thresholded by 2.5
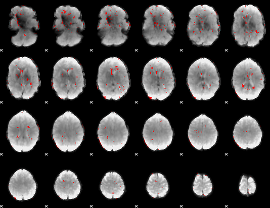 |
GLM binary result thresholded by 3
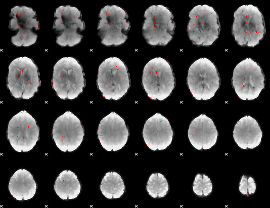 |
GLM binary result thresholded by 3.5
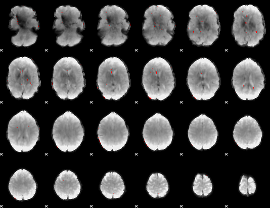 |
GLM binary result thresholded by 4
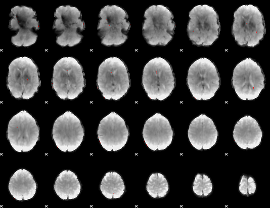 |
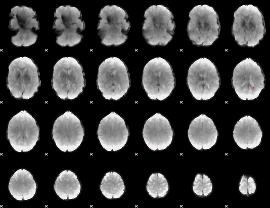
GLM Z-score result thresholded by 1
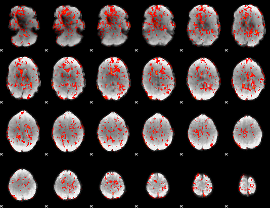 |
GLM Z-score result thresholded by 1.5
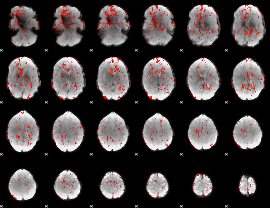 |
GLM Z-score result thresholded by 2
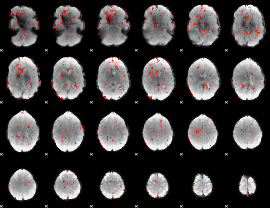 |
GLM Z-score result thresholded by 2.5
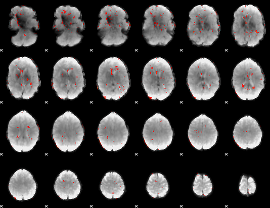 |
GLM Z-score result thresholded by 3
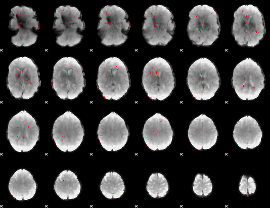 |
GLM Z-score result thresholded by 3.5
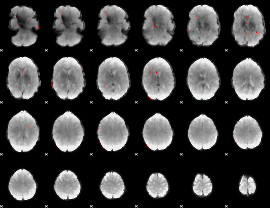 |
GLM Z-score result thresholded by 4
 |
Component:324; Jaccard:0.099107
 |
Component:140; Jaccard:0.098507
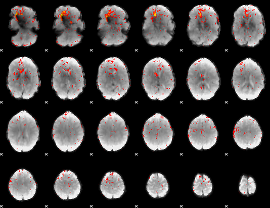 |
Component:140; Jaccard:0.084025
 |
Component:130; Jaccard:0.069054
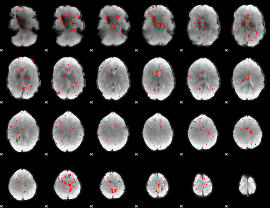 |
Component:299; Jaccard:0.044259
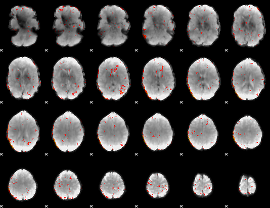 |
Component:299; Jaccard:0.026285
 |
Component:299; Jaccard:0.010636
 |
Correlation:-0.021582
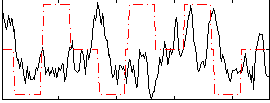 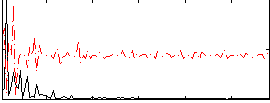 |
Correlation:0.0026987
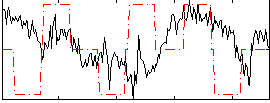  |
Correlation:0.0026987
  |
Correlation:0.079987
  |
Correlation:0.29293
 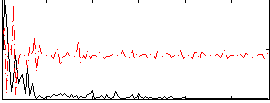 |
Correlation:0.29293
  |
Correlation:0.29293
  |
Component:140; Jaccard:0.096136
 |
Component:324; Jaccard:0.093333
 |
Component:299; Jaccard:0.079109
 |
Component:299; Jaccard:0.063692
 |
Component:130; Jaccard:0.042789
 |
Component:130; Jaccard:0.02147
 |
Component:130; Jaccard:0.007104
 |
Correlation:0.0026987
  |
Correlation:-0.021582
  |
Correlation:0.29293
  |
Correlation:0.29293
  |
Correlation:0.079987
  |
Correlation:0.079987
  |
Correlation:0.079987
  |
Component:297; Jaccard:0.075848
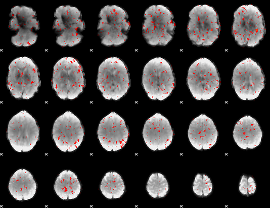 |
Component:130; Jaccard:0.081506
 |
Component:130; Jaccard:0.078772
 |
Component:104; Jaccard:0.056559
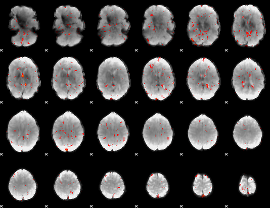 |
Component:301; Jaccard:0.033482
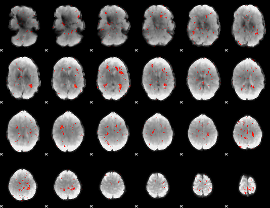 |
Component:103; Jaccard:0.014898
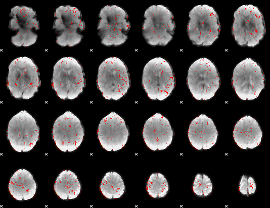 |
Component:103; Jaccard:0.006032
 |
Correlation:0.053217
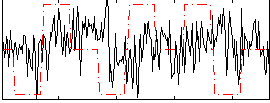 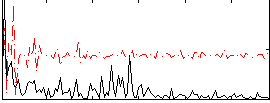 |
Correlation:0.079987
  |
Correlation:0.079987
  |
Correlation:0.10911
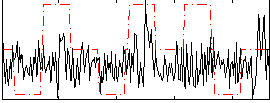 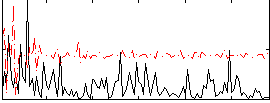 |
Correlation:0.12724
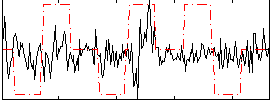 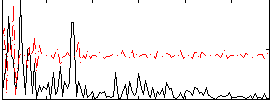 |
Correlation:0.088838
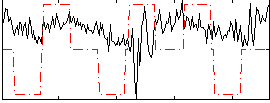 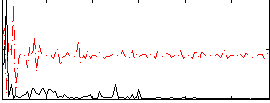 |
Correlation:0.088838
  |
Component:299; Jaccard:0.074041
 |
Component:299; Jaccard:0.080966
 |
Component:104; Jaccard:0.073893
 |
Component:140; Jaccard:0.056132
 |
Component:104; Jaccard:0.032477
 |
Component:161; Jaccard:0.014223
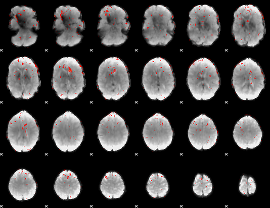 |
Component:349; Jaccard:0.005845
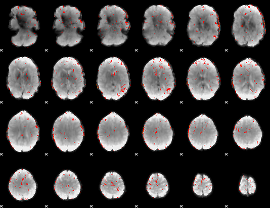 |
Correlation:0.29293
  |
Correlation:0.29293
  |
Correlation:0.10911
  |
Correlation:0.0026987
  |
Correlation:0.10911
  |
Correlation:0.24328
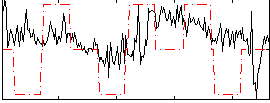  |
Correlation:0.07003
  |
Component:130; Jaccard:0.073523
 |
Component:104; Jaccard:0.075573
 |
Component:324; Jaccard:0.07049
 |
Component:301; Jaccard:0.053826
 |
Component:076; Jaccard:0.029889
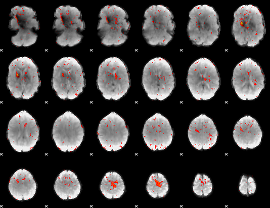 |
Component:301; Jaccard:0.014194
 |
Component:161; Jaccard:0.005254
 |
Correlation:0.079987
  |
Correlation:0.10911
  |
Correlation:-0.021582
  |
Correlation:0.12724
  |
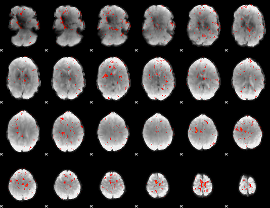
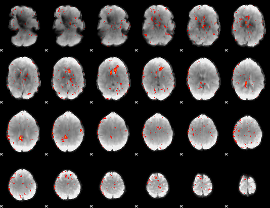
Correlation:0.034378
  |
Correlation:0.12724
  |
Correlation:0.24328
  |
Component:103; Jaccard:0.073372
 |
Component:297; Jaccard:0.074682
 |
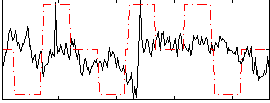
Component:162; Jaccard:0.068761
 |
Component:162; Jaccard:0.050996
 |
Component:103; Jaccard:0.02982
 |
Component:104; Jaccard:0.013972
 |
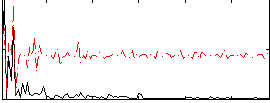
Component:102; Jaccard:0.004727
 |
Correlation:0.088838
  |
Correlation:0.053217
  |
Correlation:0.052445
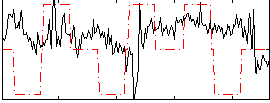 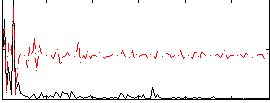 |
Correlation:0.052445
  |
Correlation:0.088838
  |
Correlation:0.10911
  |
Correlation:0.084475
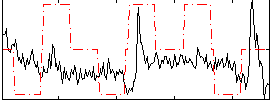 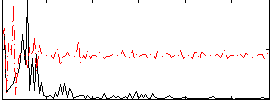 |
Component:162; Jaccard:0.073348
 |
Component:162; Jaccard:0.074667
 |
Component:103; Jaccard:0.065565
 |
Component:103; Jaccard:0.047908
 |
Component:140; Jaccard:0.029014
 |
Component:076; Jaccard:0.013446
 |
Component:301; Jaccard:0.004701
 |
Correlation:0.052445
  |
Correlation:0.052445
  |
Correlation:0.088838
  |
Correlation:0.088838
  |
Correlation:0.0026987
  |
Correlation:0.034378
  |
Correlation:0.12724
  |
Component:111; Jaccard:0.07226
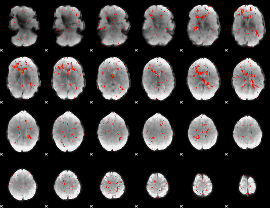 |
Component:103; Jaccard:0.074312
 |
Component:111; Jaccard:0.065463
 |
Component:076; Jaccard:0.047344
 |
Component:162; Jaccard:0.028955
 |
Component:349; Jaccard:0.012666
 |
Component:162; Jaccard:0.004486
 |
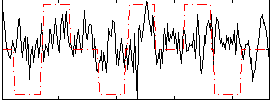
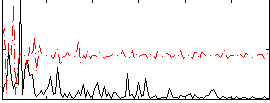
Correlation:0.1629
  |
Correlation:0.088838
  |
Correlation:0.1629
  |
Correlation:0.034378
  |
Correlation:0.052445
  |
Correlation:0.07003
  |
Correlation:0.052445
  |
Component:082; Jaccard:0.06988
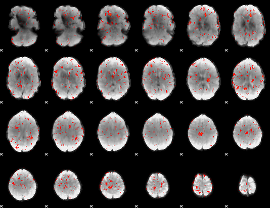 |
Component:111; Jaccard:0.072568
 |
Component:301; Jaccard:0.063509
 |
Component:111; Jaccard:0.046942
 |
Component:161; Jaccard:0.025608
 |
Component:162; Jaccard:0.012259
 |
Component:242; Jaccard:0.004346
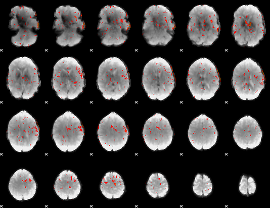 |
Correlation:0.008176
  |
Correlation:0.1629
  |
Correlation:0.12724
  |
Correlation:0.1629
  |
Correlation:0.24328
  |
Correlation:0.052445
  |
Correlation:0.0089419
  |
Component:104; Jaccard:0.069429
 |
Component:071; Jaccard:0.067843
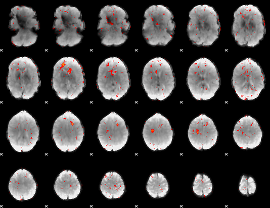 |
Component:297; Jaccard:0.063016
 |
Component:071; Jaccard:0.046599
 |
Component:349; Jaccard:0.024871
 |
Component:140; Jaccard:0.011321
 |
Component:076; Jaccard:0.004096
 |
Correlation:0.10911
  |
Correlation:0.043355
  |
Correlation:0.053217
  |
Correlation:0.043355
  |
Correlation:0.07003
  |
Correlation:0.0026987
  |
Correlation:0.034378
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
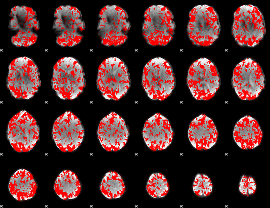 |
GLM binary result thresholded by 1.5
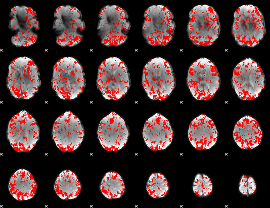 |
GLM binary result thresholded by 2
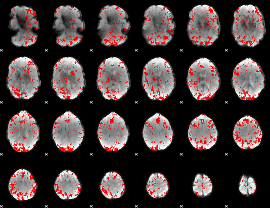 |
GLM binary result thresholded by 2.5
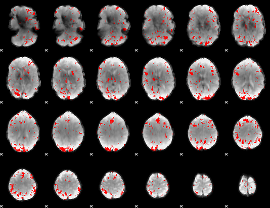 |
GLM binary result thresholded by 3
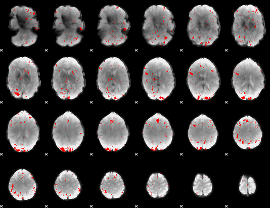 |
GLM binary result thresholded by 3.5
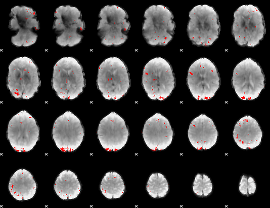 |
GLM binary result thresholded by 4
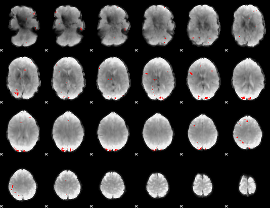 |
GLM Z-score result thresholded by 1
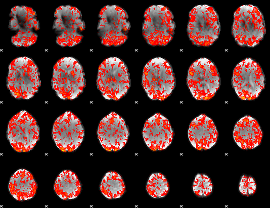 |
GLM Z-score result thresholded by 1.5
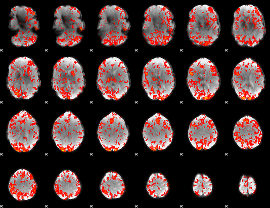 |
GLM Z-score result thresholded by 2
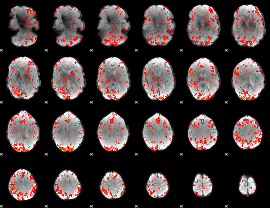 |
GLM Z-score result thresholded by 2.5
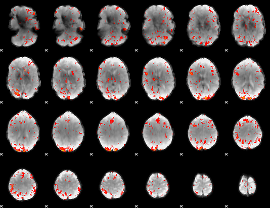 |
GLM Z-score result thresholded by 3
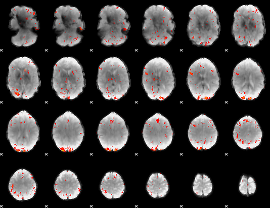 |
GLM Z-score result thresholded by 3.5
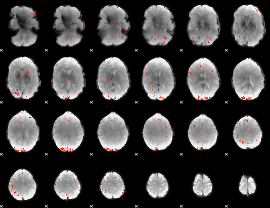 |
GLM Z-score result thresholded by 4
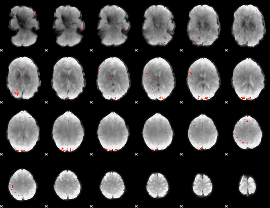 |
Component:262; Jaccard:0.11516
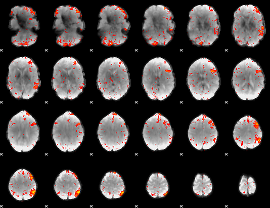 |
Component:060; Jaccard:0.13532
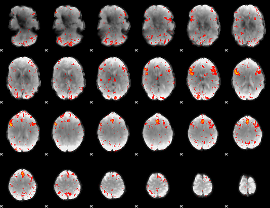 |
Component:060; Jaccard:0.15234
 |
Component:029; Jaccard:0.14963
 |
Component:029; Jaccard:0.12006
 |
Component:029; Jaccard:0.080548
 |
Component:388; Jaccard:0.047238
 |
Correlation:-0.02196
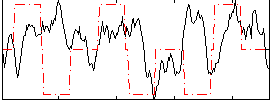 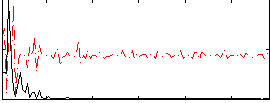 |
Correlation:0.23093
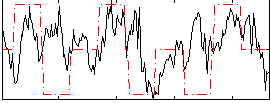 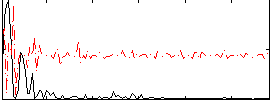 |
Correlation:0.23093
  |
Correlation:0.18265
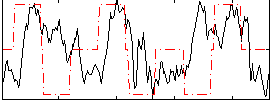 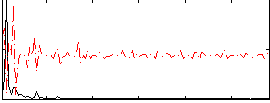 |
Correlation:0.18265
  |
Correlation:0.18265
  |
Correlation:0.0070637
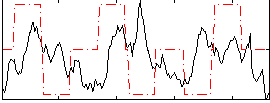 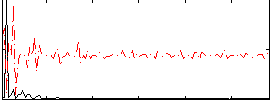 |
Component:060; Jaccard:0.11272
 |
Component:029; Jaccard:0.13368
 |
Component:029; Jaccard:0.15053
 |
Component:060; Jaccard:0.14372
 |
Component:060; Jaccard:0.10266
 |
Component:388; Jaccard:0.075096
 |
Component:029; Jaccard:0.047164
 |
Correlation:0.23093
  |
Correlation:0.18265
  |
Correlation:0.18265
  |
Correlation:0.23093
  |
Correlation:0.23093
  |
Correlation:0.0070637
  |
Correlation:0.18265
  |
Component:029; Jaccard:0.11159
 |
Component:262; Jaccard:0.13142
 |
Component:262; Jaccard:0.13656
 |
Component:262; Jaccard:0.11372
 |
Component:388; Jaccard:0.10051
 |
Component:386; Jaccard:0.063698
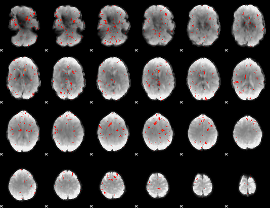 |
Component:386; Jaccard:0.043687
 |
Correlation:0.18265
  |
Correlation:-0.02196
  |
Correlation:-0.02196
  |
Correlation:-0.02196
  |
Correlation:0.0070637
  |
Correlation:0.19738
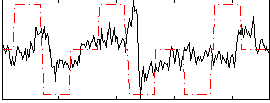 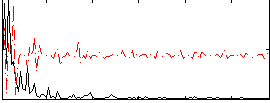 |
Correlation:0.19738
  |
Component:290; Jaccard:0.11027
 |
Component:271; Jaccard:0.11708
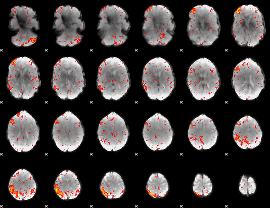 |
Component:284; Jaccard:0.1218
 |
Component:388; Jaccard:0.11263
 |
Component:190; Jaccard:0.086515
 |
Component:190; Jaccard:0.059229
 |
Component:190; Jaccard:0.038234
 |
Correlation:-0.015557
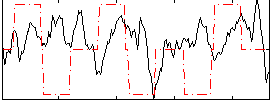 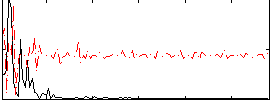 |
Correlation:0.12584
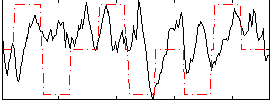 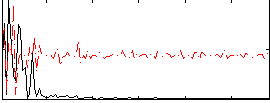 |
Correlation:0.058493
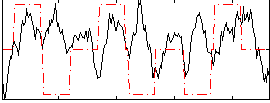 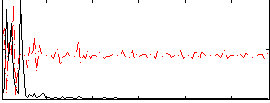 |
Correlation:0.0070637
  |
Correlation:0.18102
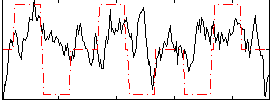 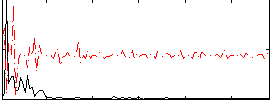 |
Correlation:0.18102
  |
Correlation:0.18102
  |
Component:271; Jaccard:0.1064
 |
Component:284; Jaccard:0.11535
 |
Component:190; Jaccard:0.11987
 |
Component:190; Jaccard:0.1113
 |
Component:386; Jaccard:0.082565
 |
Component:056; Jaccard:0.055332
 |
Component:056; Jaccard:0.034935
 |
Correlation:0.12584
  |
Correlation:0.058493
  |
Correlation:0.18102
  |
Correlation:0.18102
  |
Correlation:0.19738
  |
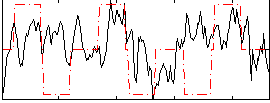
Correlation:0.080381
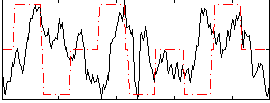  |
Correlation:0.080381
  |
Component:144; Jaccard:0.10585
 |
Component:190; Jaccard:0.11416
 |
Component:271; Jaccard:0.11773
 |
Component:284; Jaccard:0.10967
 |
Component:284; Jaccard:0.077127
 |
Component:060; Jaccard:0.054372
 |
Component:241; Jaccard:0.032143
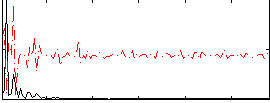
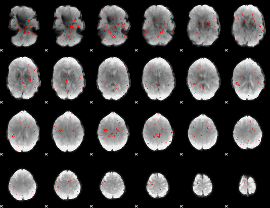 |
Correlation:0.20725
 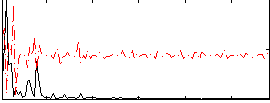 |
Correlation:0.18102
  |
Correlation:0.12584
  |
Correlation:0.058493
  |
Correlation:0.058493
  |
Correlation:0.23093
  |
Correlation:-0.046465
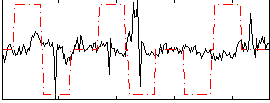 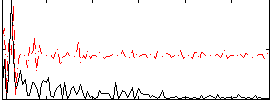 |
Component:070; Jaccard:0.10153
 |
Component:290; Jaccard:0.11391
 |
Component:144; Jaccard:0.11094
 |
Component:271; Jaccard:0.09807
 |
Component:056; Jaccard:0.07517
 |
Component:284; Jaccard:0.046649
 |
Component:060; Jaccard:0.030457
 |
Correlation:0.02192
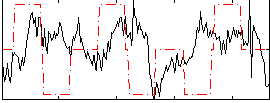  |
Correlation:-0.015557
  |
Correlation:0.20725
  |
Correlation:0.12584
  |
Correlation:0.080381
  |
Correlation:0.058493
  |
Correlation:0.23093
  |
Component:284; Jaccard:0.10142
 |
Component:144; Jaccard:0.1126
 |
Component:388; Jaccard:0.10776
 |
Component:144; Jaccard:0.095449
 |
Component:262; Jaccard:0.071497
 |
Component:322; Jaccard:0.044958
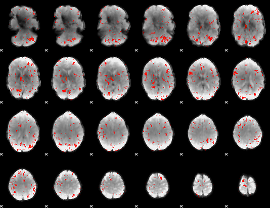 |
Component:284; Jaccard:0.027555
 |
Correlation:0.058493
  |
Correlation:0.20725
  |
Correlation:0.0070637
  |
Correlation:0.20725
  |
Correlation:-0.02196
  |
Correlation:0.23164
  |
Correlation:0.058493
  |
Component:190; Jaccard:0.10091
 |
Component:070; Jaccard:0.10974
 |
Component:251; Jaccard:0.10636
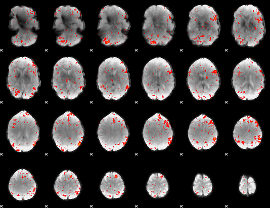 |
Component:069; Jaccard:0.094902
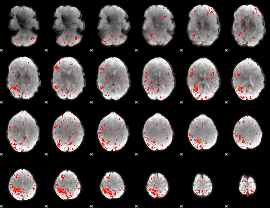 |
Component:069; Jaccard:0.066317
 |
Component:241; Jaccard:0.044821
 |
Component:322; Jaccard:0.025604
 |
Correlation:0.18102
  |
Correlation:0.02192
  |
Correlation:-0.0039458
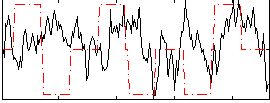 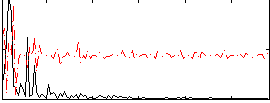 |
Correlation:0.31576
  |
Correlation:0.31576
  |
Correlation:-0.046465
  |
Correlation:0.23164
  |
Component:331; Jaccard:0.095119
 |
Component:333; Jaccard:0.10349
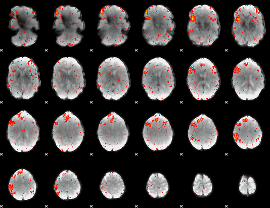 |
Component:070; Jaccard:0.10632
 |
Component:251; Jaccard:0.093635
 |
Component:144; Jaccard:0.064926
 |
Component:069; Jaccard:0.040626
 |
Component:069; Jaccard:0.023969
 |
Correlation:0.14551
  |
Correlation:0.089833
  |
Correlation:0.02192
  |
Correlation:-0.0039458
  |
Correlation:0.20725
  |
Correlation:0.31576
  |
Correlation:0.31576
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
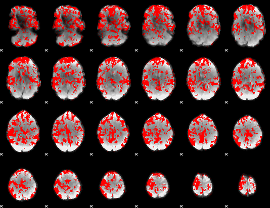 |
GLM binary result thresholded by 1.5
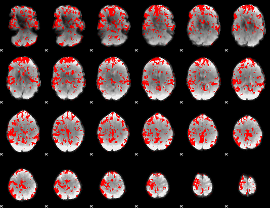 |
GLM binary result thresholded by 2
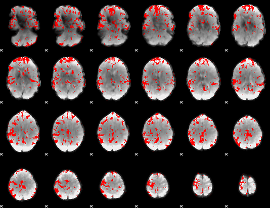 |
GLM binary result thresholded by 2.5
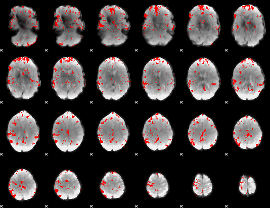 |
GLM binary result thresholded by 3
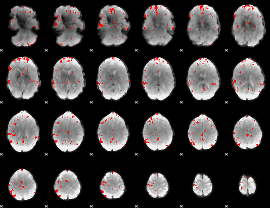 |
GLM binary result thresholded by 3.5
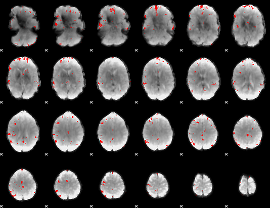 |
GLM binary result thresholded by 4
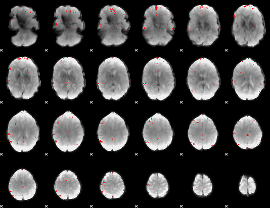 |
GLM Z-score result thresholded by 1
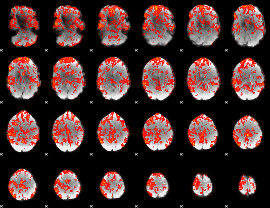 |
GLM Z-score result thresholded by 1.5
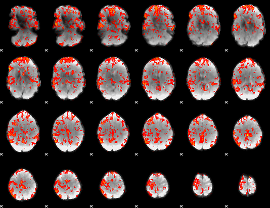 |
GLM Z-score result thresholded by 2
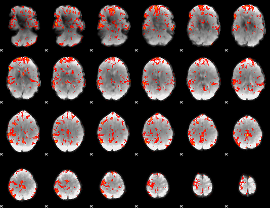 |
GLM Z-score result thresholded by 2.5
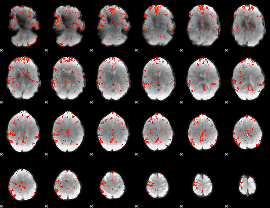 |
GLM Z-score result thresholded by 3
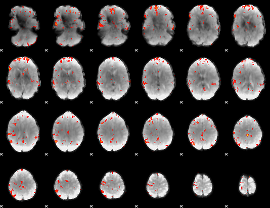 |
GLM Z-score result thresholded by 3.5
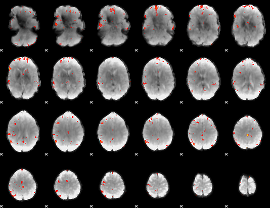 |
GLM Z-score result thresholded by 4
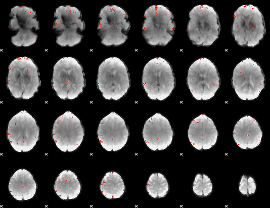 |
Component:278; Jaccard:0.12351
 |
Component:011; Jaccard:0.1464
 |
Component:011; Jaccard:0.16598
 |
Component:119; Jaccard:0.17053
 |
Component:119; Jaccard:0.15187
 |
Component:119; Jaccard:0.11125
 |
Component:119; Jaccard:0.069814
 |
Correlation:0.089096
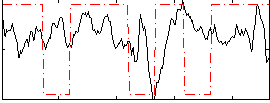 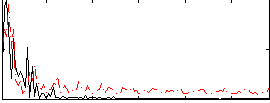 |
Correlation:0.36947
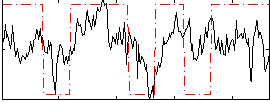  |
Correlation:0.36947
  |
Correlation:0.27482
  |
Correlation:0.27482
  |
Correlation:0.27482
  |
Correlation:0.27482
  |
Component:011; Jaccard:0.12112
 |
Component:278; Jaccard:0.14127
 |
Component:119; Jaccard:0.16229
 |
Component:011; Jaccard:0.16632
 |
Component:011; Jaccard:0.14105
 |
Component:011; Jaccard:0.095489
 |
Component:295; Jaccard:0.053566
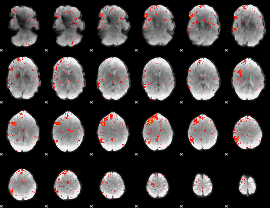 |
Correlation:0.36947
  |
Correlation:0.089096
  |
Correlation:0.27482
  |
Correlation:0.36947
  |
Correlation:0.36947
  |
Correlation:0.36947
  |
Correlation:0.16264
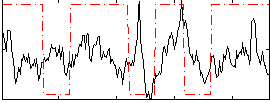 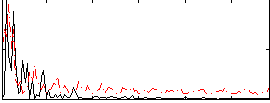 |
Component:119; Jaccard:0.11142
 |
Component:119; Jaccard:0.13715
 |
Component:278; Jaccard:0.15009
 |
Component:278; Jaccard:0.14536
 |
Component:278; Jaccard:0.121
 |
Component:295; Jaccard:0.087296
 |
Component:011; Jaccard:0.053396
 |
Correlation:0.27482
  |
Correlation:0.27482
  |
Correlation:0.089096
  |
Correlation:0.089096
  |
Correlation:0.089096
  |
Correlation:0.16264
  |
Correlation:0.36947
  |
Component:145; Jaccard:0.10708
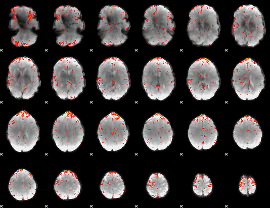 |
Component:295; Jaccard:0.12316
 |
Component:295; Jaccard:0.13878
 |
Component:295; Jaccard:0.14108
 |
Component:295; Jaccard:0.11992
 |
Component:324; Jaccard:0.08462
 |
Component:324; Jaccard:0.05299
 |
Correlation:0.15385
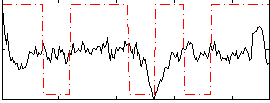 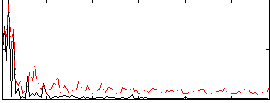 |
Correlation:0.16264
  |
Correlation:0.16264
  |
Correlation:0.16264
  |
Correlation:0.16264
  |
Correlation:0.15521
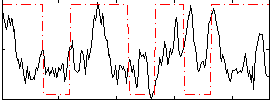 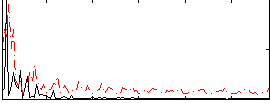 |
Correlation:0.15521
  |
Component:295; Jaccard:0.10428
 |
Component:145; Jaccard:0.1166
 |
Component:145; Jaccard:0.11895
 |
Component:324; Jaccard:0.11976
 |
Component:324; Jaccard:0.11129
 |
Component:278; Jaccard:0.084264
 |
Component:278; Jaccard:0.049335
 |
Correlation:0.16264
  |
Correlation:0.15385
  |
Correlation:0.15385
  |
Correlation:0.15521
  |
Correlation:0.15521
  |
Correlation:0.089096
  |
Correlation:0.089096
  |
Component:362; Jaccard:0.097927
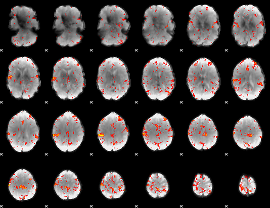 |
Component:146; Jaccard:0.1101
 |
Component:146; Jaccard:0.11708
 |
Component:146; Jaccard:0.11255
 |
Component:146; Jaccard:0.091307
 |
Component:136; Jaccard:0.064496
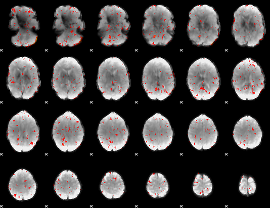 |
Component:136; Jaccard:0.04175
 |
Correlation:0.17059
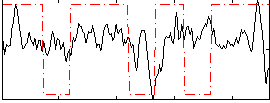 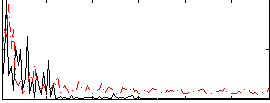 |
Correlation:0.17842
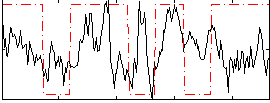  |
Correlation:0.17842
  |
Correlation:0.17842
  |
Correlation:0.17842
  |
Correlation:0.28758
  |
Correlation:0.28758
  |
Component:146; Jaccard:0.097671
 |
Component:215; Jaccard:0.10615
 |
Component:324; Jaccard:0.1154
 |
Component:145; Jaccard:0.10875
 |
Component:145; Jaccard:0.083873
 |
Component:146; Jaccard:0.062689
 |
Component:146; Jaccard:0.037909
 |
Correlation:0.17842
  |
Correlation:0.10144
  |
Correlation:0.15521
  |
Correlation:0.15385
  |
Correlation:0.15385
  |
Correlation:0.17842
  |
Correlation:0.17842
  |
Component:215; Jaccard:0.096295
 |
Component:089; Jaccard:0.1049
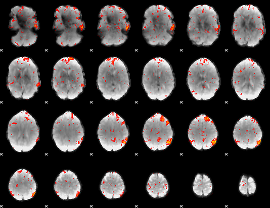 |
Component:215; Jaccard:0.10956
 |
Component:215; Jaccard:0.10162
 |
Component:136; Jaccard:0.082031
 |
Component:145; Jaccard:0.055615
 |
Component:216; Jaccard:0.031905
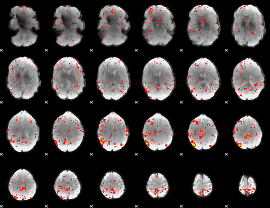 |
Correlation:0.10144
  |
Correlation:-0.0042879
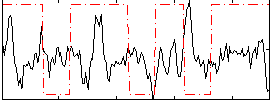 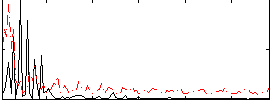 |
Correlation:0.10144
  |
Correlation:0.10144
  |
Correlation:0.28758
  |
Correlation:0.15385
  |
Correlation:0.034202
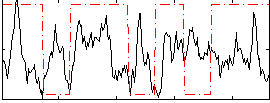 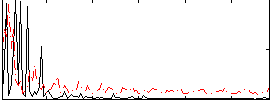 |
Component:089; Jaccard:0.095829
 |
Component:324; Jaccard:0.10459
 |
Component:089; Jaccard:0.10611
 |
Component:089; Jaccard:0.098645
 |
Component:215; Jaccard:0.081754
 |
Component:215; Jaccard:0.055594
 |
Component:215; Jaccard:0.030663
 |
Correlation:-0.0042879
  |
Correlation:0.15521
  |
Correlation:-0.0042879
  |
Correlation:-0.0042879
  |
Correlation:0.10144
  |
Correlation:0.10144
  |
Correlation:0.10144
  |
Component:324; Jaccard:0.090951
 |
Component:362; Jaccard:0.10404
 |
Component:399; Jaccard:0.10366
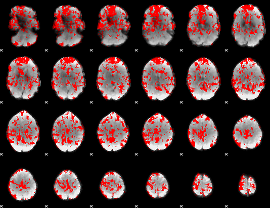
 |
Component:399; Jaccard:0.096982
 |
Component:089; Jaccard:0.07955
 |
Component:089; Jaccard:0.05339
 |
Component:145; Jaccard:0.030479
 |
Correlation:0.15521
  |
Correlation:0.17059
  |
Correlation:-0.03445
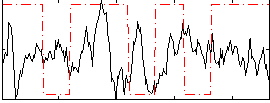 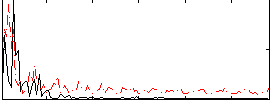 |
Correlation:-0.03445
  |
Correlation:-0.0042879
  |
Correlation:-0.0042879
  |
Correlation:0.15385
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
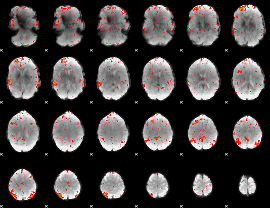
 |
GLM binary result thresholded by 1.5
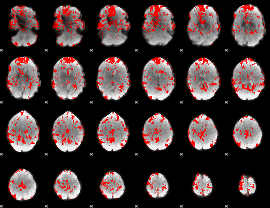 |
GLM binary result thresholded by 2
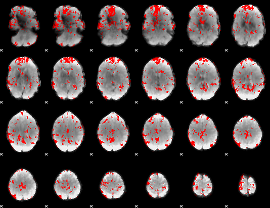 |
GLM binary result thresholded by 2.5
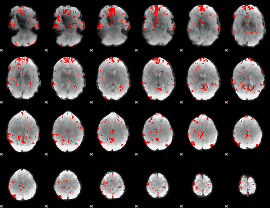 |
GLM binary result thresholded by 3
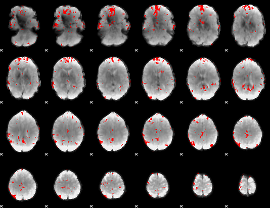 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
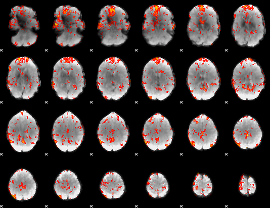 |
GLM Z-score result thresholded by 2.5
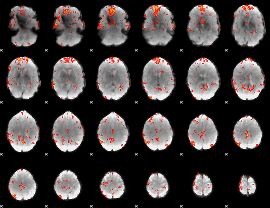 |
GLM Z-score result thresholded by 3
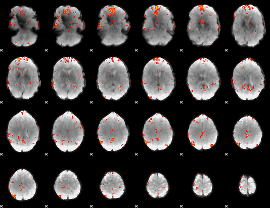 |
GLM Z-score result thresholded by 3.5
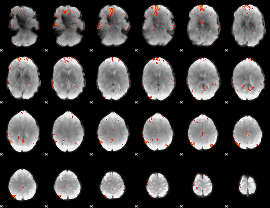 |
GLM Z-score result thresholded by 4
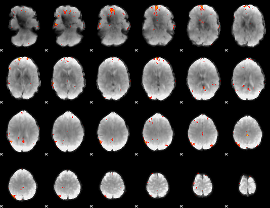 |
Component:324; Jaccard:0.12777
 |
Component:324; Jaccard:0.16014
 |
Component:324; Jaccard:0.19335
 |
Component:324; Jaccard:0.21528
 |
Component:324; Jaccard:0.21226
 |
Component:324; Jaccard:0.18021
 |
Component:324; Jaccard:0.13481
 |
Correlation:0.1188
  |
Correlation:0.1188
  |
Correlation:0.1188
  |
Correlation:0.1188
  |
Correlation:0.1188
  |
Correlation:0.1188
  |
Correlation:0.1188
  |
Component:119; Jaccard:0.1254
 |
Component:119; Jaccard:0.15288
 |
Component:119; Jaccard:0.1778
 |
Component:119; Jaccard:0.18685
 |
Component:119; Jaccard:0.17201
 |
Component:119; Jaccard:0.13965
 |
Component:119; Jaccard:0.096913
 |
Correlation:0.23363
  |
Correlation:0.23363
  |
Correlation:0.23363
  |
Correlation:0.23363
  |
Correlation:0.23363
  |
Correlation:0.23363
  |
Correlation:0.23363
  |
Component:146; Jaccard:0.11462
 |
Component:146; Jaccard:0.12859
 |
Component:146; Jaccard:0.14172
 |
Component:146; Jaccard:0.14208
 |
Component:146; Jaccard:0.1266
 |
Component:146; Jaccard:0.10307
 |
Component:141; Jaccard:0.073723
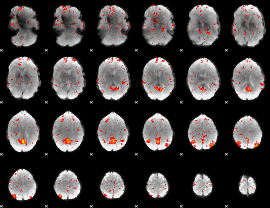 |
Correlation:0.0771
  |
Correlation:0.0771
  |
Correlation:0.0771
  |
Correlation:0.0771
  |
Correlation:0.0771
  |
Correlation:0.0771
  |
Correlation:0.034762
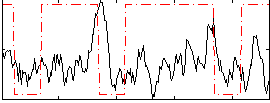 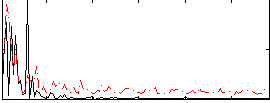 |
Component:278; Jaccard:0.10868
 |
Component:141; Jaccard:0.11649
 |
Component:141; Jaccard:0.12638
 |
Component:141; Jaccard:0.13041
 |
Component:141; Jaccard:0.11965
 |
Component:141; Jaccard:0.099763
 |
Component:146; Jaccard:0.073219
 |
Correlation:0.059597
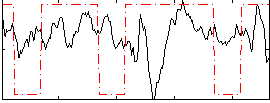 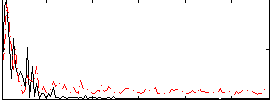 |
Correlation:0.034762
  |
Correlation:0.034762
  |
Correlation:0.034762
  |
Correlation:0.034762
  |
Correlation:0.034762
  |
Correlation:0.0771
  |
Component:141; Jaccard:0.10151
 |
Component:278; Jaccard:0.11336
 |
Component:278; Jaccard:0.11307
 |
Component:187; Jaccard:0.10778
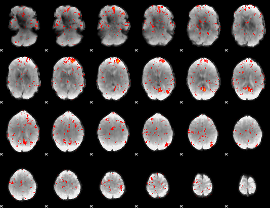 |
Component:216; Jaccard:0.094174
 |
Component:216; Jaccard:0.075579
 |
Component:216; Jaccard:0.05586
 |
Correlation:0.034762
  |
Correlation:0.059597
  |
Correlation:0.059597
  |
Correlation:0.043839
  |
Correlation:0.011942
  |
Correlation:0.011942
  |
Correlation:0.011942
  |
Component:216; Jaccard:0.097372
 |
Component:216; Jaccard:0.10719
 |
Component:216; Jaccard:0.11092
 |
Component:216; Jaccard:0.10557
 |
Component:187; Jaccard:0.091594
 |
Component:278; Jaccard:0.069709
 |
Component:278; Jaccard:0.050647
 |
Correlation:0.011942
  |
Correlation:0.011942
  |
Correlation:0.011942
  |
Correlation:0.011942
  |
Correlation:0.043839
  |
Correlation:0.059597
  |
Correlation:0.059597
  |
Component:188; Jaccard:0.096367
 |
Component:187; Jaccard:0.10523
 |
Component:187; Jaccard:0.10986
 |
Component:278; Jaccard:0.10485
 |
Component:278; Jaccard:0.089156
 |
Component:187; Jaccard:0.069333
 |
Component:187; Jaccard:0.048824
 |
Correlation:0.12968
  |
Correlation:0.043839
  |
Correlation:0.043839
  |
Correlation:0.059597
  |
Correlation:0.059597
  |
Correlation:0.043839
  |
Correlation:0.043839
  |
Component:187; Jaccard:0.095915
 |
Component:188; Jaccard:0.10267
 |
Component:188; Jaccard:0.1026
 |
Component:188; Jaccard:0.093491
 |
Component:011; Jaccard:0.074672
 |
Component:018; Jaccard:0.057179
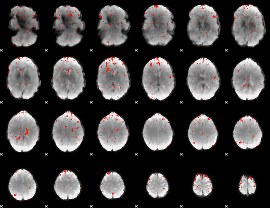 |
Component:091; Jaccard:0.047595
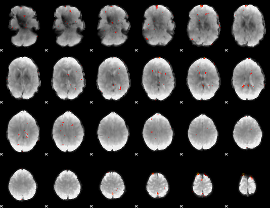 |
Correlation:0.043839
  |
Correlation:0.12968
  |
Correlation:0.12968
  |
Correlation:0.12968
  |
Correlation:-0.1268
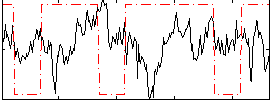 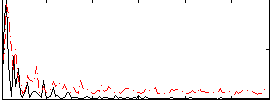 |
Correlation:0.26073
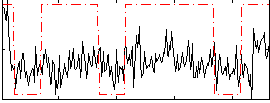 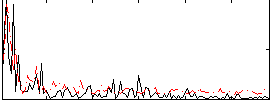 |
Correlation:0.24866
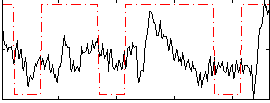 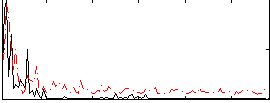 |
Component:011; Jaccard:0.092723
 |
Component:011; Jaccard:0.098036
 |
Component:011; Jaccard:0.097138
 |
Component:011; Jaccard:0.088797
 |
Component:188; Jaccard:0.073959
 |
Component:011; Jaccard:0.055394
 |
Component:018; Jaccard:0.04589
 |
Correlation:-0.1268
  |
Correlation:-0.1268
  |
Correlation:-0.1268
  |
Correlation:-0.1268
  |
Correlation:0.12968
  |
Correlation:-0.1268
  |
Correlation:0.26073
  |
Component:382; Jaccard:0.089151
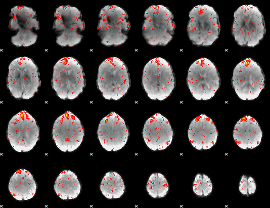 |
Component:382; Jaccard:0.093096
 |
Component:382; Jaccard:0.091045
 |
Component:039; Jaccard:0.085658
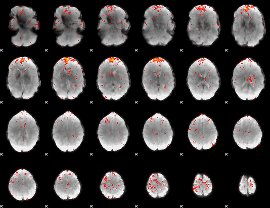 |
Component:039; Jaccard:0.069383
 |
Component:295; Jaccard:0.054557
 |
Component:295; Jaccard:0.039481
 |
Correlation:0.14093
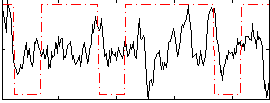 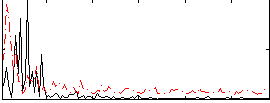 |
Correlation:0.14093
  |
Correlation:0.14093
  |
Correlation:0.1293
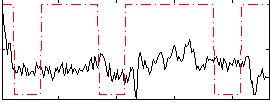 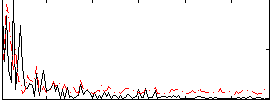 |
Correlation:0.1293
  |
Correlation:0.24572
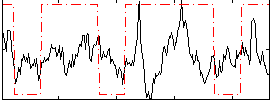 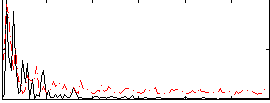 |
Correlation:0.24572
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































