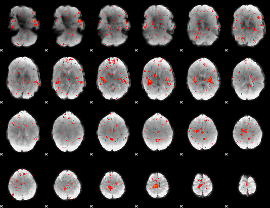
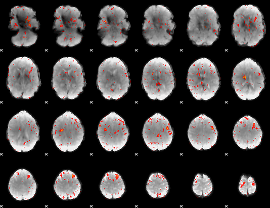
Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
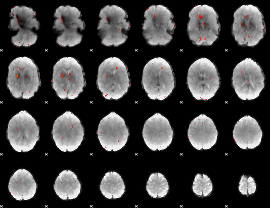
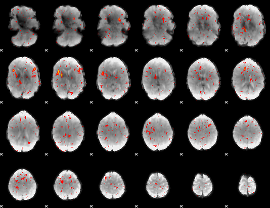
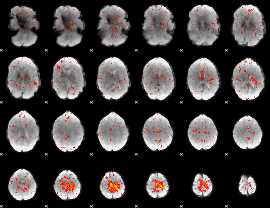
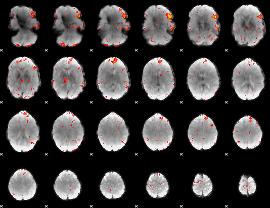
GLM result group-wise
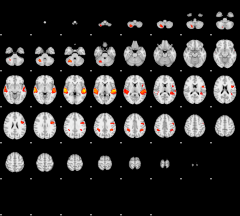 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
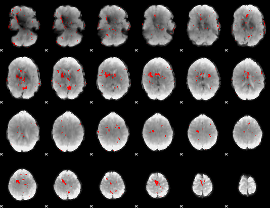 |
GLM binary result thresholded by 3
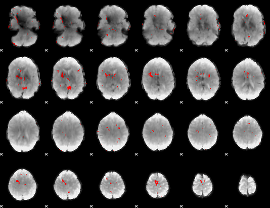 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
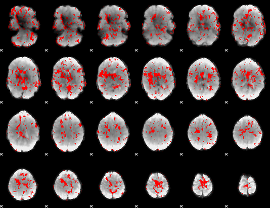
GLM Z-score result thresholded by 1
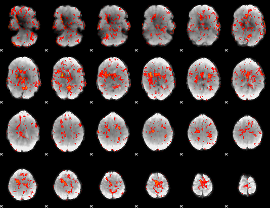 |
GLM Z-score result thresholded by 1.5
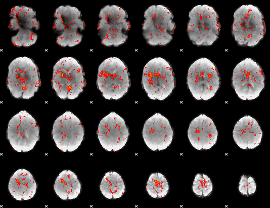 |
GLM Z-score result thresholded by 2
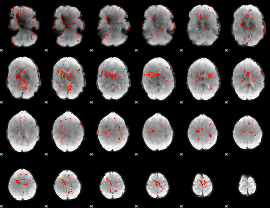 |
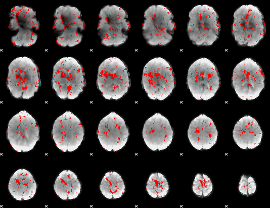
GLM Z-score result thresholded by 2.5
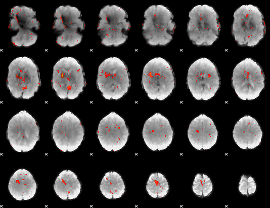 |
GLM Z-score result thresholded by 3
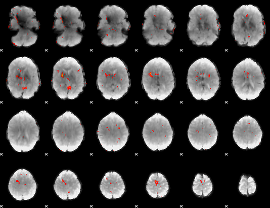 |
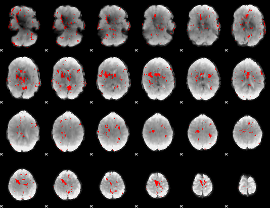
GLM Z-score result thresholded by 3.5
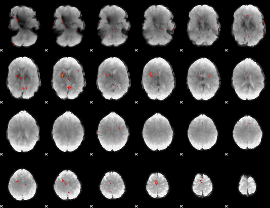 |
GLM Z-score result thresholded by 4
 |
Component:274; Jaccard:0.12986
 |
Component:274; Jaccard:0.14479
 |
Component:315; Jaccard:0.15668
 |
Component:315; Jaccard:0.14944
 |
Component:315; Jaccard:0.11078
 |
Component:315; Jaccard:0.072422
 |
Component:358; Jaccard:0.042562
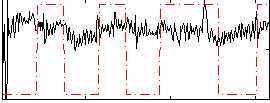
 |
Correlation:0.16348
 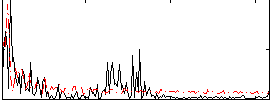 |
Correlation:0.16348
  |
Correlation:-0.12589
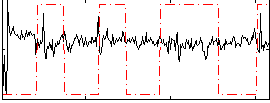  |
Correlation:-0.12589
  |
Correlation:-0.12589
  |
Correlation:-0.12589
  |
Correlation:-0.02025
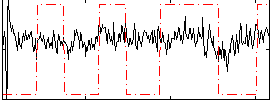
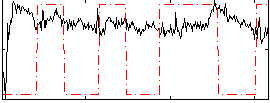  |
Component:315; Jaccard:0.11753
 |
Component:315; Jaccard:0.14384
 |
Component:274; Jaccard:0.14016
 |
Component:274; Jaccard:0.11613
 |
Component:274; Jaccard:0.079197
 |
Component:358; Jaccard:0.06531
 |
Component:315; Jaccard:0.036943
 |
Correlation:-0.12589
  |
Correlation:-0.12589
  |
Correlation:0.16348
  |
Correlation:0.16348
  |
Correlation:0.16348
  |
Correlation:-0.02025
  |
Correlation:-0.12589
  |
Component:311; Jaccard:0.10341
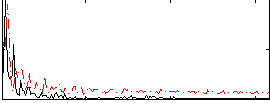
 |
Component:011; Jaccard:0.10861
 |
Component:011; Jaccard:0.10312
 |
Component:358; Jaccard:0.081564
 |
Component:358; Jaccard:0.077426
 |
Component:274; Jaccard:0.043865
 |
Component:274; Jaccard:0.019348
 |
Correlation:0.16925
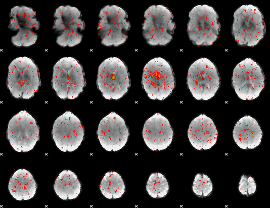
 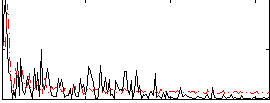 |
Correlation:-0.070152
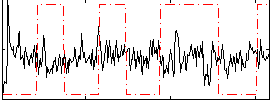 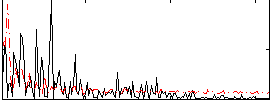 |
Correlation:-0.070152
  |
Correlation:-0.02025
  |
Correlation:-0.02025
  |
Correlation:0.16348
  |
Correlation:0.16348
  |
Component:011; Jaccard:0.10193
 |
Component:257; Jaccard:0.1061
 |
Component:257; Jaccard:0.096546
 |
Component:011; Jaccard:0.080146
 |
Component:257; Jaccard:0.056051
 |
Component:257; Jaccard:0.03244
 |
Component:008; Jaccard:0.018023
 |
Correlation:-0.070152
  |
Correlation:-0.12928
  |
Correlation:-0.12928
  |
Correlation:-0.070152
  |
Correlation:-0.12928
  |
Correlation:-0.12928
  |
Correlation:0.19064
  |
Component:257; Jaccard:0.10034
 |
Component:311; Jaccard:0.10298
 |
Component:387; Jaccard:0.093757
 |
Component:257; Jaccard:0.078078
 |
Component:011; Jaccard:0.05563
 |
Component:013; Jaccard:0.031771
 |
Component:013; Jaccard:0.017473
 |
Correlation:-0.12928
  |
Correlation:0.16925
  |
Correlation:0.036407
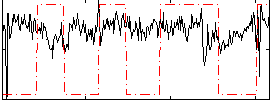 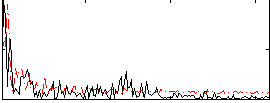 |
Correlation:-0.12928
  |
Correlation:-0.070152
  |
Correlation:-0.15548
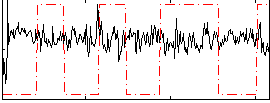 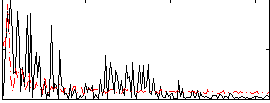 |
Correlation:-0.15548
  |
Component:125; Jaccard:0.09594
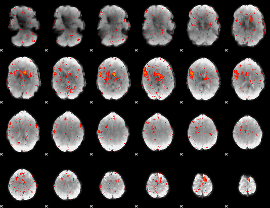 |
Component:387; Jaccard:0.10282
 |
Component:018; Jaccard:0.09201
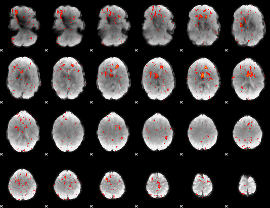 |
Component:013; Jaccard:0.074537
 |
Component:013; Jaccard:0.052284
 |
Component:011; Jaccard:0.031098
 |
Component:257; Jaccard:0.016711
 |
Correlation:0.16668
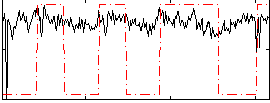 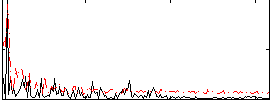 |
Correlation:0.036407
  |
Correlation:0.28577
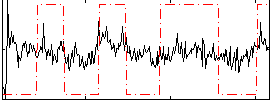 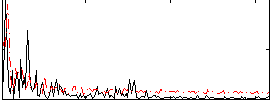 |
Correlation:-0.15548
  |
Correlation:-0.15548
  |
Correlation:-0.070152
  |
Correlation:-0.12928
  |
Component:018; Jaccard:0.095878
 |
Component:018; Jaccard:0.10025
 |
Component:311; Jaccard:0.089597
 |
Component:018; Jaccard:0.074015
 |
Component:115; Jaccard:0.052113
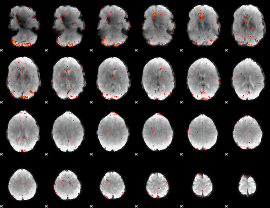 |
Component:326; Jaccard:0.030751
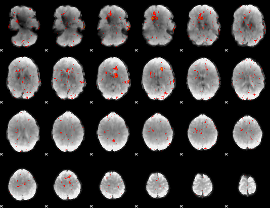 |
Component:326; Jaccard:0.01548
 |
Correlation:0.28577
  |
Correlation:0.28577
  |
Correlation:0.16925
  |
Correlation:0.28577
  |
Correlation:-0.026415
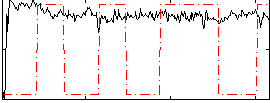 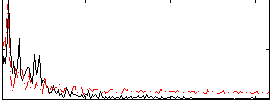 |
Correlation:0.010001
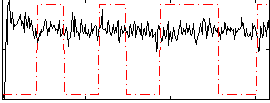  |
Correlation:0.010001
  |
Component:387; Jaccard:0.095479
 |
Component:013; Jaccard:0.093495
 |
Component:013; Jaccard:0.08792
 |
Component:387; Jaccard:0.073844
 |
Component:018; Jaccard:0.048609
 |
Component:115; Jaccard:0.029799
 |
Component:011; Jaccard:0.015224
 |
Correlation:0.036407
  |
Correlation:-0.15548
  |
Correlation:-0.15548
  |
Correlation:0.036407
  |
Correlation:0.28577
  |
Correlation:-0.026415
  |
Correlation:-0.070152
  |
Component:013; Jaccard:0.087907
 |
Component:125; Jaccard:0.091673
 |
Component:125; Jaccard:0.079776
 |
Component:115; Jaccard:0.068417
 |
Component:326; Jaccard:0.048401
 |
Component:008; Jaccard:0.02886
 |
Component:134; Jaccard:0.013408
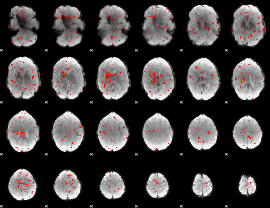 |
Correlation:-0.15548
  |
Correlation:0.16668
  |
Correlation:0.16668
  |
Correlation:-0.026415
  |
Correlation:0.010001
  |
Correlation:0.19064
  |
Correlation:0.046747
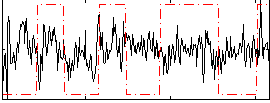  |
Component:019; Jaccard:0.086608
 |
Component:008; Jaccard:0.085861
 |
Component:115; Jaccard:0.07969
 |
Component:311; Jaccard:0.067505
 |
Component:387; Jaccard:0.047948
 |
Component:134; Jaccard:0.026503
 |
Component:311; Jaccard:0.011804
 |
Correlation:-0.049228
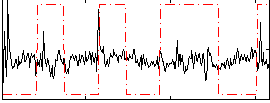 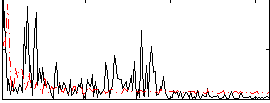 |
Correlation:0.19064
  |
Correlation:-0.026415
  |
Correlation:0.16925
  |
Correlation:0.036407
  |
Correlation:0.046747
  |
Correlation:0.16925
  |
Cope: 02:STORY BACK
|
GLM result group-wise
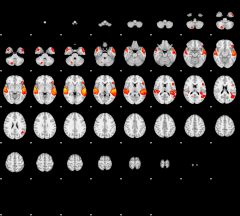 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
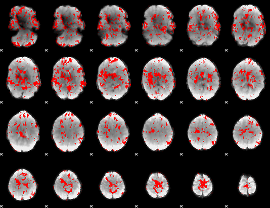
GLM binary result thresholded by 2
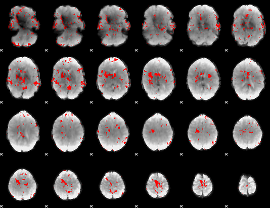 |
GLM binary result thresholded by 2.5
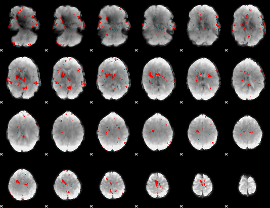 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
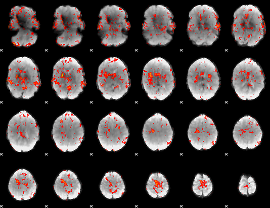 |
GLM Z-score result thresholded by 2
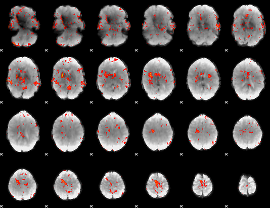 |
GLM Z-score result thresholded by 2.5
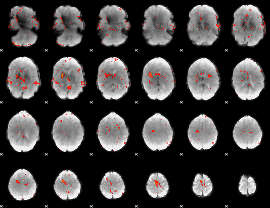 |
GLM Z-score result thresholded by 3
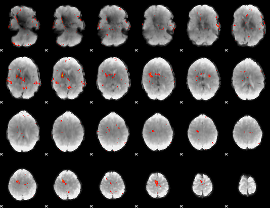 |
GLM Z-score result thresholded by 3.5
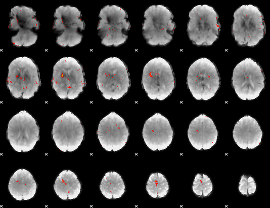 |
GLM Z-score result thresholded by 4
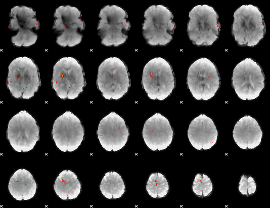 |
Component:186; Jaccard:0.13304
 |
Component:315; Jaccard:0.13512
 |
Component:315; Jaccard:0.15064
 |
Component:315; Jaccard:0.14792
 |
Component:315; Jaccard:0.12192
 |
Component:315; Jaccard:0.086009
 |
Component:315; Jaccard:0.048681
 |
Correlation:0.54271
  |
Correlation:0.090856
  |
Correlation:0.090856
  |
Correlation:0.090856
  |
Correlation:0.090856
  |
Correlation:0.090856
  |
Correlation:0.090856
  |
Component:257; Jaccard:0.11004
 |
Component:186; Jaccard:0.13356
 |
Component:186; Jaccard:0.12004
 |
Component:257; Jaccard:0.097865
 |
Component:257; Jaccard:0.069734
 |
Component:358; Jaccard:0.058227
 |
Component:358; Jaccard:0.042094
 |
Correlation:0.12331
  |
Correlation:0.54271
  |
Correlation:0.54271
  |
Correlation:0.12331
  |
Correlation:0.12331
  |
Correlation:0.021403
 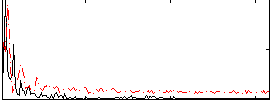 |
Correlation:0.021403
  |
Component:315; Jaccard:0.10921
 |
Component:257; Jaccard:0.11769
 |
Component:257; Jaccard:0.11344
 |
Component:186; Jaccard:0.096916
 |
Component:274; Jaccard:0.065304
 |
Component:257; Jaccard:0.043982
 |
Component:013; Jaccard:0.026036
 |
Correlation:0.090856
  |
Correlation:0.12331
  |
Correlation:0.12331
  |
Correlation:0.54271
  |
Correlation:-0.1952
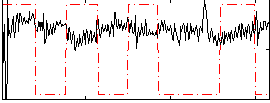 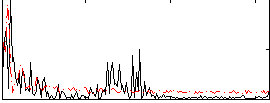 |
Correlation:0.12331
  |
Correlation:0.12664
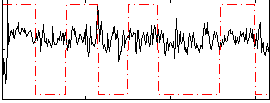  |
Component:274; Jaccard:0.10687
 |
Component:274; Jaccard:0.11374
 |
Component:274; Jaccard:0.11086
 |
Component:274; Jaccard:0.091172
 |
Component:013; Jaccard:0.06386
 |
Component:013; Jaccard:0.043612
 |
Component:257; Jaccard:0.023389
 |
Correlation:-0.1952
  |
Correlation:-0.1952
  |
Correlation:-0.1952
  |
Correlation:-0.1952
  |
Correlation:0.12664
  |
Correlation:0.12664
  |
Correlation:0.12331
  |
Component:375; Jaccard:0.10478
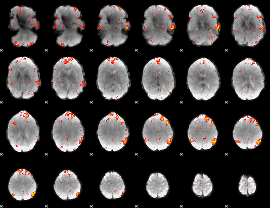 |
Component:013; Jaccard:0.10436
 |
Component:013; Jaccard:0.10272
 |
Component:013; Jaccard:0.088633
 |
Component:186; Jaccard:0.062682
 |
Component:274; Jaccard:0.041342
 |
Component:011; Jaccard:0.020918
 |
Correlation:0.5465
  |
Correlation:0.12664
  |
Correlation:0.12664
  |
Correlation:0.12664
  |
Correlation:0.54271
  |
Correlation:-0.1952
  |
Correlation:0.021355
  |
Component:013; Jaccard:0.09625
 |
Component:375; Jaccard:0.10276
 |
Component:387; Jaccard:0.09964
 |
Component:011; Jaccard:0.083246
 |
Component:358; Jaccard:0.062577
 |
Component:011; Jaccard:0.038992
 |
Component:274; Jaccard:0.02019
 |
Correlation:0.12664
  |
Correlation:0.5465
  |
Correlation:-0.016957
  |
Correlation:0.021355
  |
Correlation:0.021403
  |
Correlation:0.021355
  |
Correlation:-0.1952
  |
Component:387; Jaccard:0.093906
 |
Component:387; Jaccard:0.10174
 |
Component:011; Jaccard:0.098484
 |
Component:387; Jaccard:0.081833
 |
Component:011; Jaccard:0.059428
 |
Component:115; Jaccard:0.03634
 |
Component:008; Jaccard:0.020034
 |
Correlation:-0.016957
  |
Correlation:-0.016957
  |
Correlation:0.021355
  |
Correlation:-0.016957
  |
Correlation:0.021355
  |
Correlation:0.022722
  |
Correlation:-0.16652
  |
Component:011; Jaccard:0.093179
 |
Component:011; Jaccard:0.099568
 |
Component:375; Jaccard:0.090919
 |
Component:115; Jaccard:0.073881
 |
Component:387; Jaccard:0.059371
 |
Component:326; Jaccard:0.034926
 |
Component:115; Jaccard:0.018433
 |
Correlation:0.021355
  |
Correlation:0.021355
  |
Correlation:0.5465
  |
Correlation:0.022722
  |
Correlation:-0.016957
  |
Correlation:-0.0048294
  |
Correlation:0.022722
  |
Component:059; Jaccard:0.092686
 |
Component:059; Jaccard:0.092876
 |
Component:311; Jaccard:0.084643
 |
Component:375; Jaccard:0.070958
 |
Component:115; Jaccard:0.058241
 |
Component:186; Jaccard:0.034432
 |
Component:186; Jaccard:0.017911
 |
Correlation:0.34279
  |
Correlation:0.34279
  |
Correlation:-0.17389
  |
Correlation:0.5465
  |
Correlation:0.022722
  |
Correlation:0.54271
  |
Correlation:0.54271
  |
Component:311; Jaccard:0.091924
 |
Component:311; Jaccard:0.091616
 |
Component:059; Jaccard:0.083443
 |
Component:311; Jaccard:0.06796
 |
Component:326; Jaccard:0.050251
 |
Component:387; Jaccard:0.033696
 |
Component:326; Jaccard:0.017895
 |
Correlation:-0.17389
  |
Correlation:-0.17389
  |
Correlation:0.34279
  |
Correlation:-0.17389
  |
Correlation:-0.0048294
  |
Correlation:-0.016957
  |
Correlation:-0.0048294
  |
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
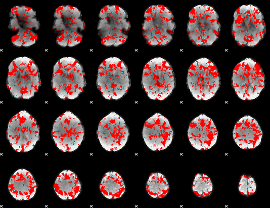 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
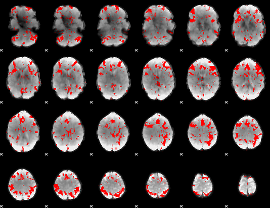 |
GLM binary result thresholded by 2.5
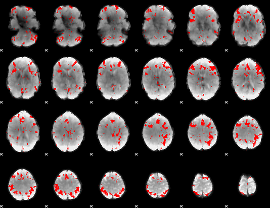 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
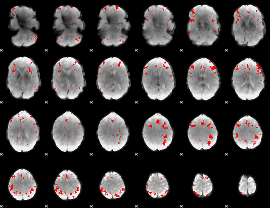 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
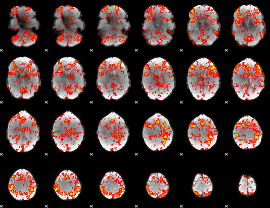 |
GLM Z-score result thresholded by 1.5
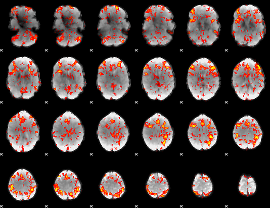 |
GLM Z-score result thresholded by 2
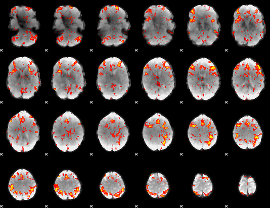 |
GLM Z-score result thresholded by 2.5
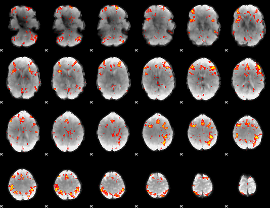 |
GLM Z-score result thresholded by 3
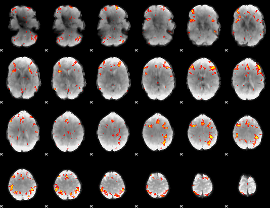 |
GLM Z-score result thresholded by 3.5
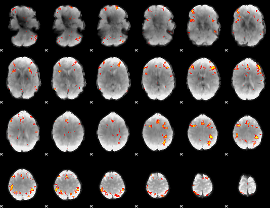 |
GLM Z-score result thresholded by 4
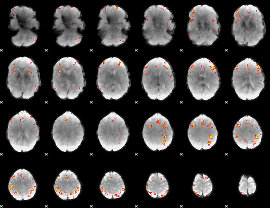 |
Component:207; Jaccard:0.17173
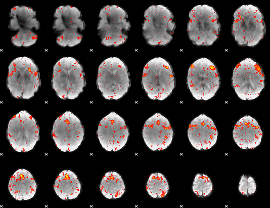 |
Component:268; Jaccard:0.20674
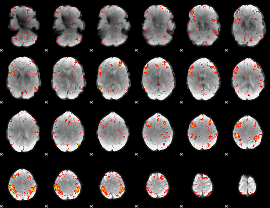 |
Component:268; Jaccard:0.23902
 |
Component:268; Jaccard:0.2534
 |
Component:268; Jaccard:0.24569
 |
Component:268; Jaccard:0.21896
 |
Component:268; Jaccard:0.18352
 |
Correlation:0.68685
  |
Correlation:0.64168
  |
Correlation:0.64168
  |
Correlation:0.64168
  |
Correlation:0.64168
  |
Correlation:0.64168
  |
Correlation:0.64168
  |
Component:268; Jaccard:0.1705
 |
Component:207; Jaccard:0.2066
 |
Component:207; Jaccard:0.23509
 |
Component:207; Jaccard:0.24606
 |
Component:207; Jaccard:0.23608
 |
Component:207; Jaccard:0.20594
 |
Component:207; Jaccard:0.1668
 |
Correlation:0.64168
  |
Correlation:0.68685
  |
Correlation:0.68685
  |
Correlation:0.68685
  |
Correlation:0.68685
  |
Correlation:0.68685
  |
Correlation:0.68685
  |
Component:239; Jaccard:0.16673
 |
Component:239; Jaccard:0.19919
 |
Component:239; Jaccard:0.22396
 |
Component:239; Jaccard:0.23191
 |
Component:239; Jaccard:0.2251
 |
Component:239; Jaccard:0.19726
 |
Component:239; Jaccard:0.16362
 |
Correlation:0.60696
  |
Correlation:0.60696
  |
Correlation:0.60696
  |
Correlation:0.60696
  |
Correlation:0.60696
  |
Correlation:0.60696
  |
Correlation:0.60696
  |
Component:279; Jaccard:0.14999
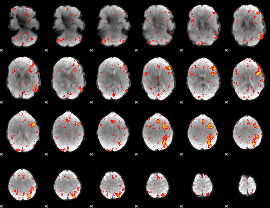 |
Component:279; Jaccard:0.17265
 |
Component:279; Jaccard:0.19036
 |
Component:279; Jaccard:0.19544
 |
Component:279; Jaccard:0.18531
 |
Component:279; Jaccard:0.16573
 |
Component:279; Jaccard:0.1372
 |
Correlation:0.63261
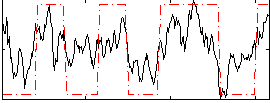 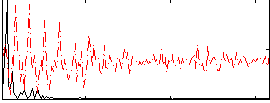 |
Correlation:0.63261
  |
Correlation:0.63261
  |
Correlation:0.63261
  |
Correlation:0.63261
  |
Correlation:0.63261
  |
Correlation:0.63261
  |
Component:169; Jaccard:0.1303
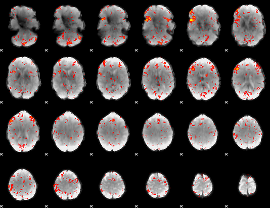 |
Component:169; Jaccard:0.1458
 |
Component:169; Jaccard:0.15931
 |
Component:169; Jaccard:0.16365
 |
Component:169; Jaccard:0.15345
 |
Component:169; Jaccard:0.13465
 |
Component:169; Jaccard:0.11241
 |
Correlation:0.53276
 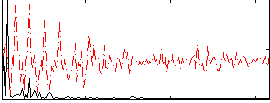 |
Correlation:0.53276
  |
Correlation:0.53276
  |
Correlation:0.53276
  |
Correlation:0.53276
  |
Correlation:0.53276
  |
Correlation:0.53276
  |
Component:204; Jaccard:0.11511
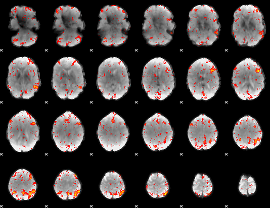 |
Component:086; Jaccard:0.12983
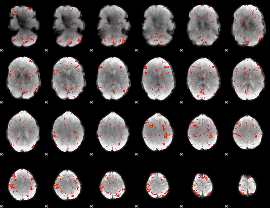 |
Component:086; Jaccard:0.14108
 |
Component:086; Jaccard:0.14509
 |
Component:173; Jaccard:0.13767
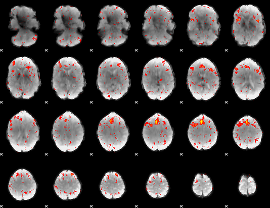 |
Component:173; Jaccard:0.12468
 |
Component:173; Jaccard:0.10638
 |
Correlation:0.34892
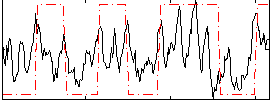 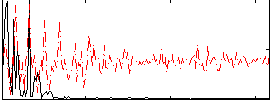 |
Correlation:0.52764
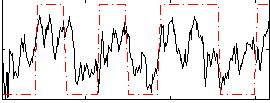 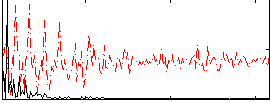 |
Correlation:0.52764
  |
Correlation:0.52764
  |
Correlation:0.39357
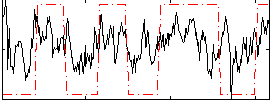 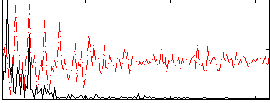 |
Correlation:0.39357
  |
Correlation:0.39357
  |
Component:086; Jaccard:0.11509
 |
Component:204; Jaccard:0.12407
 |
Component:173; Jaccard:0.13027
 |
Component:173; Jaccard:0.13788
 |
Component:086; Jaccard:0.13689
 |
Component:086; Jaccard:0.12108
 |
Component:086; Jaccard:0.098299
 |
Correlation:0.52764
  |
Correlation:0.34892
  |
Correlation:0.39357
  |
Correlation:0.39357
  |
Correlation:0.52764
  |
Correlation:0.52764
  |
Correlation:0.52764
  |
Component:269; Jaccard:0.11134
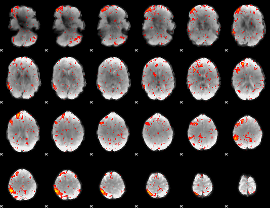 |
Component:294; Jaccard:0.1184
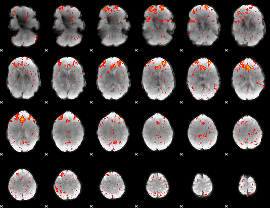 |
Component:204; Jaccard:0.12789
 |
Component:204; Jaccard:0.12293
 |
Component:294; Jaccard:0.11404
 |
Component:294; Jaccard:0.094281
 |
Component:294; Jaccard:0.074771
 |
Correlation:0.46753
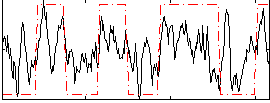 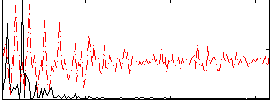 |
Correlation:0.37902
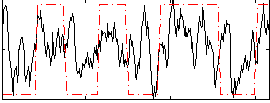 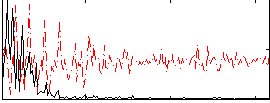 |
Correlation:0.34892
  |
Correlation:0.34892
  |
Correlation:0.37902
  |
Correlation:0.37902
  |
Correlation:0.37902
  |
Component:294; Jaccard:0.10791
 |
Component:269; Jaccard:0.11675
 |
Component:294; Jaccard:0.12381
 |
Component:294; Jaccard:0.12249
 |
Component:204; Jaccard:0.10785
 |
Component:204; Jaccard:0.08992
 |
Component:204; Jaccard:0.071219
 |
Correlation:0.37902
  |
Correlation:0.46753
  |
Correlation:0.37902
  |
Correlation:0.37902
  |
Correlation:0.34892
  |
Correlation:0.34892
  |
Correlation:0.34892
  |
Component:173; Jaccard:0.09806
 |
Component:173; Jaccard:0.114
 |
Component:269; Jaccard:0.1178
 |
Component:269; Jaccard:0.11123
 |
Component:267; Jaccard:0.098467
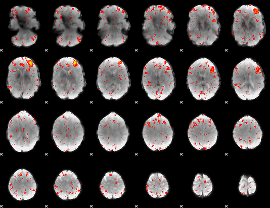 |
Component:267; Jaccard:0.08429
 |
Component:267; Jaccard:0.067084
 |
Correlation:0.39357
  |
Correlation:0.39357
  |
Correlation:0.46753
  |
Correlation:0.46753
  |
Correlation:0.32986
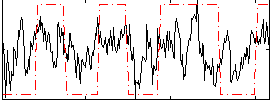 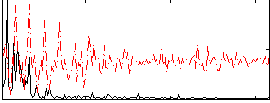 |
Correlation:0.32986
  |
Correlation:0.32986
  |
Cope: 04:STORY-MATH BACK
|
GLM result group-wise
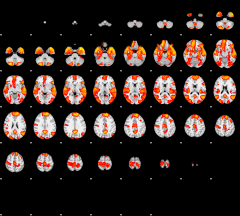 |
GLM binary result thresholded by 1
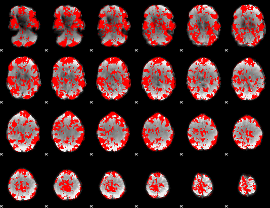 |
GLM binary result thresholded by 1.5
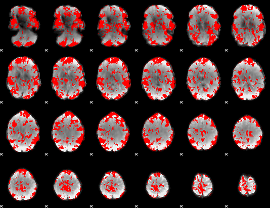 |
GLM binary result thresholded by 2
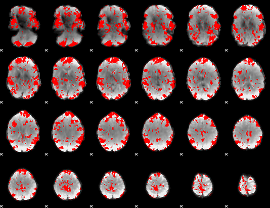 |
GLM binary result thresholded by 2.5
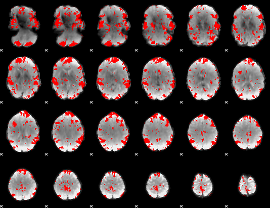 |
GLM binary result thresholded by 3
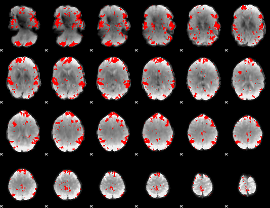 |
GLM binary result thresholded by 3.5
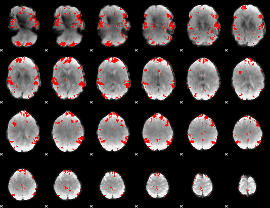 |
GLM binary result thresholded by 4
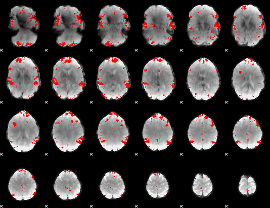 |
GLM Z-score result thresholded by 1
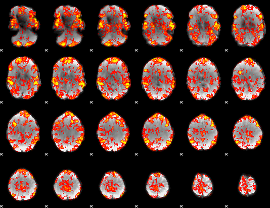 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:178; Jaccard:0.1394
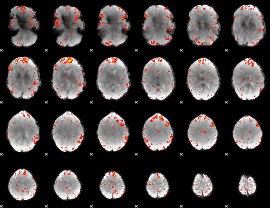 |
Component:178; Jaccard:0.16372
 |
Component:237; Jaccard:0.20102
 |
Component:186; Jaccard:0.2401
 |
Component:186; Jaccard:0.28195
 |
Component:186; Jaccard:0.31022
 |
Component:186; Jaccard:0.32261
 |
Correlation:0.45247
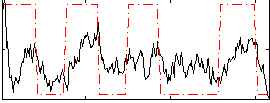 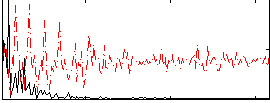 |
Correlation:0.45247
  |
Correlation:0.61721
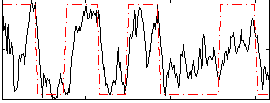 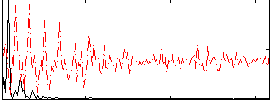 |
Correlation:0.56282
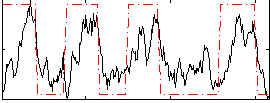 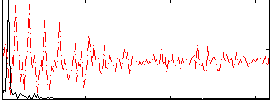 |
Correlation:0.56282
  |
Correlation:0.56282
  |
Correlation:0.56282
  |
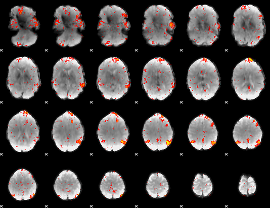
Component:237; Jaccard:0.13051
 |
Component:237; Jaccard:0.16337
 |
Component:186; Jaccard:0.19515
 |
Component:237; Jaccard:0.23657
 |
Component:237; Jaccard:0.26272
 |
Component:237; Jaccard:0.28038
 |
Component:237; Jaccard:0.28435
 |
Correlation:0.61721
  |
Correlation:0.61721
  |
Correlation:0.56282
  |
Correlation:0.61721
  |
Correlation:0.61721
  |
Correlation:0.61721
  |
Correlation:0.61721
  |
Component:186; Jaccard:0.121
 |
Component:186; Jaccard:0.15415
 |
Component:178; Jaccard:0.18783
 |
Component:293; Jaccard:0.21038
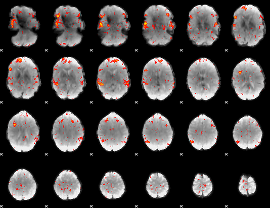 |
Component:293; Jaccard:0.24329
 |
Component:293; Jaccard:0.26606
 |
Component:293; Jaccard:0.2743
 |
Correlation:0.56282
  |
Correlation:0.56282
  |
Correlation:0.45247
  |
Correlation:0.666
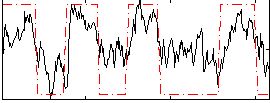 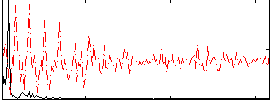 |
Correlation:0.666
  |
Correlation:0.666
  |
Correlation:0.666
  |
Component:285; Jaccard:0.11253
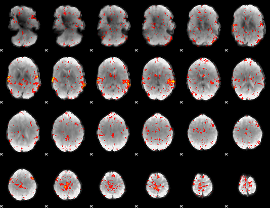 |
Component:293; Jaccard:0.13583
 |
Component:293; Jaccard:0.17241
 |
Component:178; Jaccard:0.20272
 |
Component:178; Jaccard:0.20941
 |
Component:178; Jaccard:0.20487
 |
Component:375; Jaccard:0.20447
 |
Correlation:0.24017
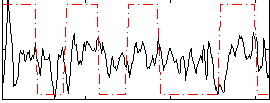 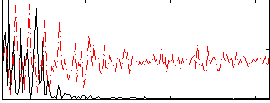 |
Correlation:0.666
  |
Correlation:0.666
  |
Correlation:0.45247
  |
Correlation:0.45247
  |
Correlation:0.45247
  |
Correlation:0.55995
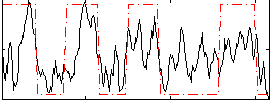 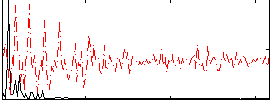 |
Component:198; Jaccard:0.11209
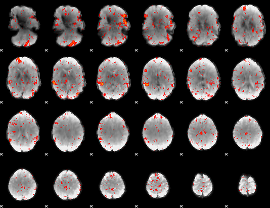 |
Component:198; Jaccard:0.12732
 |
Component:375; Jaccard:0.14896
 |
Component:375; Jaccard:0.17351
 |
Component:375; Jaccard:0.19261
 |
Component:375; Jaccard:0.20277
 |
Component:178; Jaccard:0.18832
 |
Correlation:0.27495
 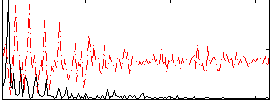 |
Correlation:0.27495
  |
Correlation:0.55995
  |
Correlation:0.55995
  |
Correlation:0.55995
  |
Correlation:0.55995
  |
Correlation:0.45247
  |
Component:357; Jaccard:0.11103
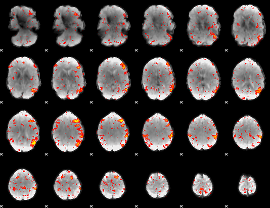 |
Component:375; Jaccard:0.12366
 |
Component:198; Jaccard:0.1418
 |
Component:135; Jaccard:0.15843
 |
Component:135; Jaccard:0.17169
 |
Component:135; Jaccard:0.17683
 |
Component:135; Jaccard:0.17146
 |
Correlation:0.18902
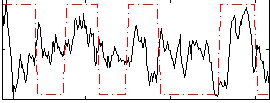  |
Correlation:0.55995
  |
Correlation:0.27495
  |
Correlation:0.50003
  |
Correlation:0.50003
  |
Correlation:0.50003
  |
Correlation:0.50003
  |
Component:160; Jaccard:0.10711
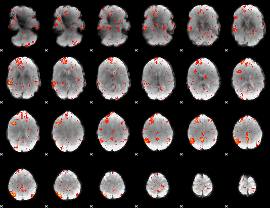 |
Component:201; Jaccard:0.12296
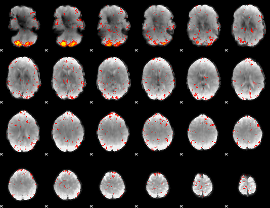 |
Component:135; Jaccard:0.14152
 |
Component:201; Jaccard:0.1521
 |
Component:201; Jaccard:0.15932
 |
Component:201; Jaccard:0.15804
 |
Component:201; Jaccard:0.14989
 |
Correlation:0.20702
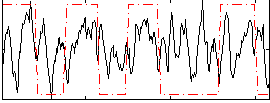 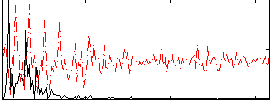 |
Correlation:0.41565
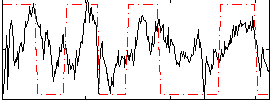  |
Correlation:0.50003
  |
Correlation:0.41565
  |
Correlation:0.41565
  |
Correlation:0.41565
  |
Correlation:0.41565
  |
Component:201; Jaccard:0.10619
 |
Component:285; Jaccard:0.12236
 |
Component:201; Jaccard:0.13889
 |
Component:198; Jaccard:0.15105
 |
Component:198; Jaccard:0.15289
 |
Component:198; Jaccard:0.15056
 |
Component:153; Jaccard:0.14186
 |
Correlation:0.41565
  |
Correlation:0.24017
  |
Correlation:0.41565
  |
Correlation:0.27495
  |
Correlation:0.27495
  |
Correlation:0.27495
  |
Correlation:0.5845
  |
Component:293; Jaccard:0.10615
 |
Component:135; Jaccard:0.12193
 |
Component:160; Jaccard:0.13104
 |
Component:160; Jaccard:0.13813
 |
Component:160; Jaccard:0.13973
 |
Component:153; Jaccard:0.14103
 |
Component:198; Jaccard:0.14127
 |
Correlation:0.666
  |
Correlation:0.50003
  |
Correlation:0.20702
  |
Correlation:0.20702
  |
Correlation:0.20702
  |
Correlation:0.5845
  |
Correlation:0.27495
  |
Component:144; Jaccard:0.10604
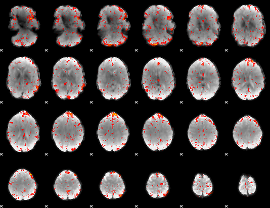 |
Component:160; Jaccard:0.11983
 |
Component:285; Jaccard:0.12904
 |
Component:285; Jaccard:0.13069
 |
Component:153; Jaccard:0.13485
 |
Component:160; Jaccard:0.1333
 |
Component:372; Jaccard:0.1302
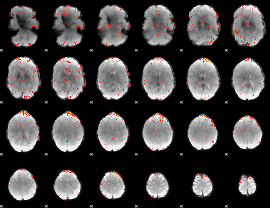 |
Correlation:0.13551
  |
Correlation:0.20702
  |
Correlation:0.24017
  |
Correlation:0.24017
  |
Correlation:0.5845
  |
Correlation:0.20702
  |
Correlation:0.45368
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
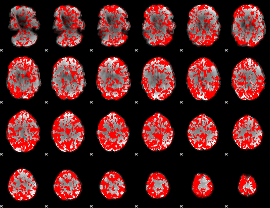 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
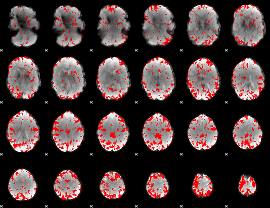 |
GLM binary result thresholded by 3
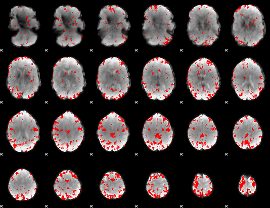 |
GLM binary result thresholded by 3.5
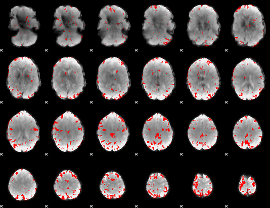 |
GLM binary result thresholded by 4
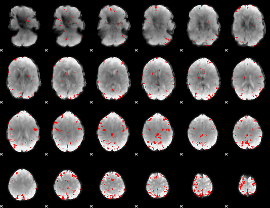 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
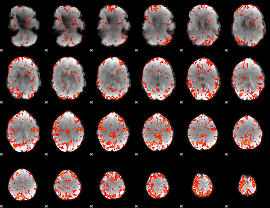 |
GLM Z-score result thresholded by 3
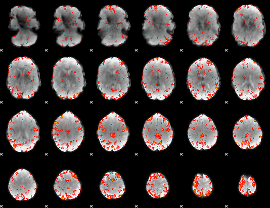 |
GLM Z-score result thresholded by 3.5
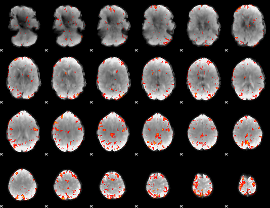 |
GLM Z-score result thresholded by 4
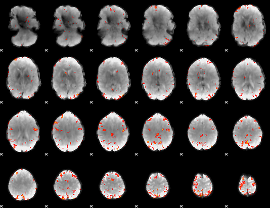 |
Component:263; Jaccard:0.11381
 |
Component:263; Jaccard:0.13011
 |
Component:263; Jaccard:0.14585
 |
Component:263; Jaccard:0.15552
 |
Component:263; Jaccard:0.15665
 |
Component:341; Jaccard:0.14356
 |
Component:341; Jaccard:0.11986
 |
Correlation:-0.15708
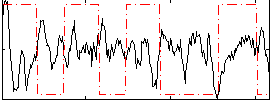 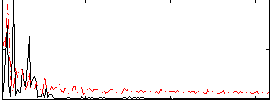 |
Correlation:-0.15708
  |
Correlation:-0.15708
  |
Correlation:-0.15708
  |
Correlation:-0.15708
  |
Correlation:-0.098861
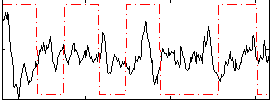 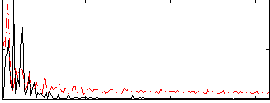 |
Correlation:-0.098861
  |
Component:042; Jaccard:0.11082
 |
Component:042; Jaccard:0.12714
 |
Component:042; Jaccard:0.14252
 |
Component:042; Jaccard:0.15114
 |
Component:341; Jaccard:0.15031
 |
Component:263; Jaccard:0.13981
 |
Component:240; Jaccard:0.11506
 |
Correlation:-0.012051
  |
Correlation:-0.012051
  |
Correlation:-0.012051
  |
Correlation:-0.012051
  |
Correlation:-0.098861
  |
Correlation:-0.15708
  |
Correlation:-0.032983
 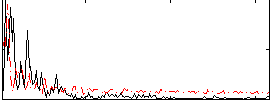 |
Component:341; Jaccard:0.10381
 |
Component:341; Jaccard:0.11831
 |
Component:341; Jaccard:0.13284
 |
Component:341; Jaccard:0.14534
 |
Component:042; Jaccard:0.14777
 |
Component:042; Jaccard:0.12933
 |
Component:263; Jaccard:0.11115
 |
Correlation:-0.098861
  |
Correlation:-0.098861
  |
Correlation:-0.098861
  |
Correlation:-0.098861
  |
Correlation:-0.012051
  |
Correlation:-0.012051
  |
Correlation:-0.15708
  |
Component:032; Jaccard:0.10017
 |
Component:389; Jaccard:0.11317
 |
Component:389; Jaccard:0.12765
 |
Component:389; Jaccard:0.13497
 |
Component:389; Jaccard:0.13144
 |
Component:240; Jaccard:0.12808
 |
Component:042; Jaccard:0.10095
 |
Correlation:-0.16272
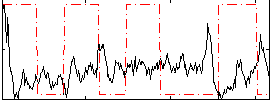 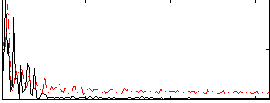 |
Correlation:-0.13685
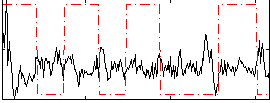  |
Correlation:-0.13685
  |
Correlation:-0.13685
  |
Correlation:-0.13685
  |
Correlation:-0.032983
  |
Correlation:-0.012051
  |
Component:316; Jaccard:0.098267
 |
Component:032; Jaccard:0.11009
 |
Component:049; Jaccard:0.11938
 |
Component:032; Jaccard:0.12481
 |
Component:240; Jaccard:0.12567
 |
Component:389; Jaccard:0.11411
 |
Component:389; Jaccard:0.088834
 |
Correlation:-0.12116
  |
Correlation:-0.16272
  |
Correlation:0.16778
  |
Correlation:-0.16272
  |
Correlation:-0.032983
  |
Correlation:-0.13685
  |
Correlation:-0.13685
  |
Component:389; Jaccard:0.09801
 |
Component:049; Jaccard:0.10833
 |
Component:032; Jaccard:0.119
 |
Component:049; Jaccard:0.12411
 |
Component:032; Jaccard:0.12207
 |
Component:032; Jaccard:0.10915
 |
Component:032; Jaccard:0.085869
 |
Correlation:-0.13685
  |
Correlation:0.16778
  |
Correlation:-0.16272
  |
Correlation:0.16778
  |
Correlation:-0.16272
  |
Correlation:-0.16272
  |
Correlation:-0.16272
  |
Component:049; Jaccard:0.096689
 |
Component:233; Jaccard:0.10665
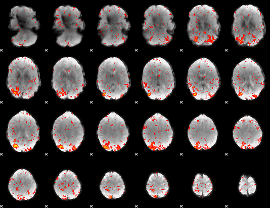 |
Component:233; Jaccard:0.11612
 |
Component:233; Jaccard:0.11963
 |
Component:049; Jaccard:0.11911
 |
Component:049; Jaccard:0.10571
 |
Component:049; Jaccard:0.084281
 |
Correlation:0.16778
  |
Correlation:0.16008
  |
Correlation:0.16008
  |
Correlation:0.16008
  |
Correlation:0.16778
  |
Correlation:0.16778
  |
Correlation:0.16778
  |
Component:233; Jaccard:0.09578
 |
Component:316; Jaccard:0.10486
 |
Component:331; Jaccard:0.1098
 |
Component:240; Jaccard:0.11299
 |
Component:233; Jaccard:0.11486
 |
Component:233; Jaccard:0.097883
 |
Component:233; Jaccard:0.076625
 |
Correlation:0.16008
  |
Correlation:-0.12116
  |
Correlation:-0.22553
  |
Correlation:-0.032983
  |
Correlation:0.16008
  |
Correlation:0.16008
  |
Correlation:0.16008
  |
Component:114; Jaccard:0.094881
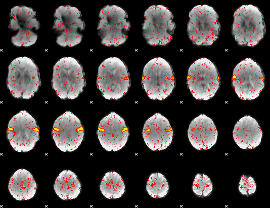 |
Component:331; Jaccard:0.10186
 |
Component:316; Jaccard:0.10772
 |
Component:331; Jaccard:0.1119
 |
Component:331; Jaccard:0.10549
 |
Component:195; Jaccard:0.090222
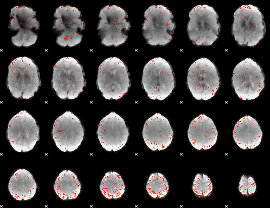 |
Component:195; Jaccard:0.074035
 |
Correlation:-0.040751
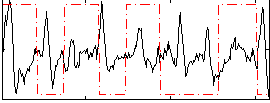 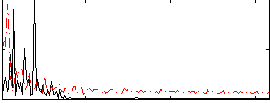 |
Correlation:-0.22553
  |
Correlation:-0.12116
  |
Correlation:-0.22553
  |
Correlation:-0.22553
  |
Correlation:-0.0087208
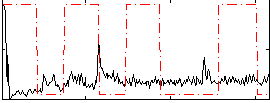  |
Correlation:-0.0087208
  |
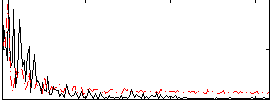
Component:111; Jaccard:0.09406
 |
Component:114; Jaccard:0.10146
 |
Component:114; Jaccard:0.10724
 |
Component:114; Jaccard:0.10651
 |
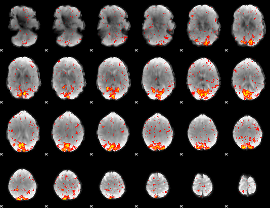
Component:361; Jaccard:0.098876
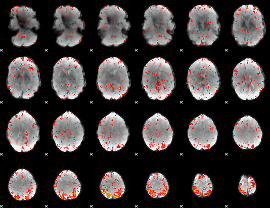 |
Component:331; Jaccard:0.090185
 |
Component:159; Jaccard:0.06998
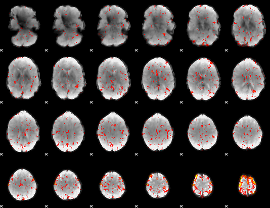 |
Correlation:-0.21807
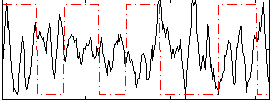 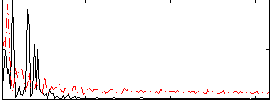 |
Correlation:-0.040751
  |
Correlation:-0.040751
  |
Correlation:-0.040751
  |
Correlation:-0.26204
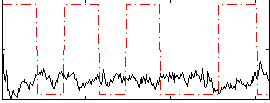  |
Correlation:-0.22553
  |
Correlation:-0.15176
  |
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
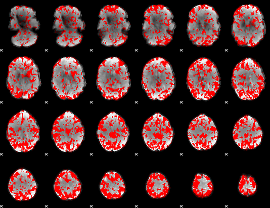 |
GLM binary result thresholded by 2
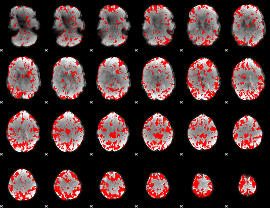 |
GLM binary result thresholded by 2.5
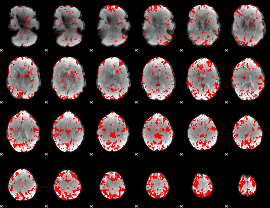 |
GLM binary result thresholded by 3
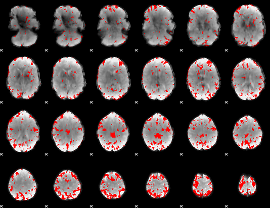 |
GLM binary result thresholded by 3.5
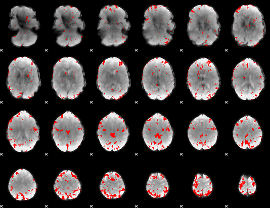 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
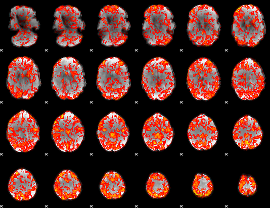 |
GLM Z-score result thresholded by 1.5
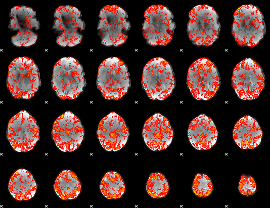 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
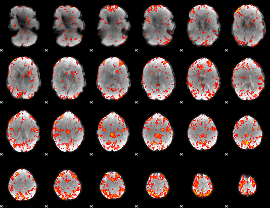 |
GLM Z-score result thresholded by 3
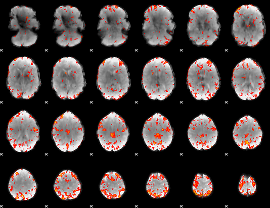 |
GLM Z-score result thresholded by 3.5
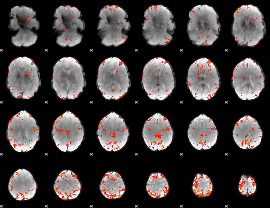 |
GLM Z-score result thresholded by 4
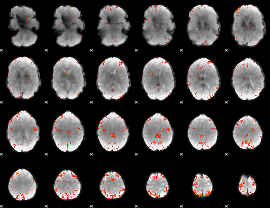 |
Component:263; Jaccard:0.11856
 |
Component:263; Jaccard:0.13466
 |
Component:263; Jaccard:0.15143
 |
Component:263; Jaccard:0.16126
 |
Component:263; Jaccard:0.16272
 |
Component:239; Jaccard:0.15292
 |
Component:239; Jaccard:0.13153
 |
Correlation:0.12186
  |
Correlation:0.12186
  |
Correlation:0.12186
  |
Correlation:0.12186
  |
Correlation:0.12186
  |
Correlation:0.60269
  |
Correlation:0.60269
  |
Component:042; Jaccard:0.11501
 |
Component:042; Jaccard:0.13189
 |
Component:042; Jaccard:0.14873
 |
Component:042; Jaccard:0.16021
 |
Component:239; Jaccard:0.15829
 |
Component:263; Jaccard:0.14625
 |
Component:341; Jaccard:0.11875
 |
Correlation:-0.01529
  |
Correlation:-0.01529
  |
Correlation:-0.01529
  |
Correlation:-0.01529
  |
Correlation:0.60269
  |
Correlation:0.12186
  |
Correlation:0.063459
  |
Component:341; Jaccard:0.10741
 |
Component:341; Jaccard:0.12192
 |
Component:341; Jaccard:0.13614
 |
Component:239; Jaccard:0.14916
 |
Component:042; Jaccard:0.15621
 |
Component:341; Jaccard:0.14319
 |
Component:263; Jaccard:0.11818
 |
Correlation:0.063459
  |
Correlation:0.063459
  |
Correlation:0.063459
  |
Correlation:0.60269
  |
Correlation:-0.01529
  |
Correlation:0.063459
  |
Correlation:0.12186
  |
Component:389; Jaccard:0.10429
 |
Component:389; Jaccard:0.12102
 |
Component:389; Jaccard:0.13539
 |
Component:341; Jaccard:0.14591
 |
Component:341; Jaccard:0.15098
 |
Component:042; Jaccard:0.13949
 |
Component:240; Jaccard:0.11354
 |
Correlation:0.14967
  |
Correlation:0.14967
  |
Correlation:0.14967
  |
Correlation:0.063459
  |
Correlation:0.063459
  |
Correlation:-0.01529
  |
Correlation:0.021074
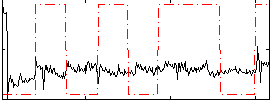 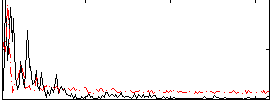 |
Component:361; Jaccard:0.10223
 |
Component:239; Jaccard:0.11639
 |
Component:239; Jaccard:0.13487
 |
Component:389; Jaccard:0.14568
 |
Component:389; Jaccard:0.14382
 |
Component:240; Jaccard:0.12579
 |
Component:042; Jaccard:0.10838
 |
Correlation:0.2627
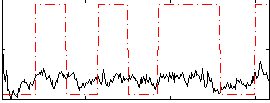 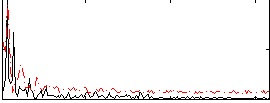 |
Correlation:0.60269
  |
Correlation:0.60269
  |
Correlation:0.14967
  |
Correlation:0.14967
  |
Correlation:0.021074
  |
Correlation:-0.01529
  |
Component:032; Jaccard:0.10161
 |
Component:361; Jaccard:0.11109
 |
Component:361; Jaccard:0.11915
 |
Component:331; Jaccard:0.12372
 |
Component:240; Jaccard:0.12442
 |
Component:389; Jaccard:0.12561
 |
Component:389; Jaccard:0.096357
 |
Correlation:0.14539
  |
Correlation:0.2627
  |
Correlation:0.2627
  |
Correlation:0.24811
  |
Correlation:0.021074
  |
Correlation:0.14967
  |
Correlation:0.14967
  |
Component:316; Jaccard:0.10025
 |
Component:032; Jaccard:0.11062
 |
Component:032; Jaccard:0.1176
 |
Component:361; Jaccard:0.12266
 |
Component:331; Jaccard:0.12281
 |
Component:331; Jaccard:0.10792
 |
Component:331; Jaccard:0.084975
 |
Correlation:0.13638
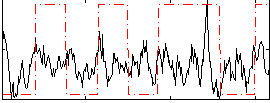 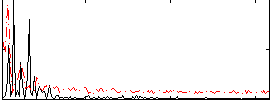 |
Correlation:0.14539
  |
Correlation:0.14539
  |
Correlation:0.2627
  |
Correlation:0.24811
  |
Correlation:0.24811
  |
Correlation:0.24811
  |
Component:111; Jaccard:0.099949
 |
Component:331; Jaccard:0.10796
 |
Component:331; Jaccard:0.11739
 |
Component:032; Jaccard:0.1198
 |
Component:361; Jaccard:0.1188
 |
Component:361; Jaccard:0.10561
 |
Component:361; Jaccard:0.080813
 |
Correlation:0.23651
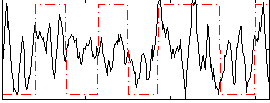 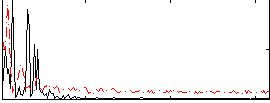 |
Correlation:0.24811
  |
Correlation:0.24811
  |
Correlation:0.14539
  |
Correlation:0.2627
  |
Correlation:0.2627
  |
Correlation:0.2627
  |
Component:239; Jaccard:0.099741
 |
Component:111; Jaccard:0.10654
 |
Component:318; Jaccard:0.10949
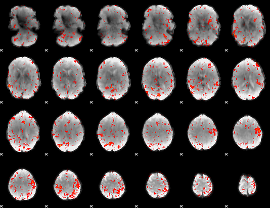 |
Component:240; Jaccard:0.11507
 |
Component:032; Jaccard:0.1155
 |
Component:032; Jaccard:0.10174
 |
Component:032; Jaccard:0.079188
 |
Correlation:0.60269
  |
Correlation:0.23651
  |
Correlation:0.32706
  |
Correlation:0.021074
  |
Correlation:0.14539
  |
Correlation:0.14539
  |
Correlation:0.14539
  |
Component:318; Jaccard:0.099227
 |
Component:316; Jaccard:0.10545
 |
Component:111; Jaccard:0.10873
 |
Component:269; Jaccard:0.10842
 |
Component:159; Jaccard:0.10634
 |
Component:159; Jaccard:0.096231
 |
Component:159; Jaccard:0.07673
 |
Correlation:0.32706
  |
Correlation:0.13638
  |
Correlation:0.23651
  |
Correlation:0.4418
 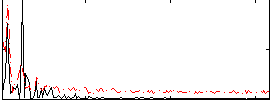 |
Correlation:0.1423
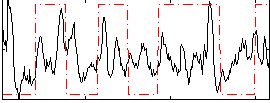 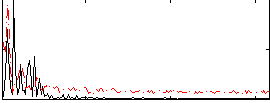 |
Correlation:0.1423
  |
Correlation:0.1423
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































