Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
GLM result group-wise
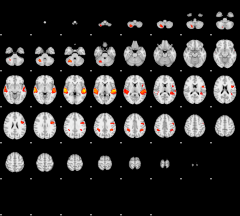 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
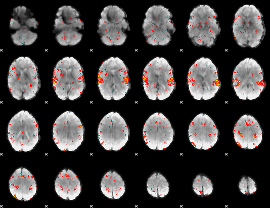 |
GLM Z-score result thresholded by 2.5
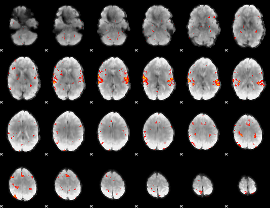 |
GLM Z-score result thresholded by 3
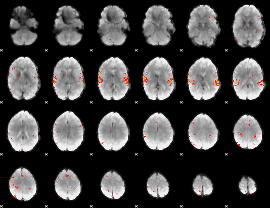 |
GLM Z-score result thresholded by 3.5
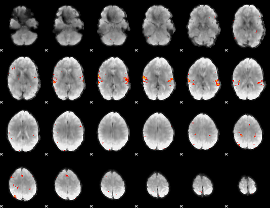 |
GLM Z-score result thresholded by 4
 |
Component:091; Jaccard:0.15855
 |
Component:091; Jaccard:0.17169
 |
Component:201; Jaccard:0.17176
 |
Component:201; Jaccard:0.15628
 |
Component:201; Jaccard:0.12953
 |
Component:201; Jaccard:0.098973
 |
Component:201; Jaccard:0.06945
 |
Correlation:-0.19976
  |
Correlation:-0.19976
  |
Correlation:-0.23209
  |
Correlation:-0.23209
  |
Correlation:-0.23209
  |
Correlation:-0.23209
  |
Correlation:-0.23209
  |
Component:201; Jaccard:0.13298
 |
Component:201; Jaccard:0.15906
 |
Component:091; Jaccard:0.16331
 |
Component:091; Jaccard:0.13769
 |
Component:091; Jaccard:0.1069
 |
Component:091; Jaccard:0.078028
 |
Component:091; Jaccard:0.050909
 |
Correlation:-0.23209
  |
Correlation:-0.23209
  |
Correlation:-0.19976
  |
Correlation:-0.19976
  |
Correlation:-0.19976
  |
Correlation:-0.19976
  |
Correlation:-0.19976
  |
Component:195; Jaccard:0.12157
 |
Component:195; Jaccard:0.1204
 |
Component:362; Jaccard:0.10537
 |
Component:362; Jaccard:0.095735
 |
Component:362; Jaccard:0.080954
 |
Component:362; Jaccard:0.064753
 |
Component:362; Jaccard:0.046379
 |
Correlation:0.653
  |
Correlation:0.653
  |
Correlation:-0.38134
  |
Correlation:-0.38134
  |
Correlation:-0.38134
  |
Correlation:-0.38134
  |
Correlation:-0.38134
  |
Component:092; Jaccard:0.10152
 |
Component:092; Jaccard:0.10934
 |
Component:092; Jaccard:0.10451
 |
Component:092; Jaccard:0.091511
 |
Component:092; Jaccard:0.071436
 |
Component:092; Jaccard:0.051935
 |
Component:092; Jaccard:0.03386
 |
Correlation:0.038137
  |
Correlation:0.038137
  |
Correlation:0.038137
  |
Correlation:0.038137
  |
Correlation:0.038137
  |
Correlation:0.038137
  |
Correlation:0.038137
  |
Component:111; Jaccard:0.1008
 |
Component:111; Jaccard:0.10172
 |
Component:195; Jaccard:0.10243
 |
Component:195; Jaccard:0.072812
 |
Component:195; Jaccard:0.046934
 |
Component:311; Jaccard:0.027722
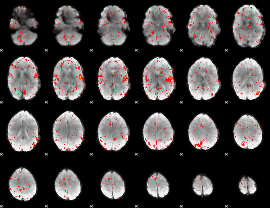 |
Component:311; Jaccard:0.019612
 |
Correlation:0.5012
  |
Correlation:0.5012
  |
Correlation:0.653
  |
Correlation:0.653
  |
Correlation:0.653
  |
Correlation:-0.25459
  |
Correlation:-0.25459
  |
Component:339; Jaccard:0.096517
 |
Component:362; Jaccard:0.1014
 |
Component:111; Jaccard:0.088775
 |
Component:111; Jaccard:0.065205
 |
Component:111; Jaccard:0.043652
 |
Component:339; Jaccard:0.027011
 |
Component:339; Jaccard:0.015992
 |
Correlation:0.26638
  |
Correlation:-0.38134
  |
Correlation:0.5012
  |
Correlation:0.5012
  |
Correlation:0.5012
  |
Correlation:0.26638
  |
Correlation:0.26638
  |
Component:351; Jaccard:0.092458
 |
Component:339; Jaccard:0.094261
 |
Component:339; Jaccard:0.079427
 |
Component:339; Jaccard:0.060477
 |
Component:339; Jaccard:0.041847
 |
Component:195; Jaccard:0.02638
 |
Component:254; Jaccard:0.015877
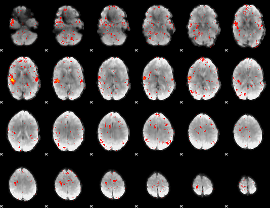 |
Correlation:0.64834
  |
Correlation:0.26638
  |
Correlation:0.26638
  |
Correlation:0.26638
  |
Correlation:0.26638
  |
Correlation:0.653
  |
Correlation:-0.24409
  |
Component:362; Jaccard:0.090885
 |
Component:351; Jaccard:0.086524
 |
Component:234; Jaccard:0.075683
 |
Component:234; Jaccard:0.056578
 |
Component:241; Jaccard:0.03875
 |
Component:111; Jaccard:0.025089
 |
Component:241; Jaccard:0.014711
 |
Correlation:-0.38134
  |
Correlation:0.64834
  |
Correlation:0.62223
  |
Correlation:0.62223
  |
Correlation:0.38266
  |
Correlation:0.5012
  |
Correlation:0.38266
  |
Component:309; Jaccard:0.089675
 |
Component:309; Jaccard:0.085175
 |
Component:036; Jaccard:0.07386
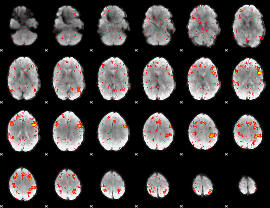 |
Component:309; Jaccard:0.054468
 |
Component:036; Jaccard:0.037649
 |
Component:241; Jaccard:0.024676
 |
Component:111; Jaccard:0.014228
 |
Correlation:0.5608
  |
Correlation:0.5608
  |
Correlation:0.54769
 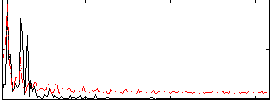 |
Correlation:0.5608
  |
Correlation:0.54769
  |
Correlation:0.38266
  |
Correlation:0.5012
  |
Component:208; Jaccard:0.088845
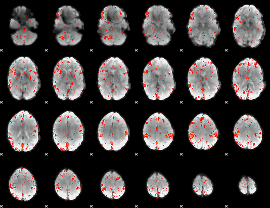 |
Component:234; Jaccard:0.083506
 |
Component:309; Jaccard:0.072841
 |
Component:241; Jaccard:0.053873
 |
Component:309; Jaccard:0.037602
 |
Component:036; Jaccard:0.023951
 |
Component:304; Jaccard:0.013889
 |
Correlation:0.37671
  |
Correlation:0.62223
  |
Correlation:0.5608
  |
Correlation:0.38266
  |
Correlation:0.5608
  |
Correlation:0.54769
  |
Correlation:-0.139
  |
Cope: 02:STORY BACK
|
GLM result group-wise
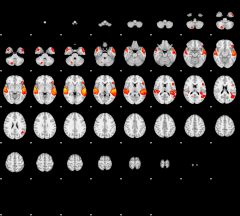 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
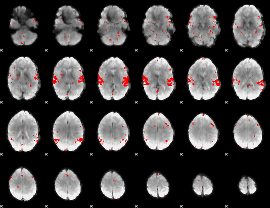 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
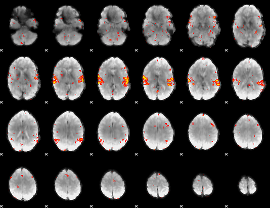 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:091; Jaccard:0.16278
 |
Component:201; Jaccard:0.18914
 |
Component:201; Jaccard:0.20979
 |
Component:201; Jaccard:0.20494
 |
Component:201; Jaccard:0.18111
 |
Component:201; Jaccard:0.14645
 |
Component:201; Jaccard:0.10815
 |
Correlation:0.12033
  |
Correlation:0.2124
  |
Correlation:0.2124
  |
Correlation:0.2124
  |
Correlation:0.2124
  |
Correlation:0.2124
  |
Correlation:0.2124
  |
Component:142; Jaccard:0.15584
 |
Component:091; Jaccard:0.17882
 |
Component:091; Jaccard:0.17314
 |
Component:362; Jaccard:0.16959
 |
Component:362; Jaccard:0.1485
 |
Component:362; Jaccard:0.12465
 |
Component:362; Jaccard:0.097283
 |
Correlation:0.55036
  |
Correlation:0.12033
  |
Correlation:0.12033
  |
Correlation:0.32764
  |
Correlation:0.32764
  |
Correlation:0.32764
  |
Correlation:0.32764
  |
Component:201; Jaccard:0.1549
 |
Component:142; Jaccard:0.16849
 |
Component:362; Jaccard:0.17201
 |
Component:091; Jaccard:0.15673
 |
Component:091; Jaccard:0.1288
 |
Component:091; Jaccard:0.10439
 |
Component:091; Jaccard:0.078662
 |
Correlation:0.2124
  |
Correlation:0.55036
  |
Correlation:0.32764
  |
Correlation:0.12033
  |
Correlation:0.12033
  |
Correlation:0.12033
  |
Correlation:0.12033
  |
Component:362; Jaccard:0.1337
 |
Component:362; Jaccard:0.15662
 |
Component:142; Jaccard:0.16547
 |
Component:142; Jaccard:0.14188
 |
Component:142; Jaccard:0.11268
 |
Component:142; Jaccard:0.083279
 |
Component:142; Jaccard:0.056246
 |
Correlation:0.32764
  |
Correlation:0.32764
  |
Correlation:0.55036
  |
Correlation:0.55036
  |
Correlation:0.55036
  |
Correlation:0.55036
  |
Correlation:0.55036
  |
Component:167; Jaccard:0.12099
 |
Component:167; Jaccard:0.11961
 |
Component:167; Jaccard:0.10264
 |
Component:311; Jaccard:0.084181
 |
Component:092; Jaccard:0.06981
 |
Component:092; Jaccard:0.057318
 |
Component:092; Jaccard:0.042341
 |
Correlation:0.17115
  |
Correlation:0.17115
  |
Correlation:0.17115
  |
Correlation:0.26428
  |
Correlation:-0.067589
  |
Correlation:-0.067589
  |
Correlation:-0.067589
  |
Component:311; Jaccard:0.10841
 |
Component:311; Jaccard:0.1092
 |
Component:311; Jaccard:0.10005
 |
Component:092; Jaccard:0.083618
 |
Component:311; Jaccard:0.06601
 |
Component:311; Jaccard:0.050381
 |
Component:311; Jaccard:0.038044
 |
Correlation:0.26428
  |
Correlation:0.26428
  |
Correlation:0.26428
  |
Correlation:-0.067589
  |
Correlation:0.26428
  |
Correlation:0.26428
  |
Correlation:0.26428
  |
Component:092; Jaccard:0.092347
 |
Component:092; Jaccard:0.096786
 |
Component:092; Jaccard:0.092812
 |
Component:167; Jaccard:0.080461
 |
Component:167; Jaccard:0.055304
 |
Component:033; Jaccard:0.043145
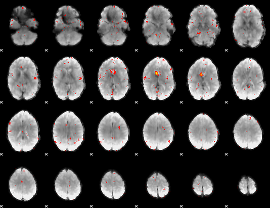 |
Component:033; Jaccard:0.033351
 |
Correlation:-0.067589
  |
Correlation:-0.067589
  |
Correlation:-0.067589
  |
Correlation:0.17115
  |
Correlation:0.17115
  |
Correlation:0.33724
  |
Correlation:0.33724
  |
Component:184; Jaccard:0.091229
 |
Component:254; Jaccard:0.087054
 |
Component:254; Jaccard:0.081859
 |
Component:254; Jaccard:0.068808
 |
Component:254; Jaccard:0.054444
 |
Component:254; Jaccard:0.041362
 |
Component:254; Jaccard:0.033058
 |
Correlation:0.022796
  |
Correlation:0.24492
  |
Correlation:0.24492
  |
Correlation:0.24492
  |
Correlation:0.24492
  |
Correlation:0.24492
  |
Correlation:0.24492
  |
Component:229; Jaccard:0.090712
 |
Component:229; Jaccard:0.082555
 |
Component:096; Jaccard:0.068636
 |
Component:033; Jaccard:0.056962
 |
Component:033; Jaccard:0.049059
 |
Component:167; Jaccard:0.034208
 |
Component:225; Jaccard:0.023191
 |
Correlation:-0.077393
  |
Correlation:-0.077393
  |
Correlation:0.20798
  |
Correlation:0.33724
  |
Correlation:0.33724
  |
Correlation:0.17115
  |
Correlation:0.34005
  |
Component:254; Jaccard:0.085793
 |
Component:184; Jaccard:0.08246
 |
Component:229; Jaccard:0.067235
 |
Component:096; Jaccard:0.056505
 |
Component:096; Jaccard:0.042657
 |
Component:096; Jaccard:0.031837
 |
Component:096; Jaccard:0.019579
 |
Correlation:0.24492
  |
Correlation:0.022796
  |
Correlation:-0.077393
  |
Correlation:0.20798
  |
Correlation:0.20798
  |
Correlation:0.20798
  |
Correlation:0.20798
  |
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
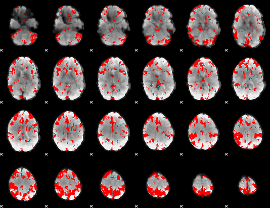 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
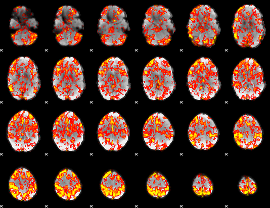 |
GLM Z-score result thresholded by 1.5
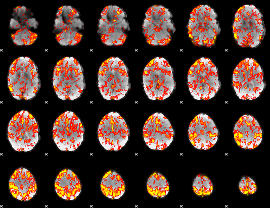 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
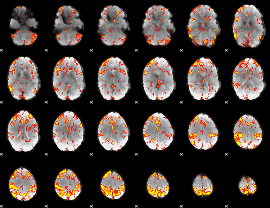 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
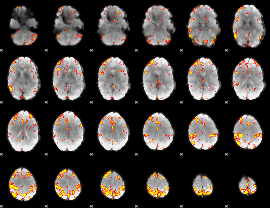 |
Component:138; Jaccard:0.15406
 |
Component:138; Jaccard:0.18262
 |
Component:138; Jaccard:0.21398
 |
Component:138; Jaccard:0.24469
 |
Component:138; Jaccard:0.27093
 |
Component:162; Jaccard:0.29322
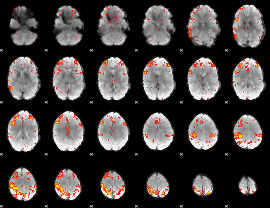 |
Component:162; Jaccard:0.31293
 |
Correlation:0.54683
  |
Correlation:0.54683
  |
Correlation:0.54683
  |
Correlation:0.54683
  |
Correlation:0.54683
  |
Correlation:0.70869
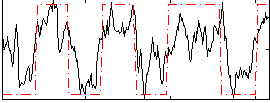 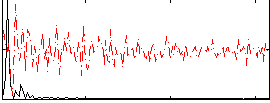 |
Correlation:0.70869
  |
Component:372; Jaccard:0.1477
 |
Component:372; Jaccard:0.17211
 |
Component:162; Jaccard:0.20199
 |
Component:162; Jaccard:0.23631
 |
Component:162; Jaccard:0.26829
 |
Component:138; Jaccard:0.28981
 |
Component:138; Jaccard:0.29904
 |
Correlation:0.53504
  |
Correlation:0.53504
  |
Correlation:0.70869
  |
Correlation:0.70869
  |
Correlation:0.70869
  |
Correlation:0.54683
  |
Correlation:0.54683
  |
Component:162; Jaccard:0.14052
 |
Component:162; Jaccard:0.16941
 |
Component:372; Jaccard:0.19704
 |
Component:372; Jaccard:0.22188
 |
Component:372; Jaccard:0.24124
 |
Component:372; Jaccard:0.25181
 |
Component:372; Jaccard:0.25842
 |
Correlation:0.70869
  |
Correlation:0.70869
  |
Correlation:0.53504
  |
Correlation:0.53504
  |
Correlation:0.53504
  |
Correlation:0.53504
  |
Correlation:0.53504
  |
Component:309; Jaccard:0.12149
 |
Component:195; Jaccard:0.14089
 |
Component:195; Jaccard:0.16084
 |
Component:195; Jaccard:0.18038
 |
Component:195; Jaccard:0.19435
 |
Component:195; Jaccard:0.20447
 |
Component:195; Jaccard:0.20687
 |
Correlation:0.58257
  |
Correlation:0.68127
  |
Correlation:0.68127
  |
Correlation:0.68127
  |
Correlation:0.68127
  |
Correlation:0.68127
  |
Correlation:0.68127
  |
Component:195; Jaccard:0.12052
 |
Component:309; Jaccard:0.13842
 |
Component:309; Jaccard:0.15471
 |
Component:350; Jaccard:0.17067
 |
Component:350; Jaccard:0.18473
 |
Component:350; Jaccard:0.19322
 |
Component:350; Jaccard:0.19419
 |
Correlation:0.68127
  |
Correlation:0.58257
  |
Correlation:0.58257
  |
Correlation:0.54011
  |
Correlation:0.54011
  |
Correlation:0.54011
  |
Correlation:0.54011
  |
Component:068; Jaccard:0.11452
 |
Component:350; Jaccard:0.13056
 |
Component:350; Jaccard:0.15087
 |
Component:309; Jaccard:0.1706
 |
Component:309; Jaccard:0.18096
 |
Component:309; Jaccard:0.18637
 |
Component:309; Jaccard:0.18581
 |
Correlation:0.4277
  |
Correlation:0.54011
  |
Correlation:0.54011
  |
Correlation:0.58257
  |
Correlation:0.58257
  |
Correlation:0.58257
  |
Correlation:0.58257
  |
Component:350; Jaccard:0.11095
 |
Component:068; Jaccard:0.12654
 |
Component:068; Jaccard:0.13748
 |
Component:068; Jaccard:0.14814
 |
Component:068; Jaccard:0.15381
 |
Component:068; Jaccard:0.1551
 |
Component:068; Jaccard:0.15227
 |
Correlation:0.54011
  |
Correlation:0.4277
  |
Correlation:0.4277
  |
Correlation:0.4277
  |
Correlation:0.4277
  |
Correlation:0.4277
  |
Correlation:0.4277
  |
Component:069; Jaccard:0.10927
 |
Component:078; Jaccard:0.1175
 |
Component:351; Jaccard:0.12536
 |
Component:351; Jaccard:0.13598
 |
Component:351; Jaccard:0.14288
 |
Component:351; Jaccard:0.14667
 |
Component:351; Jaccard:0.14456
 |
Correlation:0.33564
  |
Correlation:0.29698
  |
Correlation:0.67521
  |
Correlation:0.67521
  |
Correlation:0.67521
  |
Correlation:0.67521
  |
Correlation:0.67521
  |
Component:078; Jaccard:0.1079
 |
Component:069; Jaccard:0.11673
 |
Component:078; Jaccard:0.12399
 |
Component:279; Jaccard:0.12895
 |
Component:279; Jaccard:0.13217
 |
Component:279; Jaccard:0.1319
 |
Component:279; Jaccard:0.12956
 |
Correlation:0.29698
  |
Correlation:0.33564
  |
Correlation:0.29698
  |
Correlation:0.32835
  |
Correlation:0.32835
  |
Correlation:0.32835
  |
Correlation:0.32835
  |
Component:358; Jaccard:0.10692
 |
Component:322; Jaccard:0.11487
 |
Component:069; Jaccard:0.12296
 |
Component:078; Jaccard:0.12859
 |
Component:322; Jaccard:0.12909
 |
Component:322; Jaccard:0.12815
 |
Component:322; Jaccard:0.12298
 |
Correlation:0.35627
  |
Correlation:0.36127
  |
Correlation:0.33564
  |
Correlation:0.29698
  |
Correlation:0.36127
  |
Correlation:0.36127
  |
Correlation:0.36127
  |
Cope: 04:STORY-MATH BACK
|
GLM result group-wise
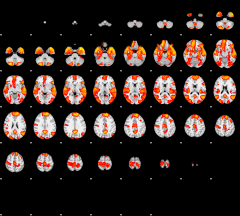 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
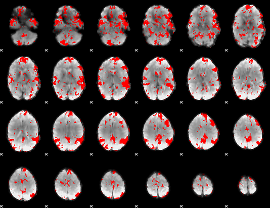 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
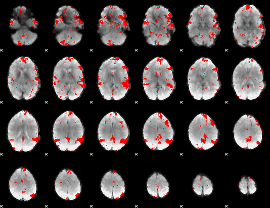 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
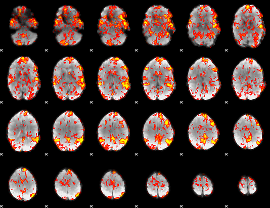 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
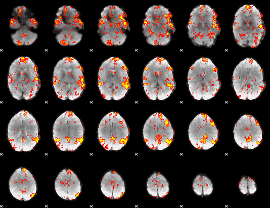 |
GLM Z-score result thresholded by 2.5
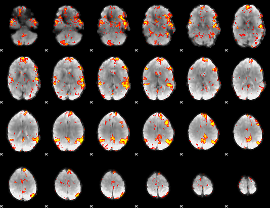 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:142; Jaccard:0.1758
 |
Component:142; Jaccard:0.22434
 |
Component:142; Jaccard:0.28085
 |
Component:142; Jaccard:0.34039
 |
Component:142; Jaccard:0.38649
 |
Component:142; Jaccard:0.41164
 |
Component:142; Jaccard:0.41351
 |
Correlation:0.5802
  |
Correlation:0.5802
  |
Correlation:0.5802
  |
Correlation:0.5802
  |
Correlation:0.5802
  |
Correlation:0.5802
  |
Correlation:0.5802
  |
Component:303; Jaccard:0.1425
 |
Component:303; Jaccard:0.16562
 |
Component:303; Jaccard:0.18635
 |
Component:303; Jaccard:0.20125
 |
Component:303; Jaccard:0.20662
 |
Component:303; Jaccard:0.20143
 |
Component:303; Jaccard:0.18724
 |
Correlation:0.45839
  |
Correlation:0.45839
  |
Correlation:0.45839
  |
Correlation:0.45839
  |
Correlation:0.45839
  |
Correlation:0.45839
  |
Correlation:0.45839
  |
Component:266; Jaccard:0.13838
 |
Component:266; Jaccard:0.15824
 |
Component:266; Jaccard:0.17493
 |
Component:266; Jaccard:0.18324
 |
Component:266; Jaccard:0.18645
 |
Component:266; Jaccard:0.17742
 |
Component:175; Jaccard:0.16939
 |
Correlation:0.46625
  |
Correlation:0.46625
  |
Correlation:0.46625
  |
Correlation:0.46625
  |
Correlation:0.46625
  |
Correlation:0.46625
  |
Correlation:0.47182
  |
Component:175; Jaccard:0.12612
 |
Component:175; Jaccard:0.14315
 |
Component:175; Jaccard:0.15879
 |
Component:175; Jaccard:0.172
 |
Component:175; Jaccard:0.17749
 |
Component:175; Jaccard:0.17667
 |
Component:266; Jaccard:0.16496
 |
Correlation:0.47182
  |
Correlation:0.47182
  |
Correlation:0.47182
  |
Correlation:0.47182
  |
Correlation:0.47182
  |
Correlation:0.47182
  |
Correlation:0.46625
  |
Component:121; Jaccard:0.12606
 |
Component:121; Jaccard:0.13714
 |
Component:390; Jaccard:0.1525
 |
Component:390; Jaccard:0.16379
 |
Component:390; Jaccard:0.16637
 |
Component:390; Jaccard:0.16125
 |
Component:390; Jaccard:0.14493
 |
Correlation:0.1791
  |
Correlation:0.1791
  |
Correlation:0.42257
  |
Correlation:0.42257
  |
Correlation:0.42257
  |
Correlation:0.42257
  |
Correlation:0.42257
  |
Component:390; Jaccard:0.11605
 |
Component:390; Jaccard:0.13411
 |
Component:121; Jaccard:0.14163
 |
Component:121; Jaccard:0.14487
 |
Component:121; Jaccard:0.1392
 |
Component:121; Jaccard:0.1282
 |
Component:121; Jaccard:0.11413
 |
Correlation:0.42257
  |
Correlation:0.42257
  |
Correlation:0.1791
  |
Correlation:0.1791
  |
Correlation:0.1791
  |
Correlation:0.1791
  |
Correlation:0.1791
  |
Component:167; Jaccard:0.10186
 |
Component:167; Jaccard:0.11382
 |
Component:167; Jaccard:0.12268
 |
Component:167; Jaccard:0.12811
 |
Component:167; Jaccard:0.12822
 |
Component:167; Jaccard:0.12064
 |
Component:167; Jaccard:0.10883
 |
Correlation:0.18529
  |
Correlation:0.18529
  |
Correlation:0.18529
  |
Correlation:0.18529
  |
Correlation:0.18529
  |
Correlation:0.18529
  |
Correlation:0.18529
  |
Component:096; Jaccard:0.094289
 |
Component:096; Jaccard:0.10305
 |
Component:096; Jaccard:0.10776
 |
Component:096; Jaccard:0.10948
 |
Component:096; Jaccard:0.10677
 |
Component:362; Jaccard:0.10255
 |
Component:362; Jaccard:0.09576
 |
Correlation:0.21197
  |
Correlation:0.21197
  |
Correlation:0.21197
  |
Correlation:0.21197
  |
Correlation:0.21197
  |
Correlation:0.36116
  |
Correlation:0.36116
  |
Component:396; Jaccard:0.091374
 |
Component:319; Jaccard:0.097875
 |
Component:319; Jaccard:0.10099
 |
Component:362; Jaccard:0.10462
 |
Component:362; Jaccard:0.10402
 |
Component:096; Jaccard:0.1011
 |
Component:051; Jaccard:0.091391
 |
Correlation:0.10715
  |
Correlation:0.22781
  |
Correlation:0.22781
  |
Correlation:0.36116
  |
Correlation:0.36116
  |
Correlation:0.21197
  |
Correlation:0.44997
  |
Component:319; Jaccard:0.090666
 |
Component:362; Jaccard:0.094568
 |
Component:362; Jaccard:0.10097
 |
Component:319; Jaccard:0.099538
 |
Component:319; Jaccard:0.09668
 |
Component:051; Jaccard:0.094931
 |
Component:096; Jaccard:0.090538
 |
Correlation:0.22781
  |
Correlation:0.36116
  |
Correlation:0.36116
  |
Correlation:0.22781
  |
Correlation:0.22781
  |
Correlation:0.44997
  |
Correlation:0.21197
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
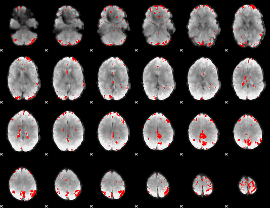 |
GLM binary result thresholded by 4
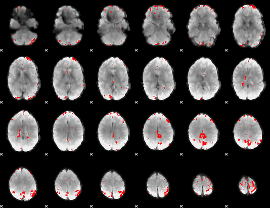 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
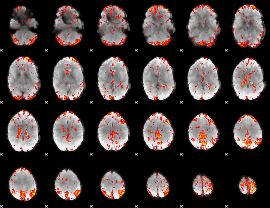 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
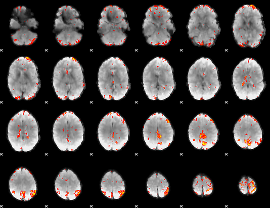 |
GLM Z-score result thresholded by 4
 |
Component:187; Jaccard:0.12648
 |
Component:222; Jaccard:0.14877
 |
Component:222; Jaccard:0.17229
 |
Component:222; Jaccard:0.18917
 |
Component:222; Jaccard:0.19087
 |
Component:222; Jaccard:0.17267
 |
Component:222; Jaccard:0.1405
 |
Correlation:-0.046251
 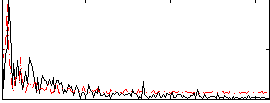 |
Correlation:-0.006601
  |
Correlation:-0.006601
  |
Correlation:-0.006601
  |
Correlation:-0.006601
  |
Correlation:-0.006601
  |
Correlation:-0.006601
  |
Component:222; Jaccard:0.12308
 |
Component:187; Jaccard:0.14458
 |
Component:291; Jaccard:0.15852
 |
Component:291; Jaccard:0.17401
 |
Component:291; Jaccard:0.16625
 |
Component:291; Jaccard:0.1352
 |
Component:291; Jaccard:0.097326
 |
Correlation:-0.006601
  |
Correlation:-0.046251
  |
Correlation:-0.25861
  |
Correlation:-0.25861
  |
Correlation:-0.25861
  |
Correlation:-0.25861
  |
Correlation:-0.25861
  |
Component:358; Jaccard:0.12115
 |
Component:358; Jaccard:0.13922
 |
Component:187; Jaccard:0.15833
 |
Component:187; Jaccard:0.16103
 |
Component:187; Jaccard:0.14951
 |
Component:187; Jaccard:0.12472
 |
Component:187; Jaccard:0.09546
 |
Correlation:-0.3287
  |
Correlation:-0.3287
  |
Correlation:-0.046251
  |
Correlation:-0.046251
  |
Correlation:-0.046251
  |
Correlation:-0.046251
  |
Correlation:-0.046251
  |
Component:344; Jaccard:0.11189
 |
Component:291; Jaccard:0.13243
 |
Component:358; Jaccard:0.15485
 |
Component:358; Jaccard:0.16084
 |
Component:358; Jaccard:0.14899
 |
Component:358; Jaccard:0.12126
 |
Component:358; Jaccard:0.090758
 |
Correlation:-0.072212
  |
Correlation:-0.25861
  |
Correlation:-0.3287
  |
Correlation:-0.3287
  |
Correlation:-0.3287
  |
Correlation:-0.3287
  |
Correlation:-0.3287
  |
Component:398; Jaccard:0.11116
 |
Component:344; Jaccard:0.12633
 |
Component:344; Jaccard:0.13794
 |
Component:372; Jaccard:0.14132
 |
Component:207; Jaccard:0.13066
 |
Component:186; Jaccard:0.1103
 |
Component:186; Jaccard:0.087552
 |
Correlation:-0.093168
 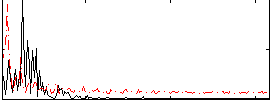 |
Correlation:-0.072212
  |
Correlation:-0.072212
  |
Correlation:-0.49479
  |
Correlation:0.22427
  |
Correlation:-0.10382
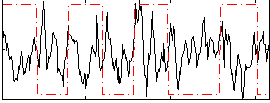  |
Correlation:-0.10382
  |
Component:372; Jaccard:0.11089
 |
Component:372; Jaccard:0.12501
 |
Component:372; Jaccard:0.13706
 |
Component:078; Jaccard:0.14062
 |
Component:372; Jaccard:0.12844
 |
Component:207; Jaccard:0.11013
 |
Component:207; Jaccard:0.083726
 |
Correlation:-0.49479
  |
Correlation:-0.49479
  |
Correlation:-0.49479
  |
Correlation:-0.28957
 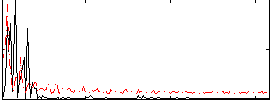 |
Correlation:-0.49479
  |
Correlation:0.22427
  |
Correlation:0.22427
  |
Component:291; Jaccard:0.10698
 |
Component:078; Jaccard:0.12075
 |
Component:078; Jaccard:0.13574
 |
Component:344; Jaccard:0.14009
 |
Component:078; Jaccard:0.12679
 |
Component:372; Jaccard:0.10513
 |
Component:065; Jaccard:0.07654
 |
Correlation:-0.25861
  |
Correlation:-0.28957
  |
Correlation:-0.28957
  |
Correlation:-0.072212
  |
Correlation:-0.28957
  |
Correlation:-0.49479
  |
Correlation:0.27008
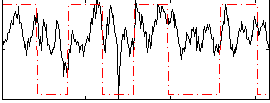 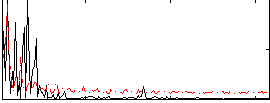 |
Component:186; Jaccard:0.10664
 |
Component:398; Jaccard:0.12048
 |
Component:207; Jaccard:0.12798
 |
Component:207; Jaccard:0.13611
 |
Component:186; Jaccard:0.12633
 |
Component:078; Jaccard:0.0994
 |
Component:394; Jaccard:0.075128
 |
Correlation:-0.10382
  |
Correlation:-0.093168
  |
Correlation:0.22427
  |
Correlation:0.22427
  |
Correlation:-0.10382
  |
Correlation:-0.28957
  |
Correlation:-0.092138
  |
Component:394; Jaccard:0.10567
 |
Component:186; Jaccard:0.11856
 |
Component:186; Jaccard:0.12781
 |
Component:186; Jaccard:0.13073
 |
Component:344; Jaccard:0.12367
 |
Component:344; Jaccard:0.095675
 |
Component:372; Jaccard:0.074036
 |
Correlation:-0.092138
  |
Correlation:-0.10382
  |
Correlation:-0.10382
  |
Correlation:-0.10382
  |
Correlation:-0.072212
  |
Correlation:-0.072212
  |
Correlation:-0.49479
  |
Component:078; Jaccard:0.10502
 |
Component:394; Jaccard:0.11361
 |
Component:398; Jaccard:0.12551
 |
Component:398; Jaccard:0.12159
 |
Component:394; Jaccard:0.10977
 |
Component:065; Jaccard:0.095296
 |
Component:322; Jaccard:0.067708
 |
Correlation:-0.28957
  |
Correlation:-0.092138
  |
Correlation:-0.093168
  |
Correlation:-0.093168
  |
Correlation:-0.092138
  |
Correlation:0.27008
  |
Correlation:-0.34379
  |
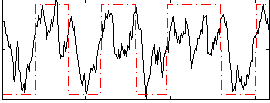
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
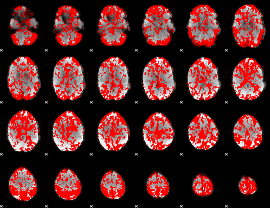 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
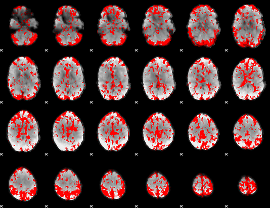 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
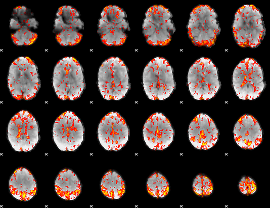 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:372; Jaccard:0.13792
 |
Component:372; Jaccard:0.16897
 |
Component:372; Jaccard:0.20098
 |
Component:372; Jaccard:0.23169
 |
Component:372; Jaccard:0.25251
 |
Component:372; Jaccard:0.25777
 |
Component:372; Jaccard:0.24374
 |
Correlation:0.55552
  |
Correlation:0.55552
  |
Correlation:0.55552
  |
Correlation:0.55552
  |
Correlation:0.55552
  |
Correlation:0.55552
  |
Correlation:0.55552
  |
Component:358; Jaccard:0.12785
 |
Component:358; Jaccard:0.15079
 |
Component:358; Jaccard:0.17338
 |
Component:291; Jaccard:0.19167
 |
Component:291; Jaccard:0.21773
 |
Component:291; Jaccard:0.22794
 |
Component:291; Jaccard:0.22053
 |
Correlation:0.37068
  |
Correlation:0.37068
  |
Correlation:0.37068
  |
Correlation:0.30485
  |
Correlation:0.30485
  |
Correlation:0.30485
  |
Correlation:0.30485
  |
Component:187; Jaccard:0.11863
 |
Component:078; Jaccard:0.13465
 |
Component:291; Jaccard:0.16168
 |
Component:358; Jaccard:0.19135
 |
Component:358; Jaccard:0.20053
 |
Component:358; Jaccard:0.19578
 |
Component:358; Jaccard:0.18156
 |
Correlation:0.038256
  |
Correlation:0.29341
  |
Correlation:0.30485
  |
Correlation:0.37068
  |
Correlation:0.37068
  |
Correlation:0.37068
  |
Correlation:0.37068
  |
Component:398; Jaccard:0.11706
 |
Component:187; Jaccard:0.13348
 |
Component:078; Jaccard:0.15648
 |
Component:078; Jaccard:0.17544
 |
Component:078; Jaccard:0.18655
 |
Component:078; Jaccard:0.18788
 |
Component:078; Jaccard:0.1745
 |
Correlation:0.14251
  |
Correlation:0.038256
  |
Correlation:0.29341
  |
Correlation:0.29341
  |
Correlation:0.29341
  |
Correlation:0.29341
  |
Correlation:0.29341
  |
Component:138; Jaccard:0.11424
 |
Component:291; Jaccard:0.13148
 |
Component:322; Jaccard:0.14794
 |
Component:350; Jaccard:0.15851
 |
Component:350; Jaccard:0.17431
 |
Component:350; Jaccard:0.1778
 |
Component:350; Jaccard:0.16878
 |
Correlation:0.55212
  |
Correlation:0.30485
  |
Correlation:0.3654
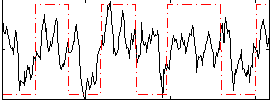  |
Correlation:0.54802
  |
Correlation:0.54802
  |
Correlation:0.54802
  |
Correlation:0.54802
  |
Component:078; Jaccard:0.11348
 |
Component:322; Jaccard:0.13104
 |
Component:138; Jaccard:0.1458
 |
Component:322; Jaccard:0.15804
 |
Component:138; Jaccard:0.16249
 |
Component:138; Jaccard:0.15987
 |
Component:138; Jaccard:0.14848
 |
Correlation:0.29341
  |
Correlation:0.3654
  |
Correlation:0.55212
  |
Correlation:0.3654
  |
Correlation:0.55212
  |
Correlation:0.55212
  |
Correlation:0.55212
  |
Component:322; Jaccard:0.11324
 |
Component:398; Jaccard:0.13087
 |
Component:187; Jaccard:0.1448
 |
Component:138; Jaccard:0.15719
 |
Component:322; Jaccard:0.1593
 |
Component:322; Jaccard:0.15603
 |
Component:322; Jaccard:0.1421
 |
Correlation:0.3654
  |
Correlation:0.14251
  |
Correlation:0.038256
  |
Correlation:0.55212
  |
Correlation:0.3654
  |
Correlation:0.3654
  |
Correlation:0.3654
  |
Component:344; Jaccard:0.10806
 |
Component:138; Jaccard:0.13036
 |
Component:398; Jaccard:0.14
 |
Component:187; Jaccard:0.1495
 |
Component:187; Jaccard:0.14547
 |
Component:222; Jaccard:0.13729
 |
Component:222; Jaccard:0.12526
 |
Correlation:0.13141
  |
Correlation:0.55212
  |
Correlation:0.14251
  |
Correlation:0.038256
  |
Correlation:0.038256
  |
Correlation:0.055111
  |
Correlation:0.055111
  |
Component:222; Jaccard:0.10697
 |
Component:222; Jaccard:0.12211
 |
Component:350; Jaccard:0.13866
 |
Component:398; Jaccard:0.1429
 |
Component:222; Jaccard:0.1437
 |
Component:187; Jaccard:0.13396
 |
Component:043; Jaccard:0.12498
 |
Correlation:0.055111
  |
Correlation:0.055111
  |
Correlation:0.54802
  |
Correlation:0.14251
  |
Correlation:0.055111
  |
Correlation:0.038256
  |
Correlation:0.26121
  |
Component:291; Jaccard:0.10624
 |
Component:344; Jaccard:0.12079
 |
Component:222; Jaccard:0.13319
 |
Component:222; Jaccard:0.14106
 |
Component:398; Jaccard:0.13925
 |
Component:043; Jaccard:0.13363
 |
Component:187; Jaccard:0.11862
 |
Correlation:0.30485
  |
Correlation:0.13141
  |
Correlation:0.055111
  |
Correlation:0.055111
  |
Correlation:0.14251
  |
Correlation:0.26121
  |
Correlation:0.038256
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































