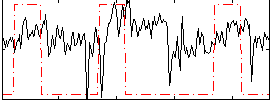
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
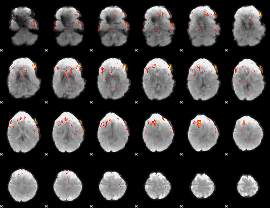
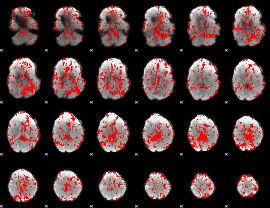
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
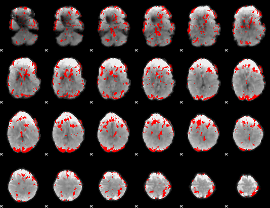 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
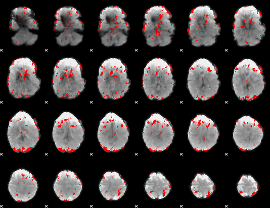 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
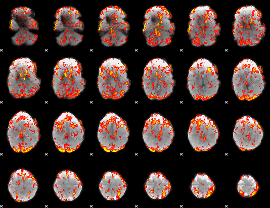 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
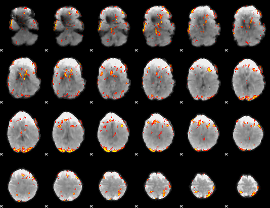 |
GLM Z-score result thresholded by 4
 |
Component:374; Jaccard:0.12588
 |
Component:363; Jaccard:0.14202
 |
Component:291; Jaccard:0.15951
 |
Component:291; Jaccard:0.1833
 |
Component:291; Jaccard:0.20495
 |
Component:291; Jaccard:0.21788
 |
Component:291; Jaccard:0.2215
 |
Correlation:0.13435
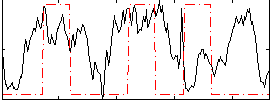  |
Correlation:0.18711
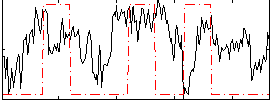  |
Correlation:0.25578
  |
Correlation:0.25578
  |
Correlation:0.25578
  |
Correlation:0.25578
  |
Correlation:0.25578
  |
Component:363; Jaccard:0.12361
 |
Component:374; Jaccard:0.13729
 |
Component:363; Jaccard:0.15847
 |
Component:363; Jaccard:0.17179
 |
Component:363; Jaccard:0.17572
 |
Component:363; Jaccard:0.1753
 |
Component:363; Jaccard:0.1658
 |
Correlation:0.18711
  |
Correlation:0.13435
  |
Correlation:0.18711
  |
Correlation:0.18711
  |
Correlation:0.18711
  |
Correlation:0.18711
  |
Correlation:0.18711
  |
Component:318; Jaccard:0.11251
 |
Component:291; Jaccard:0.13348
 |
Component:374; Jaccard:0.14609
 |
Component:170; Jaccard:0.15186
 |
Component:170; Jaccard:0.16101
 |
Component:170; Jaccard:0.16159
 |
Component:170; Jaccard:0.15848
 |
Correlation:-0.005029
  |
Correlation:0.25578
  |
Correlation:0.13435
  |
Correlation:0.33061
  |
Correlation:0.33061
  |
Correlation:0.33061
  |
Correlation:0.33061
  |
Component:291; Jaccard:0.11042
 |
Component:318; Jaccard:0.12537
 |
Component:318; Jaccard:0.13738
 |
Component:374; Jaccard:0.14782
 |
Component:198; Jaccard:0.14919
 |
Component:198; Jaccard:0.15462
 |
Component:198; Jaccard:0.15621
 |
Correlation:0.25578
  |
Correlation:-0.005029
  |
Correlation:-0.005029
  |
Correlation:0.13435
  |
Correlation:0.18079
  |
Correlation:0.18079
  |
Correlation:0.18079
  |
Component:044; Jaccard:0.10782
 |
Component:238; Jaccard:0.12205
 |
Component:170; Jaccard:0.13719
 |
Component:318; Jaccard:0.14494
 |
Component:238; Jaccard:0.14744
 |
Component:172; Jaccard:0.14706
 |
Component:172; Jaccard:0.14231
 |
Correlation:0.21097
  |
Correlation:0.22565
  |
Correlation:0.33061
  |
Correlation:-0.005029
  |
Correlation:0.22565
  |
Correlation:0.072973
  |
Correlation:0.072973
  |
Component:238; Jaccard:0.10775
 |
Component:044; Jaccard:0.12179
 |
Component:238; Jaccard:0.13589
 |
Component:238; Jaccard:0.14346
 |
Component:044; Jaccard:0.14608
 |
Component:044; Jaccard:0.14586
 |
Component:044; Jaccard:0.1395
 |
Correlation:0.22565
  |
Correlation:0.21097
  |
Correlation:0.22565
  |
Correlation:0.22565
  |
Correlation:0.21097
  |
Correlation:0.21097
  |
Correlation:0.21097
  |
Component:170; Jaccard:0.10076
 |
Component:170; Jaccard:0.11927
 |
Component:044; Jaccard:0.13357
 |
Component:044; Jaccard:0.14276
 |
Component:318; Jaccard:0.14542
 |
Component:238; Jaccard:0.14393
 |
Component:238; Jaccard:0.13549
 |
Correlation:0.33061
  |
Correlation:0.33061
  |
Correlation:0.21097
  |
Correlation:0.21097
  |
Correlation:-0.005029
  |
Correlation:0.22565
  |
Correlation:0.22565
  |
Component:148; Jaccard:0.10033
 |
Component:148; Jaccard:0.11073
 |
Component:172; Jaccard:0.1257
 |
Component:172; Jaccard:0.13593
 |
Component:172; Jaccard:0.14481
 |
Component:318; Jaccard:0.13833
 |
Component:318; Jaccard:0.12933
 |
Correlation:0.022295
  |
Correlation:0.022295
  |
Correlation:0.072973
  |
Correlation:0.072973
  |
Correlation:0.072973
  |
Correlation:-0.005029
  |
Correlation:-0.005029
  |
Component:173; Jaccard:0.098048
 |
Component:172; Jaccard:0.11017
 |
Component:148; Jaccard:0.1205
 |
Component:198; Jaccard:0.13396
 |
Component:374; Jaccard:0.1428
 |
Component:374; Jaccard:0.13628
 |
Component:374; Jaccard:0.11993
 |
Correlation:0.10212
  |
Correlation:0.072973
  |
Correlation:0.022295
  |
Correlation:0.18079
  |
Correlation:0.13435
  |
Correlation:0.13435
  |
Correlation:0.13435
  |
Component:231; Jaccard:0.097282
 |
Component:226; Jaccard:0.10831
 |
Component:226; Jaccard:0.11761
 |
Component:148; Jaccard:0.12732
 |
Component:148; Jaccard:0.12815
 |
Component:148; Jaccard:0.1259
 |
Component:148; Jaccard:0.11789
 |
Correlation:0.12044
  |
Correlation:0.14862
  |
Correlation:0.14862
  |
Correlation:0.022295
  |
Correlation:0.022295
  |
Correlation:0.022295
  |
Correlation:0.022295
  |
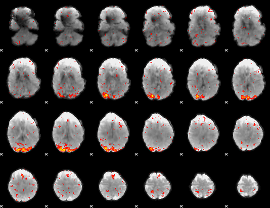
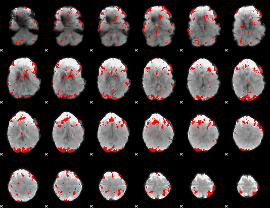
Cope: 02:REL BACK
|
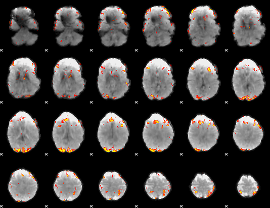
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
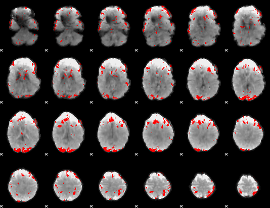 |
GLM binary result thresholded by 4
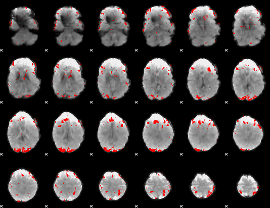 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
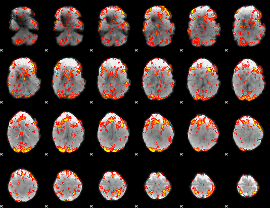 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
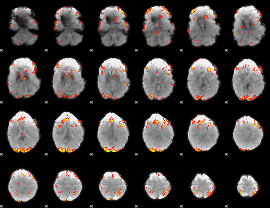 |
GLM Z-score result thresholded by 4
 |
Component:374; Jaccard:0.12864
 |
Component:374; Jaccard:0.14832
 |
Component:374; Jaccard:0.16625
 |
Component:374; Jaccard:0.17599
 |
Component:374; Jaccard:0.1779
 |
Component:363; Jaccard:0.17584
 |
Component:363; Jaccard:0.17033
 |
Correlation:-0.093882
  |
Correlation:-0.093882
  |
Correlation:-0.093882
  |
Correlation:-0.093882
  |
Correlation:-0.093882
  |
Correlation:-0.037824
  |
Correlation:-0.037824
  |
Component:363; Jaccard:0.11586
 |
Component:363; Jaccard:0.13495
 |
Component:363; Jaccard:0.1532
 |
Component:363; Jaccard:0.16924
 |
Component:363; Jaccard:0.17669
 |
Component:374; Jaccard:0.17271
 |
Component:226; Jaccard:0.16188
 |
Correlation:-0.037824
  |
Correlation:-0.037824
  |
Correlation:-0.037824
  |
Correlation:-0.037824
  |
Correlation:-0.037824
  |
Correlation:-0.093882
  |
Correlation:0.37281
  |
Component:318; Jaccard:0.11182
 |
Component:318; Jaccard:0.13031
 |
Component:318; Jaccard:0.14722
 |
Component:318; Jaccard:0.16052
 |
Component:226; Jaccard:0.16647
 |
Component:226; Jaccard:0.16654
 |
Component:044; Jaccard:0.16069
 |
Correlation:0.1747
 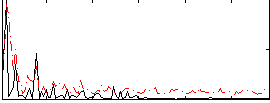 |
Correlation:0.1747
  |
Correlation:0.1747
  |
Correlation:0.1747
  |
Correlation:0.37281
  |
Correlation:0.37281
  |
Correlation:0.13476
 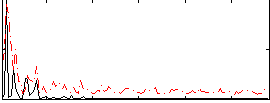 |
Component:304; Jaccard:0.11176
 |
Component:304; Jaccard:0.12922
 |
Component:226; Jaccard:0.14428
 |
Component:226; Jaccard:0.15993
 |
Component:318; Jaccard:0.16426
 |
Component:044; Jaccard:0.16483
 |
Component:374; Jaccard:0.15923
 |
Correlation:0.37018
  |
Correlation:0.37018
  |
Correlation:0.37281
  |
Correlation:0.37281
  |
Correlation:0.1747
  |
Correlation:0.13476
  |
Correlation:-0.093882
  |
Component:044; Jaccard:0.10666
 |
Component:226; Jaccard:0.1254
 |
Component:304; Jaccard:0.14295
 |
Component:044; Jaccard:0.15427
 |
Component:044; Jaccard:0.1615
 |
Component:172; Jaccard:0.16059
 |
Component:172; Jaccard:0.15874
 |
Correlation:0.13476
  |
Correlation:0.37281
  |
Correlation:0.37018
  |
Correlation:0.13476
  |
Correlation:0.13476
  |
Correlation:0.16072
 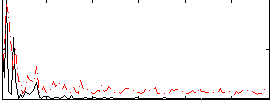 |
Correlation:0.16072
  |
Component:226; Jaccard:0.10611
 |
Component:044; Jaccard:0.12383
 |
Component:044; Jaccard:0.1411
 |
Component:304; Jaccard:0.15248
 |
Component:304; Jaccard:0.15504
 |
Component:318; Jaccard:0.15673
 |
Component:318; Jaccard:0.14259
 |
Correlation:0.37281
  |
Correlation:0.13476
  |
Correlation:0.13476
  |
Correlation:0.37018
  |
Correlation:0.37018
  |
Correlation:0.1747
  |
Correlation:0.1747
  |
Component:148; Jaccard:0.10022
 |
Component:170; Jaccard:0.11377
 |
Component:170; Jaccard:0.12901
 |
Component:172; Jaccard:0.14243
 |
Component:172; Jaccard:0.15421
 |
Component:304; Jaccard:0.15039
 |
Component:304; Jaccard:0.1412
 |
Correlation:0.22217
  |
Correlation:0.1271
  |
Correlation:0.1271
  |
Correlation:0.16072
  |
Correlation:0.16072
  |
Correlation:0.37018
  |
Correlation:0.37018
  |
Component:333; Jaccard:0.098674
 |
Component:148; Jaccard:0.1136
 |
Component:148; Jaccard:0.12641
 |
Component:170; Jaccard:0.1404
 |
Component:148; Jaccard:0.14551
 |
Component:148; Jaccard:0.14423
 |
Component:148; Jaccard:0.14105
 |
Correlation:0.039161
  |
Correlation:0.22217
  |
Correlation:0.22217
  |
Correlation:0.1271
  |
Correlation:0.22217
  |
Correlation:0.22217
  |
Correlation:0.22217
  |
Component:170; Jaccard:0.096317
 |
Component:172; Jaccard:0.10789
 |
Component:172; Jaccard:0.12583
 |
Component:148; Jaccard:0.13893
 |
Component:170; Jaccard:0.14381
 |
Component:170; Jaccard:0.14064
 |
Component:291; Jaccard:0.12959
 |
Correlation:0.1271
  |
Correlation:0.16072
  |
Correlation:0.16072
  |
Correlation:0.22217
  |
Correlation:0.1271
  |
Correlation:0.1271
  |
Correlation:-0.027978
  |
Component:172; Jaccard:0.091041
 |
Component:333; Jaccard:0.10657
 |
Component:291; Jaccard:0.12182
 |
Component:291; Jaccard:0.13468
 |
Component:291; Jaccard:0.14075
 |
Component:291; Jaccard:0.14055
 |
Component:170; Jaccard:0.12424
 |
Correlation:0.16072
  |
Correlation:0.039161
  |
Correlation:-0.027978
  |
Correlation:-0.027978
  |
Correlation:-0.027978
  |
Correlation:-0.027978
  |
Correlation:0.1271
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
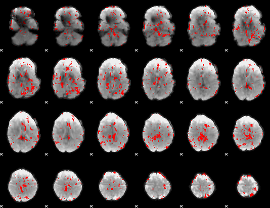 |
GLM binary result thresholded by 2.5
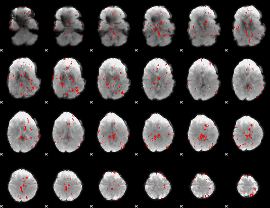 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
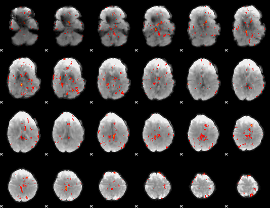 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
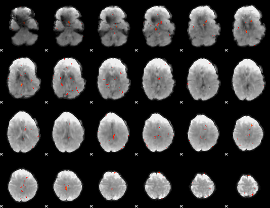 |
GLM Z-score result thresholded by 4
 |
Component:231; Jaccard:0.094935
 |
Component:209; Jaccard:0.11121
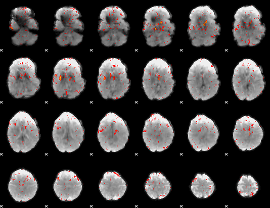 |
Component:209; Jaccard:0.12578
 |
Component:209; Jaccard:0.12187
 |
Component:209; Jaccard:0.10027
 |
Component:209; Jaccard:0.068007
 |
Component:209; Jaccard:0.03657
 |
Correlation:0.071487
  |
Correlation:0.3982
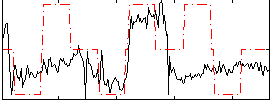 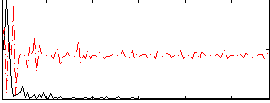 |
Correlation:0.3982
  |
Correlation:0.3982
  |
Correlation:0.3982
  |
Correlation:0.3982
  |
Correlation:0.3982
  |
Component:209; Jaccard:0.092959
 |
Component:351; Jaccard:0.10126
 |
Component:351; Jaccard:0.10518
 |
Component:351; Jaccard:0.094706
 |
Component:351; Jaccard:0.069213
 |
Component:351; Jaccard:0.043018
 |
Component:351; Jaccard:0.025169
 |
Correlation:0.3982
  |
Correlation:0.20331
  |
Correlation:0.20331
  |
Correlation:0.20331
  |
Correlation:0.20331
  |
Correlation:0.20331
  |
Correlation:0.20331
  |
Component:351; Jaccard:0.090435
 |
Component:231; Jaccard:0.098965
 |
Component:295; Jaccard:0.099296
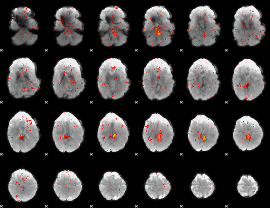 |
Component:295; Jaccard:0.091088
 |
Component:295; Jaccard:0.063708
 |
Component:295; Jaccard:0.033835
 |
Component:053; Jaccard:0.019028
 |
Correlation:0.20331
  |
Correlation:0.071487
  |
Correlation:0.14037
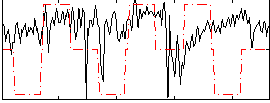  |
Correlation:0.14037
  |
Correlation:0.14037
  |
Correlation:0.14037
  |
Correlation:0.28818
 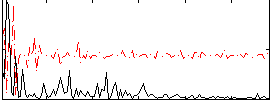 |
Component:322; Jaccard:0.085145
 |
Component:295; Jaccard:0.095529
 |
Component:231; Jaccard:0.092942
 |
Component:231; Jaccard:0.071987
 |
Component:231; Jaccard:0.045649
 |
Component:053; Jaccard:0.031075
 |
Component:360; Jaccard:0.017611
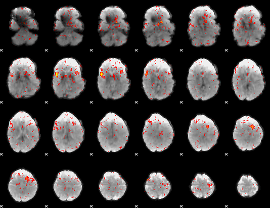 |
Correlation:0.18971
 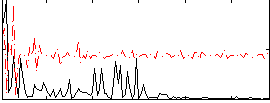 |
Correlation:0.14037
  |
Correlation:0.071487
  |
Correlation:0.071487
  |
Correlation:0.071487
  |
Correlation:0.28818
  |
Correlation:0.33302
  |
Component:348; Jaccard:0.083906
 |
Component:322; Jaccard:0.092145
 |
Component:322; Jaccard:0.085231
 |
Component:109; Jaccard:0.069387
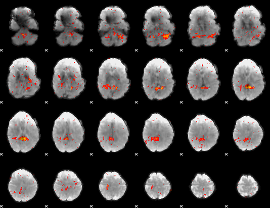 |
Component:053; Jaccard:0.045224
 |
Component:360; Jaccard:0.027866
 |
Component:295; Jaccard:0.015677
 |
Correlation:0.046741
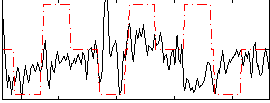 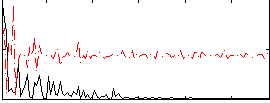 |
Correlation:0.18971
  |
Correlation:0.18971
  |
Correlation:0.20431
  |
Correlation:0.28818
  |
Correlation:0.33302
  |
Correlation:0.14037
  |
Component:150; Jaccard:0.082745
 |
Component:150; Jaccard:0.084216
 |
Component:109; Jaccard:0.084111
 |
Component:150; Jaccard:0.062669
 |
Component:360; Jaccard:0.044493
 |
Component:150; Jaccard:0.023588
 |
Component:154; Jaccard:0.012852
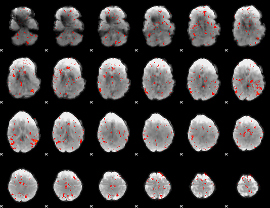 |
Correlation:0.16547
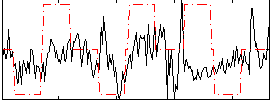  |
Correlation:0.16547
  |
Correlation:0.20431
  |
Correlation:0.16547
  |
Correlation:0.33302
  |
Correlation:0.16547
  |
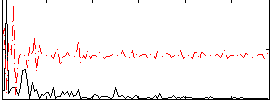
Correlation:0.19764
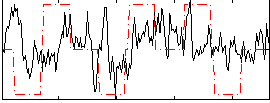  |
Component:018; Jaccard:0.079965
 |
Component:109; Jaccard:0.08368
 |
Component:150; Jaccard:0.0783
 |
Component:322; Jaccard:0.062447
 |
Component:150; Jaccard:0.041578
 |
Component:231; Jaccard:0.022713
 |
Component:060; Jaccard:0.012849
 |
Correlation:0.16671
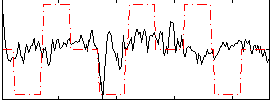  |
Correlation:0.20431
  |
Correlation:0.16547
  |
Correlation:0.18971
  |
Correlation:0.16547
  |
Correlation:0.071487
  |
Correlation:0.13912
  |
Component:295; Jaccard:0.07873
 |
Component:007; Jaccard:0.081103
 |
Component:007; Jaccard:0.074417
 |
Component:273; Jaccard:0.061124
 |
Component:291; Jaccard:0.041324
 |
Component:154; Jaccard:0.022209
 |
Component:150; Jaccard:0.012625
 |
Correlation:0.14037
  |
Correlation:0.08878
 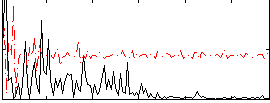 |
Correlation:0.08878
  |
Correlation:0.083798
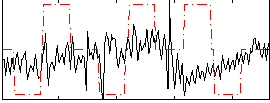  |
Correlation:0.16818
 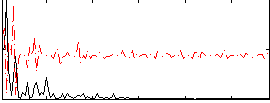 |
Correlation:0.19764
  |
Correlation:0.16547
  |
Component:007; Jaccard:0.078105
 |
Component:018; Jaccard:0.078798
 |
Component:238; Jaccard:0.073556
 |
Component:360; Jaccard:0.059781
 |
Component:238; Jaccard:0.039581
 |
Component:291; Jaccard:0.021902
 |
Component:291; Jaccard:0.012397
 |
Correlation:0.08878
  |
Correlation:0.16671
  |
Correlation:0.20438
  |
Correlation:0.33302
  |
Correlation:0.20438
  |
Correlation:0.16818
  |
Correlation:0.16818
  |
Component:131; Jaccard:0.077268
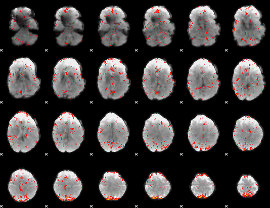 |
Component:379; Jaccard:0.077619
 |
Component:273; Jaccard:0.072441
 |
Component:238; Jaccard:0.058459
 |
Component:109; Jaccard:0.038564
 |
Component:238; Jaccard:0.020852
 |
Component:073; Jaccard:0.011812
 |
Correlation:0.13934
  |
Correlation:0.053264
  |
Correlation:0.083798
  |
Correlation:0.20438
  |
Correlation:0.20431
  |
Correlation:0.20438
  |
Correlation:0.051257
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
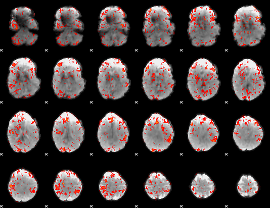 |
GLM Z-score result thresholded by 2.5
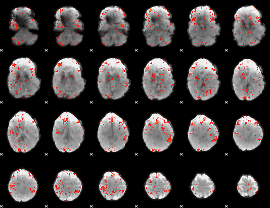 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:304; Jaccard:0.084459
 |
Component:304; Jaccard:0.10336
 |
Component:304; Jaccard:0.12475
 |
Component:304; Jaccard:0.13434
 |
Component:304; Jaccard:0.11952
 |
Component:304; Jaccard:0.080386
 |
Component:304; Jaccard:0.047233
 |
Correlation:0.22739
  |
Correlation:0.22739
  |
Correlation:0.22739
  |
Correlation:0.22739
  |
Correlation:0.22739
  |
Correlation:0.22739
  |
Correlation:0.22739
  |
Component:333; Jaccard:0.082788
 |
Component:333; Jaccard:0.092836
 |
Component:333; Jaccard:0.098199
 |
Component:333; Jaccard:0.095457
 |
Component:333; Jaccard:0.077636
 |
Component:333; Jaccard:0.047985
 |
Component:333; Jaccard:0.02495
 |
Correlation:0.066097
  |
Correlation:0.066097
  |
Correlation:0.066097
  |
Correlation:0.066097
  |
Correlation:0.066097
  |
Correlation:0.066097
  |
Correlation:0.066097
  |
Component:270; Jaccard:0.079816
 |
Component:270; Jaccard:0.0864
 |
Component:270; Jaccard:0.089819
 |
Component:270; Jaccard:0.08208
 |
Component:270; Jaccard:0.062342
 |
Component:270; Jaccard:0.038568
 |
Component:270; Jaccard:0.019505
 |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Component:096; Jaccard:0.07213
 |
Component:096; Jaccard:0.074737
 |
Component:162; Jaccard:0.073346
 |
Component:023; Jaccard:0.070451
 |
Component:023; Jaccard:0.058238
 |
Component:023; Jaccard:0.034611
 |
Component:226; Jaccard:0.01688
 |
Correlation:0.17418
  |
Correlation:0.17418
  |
Correlation:0.22538
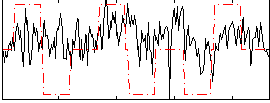  |
Correlation:0.058448
 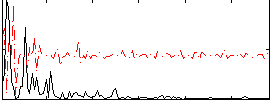 |
Correlation:0.058448
  |
Correlation:0.058448
  |
Correlation:0.13288
 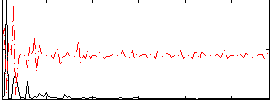 |
Component:094; Jaccard:0.071528
 |
Component:319; Jaccard:0.073525
 |
Component:050; Jaccard:0.072717
 |
Component:050; Jaccard:0.068341
 |
Component:038; Jaccard:0.052684
 |
Component:157; Jaccard:0.03378
 |
Component:038; Jaccard:0.016345
 |
Correlation:0.044596
 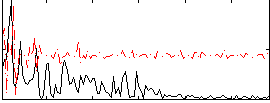 |
Correlation:0.17226
 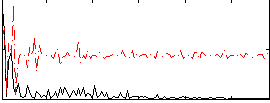 |
Correlation:0.17117
  |
Correlation:0.17117
  |
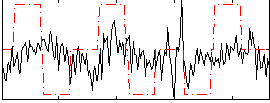
Correlation:0.037918
 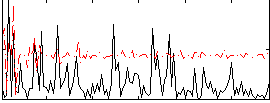 |
Correlation:0.33484
 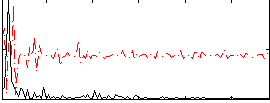 |
Correlation:0.037918
  |
Component:319; Jaccard:0.069862
 |
Component:162; Jaccard:0.07162
 |
Component:335; Jaccard:0.072698
 |
Component:162; Jaccard:0.067265
 |
Component:157; Jaccard:0.052676
 |
Component:162; Jaccard:0.032566
 |
Component:374; Jaccard:0.016252
 |
Correlation:0.17226
  |
Correlation:0.22538
  |
Correlation:0.19727
  |
Correlation:0.22538
  |
Correlation:0.33484
  |
Correlation:0.22538
  |
Correlation:-0.13527
  |
Component:145; Jaccard:0.068221
 |
Component:335; Jaccard:0.071595
 |
Component:096; Jaccard:0.072415
 |
Component:335; Jaccard:0.065179
 |
Component:162; Jaccard:0.052666
 |
Component:038; Jaccard:0.03227
 |
Component:123; Jaccard:0.015555
 |
Correlation:0.19348
  |
Correlation:0.19727
  |
Correlation:0.17418
  |
Correlation:0.19727
  |
Correlation:0.22538
  |
Correlation:0.037918
  |
Correlation:0.22016
  |
Component:162; Jaccard:0.067365
 |
Component:094; Jaccard:0.070467
 |
Component:319; Jaccard:0.071345
 |
Component:157; Jaccard:0.06502
 |
Component:123; Jaccard:0.050054
 |
Component:123; Jaccard:0.031091
 |
Component:157; Jaccard:0.015502
 |
Correlation:0.22538
  |
Correlation:0.044596
  |
Correlation:0.17226
  |
Correlation:0.33484
  |
Correlation:0.22016
  |
Correlation:0.22016
  |
Correlation:0.33484
  |
Component:283; Jaccard:0.067166
 |
Component:083; Jaccard:0.069855
 |
Component:023; Jaccard:0.070481
 |
Component:123; Jaccard:0.064231
 |
Component:050; Jaccard:0.049293
 |
Component:374; Jaccard:0.029559
 |
Component:023; Jaccard:0.015498
 |
Correlation:0.19997
  |
Correlation:-0.080257
  |
Correlation:0.058448
  |
Correlation:0.22016
  |
Correlation:0.17117
  |
Correlation:-0.13527
  |
Correlation:0.058448
  |
Component:335; Jaccard:0.066672
 |
Component:145; Jaccard:0.069565
 |
Component:083; Jaccard:0.068845
 |
Component:319; Jaccard:0.063234
 |
Component:319; Jaccard:0.048378
 |
Component:319; Jaccard:0.029177
 |
Component:168; Jaccard:0.015431
 |
Correlation:0.19727
  |
Correlation:0.19348
  |
Correlation:-0.080257
  |
Correlation:0.17226
  |
Correlation:0.17226
  |
Correlation:0.17226
  |
Correlation:0.044396
  |
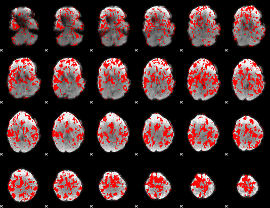
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
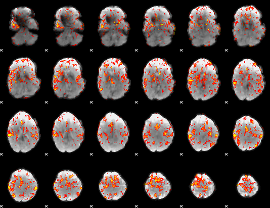
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
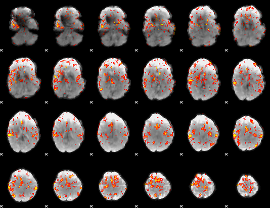 |
GLM Z-score result thresholded by 4
 |
Component:274; Jaccard:0.087376
 |
Component:274; Jaccard:0.099437
 |
Component:274; Jaccard:0.11007
 |
Component:134; Jaccard:0.1228
 |
Component:134; Jaccard:0.14372
 |
Component:134; Jaccard:0.16083
 |
Component:134; Jaccard:0.17419
 |
Correlation:0.22977
  |
Correlation:0.22977
  |
Correlation:0.22977
  |
Correlation:0.14045
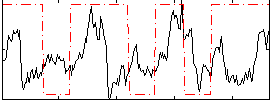 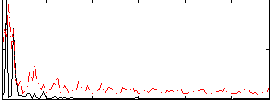 |
Correlation:0.14045
  |
Correlation:0.14045
  |
Correlation:0.14045
  |
Component:145; Jaccard:0.082269
 |
Component:193; Jaccard:0.092844
 |
Component:193; Jaccard:0.10657
 |
Component:193; Jaccard:0.12022
 |
Component:193; Jaccard:0.13205
 |
Component:204; Jaccard:0.14953
 |
Component:204; Jaccard:0.16656
 |
Correlation:0.17546
 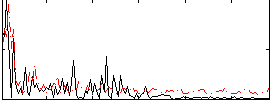 |
Correlation:0.0038091
 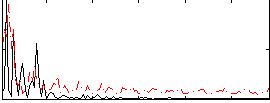 |
Correlation:0.0038091
  |
Correlation:0.0038091
  |
Correlation:0.0038091
  |
Correlation:0.32375
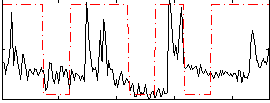  |
Correlation:0.32375
  |
Component:394; Jaccard:0.08193
 |
Component:145; Jaccard:0.091891
 |
Component:134; Jaccard:0.10336
 |
Component:274; Jaccard:0.1197
 |
Component:204; Jaccard:0.12673
 |
Component:193; Jaccard:0.13887
 |
Component:311; Jaccard:0.13856
 |
Correlation:0.1644
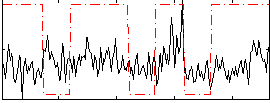 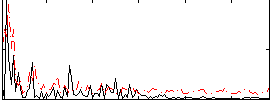 |
Correlation:0.17546
  |
Correlation:0.14045
  |
Correlation:0.22977
  |
Correlation:0.32375
  |
Correlation:0.0038091
  |
Correlation:0.020497
  |
Component:193; Jaccard:0.080586
 |
Component:394; Jaccard:0.091171
 |
Component:145; Jaccard:0.10218
 |
Component:276; Jaccard:0.11224
 |
Component:274; Jaccard:0.12518
 |
Component:311; Jaccard:0.13089
 |
Component:193; Jaccard:0.13788
 |
Correlation:0.0038091
  |
Correlation:0.1644
  |
Correlation:0.17546
  |
Correlation:0.18939
  |
Correlation:0.22977
  |
Correlation:0.020497
  |
Correlation:0.0038091
  |
Component:276; Jaccard:0.08034
 |
Component:276; Jaccard:0.090984
 |
Component:276; Jaccard:0.10216
 |
Component:145; Jaccard:0.11159
 |
Component:276; Jaccard:0.12138
 |
Component:276; Jaccard:0.12805
 |
Component:276; Jaccard:0.12972
 |
Correlation:0.18939
  |
Correlation:0.18939
  |
Correlation:0.18939
  |
Correlation:0.17546
  |
Correlation:0.18939
  |
Correlation:0.18939
  |
Correlation:0.18939
  |
Component:285; Jaccard:0.078886
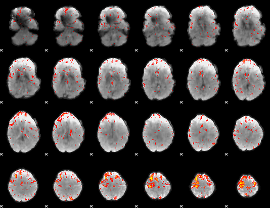 |
Component:134; Jaccard:0.086701
 |
Component:394; Jaccard:0.10005
 |
Component:394; Jaccard:0.1074
 |
Component:311; Jaccard:0.11981
 |
Component:274; Jaccard:0.12667
 |
Component:274; Jaccard:0.11886
 |
Correlation:0.19432
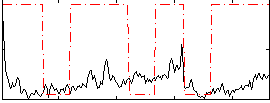  |
Correlation:0.14045
  |
Correlation:0.1644
  |
Correlation:0.1644
  |
Correlation:0.020497
  |
Correlation:0.22977
  |
Correlation:0.22977
  |
Component:331; Jaccard:0.076362
 |
Component:331; Jaccard:0.083538
 |
Component:311; Jaccard:0.093083
 |
Component:311; Jaccard:0.10692
 |
Component:145; Jaccard:0.11754
 |
Component:145; Jaccard:0.11681
 |
Component:145; Jaccard:0.11078
 |
Correlation:0.17423
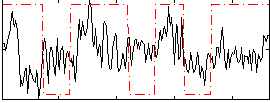 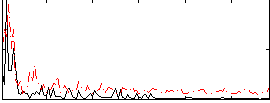 |
Correlation:0.17423
  |
Correlation:0.020497
  |
Correlation:0.020497
  |
Correlation:0.17546
  |
Correlation:0.17546
  |
Correlation:0.17546
  |
Component:362; Jaccard:0.074034
 |
Component:285; Jaccard:0.082729
 |
Component:331; Jaccard:0.088558
 |
Component:204; Jaccard:0.10404
 |
Component:394; Jaccard:0.11099
 |
Component:394; Jaccard:0.11052
 |
Component:133; Jaccard:0.10636
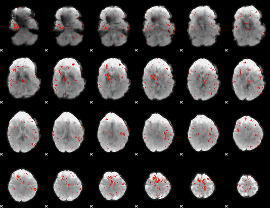 |
Correlation:0.20751
  |
Correlation:0.19432
  |
Correlation:0.17423
  |
Correlation:0.32375
  |
Correlation:0.1644
  |
Correlation:0.1644
  |
Correlation:0.12003
  |
Component:293; Jaccard:0.073731
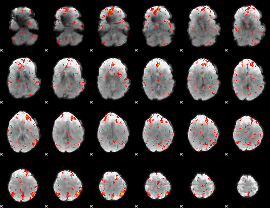 |
Component:311; Jaccard:0.080202
 |
Component:285; Jaccard:0.085894
 |
Component:331; Jaccard:0.092349
 |
Component:133; Jaccard:0.09882
 |
Component:133; Jaccard:0.10466
 |
Component:394; Jaccard:0.1008
 |
Correlation:0.10749
  |
Correlation:0.020497
  |
Correlation:0.19432
  |
Correlation:0.17423
  |
Correlation:0.12003
  |
Correlation:0.12003
  |
Correlation:0.1644
  |
Component:134; Jaccard:0.07321
 |
Component:357; Jaccard:0.078745
 |
Component:357; Jaccard:0.085337
 |
Component:286; Jaccard:0.091754
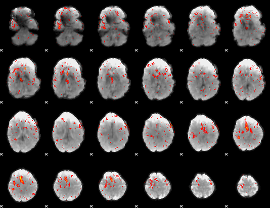 |
Component:286; Jaccard:0.097072
 |
Component:254; Jaccard:0.10039
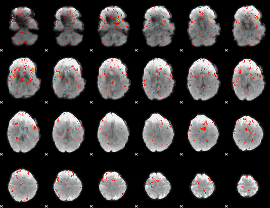 |
Component:254; Jaccard:0.098612
 |
Correlation:0.14045
  |
Correlation:0.18023
 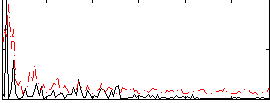 |
Correlation:0.18023
  |
Correlation:0.20586
 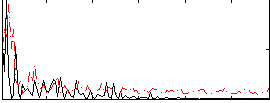 |
Correlation:0.20586
  |
Correlation:0.36541
  |
Correlation:0.36541
  |
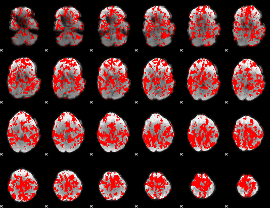
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
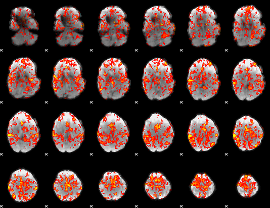 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
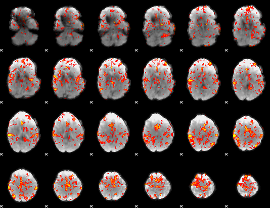 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:331; Jaccard:0.091571
 |
Component:331; Jaccard:0.1049
 |
Component:276; Jaccard:0.12108
 |
Component:134; Jaccard:0.14549
 |
Component:134; Jaccard:0.16962
 |
Component:134; Jaccard:0.18657
 |
Component:134; Jaccard:0.18995
 |
Correlation:0.2521
  |
Correlation:0.2521
  |
Correlation:0.024154
  |
Correlation:0.2692
 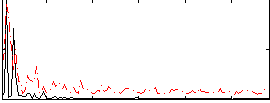 |
Correlation:0.2692
  |
Correlation:0.2692
  |
Correlation:0.2692
  |
Component:285; Jaccard:0.090747
 |
Component:276; Jaccard:0.10477
 |
Component:134; Jaccard:0.12105
 |
Component:276; Jaccard:0.13843
 |
Component:276; Jaccard:0.15476
 |
Component:276; Jaccard:0.16091
 |
Component:276; Jaccard:0.16015
 |
Correlation:0.22495
  |
Correlation:0.024154
  |
Correlation:0.2692
  |
Correlation:0.024154
  |
Correlation:0.024154
  |
Correlation:0.024154
  |
Correlation:0.024154
  |
Component:274; Jaccard:0.090252
 |
Component:193; Jaccard:0.10298
 |
Component:193; Jaccard:0.11926
 |
Component:193; Jaccard:0.13529
 |
Component:193; Jaccard:0.14765
 |
Component:193; Jaccard:0.15123
 |
Component:193; Jaccard:0.1401
 |
Correlation:0.067265
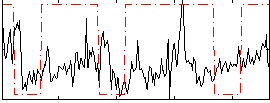 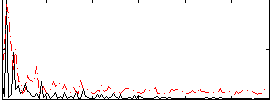 |
Correlation:0.19087
  |
Correlation:0.19087
  |
Correlation:0.19087
  |
Correlation:0.19087
  |
Correlation:0.19087
  |
Correlation:0.19087
  |
Component:276; Jaccard:0.089965
 |
Component:274; Jaccard:0.10262
 |
Component:331; Jaccard:0.11695
 |
Component:331; Jaccard:0.12624
 |
Component:311; Jaccard:0.13393
 |
Component:311; Jaccard:0.14108
 |
Component:311; Jaccard:0.13937
 |
Correlation:0.024154
  |
Correlation:0.067265
  |
Correlation:0.2521
  |
Correlation:0.2521
  |
Correlation:0.12986
 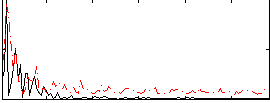 |
Correlation:0.12986
  |
Correlation:0.12986
  |
Component:394; Jaccard:0.088371
 |
Component:134; Jaccard:0.099455
 |
Component:274; Jaccard:0.1154
 |
Component:274; Jaccard:0.12581
 |
Component:274; Jaccard:0.13045
 |
Component:274; Jaccard:0.12535
 |
Component:282; Jaccard:0.1233
 |
Correlation:0.18864
  |
Correlation:0.2692
  |
Correlation:0.067265
  |
Correlation:0.067265
  |
Correlation:0.067265
  |
Correlation:0.067265
  |
Correlation:0.010581
  |
Component:193; Jaccard:0.087854
 |
Component:394; Jaccard:0.098814
 |
Component:394; Jaccard:0.10778
 |
Component:311; Jaccard:0.12084
 |
Component:331; Jaccard:0.12874
 |
Component:282; Jaccard:0.12519
 |
Component:274; Jaccard:0.11126
 |
Correlation:0.19087
  |
Correlation:0.18864
  |
Correlation:0.18864
  |
Correlation:0.12986
  |
Correlation:0.2521
  |
Correlation:0.010581
  |
Correlation:0.067265
  |
Component:134; Jaccard:0.081945
 |
Component:285; Jaccard:0.097538
 |
Component:311; Jaccard:0.10458
 |
Component:394; Jaccard:0.11231
 |
Component:282; Jaccard:0.11807
 |
Component:331; Jaccard:0.12098
 |
Component:331; Jaccard:0.10668
 |
Correlation:0.2692
  |
Correlation:0.22495
  |
Correlation:0.12986
  |
Correlation:0.18864
  |
Correlation:0.010581
  |
Correlation:0.2521
  |
Correlation:0.2521
  |
Component:357; Jaccard:0.081779
 |
Component:357; Jaccard:0.092668
 |
Component:357; Jaccard:0.10373
 |
Component:357; Jaccard:0.11184
 |
Component:357; Jaccard:0.11428
 |
Component:357; Jaccard:0.10823
 |
Component:357; Jaccard:0.092909
 |
Correlation:0.23764
  |
Correlation:0.23764
  |
Correlation:0.23764
  |
Correlation:0.23764
  |
Correlation:0.23764
  |
Correlation:0.23764
  |
Correlation:0.23764
  |
Component:303; Jaccard:0.080738
 |
Component:311; Jaccard:0.090179
 |
Component:285; Jaccard:0.10171
 |
Component:282; Jaccard:0.10693
 |
Component:394; Jaccard:0.10921
 |
Component:303; Jaccard:0.10145
 |
Component:303; Jaccard:0.09157
 |
Correlation:0.15237
  |
Correlation:0.12986
  |
Correlation:0.22495
  |
Correlation:0.010581
  |
Correlation:0.18864
  |
Correlation:0.15237
  |
Correlation:0.15237
  |
Component:277; Jaccard:0.077899
 |
Component:303; Jaccard:0.090137
 |
Component:303; Jaccard:0.097825
 |
Component:303; Jaccard:0.10368
 |
Component:303; Jaccard:0.10556
 |
Component:394; Jaccard:0.096278
 |
Component:394; Jaccard:0.077832
 |
Correlation:0.12584
  |
Correlation:0.15237
  |
Correlation:0.15237
  |
Correlation:0.15237
  |
Correlation:0.15237
  |
Correlation:0.18864
  |
Correlation:0.18864
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































