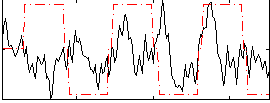
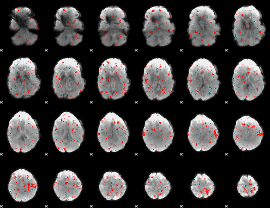
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
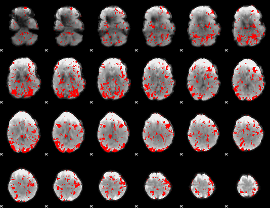 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
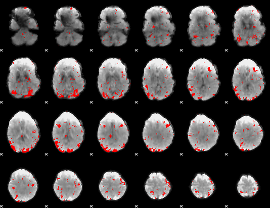
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
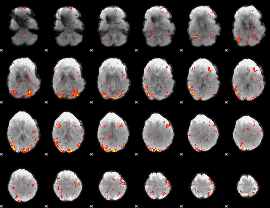 |
GLM Z-score result thresholded by 3
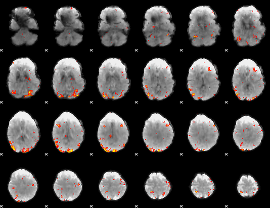 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:020; Jaccard:0.15818
 |
Component:020; Jaccard:0.21457
 |
Component:020; Jaccard:0.26827
 |
Component:020; Jaccard:0.29947
 |
Component:020; Jaccard:0.28699
 |
Component:020; Jaccard:0.24532
 |
Component:020; Jaccard:0.20272
 |
Correlation:0.58643
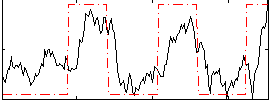 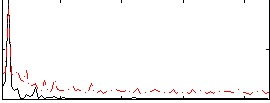 |
Correlation:0.58643
  |
Correlation:0.58643
  |
Correlation:0.58643
  |
Correlation:0.58643
  |
Correlation:0.58643
  |
Correlation:0.58643
  |
Component:248; Jaccard:0.11035
 |
Component:248; Jaccard:0.13908
 |
Component:248; Jaccard:0.16299
 |
Component:248; Jaccard:0.16033
 |
Component:035; Jaccard:0.14776
 |
Component:035; Jaccard:0.12512
 |
Component:035; Jaccard:0.10845
 |
Correlation:0.075692
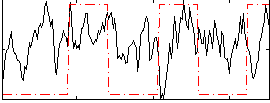  |
Correlation:0.075692
  |
Correlation:0.075692
  |
Correlation:0.075692
  |
Correlation:0.10531
  |
Correlation:0.10531
  |
Correlation:0.10531
  |
Component:035; Jaccard:0.10306
 |
Component:035; Jaccard:0.12836
 |
Component:035; Jaccard:0.15024
 |
Component:035; Jaccard:0.15562
 |
Component:248; Jaccard:0.14255
 |
Component:248; Jaccard:0.11396
 |
Component:248; Jaccard:0.090117
 |
Correlation:0.10531
  |
Correlation:0.10531
  |
Correlation:0.10531
  |
Correlation:0.10531
  |
Correlation:0.075692
  |
Correlation:0.075692
  |
Correlation:0.075692
  |
Component:372; Jaccard:0.099411
 |
Component:372; Jaccard:0.10946
 |
Component:319; Jaccard:0.11926
 |
Component:319; Jaccard:0.12047
 |
Component:319; Jaccard:0.11045
 |
Component:319; Jaccard:0.089566
 |
Component:319; Jaccard:0.069797
 |
Correlation:-0.29543
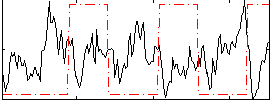  |
Correlation:-0.29543
  |
Correlation:-0.15863
 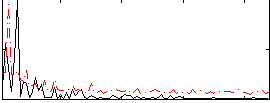 |
Correlation:-0.15863
  |
Correlation:-0.15863
  |
Correlation:-0.15863
  |
Correlation:-0.15863
  |
Component:299; Jaccard:0.093665
 |
Component:319; Jaccard:0.10641
 |
Component:372; Jaccard:0.11179
 |
Component:271; Jaccard:0.10309
 |
Component:271; Jaccard:0.095646
 |
Component:271; Jaccard:0.081833
 |
Component:271; Jaccard:0.067955
 |
Correlation:0.072996
  |
Correlation:-0.15863
  |
Correlation:-0.29543
  |
Correlation:0.37402
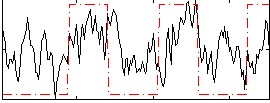  |
Correlation:0.37402
  |
Correlation:0.37402
  |
Correlation:0.37402
  |
Component:039; Jaccard:0.092699
 |
Component:039; Jaccard:0.10573
 |
Component:039; Jaccard:0.11106
 |
Component:372; Jaccard:0.102
 |
Component:299; Jaccard:0.091838
 |
Component:299; Jaccard:0.07789
 |
Component:299; Jaccard:0.06272
 |
Correlation:-0.038777
 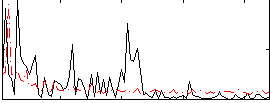 |
Correlation:-0.038777
  |
Correlation:-0.038777
  |
Correlation:-0.29543
  |
Correlation:0.072996
  |
Correlation:0.072996
  |
Correlation:0.072996
  |
Component:319; Jaccard:0.086184
 |
Component:299; Jaccard:0.10458
 |
Component:299; Jaccard:0.10664
 |
Component:299; Jaccard:0.10125
 |
Component:372; Jaccard:0.088755
 |
Component:372; Jaccard:0.073464
 |
Component:372; Jaccard:0.062461
 |
Correlation:-0.15863
  |
Correlation:0.072996
  |
Correlation:0.072996
  |
Correlation:0.072996
  |
Correlation:-0.29543
  |
Correlation:-0.29543
  |
Correlation:-0.29543
  |
Component:188; Jaccard:0.081938
 |
Component:271; Jaccard:0.094298
 |
Component:271; Jaccard:0.10321
 |
Component:039; Jaccard:0.099822
 |
Component:039; Jaccard:0.081043
 |
Component:039; Jaccard:0.061476
 |
Component:087; Jaccard:0.047427
 |
Correlation:-0.26497
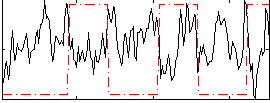  |
Correlation:0.37402
  |
Correlation:0.37402
  |
Correlation:-0.038777
  |
Correlation:-0.038777
  |
Correlation:-0.038777
  |
Correlation:-0.09715
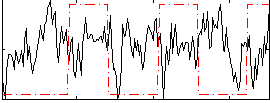  |
Component:271; Jaccard:0.081035
 |
Component:188; Jaccard:0.089811
 |
Component:188; Jaccard:0.091935
 |
Component:087; Jaccard:0.08339
 |
Component:087; Jaccard:0.074623
 |
Component:087; Jaccard:0.059046
 |
Component:039; Jaccard:0.047361
 |
Correlation:0.37402
  |
Correlation:-0.26497
  |
Correlation:-0.26497
  |
Correlation:-0.09715
  |
Correlation:-0.09715
  |
Correlation:-0.09715
  |
Correlation:-0.038777
  |
Component:059; Jaccard:0.079908
 |
Component:087; Jaccard:0.088105
 |
Component:087; Jaccard:0.09021
 |
Component:188; Jaccard:0.083328
 |
Component:188; Jaccard:0.069618
 |
Component:188; Jaccard:0.053456
 |
Component:188; Jaccard:0.039672
 |
Correlation:0.19006
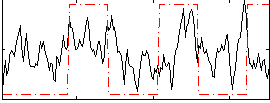 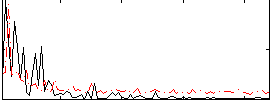 |
Correlation:-0.09715
  |
Correlation:-0.09715
  |
Correlation:-0.26497
  |
Correlation:-0.26497
  |
Correlation:-0.26497
  |
Correlation:-0.26497
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
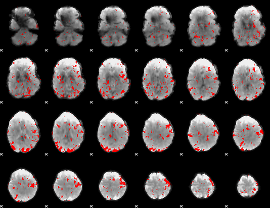 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
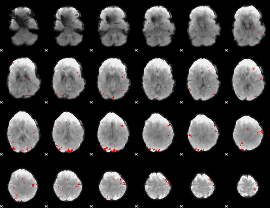 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
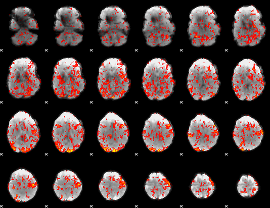 |
GLM Z-score result thresholded by 1.5
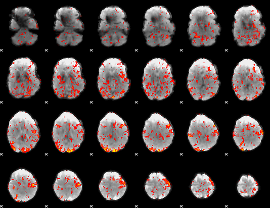 |
GLM Z-score result thresholded by 2
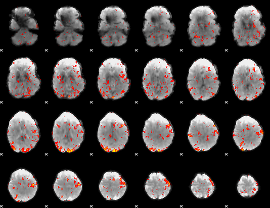 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
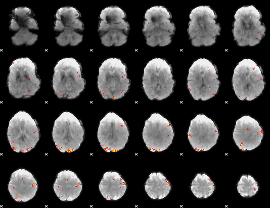 |
GLM Z-score result thresholded by 4
 |
Component:372; Jaccard:0.15199
 |
Component:372; Jaccard:0.18435
 |
Component:372; Jaccard:0.20444
 |
Component:372; Jaccard:0.19885
 |
Component:372; Jaccard:0.16031
 |
Component:372; Jaccard:0.11939
 |
Component:372; Jaccard:0.084909
 |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Correlation:0.25002
  |
Component:153; Jaccard:0.11532
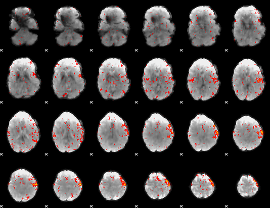 |
Component:319; Jaccard:0.13754
 |
Component:319; Jaccard:0.16214
 |
Component:319; Jaccard:0.16684
 |
Component:319; Jaccard:0.14916
 |
Component:319; Jaccard:0.11382
 |
Component:319; Jaccard:0.084767
 |
Correlation:0.1743
  |
Correlation:0.29162
  |
Correlation:0.29162
  |
Correlation:0.29162
  |
Correlation:0.29162
  |
Correlation:0.29162
  |
Correlation:0.29162
  |
Component:188; Jaccard:0.10763
 |
Component:153; Jaccard:0.12575
 |
Component:248; Jaccard:0.12944
 |
Component:248; Jaccard:0.12302
 |
Component:248; Jaccard:0.094244
 |
Component:248; Jaccard:0.064303
 |
Component:035; Jaccard:0.049244
 |
Correlation:0.20685
  |
Correlation:0.1743
  |
Correlation:-0.0049108
  |
Correlation:-0.0049108
  |
Correlation:-0.0049108
  |
Correlation:-0.0049108
  |
Correlation:-0.047722
  |
Component:319; Jaccard:0.10572
 |
Component:188; Jaccard:0.12373
 |
Component:153; Jaccard:0.12848
 |
Component:153; Jaccard:0.11083
 |
Component:035; Jaccard:0.084018
 |
Component:035; Jaccard:0.063937
 |
Component:248; Jaccard:0.04091
 |
Correlation:0.29162
  |
Correlation:0.20685
  |
Correlation:0.1743
  |
Correlation:0.1743
  |
Correlation:-0.047722
  |
Correlation:-0.047722
  |
Correlation:-0.0049108
  |
Component:248; Jaccard:0.10146
 |
Component:248; Jaccard:0.12171
 |
Component:188; Jaccard:0.12652
 |
Component:188; Jaccard:0.10952
 |
Component:153; Jaccard:0.08344
 |
Component:188; Jaccard:0.056787
 |
Component:188; Jaccard:0.035807
 |
Correlation:-0.0049108
  |
Correlation:-0.0049108
  |
Correlation:0.20685
  |
Correlation:0.20685
  |
Correlation:0.1743
  |
Correlation:0.20685
  |
Correlation:0.20685
  |
Component:039; Jaccard:0.090087
 |
Component:039; Jaccard:0.09996
 |
Component:035; Jaccard:0.10063
 |
Component:035; Jaccard:0.09974
 |
Component:188; Jaccard:0.082423
 |
Component:153; Jaccard:0.056118
 |
Component:087; Jaccard:0.03455
 |
Correlation:0.089189
  |
Correlation:0.089189
  |
Correlation:-0.047722
  |
Correlation:-0.047722
  |
Correlation:0.20685
  |
Correlation:0.1743
  |
Correlation:0.15149
  |
Component:087; Jaccard:0.083683
 |
Component:035; Jaccard:0.092333
 |
Component:039; Jaccard:0.096602
 |
Component:087; Jaccard:0.082632
 |
Component:087; Jaccard:0.06715
 |
Component:087; Jaccard:0.049537
 |
Component:153; Jaccard:0.034376
 |
Correlation:0.15149
  |
Correlation:-0.047722
  |
Correlation:0.089189
  |
Correlation:0.15149
  |
Correlation:0.15149
  |
Correlation:0.15149
  |
Correlation:0.1743
  |
Component:299; Jaccard:0.079126
 |
Component:087; Jaccard:0.091116
 |
Component:087; Jaccard:0.091921
 |
Component:039; Jaccard:0.081476
 |
Component:039; Jaccard:0.060429
 |
Component:299; Jaccard:0.044593
 |
Component:299; Jaccard:0.032401
 |
Correlation:-0.0026608
  |
Correlation:0.15149
  |
Correlation:0.15149
  |
Correlation:0.089189
  |
Correlation:0.089189
  |
Correlation:-0.0026608
  |
Correlation:-0.0026608
  |
Component:035; Jaccard:0.078659
 |
Component:299; Jaccard:0.086365
 |
Component:299; Jaccard:0.084841
 |
Component:020; Jaccard:0.0751
 |
Component:299; Jaccard:0.060104
 |
Component:039; Jaccard:0.039991
 |
Component:020; Jaccard:0.027192
 |
Correlation:-0.047722
  |
Correlation:-0.0026608
  |
Correlation:-0.0026608
  |
Correlation:-0.54089
  |
Correlation:-0.0026608
  |
Correlation:0.089189
  |
Correlation:-0.54089
  |
Component:020; Jaccard:0.077841
 |
Component:020; Jaccard:0.08444
 |
Component:020; Jaccard:0.084615
 |
Component:299; Jaccard:0.071417
 |
Component:020; Jaccard:0.057237
 |
Component:020; Jaccard:0.039873
 |
Component:018; Jaccard:0.024688
 |
Correlation:-0.54089
  |
Correlation:-0.54089
  |
Correlation:-0.54089
  |
Correlation:-0.0026608
  |
Correlation:-0.54089
  |
Correlation:-0.54089
  |
Correlation:0.06025
  |
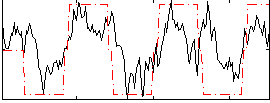
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
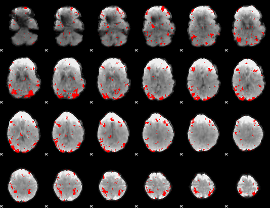 |
GLM binary result thresholded by 3
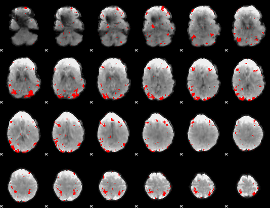 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
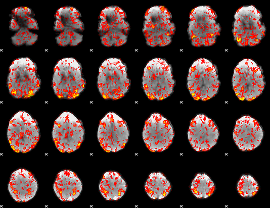 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
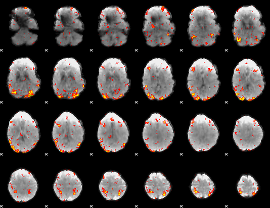 |
GLM Z-score result thresholded by 3
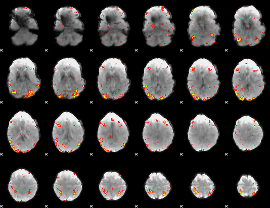 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:020; Jaccard:0.13726
 |
Component:020; Jaccard:0.2045
 |
Component:020; Jaccard:0.2863
 |
Component:020; Jaccard:0.35768
 |
Component:020; Jaccard:0.3772
 |
Component:020; Jaccard:0.35131
 |
Component:020; Jaccard:0.30088
 |
Correlation:0.61148
  |
Correlation:0.61148
  |
Correlation:0.61148
  |
Correlation:0.61148
  |
Correlation:0.61148
  |
Correlation:0.61148
  |
Correlation:0.61148
  |
Component:209; Jaccard:0.11008
 |
Component:209; Jaccard:0.14802
 |
Component:209; Jaccard:0.18116
 |
Component:209; Jaccard:0.19606
 |
Component:209; Jaccard:0.18805
 |
Component:209; Jaccard:0.17012
 |
Component:209; Jaccard:0.13867
 |
Correlation:0.61362
  |
Correlation:0.61362
  |
Correlation:0.61362
  |
Correlation:0.61362
  |
Correlation:0.61362
  |
Correlation:0.61362
  |
Correlation:0.61362
  |
Component:247; Jaccard:0.098314
 |
Component:247; Jaccard:0.12374
 |
Component:247; Jaccard:0.14461
 |
Component:271; Jaccard:0.15279
 |
Component:271; Jaccard:0.14993
 |
Component:271; Jaccard:0.14235
 |
Component:271; Jaccard:0.12418
 |
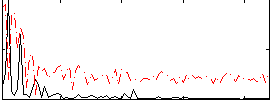
Correlation:0.5878
  |
Correlation:0.5878
  |
Correlation:0.5878
  |
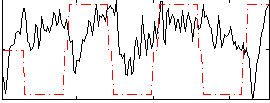
Correlation:0.34692
  |
Correlation:0.34692
  |
Correlation:0.34692
  |
Correlation:0.34692
  |
Component:320; Jaccard:0.09419
 |
Component:271; Jaccard:0.11624
 |
Component:271; Jaccard:0.13896
 |
Component:247; Jaccard:0.1522
 |
Component:247; Jaccard:0.14805
 |
Component:247; Jaccard:0.13545
 |
Component:247; Jaccard:0.11808
 |
Correlation:0.42135
  |
Correlation:0.34692
  |
Correlation:0.34692
  |
Correlation:0.5878
  |
Correlation:0.5878
  |
Correlation:0.5878
  |
Correlation:0.5878
  |
Component:335; Jaccard:0.092495
 |
Component:335; Jaccard:0.10742
 |
Component:320; Jaccard:0.11446
 |
Component:320; Jaccard:0.11301
 |
Component:320; Jaccard:0.10067
 |
Component:035; Jaccard:0.088311
 |
Component:035; Jaccard:0.075155
 |
Correlation:0.43942
  |
Correlation:0.43942
  |
Correlation:0.42135
  |
Correlation:0.42135
  |
Correlation:0.42135
  |
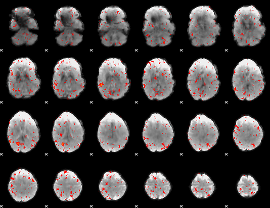
Correlation:0.083007
  |
Correlation:0.083007
  |
Component:271; Jaccard:0.090619
 |
Component:320; Jaccard:0.10653
 |
Component:335; Jaccard:0.11196
 |
Component:335; Jaccard:0.10587
 |
Component:035; Jaccard:0.098883
 |
Component:320; Jaccard:0.084902
 |
Component:320; Jaccard:0.06921
 |
Correlation:0.34692
  |
Correlation:0.42135
  |
Correlation:0.43942
  |
Correlation:0.43942
  |
Correlation:0.083007
  |
Correlation:0.42135
  |
Correlation:0.42135
  |
Component:057; Jaccard:0.083455
 |
Component:057; Jaccard:0.089238
 |
Component:035; Jaccard:0.098867
 |
Component:035; Jaccard:0.10329
 |
Component:335; Jaccard:0.087941
 |
Component:208; Jaccard:0.074564
 |
Component:208; Jaccard:0.063387
 |
Correlation:0.10849
  |
Correlation:0.10849
  |
Correlation:0.083007
  |
Correlation:0.083007
  |
Correlation:0.43942
  |
Correlation:0.40228
 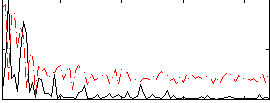 |
Correlation:0.40228
  |
Component:161; Jaccard:0.079579
 |
Component:035; Jaccard:0.085924
 |
Component:059; Jaccard:0.095196
 |
Component:059; Jaccard:0.093648
 |
Component:059; Jaccard:0.085581
 |
Component:059; Jaccard:0.072235
 |
Component:059; Jaccard:0.05797
 |
Correlation:0.22292
 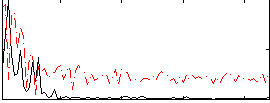 |
Correlation:0.083007
  |
Correlation:0.24697
 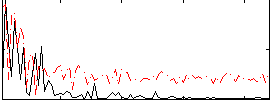 |
Correlation:0.24697
  |
Correlation:0.24697
  |
Correlation:0.24697
  |
Correlation:0.24697
  |
Component:348; Jaccard:0.077109
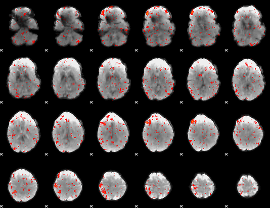 |
Component:059; Jaccard:0.085241
 |
Component:057; Jaccard:0.089297
 |
Component:208; Jaccard:0.085746
 |
Component:208; Jaccard:0.081643
 |
Component:335; Jaccard:0.068853
 |
Component:335; Jaccard:0.056789
 |
Correlation:0.41116
  |
Correlation:0.24697
  |
Correlation:0.10849
  |
Correlation:0.40228
  |
Correlation:0.40228
  |
Correlation:0.43942
  |
Correlation:0.43942
  |
Component:059; Jaccard:0.073295
 |
Component:348; Jaccard:0.084406
 |
Component:348; Jaccard:0.085603
 |
Component:348; Jaccard:0.079419
 |
Component:348; Jaccard:0.06934
 |
Component:299; Jaccard:0.06044
 |
Component:299; Jaccard:0.049676
 |
Correlation:0.24697
  |
Correlation:0.41116
  |
Correlation:0.41116
  |
Correlation:0.41116
  |
Correlation:0.41116
  |
Correlation:0.041038
  |
Correlation:0.041038
  |
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
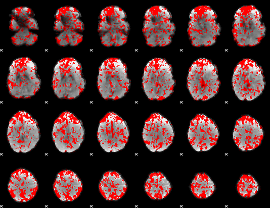 |
GLM binary result thresholded by 1.5
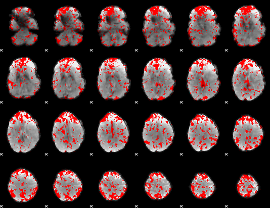 |
GLM binary result thresholded by 2
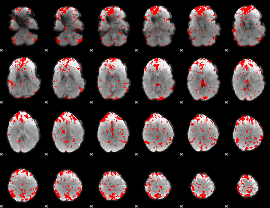 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
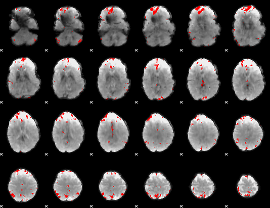 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
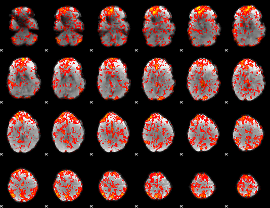 |
GLM Z-score result thresholded by 1.5
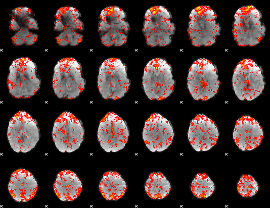 |
GLM Z-score result thresholded by 2
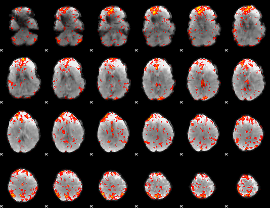 |
GLM Z-score result thresholded by 2.5
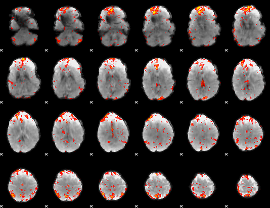 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
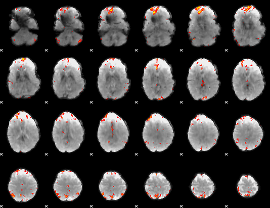 |
GLM Z-score result thresholded by 4
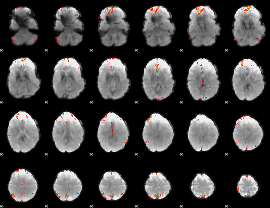 |
Component:278; Jaccard:0.11333
 |
Component:278; Jaccard:0.14352
 |
Component:278; Jaccard:0.17402
 |
Component:278; Jaccard:0.19381
 |
Component:213; Jaccard:0.18898
 |
Component:213; Jaccard:0.1666
 |
Component:213; Jaccard:0.13128
 |
Correlation:0.22379
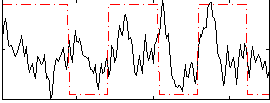  |
Correlation:0.22379
  |
Correlation:0.22379
  |
Correlation:0.22379
  |
Correlation:0.042608
  |
Correlation:0.042608
  |
Correlation:0.042608
  |
Component:213; Jaccard:0.10954
 |
Component:213; Jaccard:0.1377
 |
Component:213; Jaccard:0.16787
 |
Component:213; Jaccard:0.19033
 |
Component:278; Jaccard:0.18704
 |
Component:388; Jaccard:0.15655
 |
Component:388; Jaccard:0.12763
 |
Correlation:0.042608
  |
Correlation:0.042608
  |
Correlation:0.042608
  |
Correlation:0.042608
  |
Correlation:0.22379
  |
Correlation:-0.0041692
  |
Correlation:-0.0041692
  |
Component:245; Jaccard:0.10909
 |
Component:245; Jaccard:0.13565
 |
Component:245; Jaccard:0.15975
 |
Component:388; Jaccard:0.17421
 |
Component:388; Jaccard:0.1746
 |
Component:278; Jaccard:0.15645
 |
Component:278; Jaccard:0.11806
 |
Correlation:0.16558
  |
Correlation:0.16558
  |
Correlation:0.16558
  |
Correlation:-0.0041692
  |
Correlation:-0.0041692
  |
Correlation:0.22379
  |
Correlation:0.22379
  |
Component:219; Jaccard:0.10714
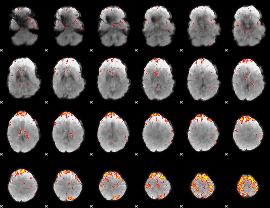 |
Component:388; Jaccard:0.12613
 |
Component:388; Jaccard:0.15287
 |
Component:245; Jaccard:0.16875
 |
Component:245; Jaccard:0.15543
 |
Component:245; Jaccard:0.12938
 |
Component:068; Jaccard:0.10139
 |
Correlation:0.052037
 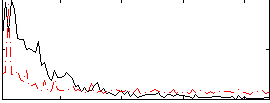 |
Correlation:-0.0041692
  |
Correlation:-0.0041692
  |
Correlation:0.16558
  |
Correlation:0.16558
  |
Correlation:0.16558
  |
Correlation:0.1419
  |
Component:388; Jaccard:0.10156
 |
Component:343; Jaccard:0.11881
 |
Component:343; Jaccard:0.14048
 |
Component:343; Jaccard:0.15024
 |
Component:343; Jaccard:0.14463
 |
Component:068; Jaccard:0.12609
 |
Component:245; Jaccard:0.10052
 |
Correlation:-0.0041692
  |
Correlation:0.25069
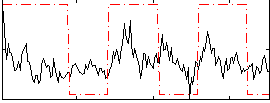  |
Correlation:0.25069
  |
Correlation:0.25069
  |
Correlation:0.25069
  |
Correlation:0.1419
  |
Correlation:0.16558
  |
Component:343; Jaccard:0.095596
 |
Component:219; Jaccard:0.1144
 |
Component:068; Jaccard:0.12488
 |
Component:068; Jaccard:0.14181
 |
Component:068; Jaccard:0.14219
 |
Component:343; Jaccard:0.12341
 |
Component:343; Jaccard:0.094791
 |
Correlation:0.25069
  |
Correlation:0.052037
  |
Correlation:0.1419
  |
Correlation:0.1419
  |
Correlation:0.1419
  |
Correlation:0.25069
  |
Correlation:0.25069
  |
Component:161; Jaccard:0.092733
 |
Component:161; Jaccard:0.10669
 |
Component:161; Jaccard:0.1178
 |
Component:044; Jaccard:0.12352
 |
Component:044; Jaccard:0.12309
 |
Component:044; Jaccard:0.10899
 |
Component:044; Jaccard:0.084038
 |
Correlation:-0.17729
  |
Correlation:-0.17729
  |
Correlation:-0.17729
  |
Correlation:0.040726
  |
Correlation:0.040726
  |
Correlation:0.040726
  |
Correlation:0.040726
  |
Component:106; Jaccard:0.089986
 |
Component:068; Jaccard:0.10392
 |
Component:044; Jaccard:0.10852
 |
Component:161; Jaccard:0.11944
 |
Component:161; Jaccard:0.1056
 |
Component:101; Jaccard:0.091955
 |
Component:101; Jaccard:0.069126
 |
Correlation:-0.055837
  |
Correlation:0.1419
  |
Correlation:0.040726
  |
Correlation:-0.17729
  |
Correlation:-0.17729
  |
Correlation:0.026639
  |
Correlation:0.026639
  |
Component:068; Jaccard:0.085891
 |
Component:106; Jaccard:0.10023
 |
Component:106; Jaccard:0.10704
 |
Component:101; Jaccard:0.10676
 |
Component:101; Jaccard:0.10477
 |
Component:161; Jaccard:0.080339
 |
Component:161; Jaccard:0.054268
 |
Correlation:0.1419
  |
Correlation:-0.055837
  |
Correlation:-0.055837
  |
Correlation:0.026639
  |
Correlation:0.026639
  |
Correlation:-0.17729
  |
Correlation:-0.17729
  |
Component:044; Jaccard:0.074014
 |
Component:044; Jaccard:0.089829
 |
Component:219; Jaccard:0.10421
 |
Component:106; Jaccard:0.10594
 |
Component:106; Jaccard:0.091718
 |
Component:106; Jaccard:0.067373
 |
Component:340; Jaccard:0.048785
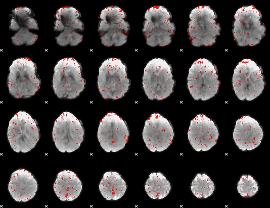 |
Correlation:0.040726
  |
Correlation:0.040726
  |
Correlation:0.052037
  |
Correlation:-0.055837
  |
Correlation:-0.055837
  |
Correlation:-0.055837
  |
Correlation:-0.23975
 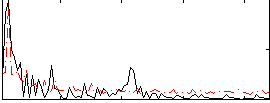 |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
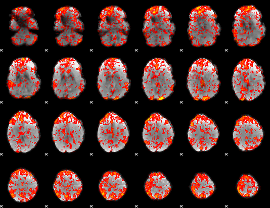 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
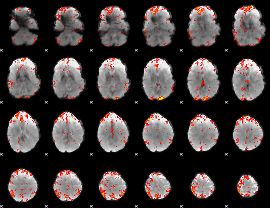 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:161; Jaccard:0.11493
 |
Component:161; Jaccard:0.1456
 |
Component:161; Jaccard:0.17845
 |
Component:161; Jaccard:0.20101
 |
Component:161; Jaccard:0.19642
 |
Component:161; Jaccard:0.16465
 |
Component:213; Jaccard:0.122
 |
Correlation:0.23369
  |
Correlation:0.23369
  |
Correlation:0.23369
  |
Correlation:0.23369
  |
Correlation:0.23369
  |
Correlation:0.23369
  |
Correlation:0.077719
  |
Component:219; Jaccard:0.11372
 |
Component:219; Jaccard:0.13319
 |
Component:213; Jaccard:0.1535
 |
Component:213; Jaccard:0.17338
 |
Component:213; Jaccard:0.1798
 |
Component:213; Jaccard:0.16267
 |
Component:161; Jaccard:0.11952
 |
Correlation:0.1474
  |
Correlation:0.1474
  |
Correlation:0.077719
  |
Correlation:0.077719
  |
Correlation:0.077719
  |
Correlation:0.077719
  |
Correlation:0.23369
  |
Component:213; Jaccard:0.10107
 |
Component:213; Jaccard:0.12633
 |
Component:278; Jaccard:0.14323
 |
Component:278; Jaccard:0.15885
 |
Component:278; Jaccard:0.1556
 |
Component:278; Jaccard:0.13529
 |
Component:278; Jaccard:0.10373
 |
Correlation:0.077719
  |
Correlation:0.077719
  |
Correlation:-0.073426
  |
Correlation:-0.073426
  |
Correlation:-0.073426
  |
Correlation:-0.073426
  |
Correlation:-0.073426
  |
Component:278; Jaccard:0.098161
 |
Component:278; Jaccard:0.12117
 |
Component:219; Jaccard:0.1417
 |
Component:388; Jaccard:0.14074
 |
Component:388; Jaccard:0.14143
 |
Component:388; Jaccard:0.12851
 |
Component:388; Jaccard:0.09801
 |
Correlation:-0.073426
  |
Correlation:-0.073426
  |
Correlation:0.1474
  |
Correlation:0.16269
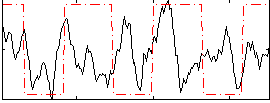  |
Correlation:0.16269
  |
Correlation:0.16269
  |
Correlation:0.16269
  |
Component:245; Jaccard:0.092021
 |
Component:245; Jaccard:0.10922
 |
Component:388; Jaccard:0.12694
 |
Component:343; Jaccard:0.13479
 |
Component:343; Jaccard:0.13146
 |
Component:343; Jaccard:0.11052
 |
Component:343; Jaccard:0.082689
 |
Correlation:-0.035974
  |
Correlation:-0.035974
  |
Correlation:0.16269
  |
Correlation:-0.16544
  |
Correlation:-0.16544
  |
Correlation:-0.16544
  |
Correlation:-0.16544
  |
Component:388; Jaccard:0.08983
 |
Component:388; Jaccard:0.1077
 |
Component:343; Jaccard:0.12474
 |
Component:245; Jaccard:0.13116
 |
Component:245; Jaccard:0.12216
 |
Component:245; Jaccard:0.10169
 |
Component:245; Jaccard:0.072796
 |
Correlation:0.16269
  |
Correlation:0.16269
  |
Correlation:-0.16544
  |
Correlation:-0.035974
  |
Correlation:-0.035974
  |
Correlation:-0.035974
  |
Correlation:-0.035974
  |
Component:106; Jaccard:0.089201
 |
Component:343; Jaccard:0.10749
 |
Component:245; Jaccard:0.12381
 |
Component:219; Jaccard:0.1258
 |
Component:247; Jaccard:0.10631
 |
Component:247; Jaccard:0.088759
 |
Component:247; Jaccard:0.067635
 |
Correlation:0.11757
  |
Correlation:-0.16544
  |
Correlation:-0.035974
  |
Correlation:0.1474
  |
Correlation:0.57997
  |
Correlation:0.57997
  |
Correlation:0.57997
  |
Component:194; Jaccard:0.088602
 |
Component:106; Jaccard:0.10236
 |
Component:106; Jaccard:0.11118
 |
Component:106; Jaccard:0.11005
 |
Component:044; Jaccard:0.10073
 |
Component:044; Jaccard:0.087804
 |
Component:044; Jaccard:0.066816
 |
Correlation:0.22075
  |
Correlation:0.11757
  |
Correlation:0.11757
  |
Correlation:0.11757
  |
Correlation:-0.0085171
  |
Correlation:-0.0085171
  |
Correlation:-0.0085171
  |
Component:343; Jaccard:0.087647
 |
Component:194; Jaccard:0.099578
 |
Component:194; Jaccard:0.10268
 |
Component:247; Jaccard:0.10583
 |
Component:106; Jaccard:0.09738
 |
Component:101; Jaccard:0.076656
 |
Component:340; Jaccard:0.059621
 |
Correlation:-0.16544
  |
Correlation:0.22075
  |
Correlation:0.22075
  |
Correlation:0.57997
  |
Correlation:0.11757
  |
Correlation:-0.14065
  |
Correlation:0.27996
  |
Component:175; Jaccard:0.078268
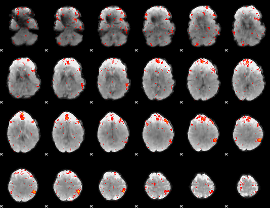 |
Component:247; Jaccard:0.087053
 |
Component:247; Jaccard:0.099361
 |
Component:044; Jaccard:0.10138
 |
Component:340; Jaccard:0.090163
 |
Component:106; Jaccard:0.075359
 |
Component:101; Jaccard:0.056455
 |
Correlation:0.17703
 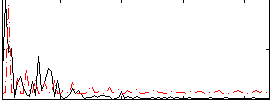 |
Correlation:0.57997
  |
Correlation:0.57997
  |
Correlation:-0.0085171
  |
Correlation:0.27996
  |
Correlation:0.11757
  |
Correlation:-0.14065
  |
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
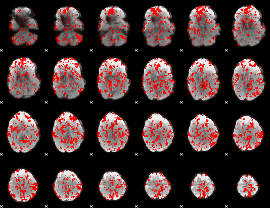 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
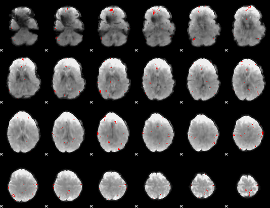 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
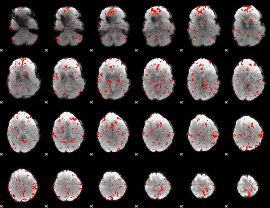 |
GLM Z-score result thresholded by 2.5
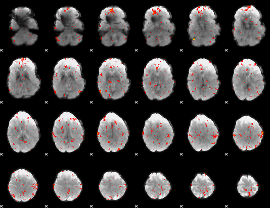 |
GLM Z-score result thresholded by 3
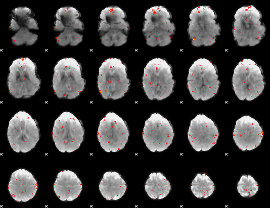 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
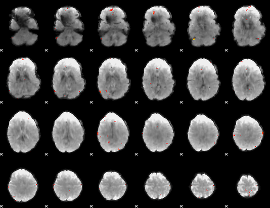 |
Component:068; Jaccard:0.091717
 |
Component:068; Jaccard:0.10312
 |
Component:068; Jaccard:0.10827
 |
Component:068; Jaccard:0.097115
 |
Component:068; Jaccard:0.071043
 |
Component:068; Jaccard:0.042921
 |
Component:068; Jaccard:0.021911
 |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Correlation:0.10092
  |
Component:245; Jaccard:0.080209
 |
Component:312; Jaccard:0.088579
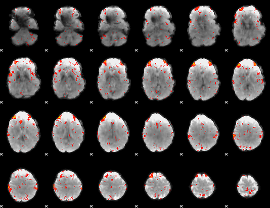 |
Component:312; Jaccard:0.088914
 |
Component:312; Jaccard:0.077762
 |
Component:312; Jaccard:0.052026
 |
Component:372; Jaccard:0.030804
 |
Component:372; Jaccard:0.018808
 |
Correlation:0.10933
  |
Correlation:0.49207
  |
Correlation:0.49207
  |
Correlation:0.49207
  |
Correlation:0.49207
  |
Correlation:0.29586
  |
Correlation:0.29586
  |
Component:312; Jaccard:0.079109
 |
Component:245; Jaccard:0.083861
 |
Component:100; Jaccard:0.084719
 |
Component:100; Jaccard:0.072682
 |
Component:100; Jaccard:0.051883
 |
Component:037; Jaccard:0.030469
 |
Component:037; Jaccard:0.015518
 |
Correlation:0.49207
  |
Correlation:0.10933
  |
Correlation:0.23934
  |
Correlation:0.23934
  |
Correlation:0.23934
  |
Correlation:0.27446
  |
Correlation:0.27446
  |
Component:388; Jaccard:0.075355
 |
Component:100; Jaccard:0.081711
 |
Component:142; Jaccard:0.080349
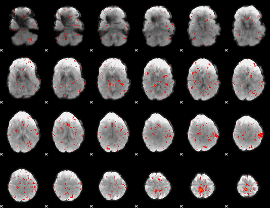 |
Component:142; Jaccard:0.065964
 |
Component:037; Jaccard:0.048344
 |
Component:100; Jaccard:0.029373
 |
Component:312; Jaccard:0.014779
 |
Correlation:-0.090507
  |
Correlation:0.23934
  |
Correlation:0.34205
  |
Correlation:0.34205
  |
Correlation:0.27446
  |
Correlation:0.23934
  |
Correlation:0.49207
  |
Component:278; Jaccard:0.074344
 |
Component:142; Jaccard:0.081546
 |
Component:245; Jaccard:0.079501
 |
Component:245; Jaccard:0.065639
 |
Component:388; Jaccard:0.045395
 |
Component:388; Jaccard:0.029114
 |
Component:388; Jaccard:0.013499
 |
Correlation:0.16122
 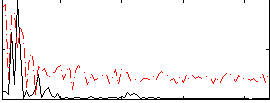 |
Correlation:0.34205
  |
Correlation:0.10933
  |
Correlation:0.10933
  |
Correlation:-0.090507
  |
Correlation:-0.090507
  |
Correlation:-0.090507
  |
Component:370; Jaccard:0.073987
 |
Component:388; Jaccard:0.080347
 |
Component:388; Jaccard:0.079217
 |
Component:388; Jaccard:0.064381
 |
Component:372; Jaccard:0.044719
 |
Component:312; Jaccard:0.028543
 |
Component:100; Jaccard:0.013267
 |
Correlation:0.2916
  |
Correlation:-0.090507
  |
Correlation:-0.090507
  |
Correlation:-0.090507
  |
Correlation:0.29586
  |
Correlation:0.49207
  |
Correlation:0.23934
  |
Component:142; Jaccard:0.073417
 |
Component:084; Jaccard:0.076424
 |
Component:084; Jaccard:0.074787
 |
Component:037; Jaccard:0.064106
 |
Component:245; Jaccard:0.044395
 |
Component:245; Jaccard:0.025192
 |
Component:142; Jaccard:0.012838
 |
Correlation:0.34205
  |
Correlation:0.47383
 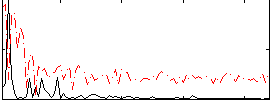 |
Correlation:0.47383
  |
Correlation:0.27446
  |
Correlation:0.10933
  |
Correlation:0.10933
  |
Correlation:0.34205
  |
Component:100; Jaccard:0.072193
 |
Component:278; Jaccard:0.075422
 |
Component:278; Jaccard:0.071885
 |
Component:084; Jaccard:0.059438
 |
Component:142; Jaccard:0.044236
 |
Component:142; Jaccard:0.024199
 |
Component:162; Jaccard:0.012234
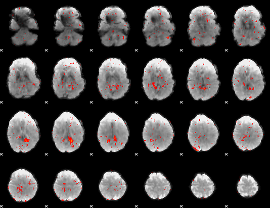 |
Correlation:0.23934
  |
Correlation:0.16122
  |
Correlation:0.16122
  |
Correlation:0.47383
  |
Correlation:0.34205
  |
Correlation:0.34205
  |
Correlation:0.2987
  |
Component:197; Jaccard:0.071774
 |
Component:370; Jaccard:0.075404
 |
Component:037; Jaccard:0.07144
 |
Component:278; Jaccard:0.057358
 |
Component:084; Jaccard:0.041198
 |
Component:153; Jaccard:0.022163
 |
Component:153; Jaccard:0.012031
 |
Correlation:0.33515
  |
Correlation:0.2916
  |
Correlation:0.27446
  |
Correlation:0.16122
  |
Correlation:0.47383
  |
Correlation:0.23475
  |
Correlation:0.23475
  |
Component:153; Jaccard:0.071077
 |
Component:197; Jaccard:0.074782
 |
Component:197; Jaccard:0.070006
 |
Component:372; Jaccard:0.055754
 |
Component:278; Jaccard:0.039907
 |
Component:079; Jaccard:0.021959
 |
Component:245; Jaccard:0.011941
 |
Correlation:0.23475
  |
Correlation:0.33515
  |
Correlation:0.33515
  |
Correlation:0.29586
  |
Correlation:0.16122
  |
Correlation:0.18834
  |
Correlation:0.10933
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































