Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
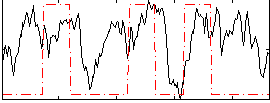
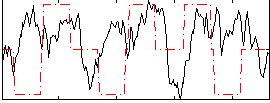
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
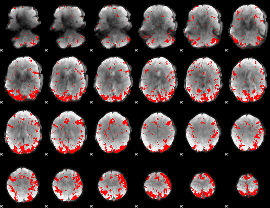 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
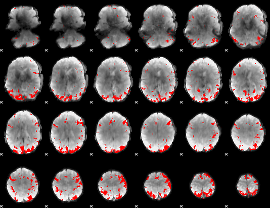 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
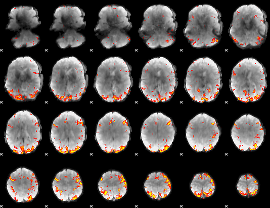 |
GLM Z-score result thresholded by 4
 |
Component:381; Jaccard:0.13975
 |
Component:381; Jaccard:0.17896
 |
Component:381; Jaccard:0.22177
 |
Component:381; Jaccard:0.25748
 |
Component:381; Jaccard:0.28023
 |
Component:381; Jaccard:0.2863
 |
Component:381; Jaccard:0.27205
 |
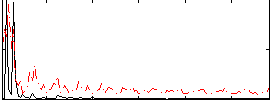
Correlation:0.14174
 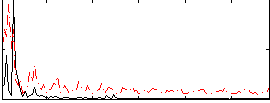 |
Correlation:0.14174
  |
Correlation:0.14174
  |
Correlation:0.14174
  |
Correlation:0.14174
  |
Correlation:0.14174
  |
Correlation:0.14174
  |
Component:364; Jaccard:0.13587
 |
Component:364; Jaccard:0.15986
 |
Component:255; Jaccard:0.19157
 |
Component:255; Jaccard:0.22312
 |
Component:213; Jaccard:0.24822
 |
Component:213; Jaccard:0.25842
 |
Component:213; Jaccard:0.24936
 |
Correlation:0.32007
  |
Correlation:0.32007
  |
Correlation:0.14696
  |
Correlation:0.14696
  |
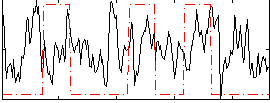
Correlation:0.28131
  |
Correlation:0.28131
  |
Correlation:0.28131
  |
Component:255; Jaccard:0.12257
 |
Component:255; Jaccard:0.15576
 |
Component:213; Jaccard:0.18409
 |
Component:213; Jaccard:0.22178
 |
Component:255; Jaccard:0.24606
 |
Component:255; Jaccard:0.25411
 |
Component:255; Jaccard:0.24515
 |
Correlation:0.14696
  |
Correlation:0.14696
  |
Correlation:0.28131
  |
Correlation:0.28131
  |
Correlation:0.14696
  |
Correlation:0.14696
  |
Correlation:0.14696
  |
Component:267; Jaccard:0.11085
 |
Component:213; Jaccard:0.14524
 |
Component:364; Jaccard:0.17748
 |
Component:364; Jaccard:0.18327
 |
Component:364; Jaccard:0.17674
 |
Component:374; Jaccard:0.16659
 |
Component:374; Jaccard:0.15177
 |
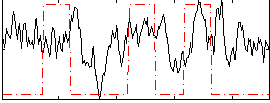
Correlation:0.098341
  |
Correlation:0.28131
  |
Correlation:0.32007
  |
Correlation:0.32007
  |
Correlation:0.32007
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Component:213; Jaccard:0.11039
 |
Component:374; Jaccard:0.12983
 |
Component:374; Jaccard:0.15188
 |
Component:374; Jaccard:0.1688
 |
Component:374; Jaccard:0.17306
 |
Component:364; Jaccard:0.16206
 |
Component:364; Jaccard:0.14303
 |
Correlation:0.28131
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.32007
  |
Correlation:0.32007
  |
Component:374; Jaccard:0.10712
 |
Component:267; Jaccard:0.12823
 |
Component:267; Jaccard:0.14098
 |
Component:267; Jaccard:0.14672
 |
Component:267; Jaccard:0.14155
 |
Component:248; Jaccard:0.13357
 |
Component:248; Jaccard:0.12208
 |
Correlation:0.13895
  |
Correlation:0.098341
  |
Correlation:0.098341
  |
Correlation:0.098341
  |
Correlation:0.098341
  |
Correlation:0.21038
  |
Correlation:0.21038
  |
Component:052; Jaccard:0.10221
 |
Component:052; Jaccard:0.1167
 |
Component:052; Jaccard:0.12756
 |
Component:248; Jaccard:0.13595
 |
Component:248; Jaccard:0.13759
 |
Component:267; Jaccard:0.12883
 |
Component:241; Jaccard:0.11464
 |
Correlation:0.17209
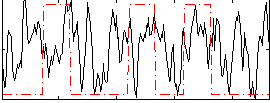  |
Correlation:0.17209
  |
Correlation:0.17209
  |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:0.098341
  |
Correlation:0.2116
  |
Component:248; Jaccard:0.097332
 |
Component:248; Jaccard:0.11223
 |
Component:248; Jaccard:0.12618
 |
Component:241; Jaccard:0.13409
 |
Component:241; Jaccard:0.13352
 |
Component:241; Jaccard:0.1283
 |
Component:267; Jaccard:0.11209
 |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:0.2116
  |
Correlation:0.2116
  |
Correlation:0.2116
  |
Correlation:0.098341
  |
Component:156; Jaccard:0.09704
 |
Component:241; Jaccard:0.10947
 |
Component:241; Jaccard:0.12489
 |
Component:052; Jaccard:0.13405
 |
Component:052; Jaccard:0.13122
 |
Component:052; Jaccard:0.12204
 |
Component:052; Jaccard:0.11062
 |
Correlation:-0.081576
  |
Correlation:0.2116
  |
Correlation:0.2116
  |
Correlation:0.17209
  |
Correlation:0.17209
  |
Correlation:0.17209
  |
Correlation:0.17209
  |
Component:389; Jaccard:0.096836
 |
Component:389; Jaccard:0.10925
 |
Component:389; Jaccard:0.11768
 |
Component:086; Jaccard:0.12535
 |
Component:086; Jaccard:0.12257
 |
Component:048; Jaccard:0.11276
 |
Component:048; Jaccard:0.099147
 |
Correlation:0.24589
  |
Correlation:0.24589
  |
Correlation:0.24589
  |
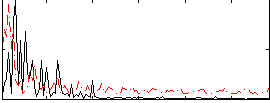
Correlation:0.04534
  |
Correlation:0.04534
  |
Correlation:0.21637
  |
Correlation:0.21637
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
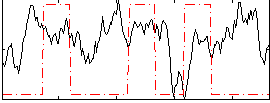
GLM binary result thresholded by 1.5
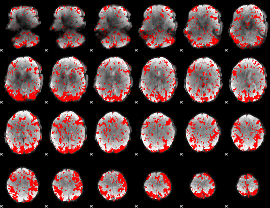 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
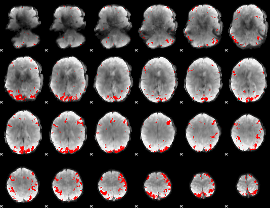 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
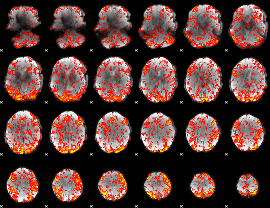 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
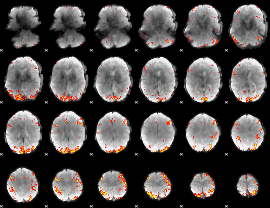 |
GLM Z-score result thresholded by 4
 |
Component:255; Jaccard:0.13881
 |
Component:255; Jaccard:0.18044
 |
Component:255; Jaccard:0.22492
 |
Component:255; Jaccard:0.26187
 |
Component:255; Jaccard:0.27916
 |
Component:255; Jaccard:0.26846
 |
Component:255; Jaccard:0.2371
 |
Correlation:0.19433
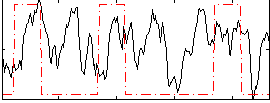  |
Correlation:0.19433
  |
Correlation:0.19433
  |
Correlation:0.19433
  |
Correlation:0.19433
  |
Correlation:0.19433
  |
Correlation:0.19433
  |
Component:381; Jaccard:0.13697
 |
Component:381; Jaccard:0.17003
 |
Component:381; Jaccard:0.20494
 |
Component:381; Jaccard:0.23131
 |
Component:381; Jaccard:0.24047
 |
Component:381; Jaccard:0.23009
 |
Component:381; Jaccard:0.20643
 |
Correlation:0.01985
  |
Correlation:0.01985
  |
Correlation:0.01985
  |
Correlation:0.01985
  |
Correlation:0.01985
  |
Correlation:0.01985
  |
Correlation:0.01985
  |
Component:364; Jaccard:0.11835
 |
Component:213; Jaccard:0.14458
 |
Component:213; Jaccard:0.17882
 |
Component:213; Jaccard:0.20639
 |
Component:213; Jaccard:0.22111
 |
Component:213; Jaccard:0.21582
 |
Component:213; Jaccard:0.19695
 |
Correlation:-0.033898
  |
Correlation:0.23884
  |
Correlation:0.23884
  |
Correlation:0.23884
  |
Correlation:0.23884
  |
Correlation:0.23884
  |
Correlation:0.23884
  |
Component:374; Jaccard:0.1146
 |
Component:086; Jaccard:0.14429
 |
Component:086; Jaccard:0.17483
 |
Component:086; Jaccard:0.19776
 |
Component:086; Jaccard:0.20072
 |
Component:086; Jaccard:0.18542
 |
Component:086; Jaccard:0.1578
 |
Correlation:0.23951
  |
Correlation:0.28296
  |
Correlation:0.28296
  |
Correlation:0.28296
  |
Correlation:0.28296
  |
Correlation:0.28296
  |
Correlation:0.28296
  |
Component:086; Jaccard:0.11376
 |
Component:374; Jaccard:0.13931
 |
Component:374; Jaccard:0.16001
 |
Component:374; Jaccard:0.17398
 |
Component:374; Jaccard:0.17358
 |
Component:374; Jaccard:0.16077
 |
Component:374; Jaccard:0.14135
 |
Correlation:0.28296
  |
Correlation:0.23951
  |
Correlation:0.23951
  |
Correlation:0.23951
  |
Correlation:0.23951
  |
Correlation:0.23951
  |
Correlation:0.23951
  |
Component:213; Jaccard:0.11196
 |
Component:364; Jaccard:0.13321
 |
Component:364; Jaccard:0.14267
 |
Component:364; Jaccard:0.14262
 |
Component:364; Jaccard:0.13348
 |
Component:364; Jaccard:0.11443
 |
Component:241; Jaccard:0.099035
 |
Correlation:0.23884
  |
Correlation:-0.033898
  |
Correlation:-0.033898
  |
Correlation:-0.033898
  |
Correlation:-0.033898
  |
Correlation:-0.033898
  |
Correlation:0.29899
  |
Component:267; Jaccard:0.10641
 |
Component:267; Jaccard:0.11829
 |
Component:267; Jaccard:0.12413
 |
Component:241; Jaccard:0.12747
 |
Component:241; Jaccard:0.12523
 |
Component:241; Jaccard:0.11436
 |
Component:364; Jaccard:0.095546
 |
Correlation:0.10115
  |
Correlation:0.10115
  |
Correlation:0.10115
  |
Correlation:0.29899
  |
Correlation:0.29899
  |
Correlation:0.29899
  |
Correlation:-0.033898
  |
Component:156; Jaccard:0.10256
 |
Component:156; Jaccard:0.11419
 |
Component:241; Jaccard:0.12123
 |
Component:267; Jaccard:0.12382
 |
Component:267; Jaccard:0.11756
 |
Component:267; Jaccard:0.10424
 |
Component:267; Jaccard:0.087124
 |
Correlation:0.15953
  |
Correlation:0.15953
  |
Correlation:0.29899
  |
Correlation:0.10115
  |
Correlation:0.10115
  |
Correlation:0.10115
  |
Correlation:0.10115
  |
Component:349; Jaccard:0.093981
 |
Component:241; Jaccard:0.10798
 |
Component:156; Jaccard:0.11929
 |
Component:288; Jaccard:0.11694
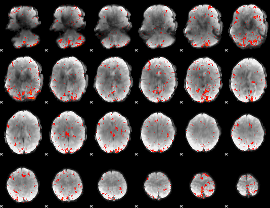 |
Component:288; Jaccard:0.11238
 |
Component:288; Jaccard:0.10049
 |
Component:288; Jaccard:0.085696
 |
Correlation:0.073257
 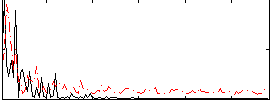 |
Correlation:0.29899
  |
Correlation:0.15953
  |
Correlation:0.226
  |
Correlation:0.226
  |
Correlation:0.226
  |
Correlation:0.226
  |
Component:052; Jaccard:0.092912
 |
Component:288; Jaccard:0.10327
 |
Component:288; Jaccard:0.11303
 |
Component:156; Jaccard:0.11633
 |
Component:156; Jaccard:0.10391
 |
Component:052; Jaccard:0.089067
 |
Component:052; Jaccard:0.075405
 |
Correlation:0.010086
  |
Correlation:0.226
  |
Correlation:0.226
  |
Correlation:0.15953
  |
Correlation:0.15953
  |
Correlation:0.010086
  |
Correlation:0.010086
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
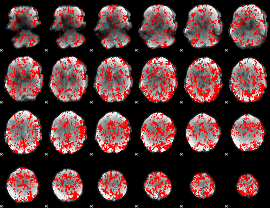 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
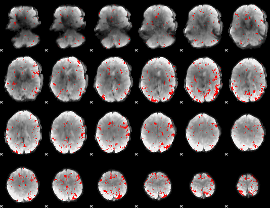 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
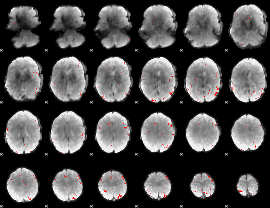 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
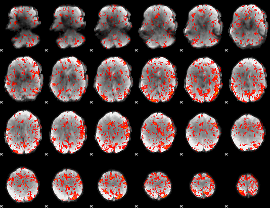 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
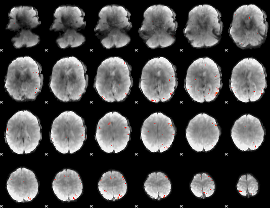 |
Component:364; Jaccard:0.096515
 |
Component:157; Jaccard:0.11083
 |
Component:157; Jaccard:0.11369
 |
Component:157; Jaccard:0.10079
 |
Component:157; Jaccard:0.07151
 |
Component:157; Jaccard:0.042205
 |
Component:157; Jaccard:0.021482
 |
Correlation:0.2098
  |
Correlation:-0.0078425
  |
Correlation:-0.0078425
  |
Correlation:-0.0078425
  |
Correlation:-0.0078425
  |
Correlation:-0.0078425
  |
Correlation:-0.0078425
  |
Component:157; Jaccard:0.095955
 |
Component:338; Jaccard:0.10527
 |
Component:338; Jaccard:0.10607
 |
Component:338; Jaccard:0.095157
 |
Component:338; Jaccard:0.067124
 |
Component:213; Jaccard:0.040109
 |
Component:213; Jaccard:0.02093
 |
Correlation:-0.0078425
  |
Correlation:0.18727
 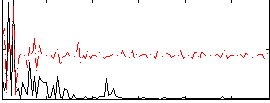 |
Correlation:0.18727
  |
Correlation:0.18727
  |
Correlation:0.18727
  |
Correlation:0.025168
  |
Correlation:0.025168
  |
Component:338; Jaccard:0.093934
 |
Component:022; Jaccard:0.1046
 |
Component:022; Jaccard:0.10426
 |
Component:248; Jaccard:0.090392
 |
Component:248; Jaccard:0.064238
 |
Component:338; Jaccard:0.039149
 |
Component:338; Jaccard:0.020733
 |
Correlation:0.18727
  |
Correlation:0.17871
  |
Correlation:0.17871
  |
Correlation:0.15796
  |
Correlation:0.15796
  |
Correlation:0.18727
  |
Correlation:0.18727
  |
Component:022; Jaccard:0.093057
 |
Component:364; Jaccard:0.10139
 |
Component:248; Jaccard:0.10111
 |
Component:381; Jaccard:0.087583
 |
Component:213; Jaccard:0.063275
 |
Component:381; Jaccard:0.038865
 |
Component:381; Jaccard:0.019917
 |
Correlation:0.17871
  |
Correlation:0.2098
  |
Correlation:0.15796
  |
Correlation:0.072247
  |
Correlation:0.025168
  |
Correlation:0.072247
  |
Correlation:0.072247
  |
Component:025; Jaccard:0.090505
 |
Component:248; Jaccard:0.098482
 |
Component:048; Jaccard:0.10049
 |
Component:048; Jaccard:0.086985
 |
Component:381; Jaccard:0.06287
 |
Component:248; Jaccard:0.037515
 |
Component:248; Jaccard:0.017897
 |
Correlation:0.20681
  |
Correlation:0.15796
  |
Correlation:0.11226
  |
Correlation:0.11226
  |
Correlation:0.072247
  |
Correlation:0.15796
  |
Correlation:0.15796
  |
Component:381; Jaccard:0.089835
 |
Component:048; Jaccard:0.0976
 |
Component:381; Jaccard:0.099087
 |
Component:022; Jaccard:0.085649
 |
Component:048; Jaccard:0.062187
 |
Component:022; Jaccard:0.035133
 |
Component:022; Jaccard:0.017182
 |
Correlation:0.072247
  |
Correlation:0.11226
  |
Correlation:0.072247
  |
Correlation:0.17871
  |
Correlation:0.11226
  |
Correlation:0.17871
  |
Correlation:0.17871
  |
Component:248; Jaccard:0.088787
 |
Component:381; Jaccard:0.097385
 |
Component:364; Jaccard:0.094613
 |
Component:213; Jaccard:0.079498
 |
Component:022; Jaccard:0.057761
 |
Component:048; Jaccard:0.034028
 |
Component:102; Jaccard:0.017025
 |
Correlation:0.15796
  |
Correlation:0.072247
  |
Correlation:0.2098
  |
Correlation:0.025168
  |
Correlation:0.17871
  |
Correlation:0.11226
  |
Correlation:0.093804
  |
Component:048; Jaccard:0.087438
 |
Component:025; Jaccard:0.089954
 |
Component:213; Jaccard:0.085095
 |
Component:364; Jaccard:0.074951
 |
Component:102; Jaccard:0.053416
 |
Component:102; Jaccard:0.033633
 |
Component:048; Jaccard:0.017009
 |
Correlation:0.11226
  |
Correlation:0.20681
  |
Correlation:0.025168
  |
Correlation:0.2098
  |
Correlation:0.093804
  |
Correlation:0.093804
  |
Correlation:0.11226
  |
Component:334; Jaccard:0.077813
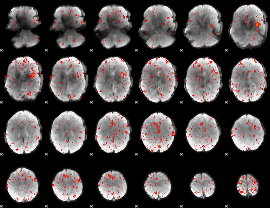 |
Component:334; Jaccard:0.084844
 |
Component:334; Jaccard:0.079956
 |
Component:102; Jaccard:0.07069
 |
Component:364; Jaccard:0.047165
 |
Component:389; Jaccard:0.025439
 |
Component:389; Jaccard:0.012784
 |
Correlation:-0.064184
  |
Correlation:-0.064184
  |
Correlation:-0.064184
  |
Correlation:0.093804
  |
Correlation:0.2098
  |
Correlation:-0.003972
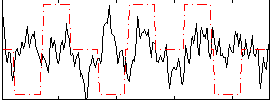  |
Correlation:-0.003972
  |
Component:389; Jaccard:0.075944
 |
Component:389; Jaccard:0.079208
 |
Component:102; Jaccard:0.079114
 |
Component:015; Jaccard:0.064624
 |
Component:015; Jaccard:0.044056
 |
Component:364; Jaccard:0.024587
 |
Component:306; Jaccard:0.012052
 |
Correlation:-0.003972
  |
Correlation:-0.003972
  |
Correlation:0.093804
  |
Correlation:0.0706
  |
Correlation:0.0706
  |
Correlation:0.2098
  |
Correlation:0.024951
 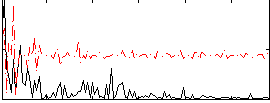 |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
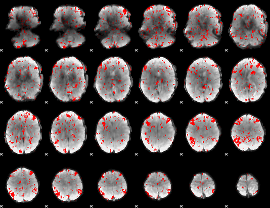 |
GLM binary result thresholded by 2
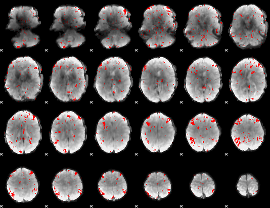 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
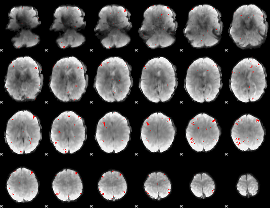 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
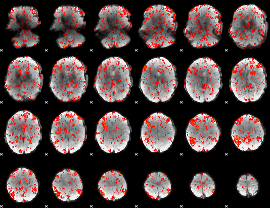 |
GLM Z-score result thresholded by 1.5
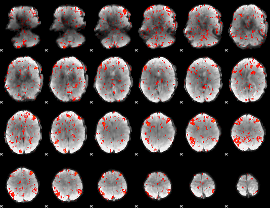 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
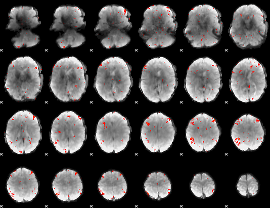 |
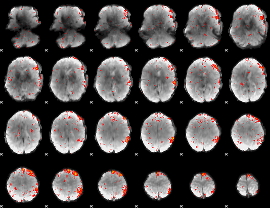
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:026; Jaccard:0.11404
 |
Component:026; Jaccard:0.13131
 |
Component:026; Jaccard:0.12398
 |
Component:026; Jaccard:0.091319
 |
Component:026; Jaccard:0.05611
 |
Component:026; Jaccard:0.026438
 |
Component:026; Jaccard:0.008911
 |
Correlation:0.067894
  |
Correlation:0.067894
  |
Correlation:0.067894
  |
Correlation:0.067894
  |
Correlation:0.067894
  |
Correlation:0.067894
  |
Correlation:0.067894
  |
Component:349; Jaccard:0.11021
 |
Component:349; Jaccard:0.11531
 |
Component:349; Jaccard:0.10063
 |
Component:349; Jaccard:0.071236
 |
Component:349; Jaccard:0.040714
 |
Component:349; Jaccard:0.018508
 |
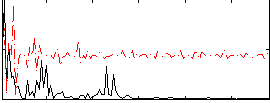
Component:321; Jaccard:0.008244
 |
Correlation:0.080967
  |
Correlation:0.080967
  |
Correlation:0.080967
  |
Correlation:0.080967
  |
Correlation:0.080967
  |
Correlation:0.080967
  |
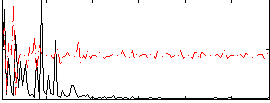
Correlation:0.14198
  |
Component:173; Jaccard:0.098948
 |
Component:173; Jaccard:0.10349
 |
Component:173; Jaccard:0.095455
 |
Component:173; Jaccard:0.069037
 |
Component:173; Jaccard:0.039452
 |
Component:173; Jaccard:0.018308
 |
Component:349; Jaccard:0.006451
 |
Correlation:0.14285
  |
Correlation:0.14285
  |
Correlation:0.14285
  |
Correlation:0.14285
  |
Correlation:0.14285
  |
Correlation:0.14285
  |
Correlation:0.080967
  |
Component:090; Jaccard:0.083848
 |
Component:090; Jaccard:0.090257
 |
Component:090; Jaccard:0.080392
 |
Component:090; Jaccard:0.058665
 |
Component:090; Jaccard:0.03716
 |
Component:321; Jaccard:0.018157
 |
Component:173; Jaccard:0.00626
 |
Correlation:0.051058
  |
Correlation:0.051058
  |
Correlation:0.051058
  |
Correlation:0.051058
  |
Correlation:0.051058
  |
Correlation:0.14198
  |
Correlation:0.14285
  |
Component:330; Jaccard:0.078598
 |
Component:330; Jaccard:0.08344
 |
Component:330; Jaccard:0.075404
 |
Component:330; Jaccard:0.054672
 |
Component:321; Jaccard:0.036866
 |
Component:090; Jaccard:0.016165
 |
Component:090; Jaccard:0.005566
 |
Correlation:0.23797
  |
Correlation:0.23797
  |
Correlation:0.23797
  |
Correlation:0.23797
  |
Correlation:0.14198
  |
Correlation:0.051058
  |
Correlation:0.051058
  |
Component:140; Jaccard:0.076723
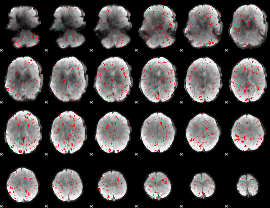 |
Component:140; Jaccard:0.077202
 |
Component:321; Jaccard:0.069567
 |
Component:321; Jaccard:0.054211
 |
Component:330; Jaccard:0.032615
 |
Component:330; Jaccard:0.015143
 |
Component:330; Jaccard:0.005148
 |
Correlation:0.14437
  |
Correlation:0.14437
  |
Correlation:0.14198
  |
Correlation:0.14198
  |
Correlation:0.23797
  |
Correlation:0.23797
  |
Correlation:0.23797
  |
Component:352; Jaccard:0.076572
 |
Component:321; Jaccard:0.072659
 |
Component:140; Jaccard:0.064166
 |
Component:086; Jaccard:0.043135
 |
Component:087; Jaccard:0.024529
 |
Component:087; Jaccard:0.012661
 |
Component:086; Jaccard:0.004542
 |
Correlation:0.20367
  |
Correlation:0.14198
  |
Correlation:0.14437
  |
Correlation:0.14084
  |
Correlation:0.14817
  |
Correlation:0.14817
  |
Correlation:0.14084
  |
Component:109; Jaccard:0.072089
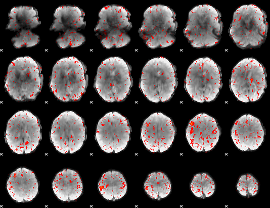 |
Component:352; Jaccard:0.069623
 |
Component:086; Jaccard:0.060222
 |
Component:140; Jaccard:0.040406
 |
Component:086; Jaccard:0.023978
 |
Component:086; Jaccard:0.012317
 |
Component:140; Jaccard:0.004078
 |
Correlation:0.12952
  |
Correlation:0.20367
  |
Correlation:0.14084
  |
Correlation:0.14437
  |
Correlation:0.14084
  |
Correlation:0.14084
  |
Correlation:0.14437
  |
Component:371; Jaccard:0.066916
 |
Component:086; Jaccard:0.069094
 |
Component:318; Jaccard:0.05451
 |
Component:318; Jaccard:0.03704
 |
Component:140; Jaccard:0.020372
 |
Component:140; Jaccard:0.009744
 |
Component:087; Jaccard:0.003983
 |
Correlation:0.16581
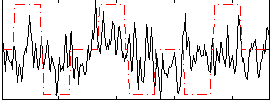 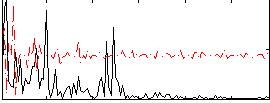 |
Correlation:0.14084
  |
Correlation:0.13186
  |
Correlation:0.13186
  |
Correlation:0.14437
  |
Correlation:0.14437
  |
Correlation:0.14817
  |
Component:086; Jaccard:0.066859
 |
Component:109; Jaccard:0.065154
 |
Component:087; Jaccard:0.050931
 |
Component:087; Jaccard:0.036938
 |
Component:318; Jaccard:0.019431
 |
Component:318; Jaccard:0.008098
 |
Component:150; Jaccard:0.003429
 |
Correlation:0.14084
  |
Correlation:0.12952
  |
Correlation:0.14817
  |
Correlation:0.14817
  |
Correlation:0.13186
  |
Correlation:0.13186
  |
Correlation:0.14738
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
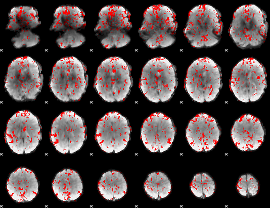 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
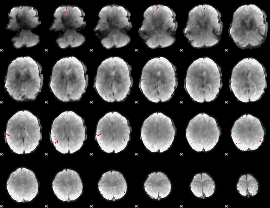 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
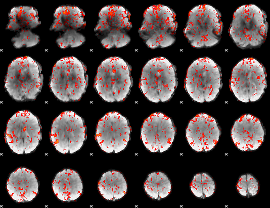 |
GLM Z-score result thresholded by 2
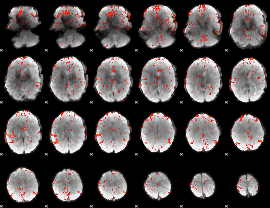 |
GLM Z-score result thresholded by 2.5
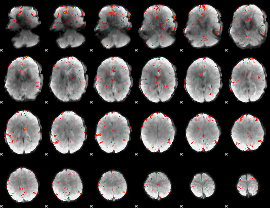 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:119; Jaccard:0.10398
 |
Component:392; Jaccard:0.11375
 |
Component:392; Jaccard:0.11642
 |
Component:191; Jaccard:0.11434
 |
Component:191; Jaccard:0.090946
 |
Component:191; Jaccard:0.057329
 |
Component:191; Jaccard:0.031291
 |
Correlation:0.26012
  |
Correlation:0.16129
  |
Correlation:0.16129
  |
Correlation:0.056169
  |
Correlation:0.056169
  |
Correlation:0.056169
  |
Correlation:0.056169
  |
Component:392; Jaccard:0.10017
 |
Component:119; Jaccard:0.11277
 |
Component:191; Jaccard:0.10958
 |
Component:392; Jaccard:0.10028
 |
Component:392; Jaccard:0.068892
 |
Component:090; Jaccard:0.042363
 |
Component:090; Jaccard:0.023503
 |
Correlation:0.16129
  |
Correlation:0.26012
  |
Correlation:0.056169
  |
Correlation:0.16129
  |
Correlation:0.16129
  |
Correlation:0.12803
 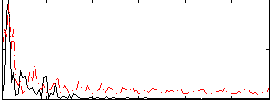 |
Correlation:0.12803
  |
Component:090; Jaccard:0.093587
 |
Component:090; Jaccard:0.10298
 |
Component:119; Jaccard:0.10667
 |
Component:090; Jaccard:0.092856
 |
Component:090; Jaccard:0.068694
 |
Component:392; Jaccard:0.040163
 |
Component:392; Jaccard:0.020676
 |
Correlation:0.12803
  |
Correlation:0.12803
  |
Correlation:0.26012
  |
Correlation:0.12803
  |
Correlation:0.12803
  |
Correlation:0.16129
  |
Correlation:0.16129
  |
Component:121; Jaccard:0.084305
 |
Component:191; Jaccard:0.096766
 |
Component:090; Jaccard:0.10507
 |
Component:119; Jaccard:0.086183
 |
Component:119; Jaccard:0.054449
 |
Component:119; Jaccard:0.030089
 |
Component:041; Jaccard:0.016846
 |
Correlation:0.13621
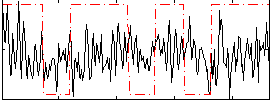  |
Correlation:0.056169
  |
Correlation:0.12803
  |
Correlation:0.26012
  |
Correlation:0.26012
  |
Correlation:0.26012
  |
Correlation:0.18416
  |
Component:219; Jaccard:0.07983
 |
Component:219; Jaccard:0.085089
 |
Component:219; Jaccard:0.081079
 |
Component:012; Jaccard:0.068436
 |
Component:012; Jaccard:0.049265
 |
Component:012; Jaccard:0.028921
 |
Component:119; Jaccard:0.014963
 |
Correlation:0.14472
 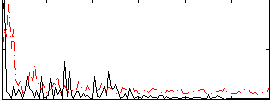 |
Correlation:0.14472
  |
Correlation:0.14472
  |
Correlation:0.17713
  |
Correlation:0.17713
  |
Correlation:0.17713
  |
Correlation:0.26012
  |
Component:259; Jaccard:0.079683
 |
Component:012; Jaccard:0.082535
 |
Component:012; Jaccard:0.080856
 |
Component:219; Jaccard:0.067907
 |
Component:289; Jaccard:0.047932
 |
Component:041; Jaccard:0.027559
 |
Component:315; Jaccard:0.014357
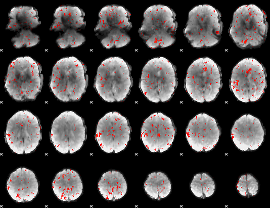 |
Correlation:0.21494
  |
Correlation:0.17713
  |
Correlation:0.17713
  |
Correlation:0.14472
  |
Correlation:0.033695
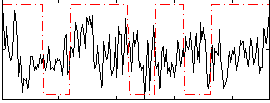  |
Correlation:0.18416
  |
Correlation:0.22988
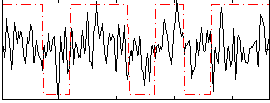  |
Component:191; Jaccard:0.079125
 |
Component:121; Jaccard:0.082485
 |
Component:009; Jaccard:0.075352
 |
Component:289; Jaccard:0.065911
 |
Component:219; Jaccard:0.042906
 |
Component:289; Jaccard:0.026742
 |
Component:012; Jaccard:0.01287
 |
Correlation:0.056169
  |
Correlation:0.13621
  |
Correlation:0.069641
 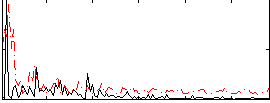 |
Correlation:0.033695
  |
Correlation:0.14472
  |
Correlation:0.033695
  |
Correlation:0.17713
  |
Component:012; Jaccard:0.077366
 |
Component:009; Jaccard:0.079784
 |
Component:121; Jaccard:0.073262
 |
Component:009; Jaccard:0.059612
 |
Component:041; Jaccard:0.042394
 |
Component:339; Jaccard:0.024531
 |
Component:339; Jaccard:0.012513
 |
Correlation:0.17713
  |
Correlation:0.069641
  |
Correlation:0.13621
  |
Correlation:0.069641
  |
Correlation:0.18416
  |
Correlation:0.035322
 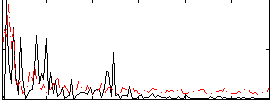 |
Correlation:0.035322
  |
Component:009; Jaccard:0.075505
 |
Component:259; Jaccard:0.079631
 |
Component:259; Jaccard:0.072701
 |
Component:041; Jaccard:0.05837
 |
Component:340; Jaccard:0.040623
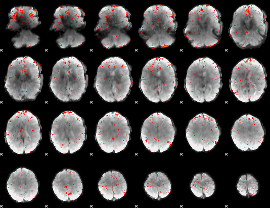 |
Component:342; Jaccard:0.02366
 |
Component:317; Jaccard:0.011924
 |
Correlation:0.069641
  |
Correlation:0.21494
  |
Correlation:0.21494
  |
Correlation:0.18416
  |
Correlation:0.025393
 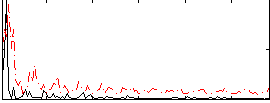 |
Correlation:0.13108
  |
Correlation:0.041541
 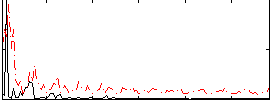 |
Component:094; Jaccard:0.074405
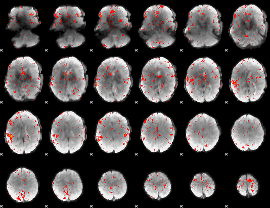 |
Component:094; Jaccard:0.076405
 |
Component:289; Jaccard:0.07244
 |
Component:259; Jaccard:0.057285
 |
Component:342; Jaccard:0.039831
 |
Component:163; Jaccard:0.023138
 |
Component:289; Jaccard:0.01184
 |
Correlation:0.069103
  |
Correlation:0.069103
  |
Correlation:0.033695
  |
Correlation:0.21494
  |
Correlation:0.13108
  |
Correlation:0.10601
  |
Correlation:0.033695
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
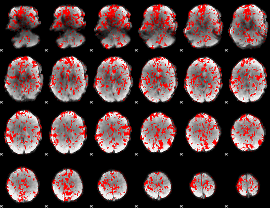 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
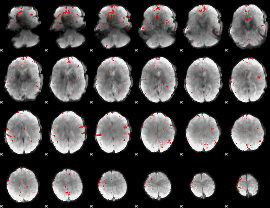 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
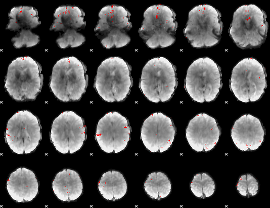 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
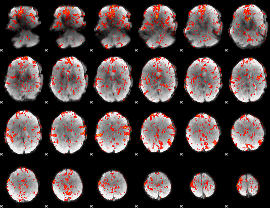 |
GLM Z-score result thresholded by 2
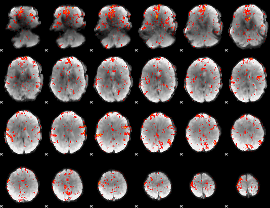 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:119; Jaccard:0.10096
 |
Component:191; Jaccard:0.11636
 |
Component:191; Jaccard:0.14491
 |
Component:191; Jaccard:0.16064
 |
Component:191; Jaccard:0.15818
 |
Component:191; Jaccard:0.13099
 |
Component:191; Jaccard:0.085623
 |
Correlation:-0.023706
  |
Correlation:0.28617
  |
Correlation:0.28617
  |
Correlation:0.28617
  |
Correlation:0.28617
  |
Correlation:0.28617
  |
Correlation:0.28617
  |
Component:317; Jaccard:0.095414
 |
Component:119; Jaccard:0.10988
 |
Component:317; Jaccard:0.1132
 |
Component:340; Jaccard:0.10714
 |
Component:340; Jaccard:0.088386
 |
Component:340; Jaccard:0.064219
 |
Component:340; Jaccard:0.039968
 |
Correlation:0.18837
  |
Correlation:-0.023706
  |
Correlation:0.18837
  |
Correlation:0.28917
  |
Correlation:0.28917
  |
Correlation:0.28917
  |
Correlation:0.28917
  |
Component:094; Jaccard:0.093941
 |
Component:317; Jaccard:0.10664
 |
Component:392; Jaccard:0.10924
 |
Component:317; Jaccard:0.10515
 |
Component:317; Jaccard:0.083093
 |
Component:392; Jaccard:0.05631
 |
Component:392; Jaccard:0.035003
 |
Correlation:0.037184
  |
Correlation:0.18837
  |
Correlation:0.15862
  |
Correlation:0.18837
  |
Correlation:0.18837
  |
Correlation:0.15862
  |
Correlation:0.15862
  |
Component:392; Jaccard:0.091498
 |
Component:392; Jaccard:0.10389
 |
Component:119; Jaccard:0.10883
 |
Component:392; Jaccard:0.10113
 |
Component:392; Jaccard:0.082408
 |
Component:317; Jaccard:0.056055
 |
Component:012; Jaccard:0.034588
 |
Correlation:0.15862
  |
Correlation:0.15862
  |
Correlation:-0.023706
  |
Correlation:0.15862
  |
Correlation:0.15862
  |
Correlation:0.18837
  |
Correlation:0.1007
  |
Component:191; Jaccard:0.090085
 |
Component:094; Jaccard:0.10316
 |
Component:340; Jaccard:0.10709
 |
Component:119; Jaccard:0.096344
 |
Component:012; Jaccard:0.074077
 |
Component:012; Jaccard:0.053383
 |
Component:317; Jaccard:0.034587
 |
Correlation:0.28617
  |
Correlation:0.037184
  |
Correlation:0.28917
  |
Correlation:-0.023706
  |
Correlation:0.1007
  |
Correlation:0.1007
  |
Correlation:0.18837
  |
Component:298; Jaccard:0.086222
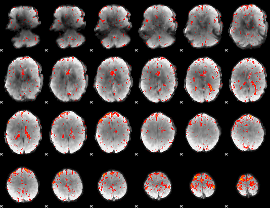 |
Component:340; Jaccard:0.097536
 |
Component:094; Jaccard:0.10442
 |
Component:094; Jaccard:0.094625
 |
Component:094; Jaccard:0.0712
 |
Component:076; Jaccard:0.047542
 |
Component:076; Jaccard:0.02973
 |
Correlation:0.15534
  |
Correlation:0.28917
  |
Correlation:0.037184
  |
Correlation:0.037184
  |
Correlation:0.037184
  |
Correlation:0.28171
  |
Correlation:0.28171
  |
Component:340; Jaccard:0.085009
 |
Component:009; Jaccard:0.092336
 |
Component:012; Jaccard:0.096067
 |
Component:012; Jaccard:0.091233
 |
Component:119; Jaccard:0.069264
 |
Component:041; Jaccard:0.04588
 |
Component:094; Jaccard:0.027705
 |
Correlation:0.28917
  |
Correlation:0.14343
  |
Correlation:0.1007
  |
Correlation:0.1007
  |
Correlation:-0.023706
  |
Correlation:0.197
  |
Correlation:0.037184
  |
Component:009; Jaccard:0.084501
 |
Component:012; Jaccard:0.090243
 |
Component:009; Jaccard:0.093289
 |
Component:009; Jaccard:0.085529
 |
Component:041; Jaccard:0.064216
 |
Component:094; Jaccard:0.045364
 |
Component:041; Jaccard:0.026991
 |
Correlation:0.14343
  |
Correlation:0.1007
  |
Correlation:0.14343
  |
Correlation:0.14343
  |
Correlation:0.197
  |
Correlation:0.037184
  |
Correlation:0.197
  |
Component:379; Jaccard:0.083168
 |
Component:379; Jaccard:0.087321
 |
Component:379; Jaccard:0.08522
 |
Component:041; Jaccard:0.07835
 |
Component:009; Jaccard:0.062887
 |
Component:119; Jaccard:0.044237
 |
Component:119; Jaccard:0.023888
 |
Correlation:0.19532
  |
Correlation:0.19532
  |
Correlation:0.19532
  |
Correlation:0.197
  |
Correlation:0.14343
  |
Correlation:-0.023706
  |
Correlation:-0.023706
  |
Component:025; Jaccard:0.080712
 |
Component:356; Jaccard:0.086559
 |
Component:041; Jaccard:0.082607
 |
Component:379; Jaccard:0.074356
 |
Component:076; Jaccard:0.062495
 |
Component:009; Jaccard:0.039734
 |
Component:009; Jaccard:0.02337
 |
Correlation:0.31271
  |
Correlation:-0.10115
  |
Correlation:0.197
  |
Correlation:0.19532
  |
Correlation:0.28171
  |
Correlation:0.14343
  |
Correlation:0.14343
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































