Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
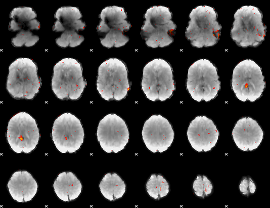
Cope: 01:FACES BACK
|
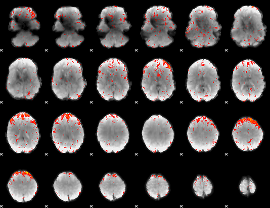
GLM result group-wise
 |
GLM binary result thresholded by 1
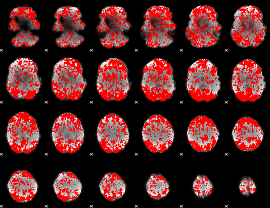 |
GLM binary result thresholded by 1.5
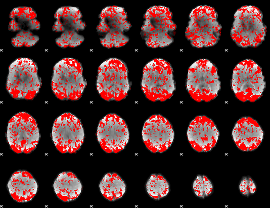 |
GLM binary result thresholded by 2
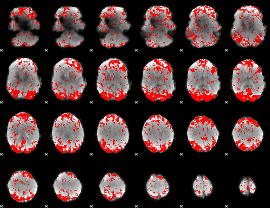 |
GLM binary result thresholded by 2.5
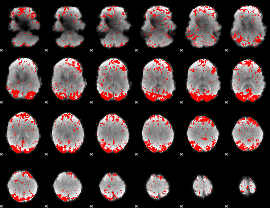 |
GLM binary result thresholded by 3
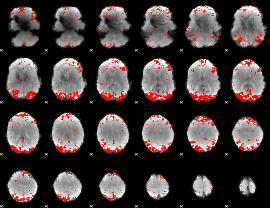 |
GLM binary result thresholded by 3.5
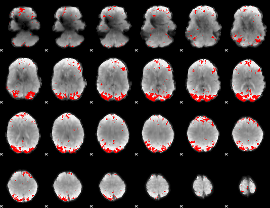 |
GLM binary result thresholded by 4
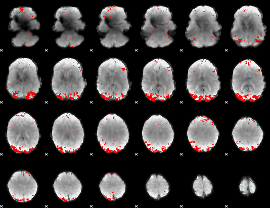 |
GLM Z-score result thresholded by 1
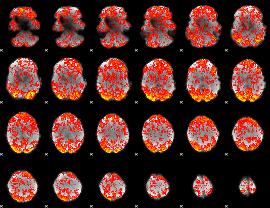 |
GLM Z-score result thresholded by 1.5
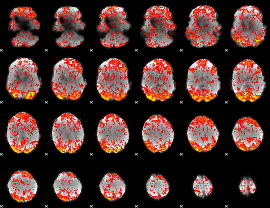 |
GLM Z-score result thresholded by 2
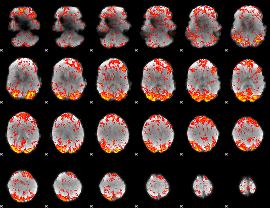 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
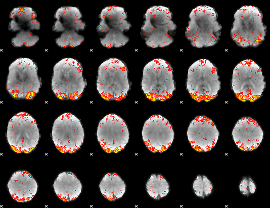 |
GLM Z-score result thresholded by 3.5
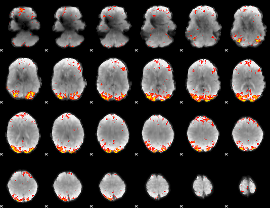 |
GLM Z-score result thresholded by 4
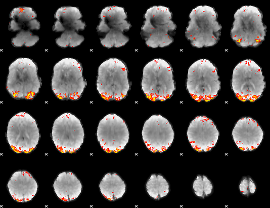 |
Component:326; Jaccard:0.088066
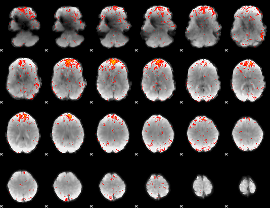 |
Component:350; Jaccard:0.11798
 |
Component:350; Jaccard:0.16064
 |
Component:350; Jaccard:0.21237
 |
Component:350; Jaccard:0.25794
 |
Component:350; Jaccard:0.28271
 |
Component:350; Jaccard:0.28343
 |
Correlation:0.26367
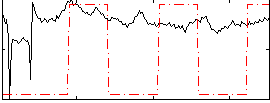  |
Correlation:0.43957
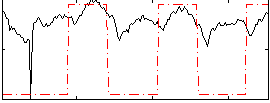 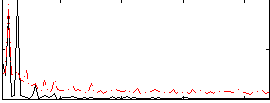 |
Correlation:0.43957
  |
Correlation:0.43957
  |
Correlation:0.43957
  |
Correlation:0.43957
  |
Correlation:0.43957
  |
Component:350; Jaccard:0.086637
 |
Component:288; Jaccard:0.10308
 |
Component:288; Jaccard:0.11942
 |
Component:253; Jaccard:0.13453
 |
Component:253; Jaccard:0.15084
 |
Component:253; Jaccard:0.15762
 |
Component:253; Jaccard:0.1545
 |
Correlation:0.43957
  |
Correlation:0.1214
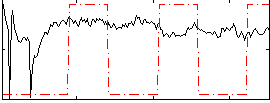  |
Correlation:0.1214
  |
Correlation:0.51858
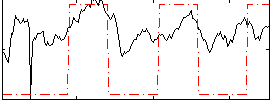  |
Correlation:0.51858
  |
Correlation:0.51858
  |
Correlation:0.51858
  |
Component:288; Jaccard:0.084072
 |
Component:326; Jaccard:0.10231
 |
Component:310; Jaccard:0.11386
 |
Component:288; Jaccard:0.12804
 |
Component:310; Jaccard:0.13296
 |
Component:310; Jaccard:0.13394
 |
Component:310; Jaccard:0.12818
 |
Correlation:0.1214
  |
Correlation:0.26367
  |
Correlation:0.56462
  |
Correlation:0.1214
  |
Correlation:0.56462
  |
Correlation:0.56462
  |
Correlation:0.56462
  |
Component:303; Jaccard:0.080904
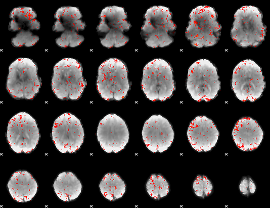 |
Component:310; Jaccard:0.096869
 |
Component:253; Jaccard:0.11314
 |
Component:310; Jaccard:0.12658
 |
Component:392; Jaccard:0.12193
 |
Component:392; Jaccard:0.11999
 |
Component:274; Jaccard:0.11578
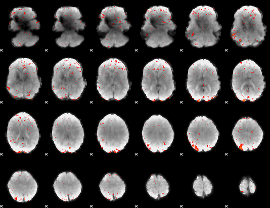 |
Correlation:0.36452
 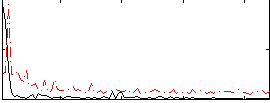 |
Correlation:0.56462
  |
Correlation:0.51858
  |
Correlation:0.56462
  |
Correlation:0.64645
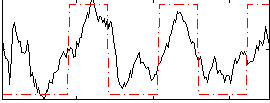 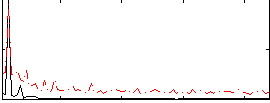 |
Correlation:0.64645
  |
Correlation:0.20574
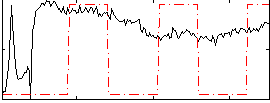 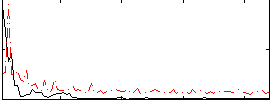 |
Component:387; Jaccard:0.080687
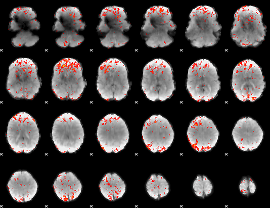 |
Component:369; Jaccard:0.092907
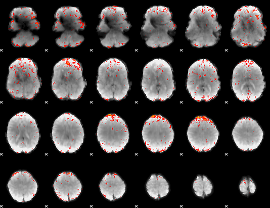 |
Component:326; Jaccard:0.11208
 |
Component:369; Jaccard:0.11969
 |
Component:369; Jaccard:0.12126
 |
Component:293; Jaccard:0.11807
 |
Component:133; Jaccard:0.11495
 |
Correlation:0.26289
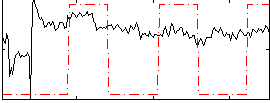  |
Correlation:0.27026
  |
Correlation:0.26367
  |
Correlation:0.27026
  |
Correlation:0.27026
  |
Correlation:-0.022477
  |
Correlation:0.14006
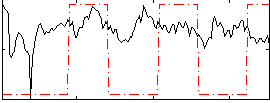  |
Component:310; Jaccard:0.079519
 |
Component:303; Jaccard:0.091773
 |
Component:369; Jaccard:0.10861
 |
Component:392; Jaccard:0.11398
 |
Component:288; Jaccard:0.11831
 |
Component:133; Jaccard:0.11789
 |
Component:293; Jaccard:0.11299
 |
Correlation:0.56462
  |
Correlation:0.36452
  |
Correlation:0.27026
  |
Correlation:0.64645
  |
Correlation:0.1214
  |
Correlation:0.14006
  |
Correlation:-0.022477
  |
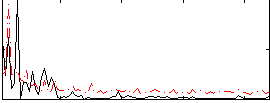
Component:194; Jaccard:0.077655
 |
Component:253; Jaccard:0.091772
 |
Component:392; Jaccard:0.10259
 |
Component:326; Jaccard:0.11188
 |
Component:133; Jaccard:0.11777
 |
Component:274; Jaccard:0.11664
 |
Component:392; Jaccard:0.11049
 |
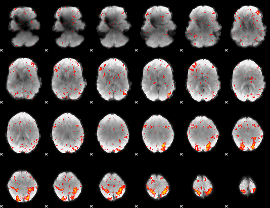
Correlation:0.17883
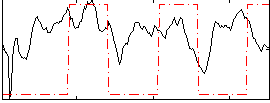  |
Correlation:0.51858
  |
Correlation:0.64645
  |
Correlation:0.26367
  |
Correlation:0.14006
  |
Correlation:0.20574
  |
Correlation:0.64645
  |
Component:369; Jaccard:0.076384
 |
Component:392; Jaccard:0.089487
 |
Component:133; Jaccard:0.099155
 |
Component:133; Jaccard:0.11024
 |
Component:293; Jaccard:0.11483
 |
Component:369; Jaccard:0.10973
 |
Component:158; Jaccard:0.10316
 |
Correlation:0.27026
  |
Correlation:0.64645
  |
Correlation:0.14006
  |
Correlation:0.14006
  |
Correlation:-0.022477
  |
Correlation:0.27026
  |
Correlation:0.16101
  |
Component:219; Jaccard:0.076283
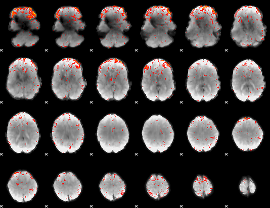 |
Component:387; Jaccard:0.08823
 |
Component:303; Jaccard:0.098309
 |
Component:293; Jaccard:0.10645
 |
Component:305; Jaccard:0.10936
 |
Component:305; Jaccard:0.1075
 |
Component:305; Jaccard:0.09886
 |
Correlation:0.11149
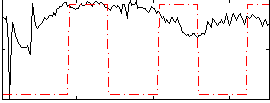 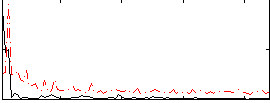 |
Correlation:0.26289
  |
Correlation:0.36452
  |
Correlation:-0.022477
  |
Correlation:0.19379
 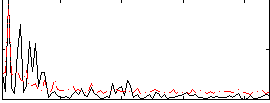 |
Correlation:0.19379
  |
Correlation:0.19379
  |
Component:035; Jaccard:0.07624
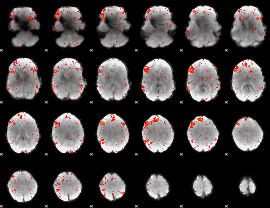 |
Component:133; Jaccard:0.086529
 |
Component:305; Jaccard:0.095718
 |
Component:305; Jaccard:0.10622
 |
Component:274; Jaccard:0.10829
 |
Component:158; Jaccard:0.10711
 |
Component:174; Jaccard:0.096084
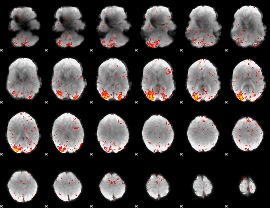 |
Correlation:0.076632
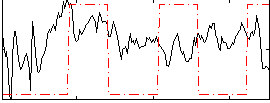  |
Correlation:0.14006
  |
Correlation:0.19379
  |
Correlation:0.19379
  |
Correlation:0.20574
  |
Correlation:0.16101
  |
Correlation:0.1931
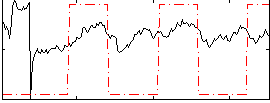  |
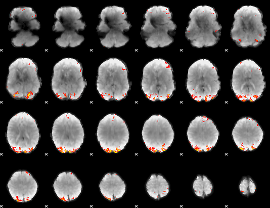
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
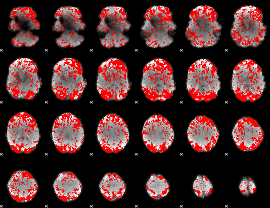 |
GLM binary result thresholded by 1.5
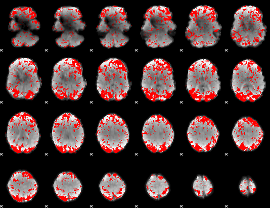 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
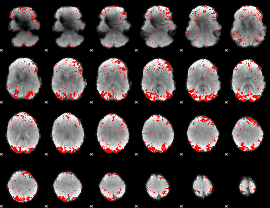 |
GLM binary result thresholded by 3
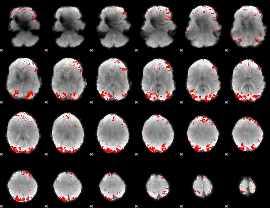 |
GLM binary result thresholded by 3.5
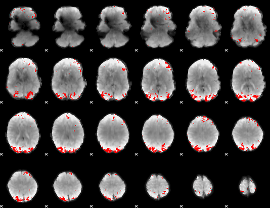 |
GLM binary result thresholded by 4
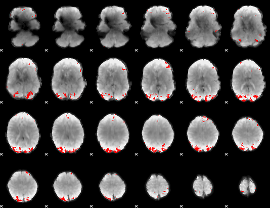 |
GLM Z-score result thresholded by 1
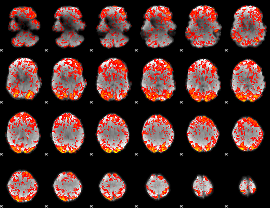 |
GLM Z-score result thresholded by 1.5
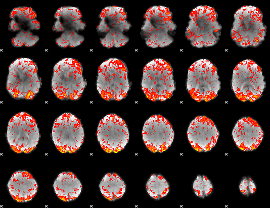 |
GLM Z-score result thresholded by 2
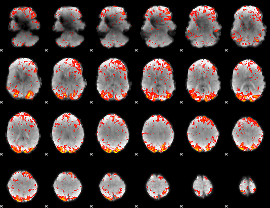 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
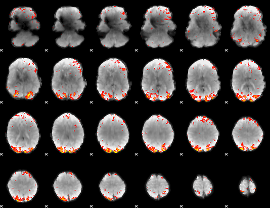 |
GLM Z-score result thresholded by 3.5
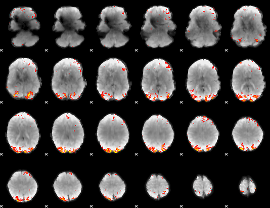 |
GLM Z-score result thresholded by 4
 |
Component:194; Jaccard:0.11405
 |
Component:350; Jaccard:0.13804
 |
Component:350; Jaccard:0.1717
 |
Component:350; Jaccard:0.20104
 |
Component:350; Jaccard:0.20986
 |
Component:350; Jaccard:0.19703
 |
Component:350; Jaccard:0.17347
 |
Correlation:0.017379
  |
Correlation:-0.37589
  |
Correlation:-0.37589
  |
Correlation:-0.37589
  |
Correlation:-0.37589
  |
Correlation:-0.37589
  |
Correlation:-0.37589
  |
Component:350; Jaccard:0.10626
 |
Component:194; Jaccard:0.13107
 |
Component:194; Jaccard:0.13898
 |
Component:194; Jaccard:0.13871
 |
Component:158; Jaccard:0.13567
 |
Component:274; Jaccard:0.13301
 |
Component:274; Jaccard:0.12272
 |
Correlation:-0.37589
  |
Correlation:0.017379
  |
Correlation:0.017379
  |
Correlation:0.017379
  |
Correlation:0.086956
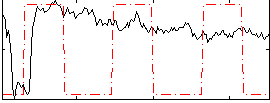 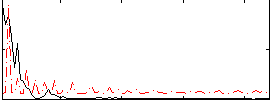 |
Correlation:0.11216
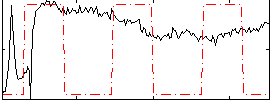 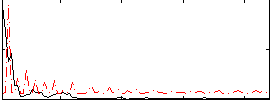 |
Correlation:0.11216
  |
Component:288; Jaccard:0.094934
 |
Component:288; Jaccard:0.11089
 |
Component:133; Jaccard:0.11907
 |
Component:293; Jaccard:0.12893
 |
Component:274; Jaccard:0.13264
 |
Component:158; Jaccard:0.13036
 |
Component:158; Jaccard:0.11827
 |
Correlation:-0.12678
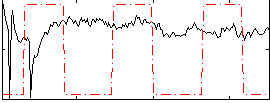 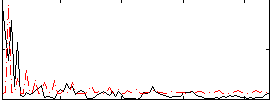 |
Correlation:-0.12678
  |
Correlation:-0.13954
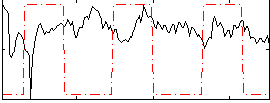 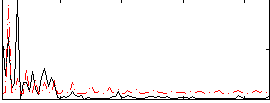 |
Correlation:-0.10102
  |
Correlation:0.11216
  |
Correlation:0.086956
  |
Correlation:0.086956
  |
Component:219; Jaccard:0.092821
 |
Component:133; Jaccard:0.10609
 |
Component:293; Jaccard:0.11832
 |
Component:158; Jaccard:0.12887
 |
Component:293; Jaccard:0.13255
 |
Component:293; Jaccard:0.12622
 |
Component:133; Jaccard:0.11475
 |
Correlation:0.089985
  |
Correlation:-0.13954
  |
Correlation:-0.10102
  |
Correlation:0.086956
  |
Correlation:-0.10102
  |
Correlation:-0.10102
  |
Correlation:-0.13954
  |
Component:326; Jaccard:0.091962
 |
Component:293; Jaccard:0.10448
 |
Component:288; Jaccard:0.11722
 |
Component:133; Jaccard:0.12764
 |
Component:133; Jaccard:0.13029
 |
Component:133; Jaccard:0.12584
 |
Component:293; Jaccard:0.11012
 |
Correlation:-0.064723
  |
Correlation:-0.10102
  |
Correlation:-0.12678
  |
Correlation:-0.13954
  |
Correlation:-0.13954
  |
Correlation:-0.13954
  |
Correlation:-0.10102
  |
Component:133; Jaccard:0.09085
 |
Component:305; Jaccard:0.10262
 |
Component:305; Jaccard:0.11664
 |
Component:305; Jaccard:0.12376
 |
Component:194; Jaccard:0.12798
 |
Component:194; Jaccard:0.1125
 |
Component:305; Jaccard:0.096691
 |
Correlation:-0.13954
  |
Correlation:-0.11115
  |
Correlation:-0.11115
  |
Correlation:-0.11115
  |
Correlation:0.017379
  |
Correlation:0.017379
  |
Correlation:-0.11115
  |
Component:387; Jaccard:0.089101
 |
Component:291; Jaccard:0.10207
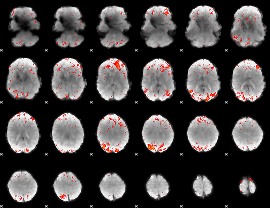 |
Component:291; Jaccard:0.1163
 |
Component:274; Jaccard:0.12185
 |
Component:305; Jaccard:0.12346
 |
Component:305; Jaccard:0.11239
 |
Component:194; Jaccard:0.09347
 |
Correlation:0.043719
  |
Correlation:-0.00077982
  |
Correlation:-0.00077982
  |
Correlation:0.11216
  |
Correlation:-0.11115
  |
Correlation:-0.11115
  |
Correlation:0.017379
  |
Component:293; Jaccard:0.088924
 |
Component:219; Jaccard:0.10068
 |
Component:158; Jaccard:0.11171
 |
Component:291; Jaccard:0.11986
 |
Component:291; Jaccard:0.1101
 |
Component:291; Jaccard:0.093912
 |
Component:291; Jaccard:0.077571
 |
Correlation:-0.10102
  |
Correlation:0.089985
  |
Correlation:0.086956
  |
Correlation:-0.00077982
  |
Correlation:-0.00077982
  |
Correlation:-0.00077982
  |
Correlation:-0.00077982
  |
Component:328; Jaccard:0.086965
 |
Component:326; Jaccard:0.098616
 |
Component:369; Jaccard:0.1046
 |
Component:288; Jaccard:0.11125
 |
Component:369; Jaccard:0.093685
 |
Component:253; Jaccard:0.084235
 |
Component:253; Jaccard:0.071422
 |
Correlation:0.12813
  |
Correlation:-0.064723
  |
Correlation:-0.089488
  |
Correlation:-0.12678
  |
Correlation:-0.089488
  |
Correlation:-0.46342
  |
Correlation:-0.46342
  |
Component:305; Jaccard:0.086919
 |
Component:063; Jaccard:0.097747
 |
Component:063; Jaccard:0.10349
 |
Component:369; Jaccard:0.10354
 |
Component:253; Jaccard:0.089854
 |
Component:369; Jaccard:0.077285
 |
Component:310; Jaccard:0.06069
 |
Correlation:-0.11115
  |
Correlation:-0.023079
  |
Correlation:-0.023079
  |
Correlation:-0.089488
  |
Correlation:-0.46342
  |
Correlation:-0.089488
  |
Correlation:-0.33097
  |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
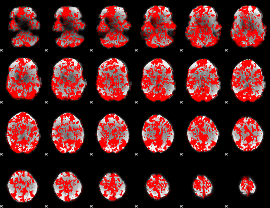 |
GLM binary result thresholded by 1.5
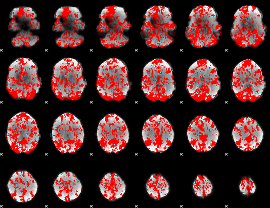 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
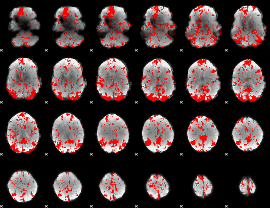 |
GLM binary result thresholded by 3
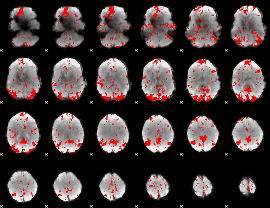 |
GLM binary result thresholded by 3.5
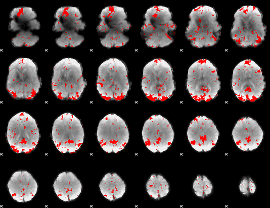 |
GLM binary result thresholded by 4
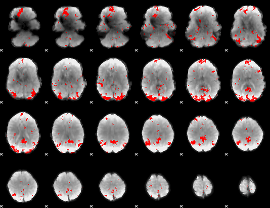 |
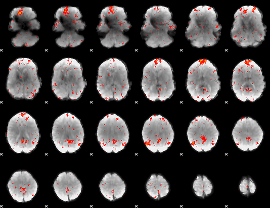
GLM Z-score result thresholded by 1
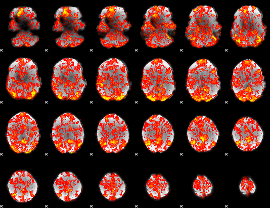 |
GLM Z-score result thresholded by 1.5
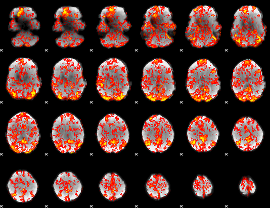 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:392; Jaccard:0.083824
 |
Component:392; Jaccard:0.10857
 |
Component:392; Jaccard:0.14376
 |
Component:392; Jaccard:0.1857
 |
Component:392; Jaccard:0.22327
 |
Component:392; Jaccard:0.24169
 |
Component:392; Jaccard:0.24269
 |
Correlation:0.72177
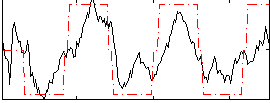  |
Correlation:0.72177
  |
Correlation:0.72177
  |
Correlation:0.72177
  |
Correlation:0.72177
  |
Correlation:0.72177
  |
Correlation:0.72177
  |
Component:385; Jaccard:0.077107
 |
Component:310; Jaccard:0.087398
 |
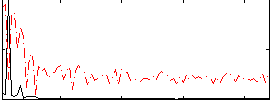
Component:193; Jaccard:0.11026
 |
Component:193; Jaccard:0.14151
 |
Component:193; Jaccard:0.17362
 |
Component:193; Jaccard:0.19553
 |
Component:193; Jaccard:0.19995
 |
Correlation:0.38045
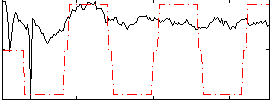  |
Correlation:0.48578
  |
Correlation:0.49532
 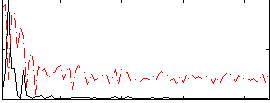 |
Correlation:0.49532
  |
Correlation:0.49532
  |
Correlation:0.49532
  |
Correlation:0.49532
  |
Component:310; Jaccard:0.073286
 |
Component:385; Jaccard:0.085611
 |
Component:310; Jaccard:0.10316
 |
Component:310; Jaccard:0.11934
 |
Component:209; Jaccard:0.13396
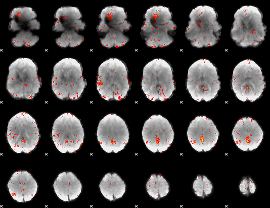 |
Component:209; Jaccard:0.13779
 |
Component:209; Jaccard:0.1376
 |
Correlation:0.48578
  |
Correlation:0.38045
  |
Correlation:0.48578
  |
Correlation:0.48578
  |
Correlation:0.39036
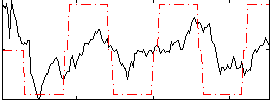 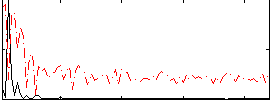 |
Correlation:0.39036
  |
Correlation:0.39036
  |
Component:371; Jaccard:0.068691
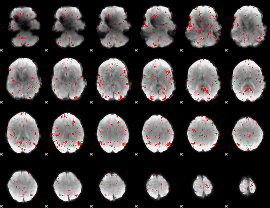 |
Component:193; Jaccard:0.085219
 |
Component:209; Jaccard:0.097317
 |
Component:209; Jaccard:0.1172
 |
Component:310; Jaccard:0.13062
 |
Component:253; Jaccard:0.13438
 |
Component:253; Jaccard:0.13315
 |
Correlation:0.49486
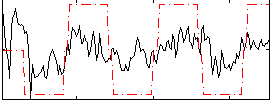  |
Correlation:0.49532
  |
Correlation:0.39036
  |
Correlation:0.39036
  |
Correlation:0.48578
  |
Correlation:0.53265
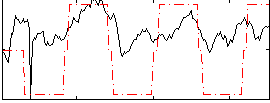  |
Correlation:0.53265
  |
Component:174; Jaccard:0.068649
 |
Component:253; Jaccard:0.078855
 |
Component:253; Jaccard:0.095569
 |
Component:253; Jaccard:0.11284
 |
Component:253; Jaccard:0.12709
 |
Component:310; Jaccard:0.13426
 |
Component:310; Jaccard:0.12935
 |
Correlation:0.33711
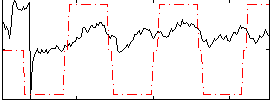 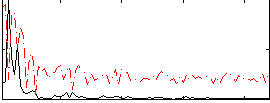 |
Correlation:0.53265
  |
Correlation:0.53265
  |
Correlation:0.53265
  |
Correlation:0.53265
  |
Correlation:0.48578
  |
Correlation:0.48578
  |
Component:303; Jaccard:0.067236
 |
Component:209; Jaccard:0.07833
 |
Component:385; Jaccard:0.092225
 |
Component:129; Jaccard:0.10461
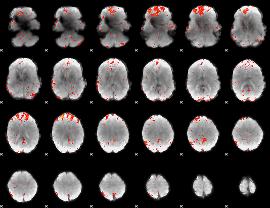 |
Component:009; Jaccard:0.11673
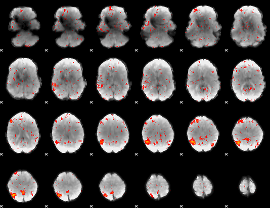 |
Component:009; Jaccard:0.12536
 |
Component:009; Jaccard:0.12117
 |
Correlation:0.27041
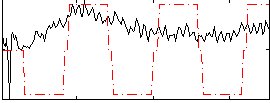 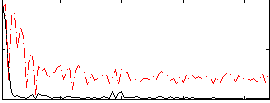 |
Correlation:0.39036
  |
Correlation:0.38045
  |
Correlation:0.34262
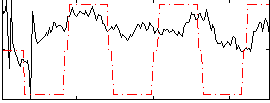 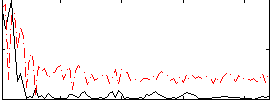 |
Correlation:0.42166
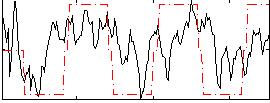  |
Correlation:0.42166
  |
Correlation:0.42166
  |
Component:193; Jaccard:0.067053
 |
Component:129; Jaccard:0.07768
 |
Component:129; Jaccard:0.091902
 |
Component:009; Jaccard:0.10204
 |
Component:129; Jaccard:0.1125
 |
Component:129; Jaccard:0.11536
 |
Component:350; Jaccard:0.11442
 |
Correlation:0.49532
  |
Correlation:0.34262
  |
Correlation:0.34262
  |
Correlation:0.42166
  |
Correlation:0.34262
  |
Correlation:0.34262
  |
Correlation:0.44232
  |
Component:129; Jaccard:0.065527
 |
Component:371; Jaccard:0.077407
 |
Component:294; Jaccard:0.089315
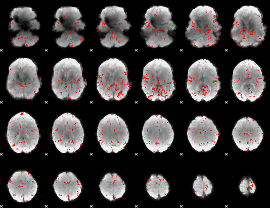 |
Component:350; Jaccard:0.098518
 |
Component:029; Jaccard:0.10899
 |
Component:350; Jaccard:0.11482
 |
Component:029; Jaccard:0.11012
 |
Correlation:0.34262
  |
Correlation:0.49486
  |
Correlation:0.44956
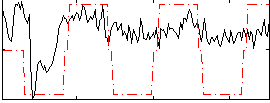 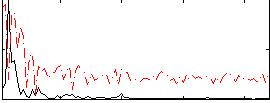 |
Correlation:0.44232
  |
Correlation:0.45311
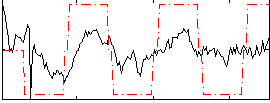 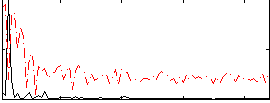 |
Correlation:0.44232
  |
Correlation:0.45311
  |
Component:294; Jaccard:0.065202
 |
Component:174; Jaccard:0.077399
 |
Component:009; Jaccard:0.08788
 |
Component:294; Jaccard:0.098189
 |
Component:350; Jaccard:0.10798
 |
Component:029; Jaccard:0.11256
 |
Component:256; Jaccard:0.10707
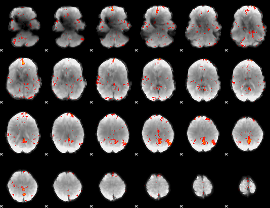 |
Correlation:0.44956
  |
Correlation:0.33711
  |
Correlation:0.42166
  |
Correlation:0.44956
  |
Correlation:0.44232
  |
Correlation:0.45311
  |
Correlation:0.39179
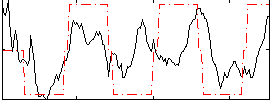 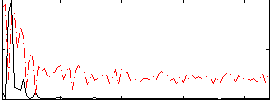 |
Component:028; Jaccard:0.06493
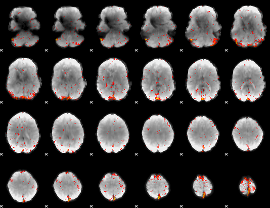 |
Component:294; Jaccard:0.077294
 |
Component:350; Jaccard:0.087267
 |
Component:256; Jaccard:0.098052
 |
Component:256; Jaccard:0.10777
 |
Component:256; Jaccard:0.11059
 |
Component:129; Jaccard:0.10436
 |
Correlation:0.48529
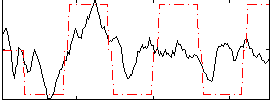 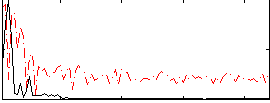 |
Correlation:0.44956
  |
Correlation:0.44232
  |
Correlation:0.39179
  |
Correlation:0.39179
  |
Correlation:0.39179
  |
Correlation:0.34262
  |
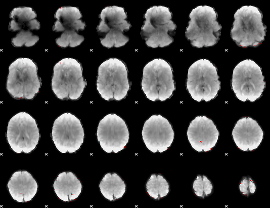
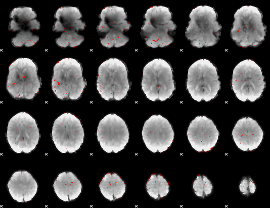
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
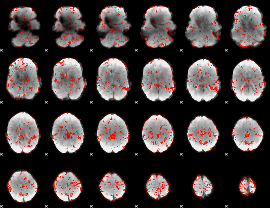
GLM binary result thresholded by 1
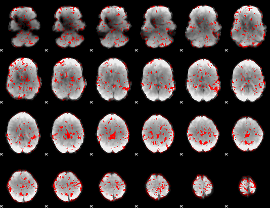 |
GLM binary result thresholded by 1.5
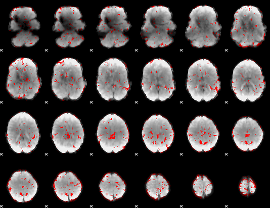 |
GLM binary result thresholded by 2
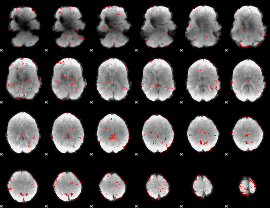 |
GLM binary result thresholded by 2.5
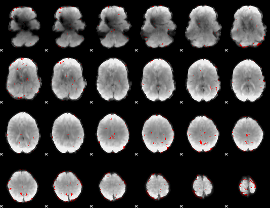 |
GLM binary result thresholded by 3
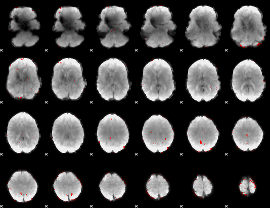 |
GLM binary result thresholded by 3.5
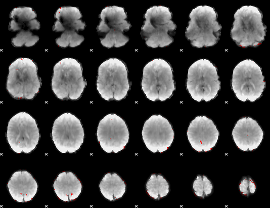 |
GLM binary result thresholded by 4
 |
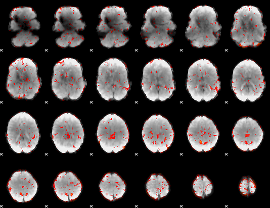
GLM Z-score result thresholded by 1
 |
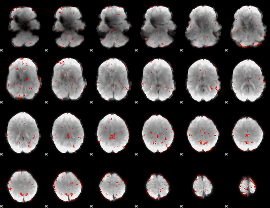
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
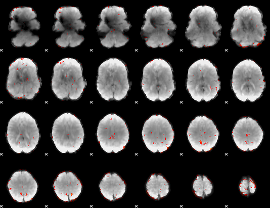 |
GLM Z-score result thresholded by 3
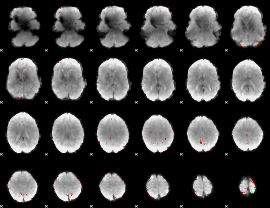 |
GLM Z-score result thresholded by 3.5
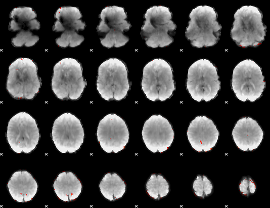 |
GLM Z-score result thresholded by 4
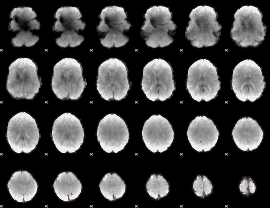 |
Component:354; Jaccard:0.13174
 |
Component:354; Jaccard:0.159
 |
Component:354; Jaccard:0.16466
 |
Component:354; Jaccard:0.14315
 |
Component:354; Jaccard:0.096361
 |
Component:162; Jaccard:0.065898
 |
Component:162; Jaccard:0.037537
 |
Correlation:0.18543
  |
Correlation:0.18543
  |
Correlation:0.18543
  |
Correlation:0.18543
  |
Correlation:0.18543
  |
Correlation:0.26332
 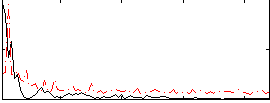 |
Correlation:0.26332
  |
Component:229; Jaccard:0.12056
 |
Component:229; Jaccard:0.14686
 |
Component:229; Jaccard:0.155
 |
Component:229; Jaccard:0.13568
 |
Component:229; Jaccard:0.092972
 |
Component:354; Jaccard:0.054811
 |
Component:314; Jaccard:0.027737
 |
Correlation:0.17925
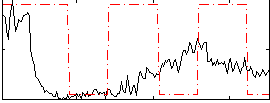 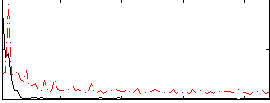 |
Correlation:0.17925
  |
Correlation:0.17925
  |
Correlation:0.17925
  |
Correlation:0.17925
  |
Correlation:0.18543
  |
Correlation:-0.025697
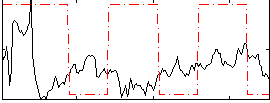 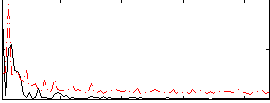 |
Component:314; Jaccard:0.10361
 |
Component:314; Jaccard:0.11599
 |
Component:314; Jaccard:0.11651
 |
Component:162; Jaccard:0.10798
 |
Component:162; Jaccard:0.090943
 |
Component:229; Jaccard:0.050999
 |
Component:229; Jaccard:0.026545
 |
Correlation:-0.025697
  |
Correlation:-0.025697
  |
Correlation:-0.025697
  |
Correlation:0.26332
  |
Correlation:0.26332
  |
Correlation:0.17925
  |
Correlation:0.17925
  |
Component:317; Jaccard:0.102
 |
Component:317; Jaccard:0.10986
 |
Component:317; Jaccard:0.1091
 |
Component:314; Jaccard:0.1007
 |
Component:314; Jaccard:0.076818
 |
Component:314; Jaccard:0.04881
 |
Component:354; Jaccard:0.026273
 |
Correlation:0.26034
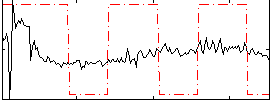 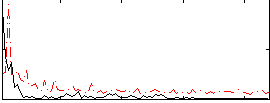 |
Correlation:0.26034
  |
Correlation:0.26034
  |
Correlation:-0.025697
  |
Correlation:-0.025697
  |
Correlation:-0.025697
  |
Correlation:0.18543
  |
Component:115; Jaccard:0.091944
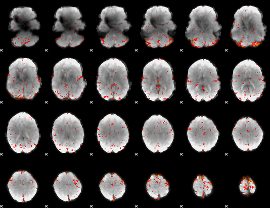 |
Component:198; Jaccard:0.098512
 |
Component:162; Jaccard:0.10496
 |
Component:183; Jaccard:0.091927
 |
Component:183; Jaccard:0.07259
 |
Component:183; Jaccard:0.047089
 |
Component:183; Jaccard:0.024327
 |
Correlation:-0.147
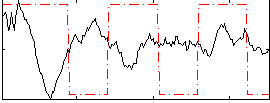 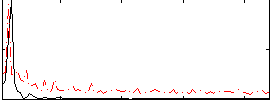 |
Correlation:0.16304
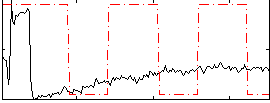 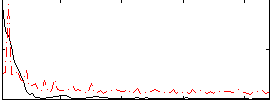 |
Correlation:0.26332
  |
Correlation:0.17652
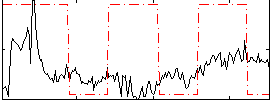 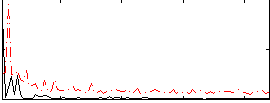 |
Correlation:0.17652
  |
Correlation:0.17652
  |
Correlation:0.17652
  |
Component:198; Jaccard:0.086576
 |
Component:183; Jaccard:0.093358
 |
Component:183; Jaccard:0.10001
 |
Component:317; Jaccard:0.091596
 |
Component:083; Jaccard:0.069836
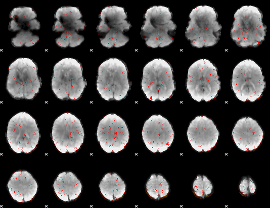 |
Component:083; Jaccard:0.046213
 |
Component:083; Jaccard:0.023084
 |
Correlation:0.16304
  |
Correlation:0.17652
  |
Correlation:0.17652
  |
Correlation:0.26034
  |
Correlation:0.13297
 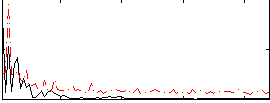 |
Correlation:0.13297
  |
Correlation:0.13297
  |
Component:160; Jaccard:0.083551
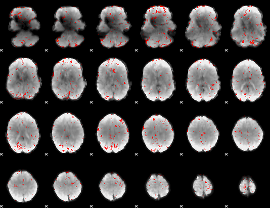 |
Component:115; Jaccard:0.087858
 |
Component:198; Jaccard:0.099583
 |
Component:198; Jaccard:0.088655
 |
Component:317; Jaccard:0.066094
 |
Component:198; Jaccard:0.040389
 |
Component:204; Jaccard:0.022635
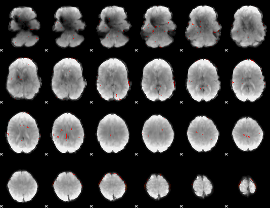 |
Correlation:-0.0052617
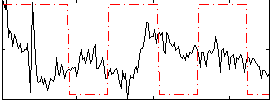 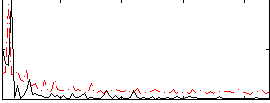 |
Correlation:-0.147
  |
Correlation:0.16304
  |
Correlation:0.16304
  |
Correlation:0.26034
  |
Correlation:0.16304
  |
Correlation:0.098555
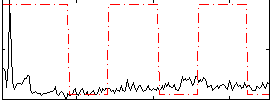 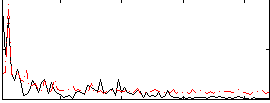 |
Component:393; Jaccard:0.080711
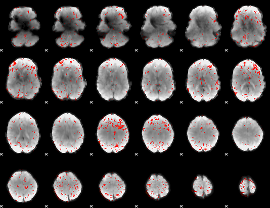 |
Component:162; Jaccard:0.087145
 |
Component:362; Jaccard:0.086809
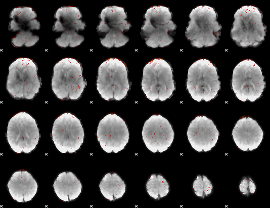 |
Component:083; Jaccard:0.083365
 |
Component:198; Jaccard:0.065311
 |
Component:102; Jaccard:0.036859
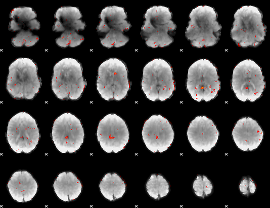 |
Component:102; Jaccard:0.022542
 |
Correlation:-0.043207
  |
Correlation:0.26332
  |
Correlation:0.12651
  |
Correlation:0.13297
  |
Correlation:0.16304
  |
Correlation:0.14748
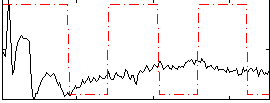  |
Correlation:0.14748
  |
Component:183; Jaccard:0.079202
 |
Component:160; Jaccard:0.083491
 |
Component:083; Jaccard:0.0849
 |
Component:362; Jaccard:0.073852
 |
Component:102; Jaccard:0.05169
 |
Component:317; Jaccard:0.036522
 |
Component:198; Jaccard:0.021357
 |
Correlation:0.17652
  |
Correlation:-0.0052617
  |
Correlation:0.13297
  |
Correlation:0.12651
  |
Correlation:0.14748
  |
Correlation:0.26034
  |
Correlation:0.16304
  |
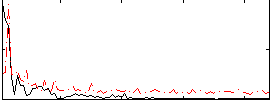
Component:324; Jaccard:0.077885
 |
Component:362; Jaccard:0.082835
 |
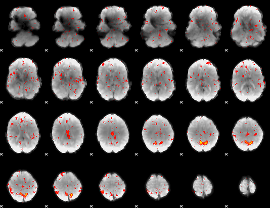
Component:276; Jaccard:0.077841
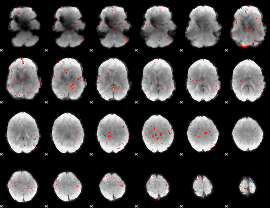 |
Component:276; Jaccard:0.06893
 |
Component:276; Jaccard:0.049561
 |
Component:204; Jaccard:0.033102
 |
Component:095; Jaccard:0.017869
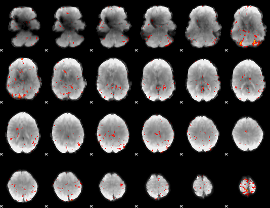 |
Correlation:0.038049
 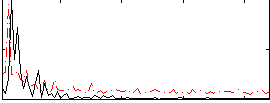 |
Correlation:0.12651
  |
Correlation:-0.013359
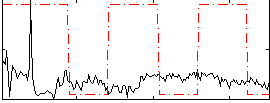 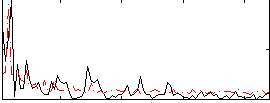 |
Correlation:-0.013359
  |
Correlation:-0.013359
  |
Correlation:0.098555
  |
Correlation:-0.16808
  |
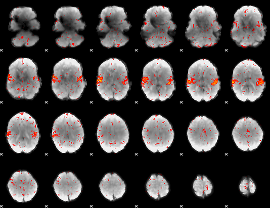
Cope: 05:neg_SHAPES BACK
|
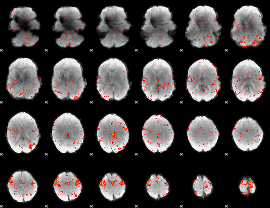
GLM result group-wise
 |
GLM binary result thresholded by 1
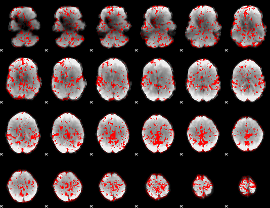 |
GLM binary result thresholded by 1.5
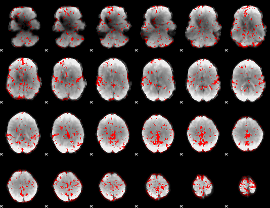 |
GLM binary result thresholded by 2
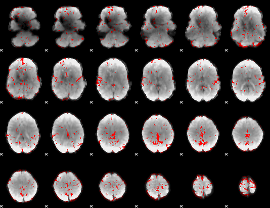 |
GLM binary result thresholded by 2.5
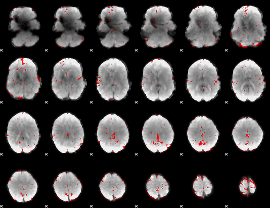 |
GLM binary result thresholded by 3
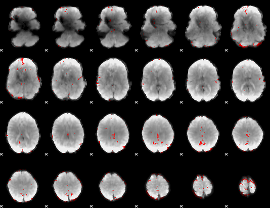 |
GLM binary result thresholded by 3.5
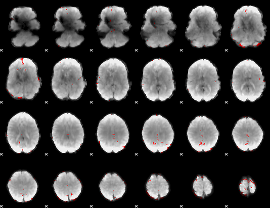 |
GLM binary result thresholded by 4
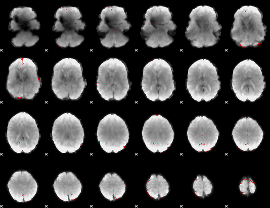 |
GLM Z-score result thresholded by 1
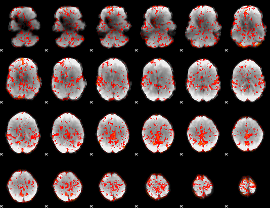 |
GLM Z-score result thresholded by 1.5
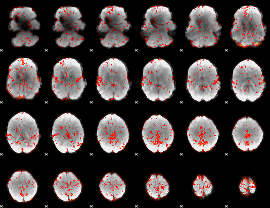 |
GLM Z-score result thresholded by 2
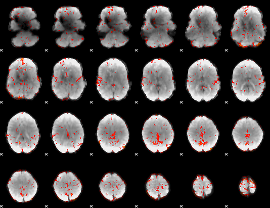 |
GLM Z-score result thresholded by 2.5
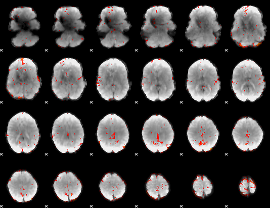 |
GLM Z-score result thresholded by 3
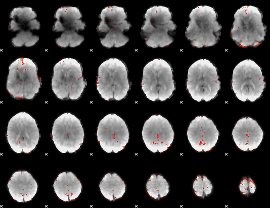 |
GLM Z-score result thresholded by 3.5
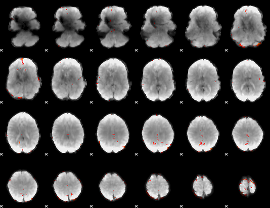 |
GLM Z-score result thresholded by 4
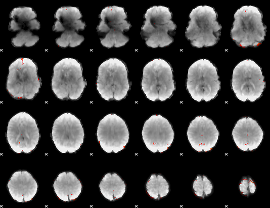 |
Component:115; Jaccard:0.12964
 |
Component:115; Jaccard:0.14966
 |
Component:115; Jaccard:0.15266
 |
Component:314; Jaccard:0.14792
 |
Component:314; Jaccard:0.12699
 |
Component:314; Jaccard:0.089728
 |
Component:314; Jaccard:0.056929
 |
Correlation:0.41871
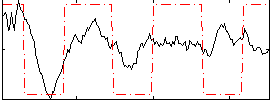 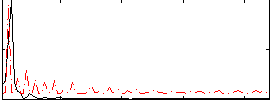 |
Correlation:0.41871
  |
Correlation:0.41871
  |
Correlation:0.25658
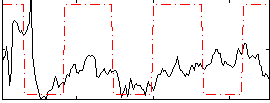  |
Correlation:0.25658
  |
Correlation:0.25658
  |
Correlation:0.25658
  |
Component:314; Jaccard:0.11139
 |
Component:314; Jaccard:0.13326
 |
Component:314; Jaccard:0.15027
 |
Component:229; Jaccard:0.13297
 |
Component:229; Jaccard:0.1062
 |
Component:083; Jaccard:0.074635
 |
Component:083; Jaccard:0.049085
 |
Correlation:0.25658
  |
Correlation:0.25658
  |
Correlation:0.25658
  |
Correlation:0.14973
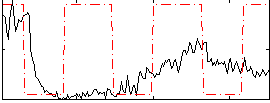 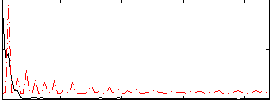 |
Correlation:0.14973
  |
Correlation:-0.027951
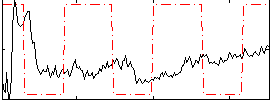 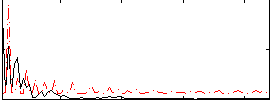 |
Correlation:-0.027951
  |
Component:354; Jaccard:0.095543
 |
Component:229; Jaccard:0.11837
 |
Component:229; Jaccard:0.13517
 |
Component:115; Jaccard:0.13142
 |
Component:115; Jaccard:0.097927
 |
Component:229; Jaccard:0.06754
 |
Component:095; Jaccard:0.041903
 |
Correlation:0.11027
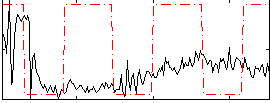 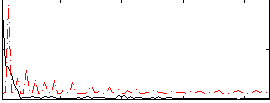 |
Correlation:0.14973
  |
Correlation:0.14973
  |
Correlation:0.41871
  |
Correlation:0.41871
  |
Correlation:0.14973
  |
Correlation:0.26591
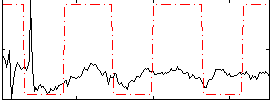 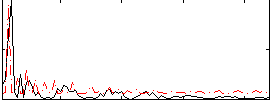 |
Component:229; Jaccard:0.094749
 |
Component:354; Jaccard:0.11566
 |
Component:354; Jaccard:0.12849
 |
Component:354; Jaccard:0.12427
 |
Component:354; Jaccard:0.094812
 |
Component:354; Jaccard:0.062275
 |
Component:162; Jaccard:0.040754
 |
Correlation:0.14973
  |
Correlation:0.11027
  |
Correlation:0.11027
  |
Correlation:0.11027
  |
Correlation:0.11027
  |
Correlation:0.11027
  |
Correlation:-0.017579
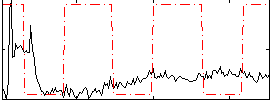  |
Component:311; Jaccard:0.091216
 |
Component:311; Jaccard:0.09681
 |
Component:276; Jaccard:0.097939
 |
Component:276; Jaccard:0.096614
 |
Component:083; Jaccard:0.091297
 |
Component:095; Jaccard:0.060606
 |
Component:229; Jaccard:0.038593
 |
Correlation:0.31204
  |
Correlation:0.31204
  |
Correlation:0.19717
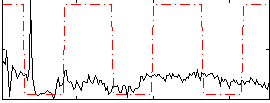 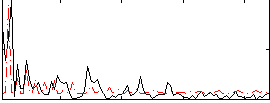 |
Correlation:0.19717
  |
Correlation:-0.027951
  |
Correlation:0.26591
  |
Correlation:0.14973
  |
Component:160; Jaccard:0.080469
 |
Component:317; Jaccard:0.087265
 |
Component:311; Jaccard:0.094775
 |
Component:083; Jaccard:0.093632
 |
Component:095; Jaccard:0.08394
 |
Component:162; Jaccard:0.059494
 |
Component:183; Jaccard:0.038017
 |
Correlation:0.24637
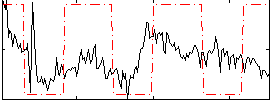 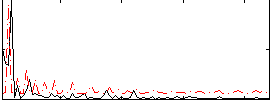 |
Correlation:0.005583
  |
Correlation:0.31204
  |
Correlation:-0.027951
  |
Correlation:0.26591
  |
Correlation:-0.017579
  |
Correlation:-0.10401
  |
Component:393; Jaccard:0.078737
 |
Component:198; Jaccard:0.086156
 |
Component:095; Jaccard:0.093979
 |
Component:198; Jaccard:0.09213
 |
Component:276; Jaccard:0.083811
 |
Component:115; Jaccard:0.058417
 |
Component:354; Jaccard:0.034817
 |
Correlation:0.099956
  |
Correlation:0.14238
  |
Correlation:0.26591
  |
Correlation:0.14238
  |
Correlation:0.19717
  |
Correlation:0.41871
  |
Correlation:0.11027
  |
Component:374; Jaccard:0.078714
 |
Component:160; Jaccard:0.085273
 |
Component:198; Jaccard:0.093481
 |
Component:095; Jaccard:0.091294
 |
Component:198; Jaccard:0.076667
 |
Component:183; Jaccard:0.057836
 |
Component:198; Jaccard:0.034737
 |
Correlation:0.28293
  |
Correlation:0.24637
  |
Correlation:0.14238
  |
Correlation:0.26591
  |
Correlation:0.14238
  |
Correlation:-0.10401
  |
Correlation:0.14238
  |
Component:317; Jaccard:0.077752
 |
Component:095; Jaccard:0.085218
 |
Component:317; Jaccard:0.091602
 |
Component:183; Jaccard:0.086475
 |
Component:183; Jaccard:0.076119
 |
Component:276; Jaccard:0.056344
 |
Component:276; Jaccard:0.03421
 |
Correlation:0.005583
  |
Correlation:0.26591
  |
Correlation:0.005583
  |
Correlation:-0.10401
  |
Correlation:-0.10401
  |
Correlation:0.19717
  |
Correlation:0.19717
  |
Component:329; Jaccard:0.076624
 |
Component:276; Jaccard:0.085049
 |
Component:329; Jaccard:0.08626
 |
Component:317; Jaccard:0.086068
 |
Component:317; Jaccard:0.070544
 |
Component:198; Jaccard:0.055455
 |
Component:115; Jaccard:0.031378
 |
Correlation:0.29858
  |
Correlation:0.19717
  |
Correlation:0.29858
  |
Correlation:0.005583
  |
Correlation:0.005583
  |
Correlation:0.14238
  |
Correlation:0.41871
  |
Cope: 06:SHAPES-FACES BACK
|
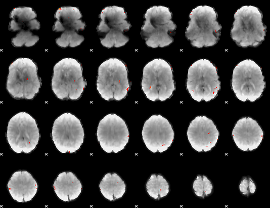
GLM result group-wise
 |
GLM binary result thresholded by 1
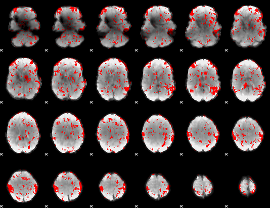 |
GLM binary result thresholded by 1.5
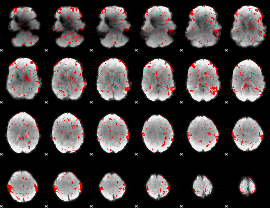 |
GLM binary result thresholded by 2
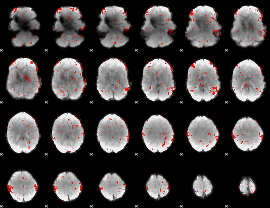 |
GLM binary result thresholded by 2.5
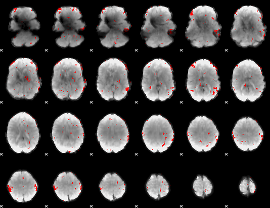 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
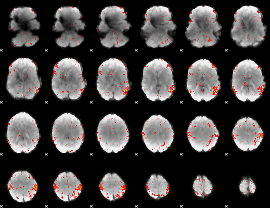
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
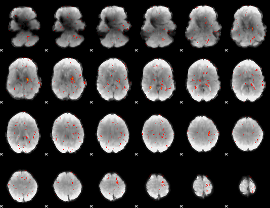
Component:108; Jaccard:0.12931
 |
Component:108; Jaccard:0.13914
 |
Component:108; Jaccard:0.13315
 |
Component:108; Jaccard:0.10719
 |
Component:108; Jaccard:0.069939
 |
Component:257; Jaccard:0.042402
 |
Component:257; Jaccard:0.026067
 |
Correlation:0.13229
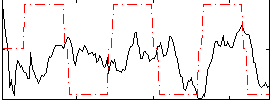 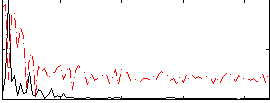 |
Correlation:0.13229
  |
Correlation:0.13229
  |
Correlation:0.13229
  |
Correlation:0.13229
  |
Correlation:0.18459
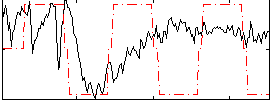 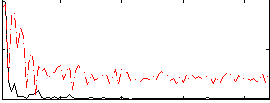 |
Correlation:0.18459
  |
Component:249; Jaccard:0.085981
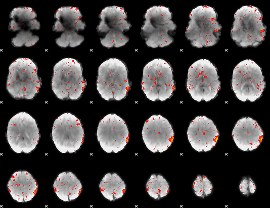 |
Component:249; Jaccard:0.087666
 |
Component:257; Jaccard:0.085712
 |
Component:257; Jaccard:0.076836
 |
Component:076; Jaccard:0.061097
 |
Component:108; Jaccard:0.041132
 |
Component:076; Jaccard:0.023598
 |
Correlation:0.25962
  |
Correlation:0.25962
  |
Correlation:0.18459
  |
Correlation:0.18459
  |
Correlation:0.36732
  |
Correlation:0.13229
  |
Correlation:0.36732
  |
Component:343; Jaccard:0.085972
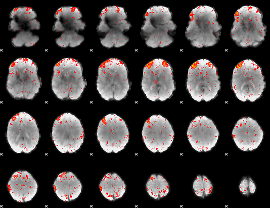 |
Component:048; Jaccard:0.082216
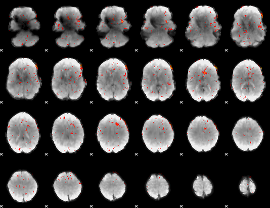 |
Component:048; Jaccard:0.083921
 |
Component:048; Jaccard:0.07609
 |
Component:048; Jaccard:0.057683
 |
Component:076; Jaccard:0.040972
 |
Component:048; Jaccard:0.02237
 |
Correlation:0.10141
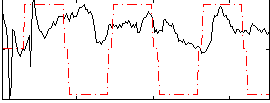 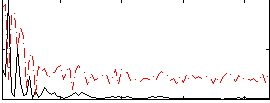 |
Correlation:0.090378
  |
Correlation:0.090378
  |
Correlation:0.090378
  |
Correlation:0.090378
  |
Correlation:0.36732
  |
Correlation:0.090378
  |
Component:222; Jaccard:0.081026
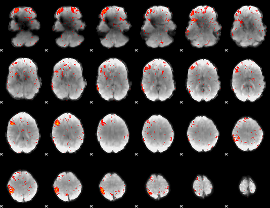 |
Component:343; Jaccard:0.081186
 |
Component:249; Jaccard:0.080728
 |
Component:076; Jaccard:0.066622
 |
Component:257; Jaccard:0.057281
 |
Component:348; Jaccard:0.038626
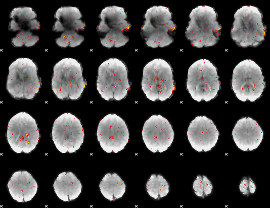 |
Component:348; Jaccard:0.020675
 |
Correlation:0.16105
  |
Correlation:0.10141
  |
Correlation:0.25962
  |
Correlation:0.36732
  |
Correlation:0.18459
  |
Correlation:0.35219
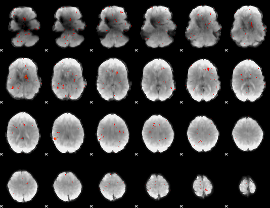
  |
Correlation:0.35219
  |
Component:284; Jaccard:0.079316
 |
Component:222; Jaccard:0.079798
 |
Component:222; Jaccard:0.071642
 |
Component:249; Jaccard:0.063844
 |
Component:348; Jaccard:0.05428
 |
Component:048; Jaccard:0.035942
 |
Component:108; Jaccard:0.020593
 |
Correlation:-0.026669
  |
Correlation:0.16105
  |
Correlation:0.16105
  |
Correlation:0.25962
  |
Correlation:0.35219
  |
Correlation:0.090378
  |
Correlation:0.13229
  |
Component:318; Jaccard:0.07764
 |
Component:257; Jaccard:0.079559
 |
Component:232; Jaccard:0.070658
 |
Component:348; Jaccard:0.063531
 |
Component:249; Jaccard:0.044565
 |
Component:232; Jaccard:0.02827
 |
Component:270; Jaccard:0.018046
 |
Correlation:0.1894
  |
Correlation:0.18459
  |
Correlation:0.40784
  |
Correlation:0.35219
  |
Correlation:0.25962
  |
Correlation:0.40784
  |
Correlation:0.3755
 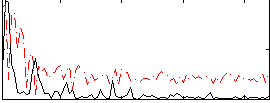 |
Component:048; Jaccard:0.073568
 |
Component:284; Jaccard:0.076806
 |
Component:261; Jaccard:0.070573
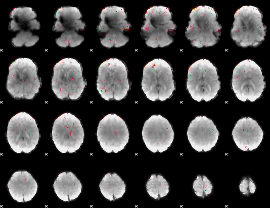 |
Component:232; Jaccard:0.061876
 |
Component:261; Jaccard:0.04427
 |
Component:249; Jaccard:0.027387
 |
Component:185; Jaccard:0.016194
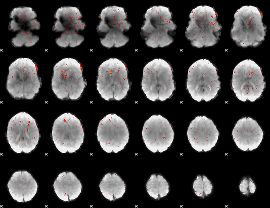 |
Correlation:0.090378
  |
Correlation:-0.026669
  |
Correlation:0.20963
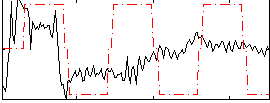  |
Correlation:0.40784
  |
Correlation:0.20963
  |
Correlation:0.25962
  |
Correlation:0.093056
  |
Component:257; Jaccard:0.071139
 |
Component:232; Jaccard:0.073145
 |
Component:343; Jaccard:0.070206
 |
Component:261; Jaccard:0.06065
 |
Component:232; Jaccard:0.043129
 |
Component:261; Jaccard:0.025068
 |
Component:232; Jaccard:0.016061
 |
Correlation:0.18459
  |
Correlation:0.40784
  |
Correlation:0.10141
  |
Correlation:0.20963
  |
Correlation:0.40784
  |
Correlation:0.20963
  |
Correlation:0.40784
  |
Component:194; Jaccard:0.069895
 |
Component:318; Jaccard:0.073018
 |
Component:348; Jaccard:0.068064
 |
Component:222; Jaccard:0.05516
 |
Component:222; Jaccard:0.035479
 |
Component:185; Jaccard:0.024579
 |
Component:249; Jaccard:0.015145
 |
Correlation:-0.087575
  |
Correlation:0.1894
  |
Correlation:0.35219
  |
Correlation:0.16105
  |
Correlation:0.16105
  |
Correlation:0.093056
  |
Correlation:0.25962
  |
Component:232; Jaccard:0.069315
 |
Component:261; Jaccard:0.067231
 |
Component:284; Jaccard:0.065621
 |
Component:343; Jaccard:0.054517
 |
Component:185; Jaccard:0.033717
 |
Component:270; Jaccard:0.024503
 |
Component:254; Jaccard:0.011489
 |
Correlation:0.40784
  |
Correlation:0.20963
  |
Correlation:-0.026669
  |
Correlation:0.10141
  |
Correlation:0.093056
  |
Correlation:0.3755
  |
Correlation:0.19356
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































