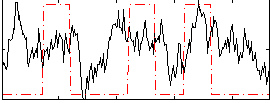
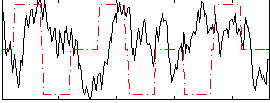
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
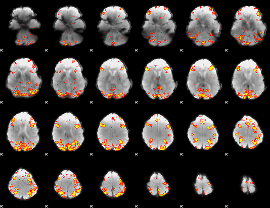
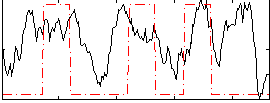
GLM result group-wise
 |
GLM binary result thresholded by 1
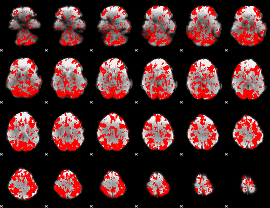 |
GLM binary result thresholded by 1.5
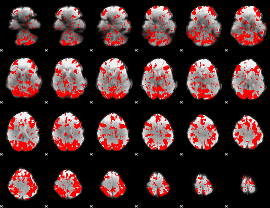 |
GLM binary result thresholded by 2
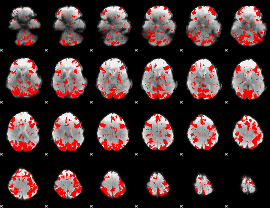 |
GLM binary result thresholded by 2.5
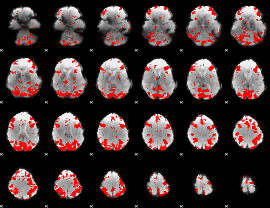 |
GLM binary result thresholded by 3
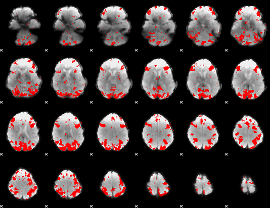 |
GLM binary result thresholded by 3.5
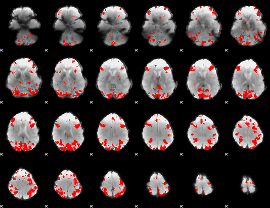 |
GLM binary result thresholded by 4
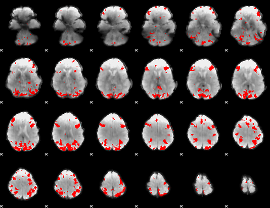 |
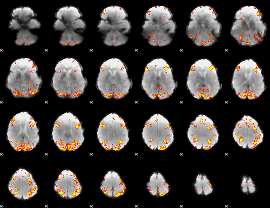
GLM Z-score result thresholded by 1
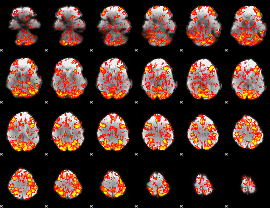 |
GLM Z-score result thresholded by 1.5
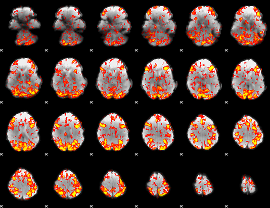 |
GLM Z-score result thresholded by 2
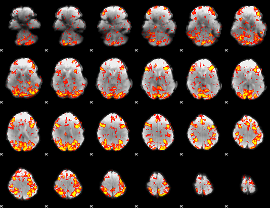 |
GLM Z-score result thresholded by 2.5
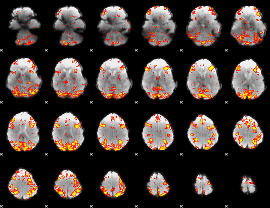 |
GLM Z-score result thresholded by 3
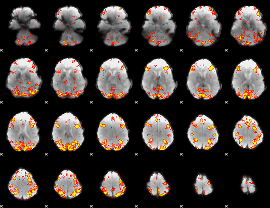 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
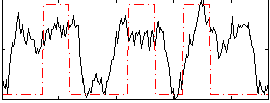
Component:114; Jaccard:0.13001
 |
Component:114; Jaccard:0.16908
 |
Component:114; Jaccard:0.21372
 |
Component:114; Jaccard:0.26251
 |
Component:114; Jaccard:0.30323
 |
Component:114; Jaccard:0.33193
 |
Component:114; Jaccard:0.34503
 |
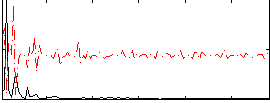
Correlation:0.394
 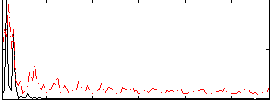 |
Correlation:0.394
  |
Correlation:0.394
  |
Correlation:0.394
  |
Correlation:0.394
  |
Correlation:0.394
  |
Correlation:0.394
  |
Component:187; Jaccard:0.12965
 |
Component:187; Jaccard:0.15787
 |
Component:186; Jaccard:0.19241
 |
Component:186; Jaccard:0.23093
 |
Component:186; Jaccard:0.26464
 |
Component:186; Jaccard:0.28843
 |
Component:186; Jaccard:0.30058
 |
Correlation:0.22348
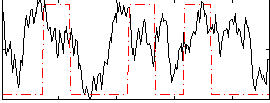 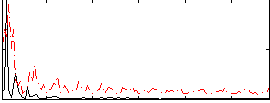 |
Correlation:0.22348
  |
Correlation:0.28191
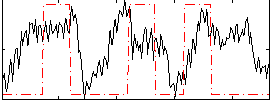 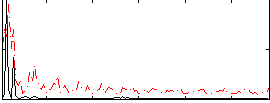 |
Correlation:0.28191
  |
Correlation:0.28191
  |
Correlation:0.28191
  |
Correlation:0.28191
  |
Component:348; Jaccard:0.12438
 |
Component:348; Jaccard:0.15604
 |
Component:348; Jaccard:0.18846
 |
Component:348; Jaccard:0.21914
 |
Component:348; Jaccard:0.24009
 |
Component:348; Jaccard:0.24979
 |
Component:348; Jaccard:0.24789
 |
Correlation:0.30568
 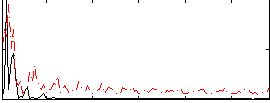 |
Correlation:0.30568
  |
Correlation:0.30568
  |
Correlation:0.30568
  |
Correlation:0.30568
  |
Correlation:0.30568
  |
Correlation:0.30568
  |
Component:231; Jaccard:0.12379
 |
Component:186; Jaccard:0.15459
 |
Component:187; Jaccard:0.18731
 |
Component:187; Jaccard:0.21428
 |
Component:231; Jaccard:0.23206
 |
Component:231; Jaccard:0.24271
 |
Component:187; Jaccard:0.24133
 |
Correlation:0.21848
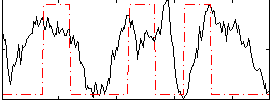 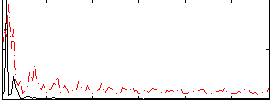 |
Correlation:0.28191
  |
Correlation:0.22348
  |
Correlation:0.22348
  |
Correlation:0.21848
  |
Correlation:0.21848
  |
Correlation:0.22348
  |
Component:186; Jaccard:0.12032
 |
Component:231; Jaccard:0.15242
 |
Component:231; Jaccard:0.18323
 |
Component:231; Jaccard:0.21144
 |
Component:187; Jaccard:0.23204
 |
Component:187; Jaccard:0.24233
 |
Component:231; Jaccard:0.24128
 |
Correlation:0.28191
  |
Correlation:0.21848
  |
Correlation:0.21848
  |
Correlation:0.21848
  |
Correlation:0.22348
  |
Correlation:0.22348
  |
Correlation:0.21848
  |
Component:346; Jaccard:0.11999
 |
Component:050; Jaccard:0.1395
 |
Component:050; Jaccard:0.16377
 |
Component:050; Jaccard:0.18499
 |
Component:050; Jaccard:0.1999
 |
Component:050; Jaccard:0.20659
 |
Component:050; Jaccard:0.20696
 |
Correlation:0.25829
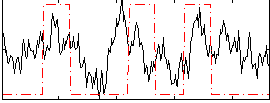 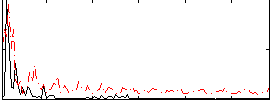 |
Correlation:0.22013
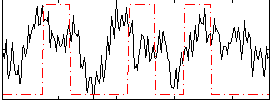 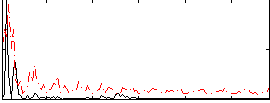 |
Correlation:0.22013
  |
Correlation:0.22013
  |
Correlation:0.22013
  |
Correlation:0.22013
  |
Correlation:0.22013
  |
Component:050; Jaccard:0.11523
 |
Component:346; Jaccard:0.13829
 |
Component:346; Jaccard:0.15394
 |
Component:346; Jaccard:0.16437
 |
Component:397; Jaccard:0.17247
 |
Component:397; Jaccard:0.17588
 |
Component:397; Jaccard:0.17311
 |
Correlation:0.22013
  |
Correlation:0.25829
  |
Correlation:0.25829
  |
Correlation:0.25829
  |
Correlation:0.28509
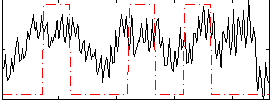 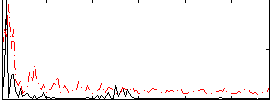 |
Correlation:0.28509
  |
Correlation:0.28509
  |
Component:218; Jaccard:0.10568
 |
Component:249; Jaccard:0.12563
 |
Component:249; Jaccard:0.14614
 |
Component:249; Jaccard:0.16208
 |
Component:249; Jaccard:0.17114
 |
Component:249; Jaccard:0.17312
 |
Component:249; Jaccard:0.16953
 |
Correlation:0.25671
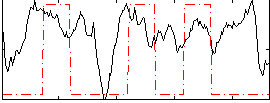 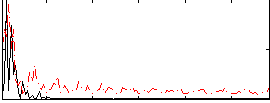 |
Correlation:0.27024
 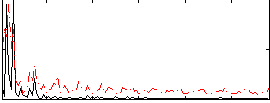 |
Correlation:0.27024
  |
Correlation:0.27024
  |
Correlation:0.27024
  |
Correlation:0.27024
  |
Correlation:0.27024
  |
Component:249; Jaccard:0.10475
 |
Component:397; Jaccard:0.12374
 |
Component:397; Jaccard:0.14417
 |
Component:397; Jaccard:0.16098
 |
Component:346; Jaccard:0.16667
 |
Component:346; Jaccard:0.16293
 |
Component:346; Jaccard:0.15384
 |
Correlation:0.27024
  |
Correlation:0.28509
  |
Correlation:0.28509
  |
Correlation:0.28509
  |
Correlation:0.25829
  |
Correlation:0.25829
  |
Correlation:0.25829
  |
Component:397; Jaccard:0.10186
 |
Component:218; Jaccard:0.11739
 |
Component:218; Jaccard:0.12634
 |
Component:354; Jaccard:0.13369
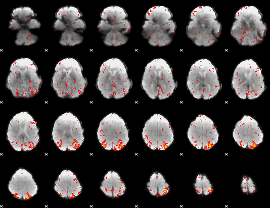 |
Component:354; Jaccard:0.13814
 |
Component:354; Jaccard:0.13899
 |
Component:354; Jaccard:0.13535
 |
Correlation:0.28509
  |
Correlation:0.25671
  |
Correlation:0.25671
  |
Correlation:0.30601
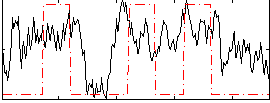 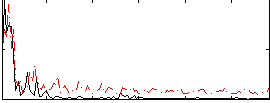 |
Correlation:0.30601
  |
Correlation:0.30601
  |
Correlation:0.30601
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
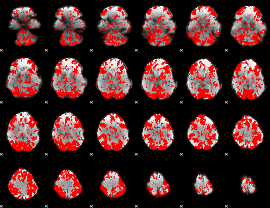 |
GLM binary result thresholded by 1.5
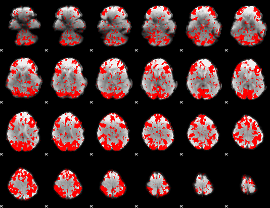 |
GLM binary result thresholded by 2
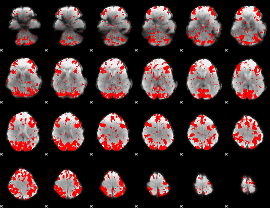 |
GLM binary result thresholded by 2.5
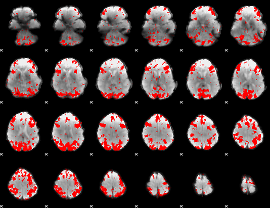 |
GLM binary result thresholded by 3
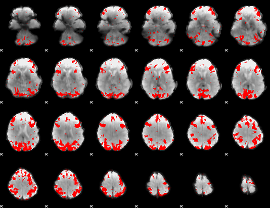 |
GLM binary result thresholded by 3.5
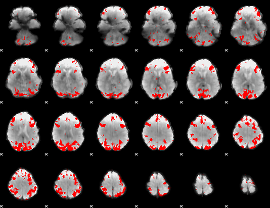 |
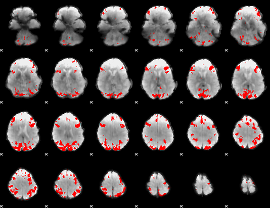
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
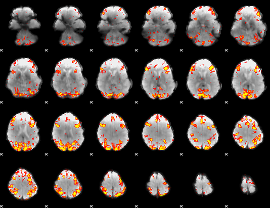 |
GLM Z-score result thresholded by 3.5
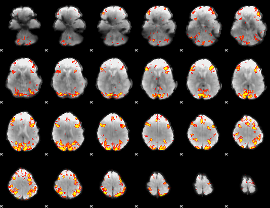 |
GLM Z-score result thresholded by 4
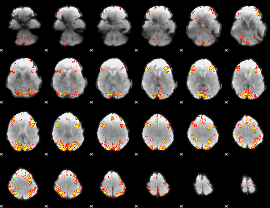 |
Component:187; Jaccard:0.13169
 |
Component:187; Jaccard:0.16408
 |
Component:348; Jaccard:0.20109
 |
Component:348; Jaccard:0.2426
 |
Component:348; Jaccard:0.2754
 |
Component:348; Jaccard:0.29178
 |
Component:348; Jaccard:0.29428
 |
Correlation:0.20795
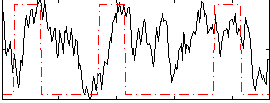  |
Correlation:0.20795
  |
Correlation:0.081067
  |
Correlation:0.081067
  |
Correlation:0.081067
  |
Correlation:0.081067
  |
Correlation:0.081067
  |
Component:348; Jaccard:0.12325
 |
Component:348; Jaccard:0.15913
 |
Component:187; Jaccard:0.19905
 |
Component:187; Jaccard:0.23278
 |
Component:187; Jaccard:0.2573
 |
Component:186; Jaccard:0.2707
 |
Component:186; Jaccard:0.28093
 |
Correlation:0.081067
  |
Correlation:0.081067
  |
Correlation:0.20795
  |
Correlation:0.20795
  |
Correlation:0.20795
  |
Correlation:0.20949
  |
Correlation:0.20949
  |
Component:346; Jaccard:0.12107
 |
Component:114; Jaccard:0.15282
 |
Component:114; Jaccard:0.1887
 |
Component:114; Jaccard:0.22357
 |
Component:186; Jaccard:0.25232
 |
Component:187; Jaccard:0.26819
 |
Component:187; Jaccard:0.27119
 |
Correlation:0.13048
  |
Correlation:0.052084
  |
Correlation:0.052084
  |
Correlation:0.052084
  |
Correlation:0.20949
  |
Correlation:0.20795
  |
Correlation:0.20795
  |
Component:114; Jaccard:0.12049
 |
Component:231; Jaccard:0.14951
 |
Component:186; Jaccard:0.18533
 |
Component:186; Jaccard:0.22179
 |
Component:114; Jaccard:0.24812
 |
Component:114; Jaccard:0.26108
 |
Component:114; Jaccard:0.26425
 |
Correlation:0.052084
  |
Correlation:0.085033
  |
Correlation:0.20949
  |
Correlation:0.20949
  |
Correlation:0.052084
  |
Correlation:0.052084
  |
Correlation:0.052084
  |
Component:231; Jaccard:0.12041
 |
Component:050; Jaccard:0.14885
 |
Component:231; Jaccard:0.18067
 |
Component:231; Jaccard:0.20744
 |
Component:231; Jaccard:0.22715
 |
Component:231; Jaccard:0.23574
 |
Component:050; Jaccard:0.23342
 |
Correlation:0.085033
  |
Correlation:0.15636
 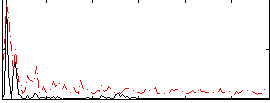 |
Correlation:0.085033
  |
Correlation:0.085033
  |
Correlation:0.085033
  |
Correlation:0.085033
  |
Correlation:0.15636
  |
Component:050; Jaccard:0.11951
 |
Component:186; Jaccard:0.14799
 |
Component:050; Jaccard:0.17979
 |
Component:050; Jaccard:0.20646
 |
Component:050; Jaccard:0.22564
 |
Component:050; Jaccard:0.23487
 |
Component:231; Jaccard:0.2331
 |
Correlation:0.15636
  |
Correlation:0.20949
  |
Correlation:0.15636
  |
Correlation:0.15636
  |
Correlation:0.15636
  |
Correlation:0.15636
  |
Correlation:0.085033
  |
Component:218; Jaccard:0.11803
 |
Component:218; Jaccard:0.13953
 |
Component:218; Jaccard:0.16004
 |
Component:218; Jaccard:0.17171
 |
Component:397; Jaccard:0.17753
 |
Component:397; Jaccard:0.18187
 |
Component:397; Jaccard:0.17686
 |
Correlation:-0.1547
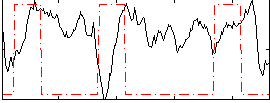 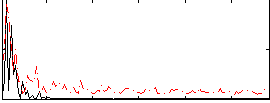 |
Correlation:-0.1547
  |
Correlation:-0.1547
  |
Correlation:-0.1547
  |
Correlation:-0.0092252
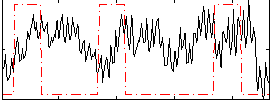 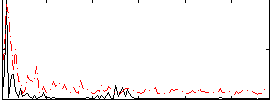 |
Correlation:-0.0092252
  |
Correlation:-0.0092252
  |
Component:186; Jaccard:0.11544
 |
Component:346; Jaccard:0.1395
 |
Component:346; Jaccard:0.15655
 |
Component:346; Jaccard:0.16681
 |
Component:218; Jaccard:0.1742
 |
Component:346; Jaccard:0.16851
 |
Component:346; Jaccard:0.15566
 |
Correlation:0.20949
  |
Correlation:0.13048
  |
Correlation:0.13048
  |
Correlation:0.13048
  |
Correlation:-0.1547
  |
Correlation:0.13048
  |
Correlation:0.13048
  |
Component:397; Jaccard:0.099303
 |
Component:397; Jaccard:0.12235
 |
Component:397; Jaccard:0.14572
 |
Component:397; Jaccard:0.1647
 |
Component:346; Jaccard:0.17042
 |
Component:218; Jaccard:0.16482
 |
Component:218; Jaccard:0.14829
 |
Correlation:-0.0092252
  |
Correlation:-0.0092252
  |
Correlation:-0.0092252
  |
Correlation:-0.0092252
  |
Correlation:0.13048
  |
Correlation:-0.1547
  |
Correlation:-0.1547
  |
Component:206; Jaccard:0.092763
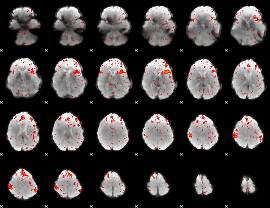 |
Component:249; Jaccard:0.10681
 |
Component:249; Jaccard:0.1202
 |
Component:249; Jaccard:0.13042
 |
Component:249; Jaccard:0.13579
 |
Component:249; Jaccard:0.13552
 |
Component:249; Jaccard:0.12951
 |
Correlation:0.17667
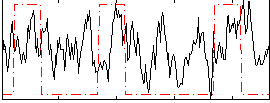 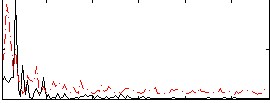 |
Correlation:0.21097
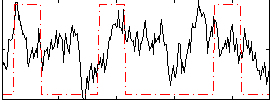 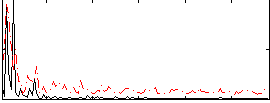 |
Correlation:0.21097
  |
Correlation:0.21097
  |
Correlation:0.21097
  |
Correlation:0.21097
  |
Correlation:0.21097
  |
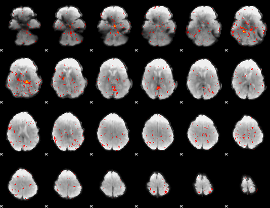
Cope: 03:MATCH-REL BACK
|
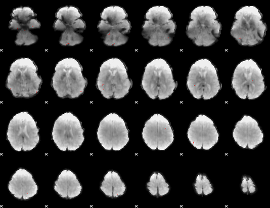
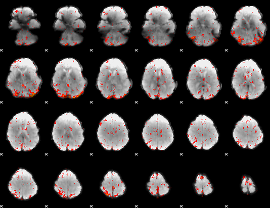
GLM result group-wise
 |
GLM binary result thresholded by 1
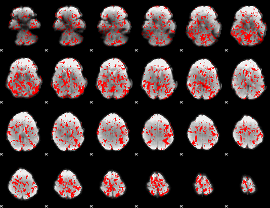 |
GLM binary result thresholded by 1.5
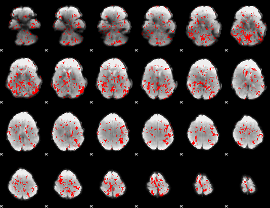 |
GLM binary result thresholded by 2
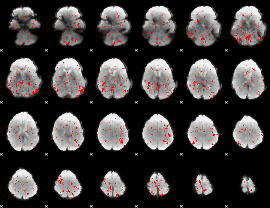 |
GLM binary result thresholded by 2.5
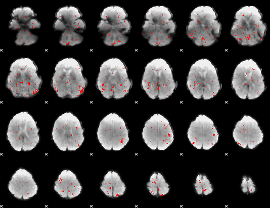 |
GLM binary result thresholded by 3
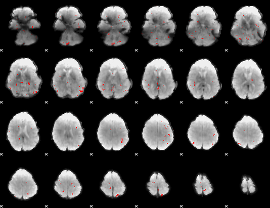 |
GLM binary result thresholded by 3.5
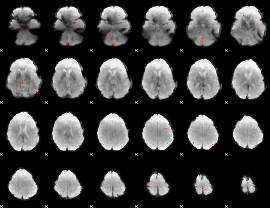 |
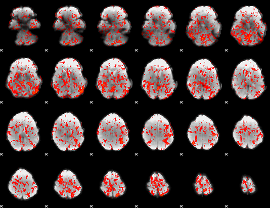
GLM binary result thresholded by 4
 |
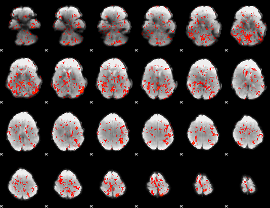
GLM Z-score result thresholded by 1
 |
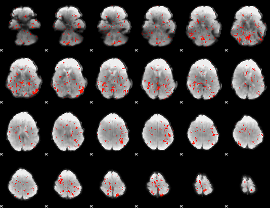
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
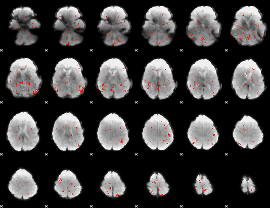 |
GLM Z-score result thresholded by 3
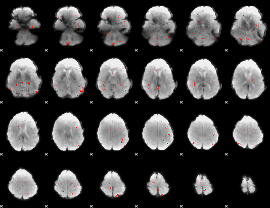 |
GLM Z-score result thresholded by 3.5
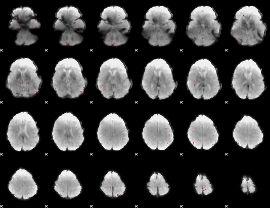 |
GLM Z-score result thresholded by 4
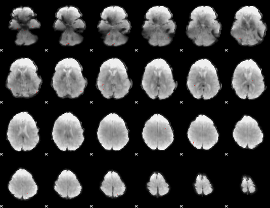 |
Component:161; Jaccard:0.09217
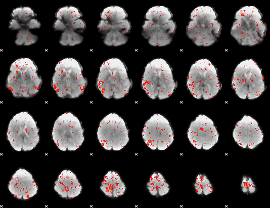 |
Component:053; Jaccard:0.096334
 |
Component:053; Jaccard:0.094228
 |
Component:053; Jaccard:0.070821
 |
Component:053; Jaccard:0.036666
 |
Component:053; Jaccard:0.014857
 |
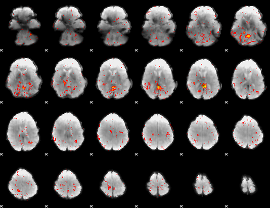
Component:393; Jaccard:0.004722
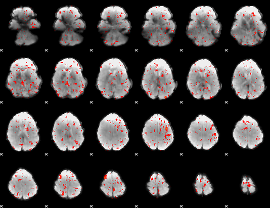 |
Correlation:0.17972
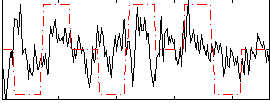 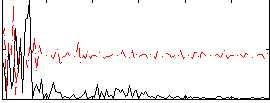 |
Correlation:0.21676
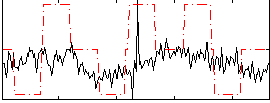 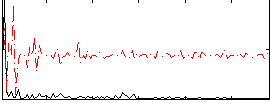 |
Correlation:0.21676
  |
Correlation:0.21676
  |
Correlation:0.21676
  |
Correlation:0.21676
  |
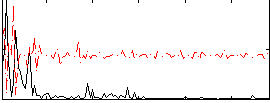
Correlation:0.18878
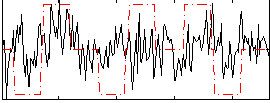 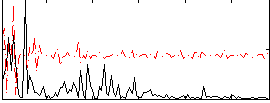 |
Component:033; Jaccard:0.090718
 |
Component:161; Jaccard:0.094662
 |
Component:393; Jaccard:0.077265
 |
Component:114; Jaccard:0.051793
 |
Component:393; Jaccard:0.027996
 |
Component:393; Jaccard:0.011594
 |
Component:053; Jaccard:0.004405
 |
Correlation:0.22384
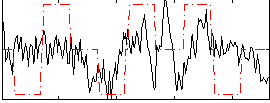  |
Correlation:0.17972
  |
Correlation:0.18878
  |
Correlation:0.20265
  |
Correlation:0.18878
  |
Correlation:0.18878
  |
Correlation:0.21676
  |
Component:393; Jaccard:0.088999
 |
Component:393; Jaccard:0.090194
 |
Component:161; Jaccard:0.076409
 |
Component:393; Jaccard:0.051612
 |
Component:114; Jaccard:0.027427
 |
Component:114; Jaccard:0.010743
 |
Component:332; Jaccard:0.003991
 |
Correlation:0.18878
  |
Correlation:0.18878
  |
Correlation:0.17972
  |
Correlation:0.18878
  |
Correlation:0.20265
  |
Correlation:0.20265
  |
Correlation:0.10071
 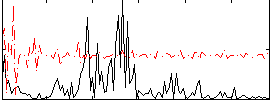 |
Component:114; Jaccard:0.08748
 |
Component:114; Jaccard:0.087994
 |
Component:114; Jaccard:0.075764
 |
Component:161; Jaccard:0.048624
 |
Component:185; Jaccard:0.024568
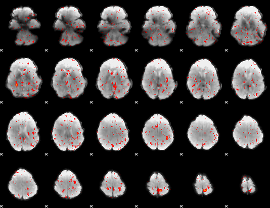 |
Component:204; Jaccard:0.009829
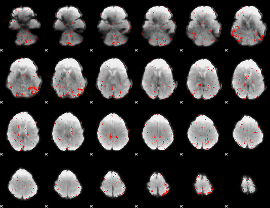 |
Component:204; Jaccard:0.003913
 |
Correlation:0.20265
  |
Correlation:0.20265
  |
Correlation:0.20265
  |
Correlation:0.17972
  |
Correlation:0.20514
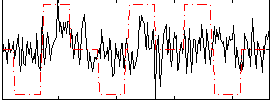 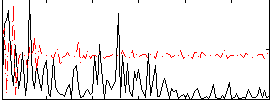 |
Correlation:0.33042
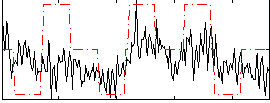  |
Correlation:0.33042
  |
Component:053; Jaccard:0.081628
 |
Component:033; Jaccard:0.086392
 |
Component:185; Jaccard:0.067604
 |
Component:185; Jaccard:0.047937
 |
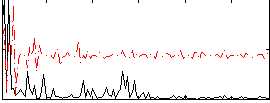
Component:305; Jaccard:0.02374
 |
Component:185; Jaccard:0.00981
 |
Component:198; Jaccard:0.003692
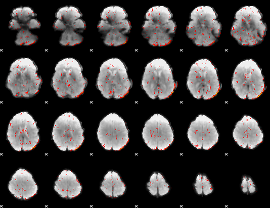 |
Correlation:0.21676
  |
Correlation:0.22384
  |
Correlation:0.20514
  |
Correlation:0.20514
  |
Correlation:0.1347
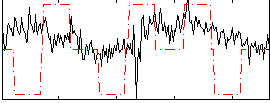 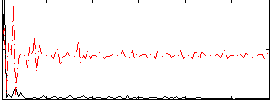 |
Correlation:0.20514
  |
Correlation:0.10321
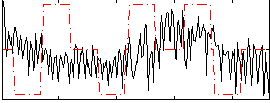 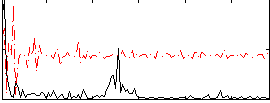 |
Component:185; Jaccard:0.076501
 |
Component:185; Jaccard:0.077397
 |
Component:033; Jaccard:0.066182
 |
Component:204; Jaccard:0.046878
 |
Component:204; Jaccard:0.022929
 |
Component:305; Jaccard:0.009709
 |
Component:114; Jaccard:0.00344
 |
Correlation:0.20514
  |
Correlation:0.20514
  |
Correlation:0.22384
  |
Correlation:0.33042
  |
Correlation:0.33042
  |
Correlation:0.1347
  |
Correlation:0.20265
  |
Component:128; Jaccard:0.07067
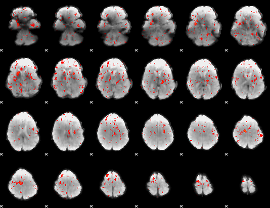 |
Component:128; Jaccard:0.070576
 |
Component:204; Jaccard:0.063621
 |
Component:305; Jaccard:0.042094
 |
Component:161; Jaccard:0.022261
 |
Component:332; Jaccard:0.009692
 |
Component:060; Jaccard:0.00334
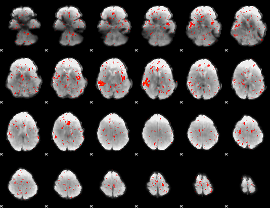 |
Correlation:0.15373
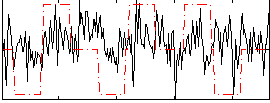 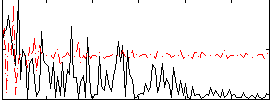 |
Correlation:0.15373
  |
Correlation:0.33042
  |
Correlation:0.1347
  |
Correlation:0.17972
  |
Correlation:0.10071
  |
Correlation:-0.0064248
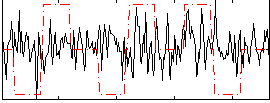 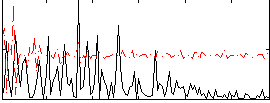 |
Component:204; Jaccard:0.067732
 |
Component:204; Jaccard:0.070462
 |
Component:128; Jaccard:0.059462
 |
Component:033; Jaccard:0.040978
 |
Component:332; Jaccard:0.020004
 |
Component:161; Jaccard:0.008464
 |
Component:185; Jaccard:0.003313
 |
Correlation:0.33042
  |
Correlation:0.33042
  |
Correlation:0.15373
  |
Correlation:0.22384
  |
Correlation:0.10071
  |
Correlation:0.17972
  |
Correlation:0.20514
  |
Component:298; Jaccard:0.067379
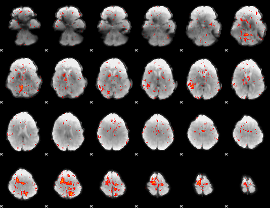 |
Component:298; Jaccard:0.064274
 |
Component:305; Jaccard:0.058891
 |
Component:128; Jaccard:0.040913
 |
Component:128; Jaccard:0.019521
 |
Component:033; Jaccard:0.007789
 |
Component:305; Jaccard:0.003248
 |
Correlation:0.13435
  |
Correlation:0.13435
  |
Correlation:0.1347
  |
Correlation:0.15373
  |
Correlation:0.15373
  |
Correlation:0.22384
  |
Correlation:0.1347
  |
Component:249; Jaccard:0.066247
 |
Component:173; Jaccard:0.064146
 |
Component:051; Jaccard:0.052681
 |
Component:051; Jaccard:0.034039
 |
Component:033; Jaccard:0.0193
 |
Component:379; Jaccard:0.007756
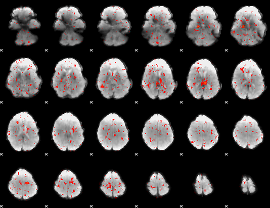 |
Component:033; Jaccard:0.003123
 |
Correlation:0.035131
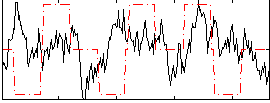 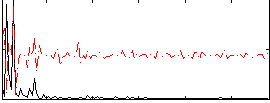 |
Correlation:0.16873
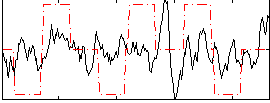  |
Correlation:0.21163
  |
Correlation:0.21163
  |
Correlation:0.22384
  |
Correlation:0.043408
 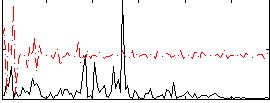 |
Correlation:0.22384
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
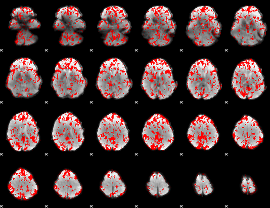 |
GLM binary result thresholded by 1.5
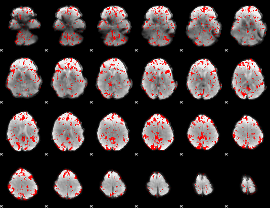 |
GLM binary result thresholded by 2
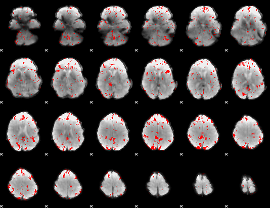 |
GLM binary result thresholded by 2.5
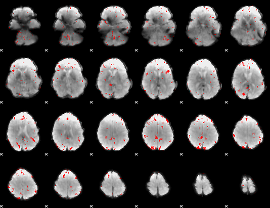 |
GLM binary result thresholded by 3
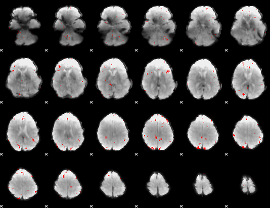 |
GLM binary result thresholded by 3.5
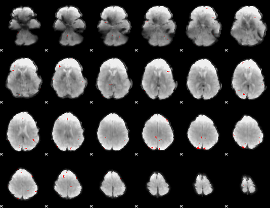 |
GLM binary result thresholded by 4
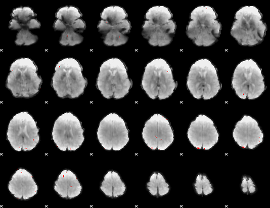 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
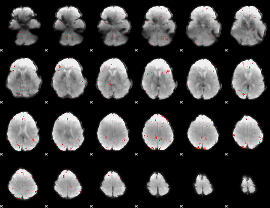 |
GLM Z-score result thresholded by 3.5
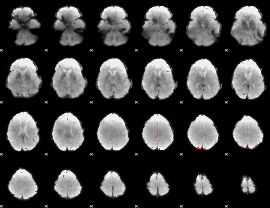 |
GLM Z-score result thresholded by 4
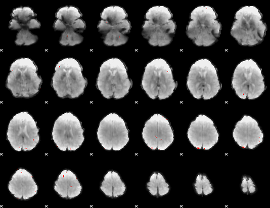 |
Component:206; Jaccard:0.11144
 |
Component:206; Jaccard:0.12382
 |
Component:206; Jaccard:0.11936
 |
Component:345; Jaccard:0.088
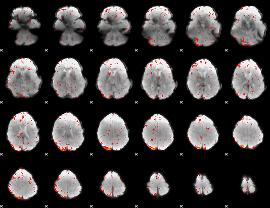 |
Component:345; Jaccard:0.055671
 |
Component:345; Jaccard:0.030551
 |
Component:345; Jaccard:0.012864
 |
Correlation:0.20809
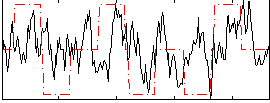 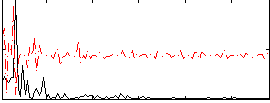 |
Correlation:0.20809
  |
Correlation:0.20809
  |
Correlation:0.092111
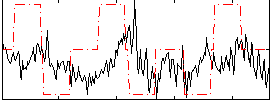 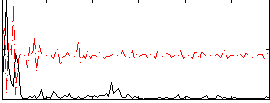 |
Correlation:0.092111
  |
Correlation:0.092111
  |
Correlation:0.092111
  |
Component:345; Jaccard:0.09486
 |
Component:345; Jaccard:0.10949
 |
Component:345; Jaccard:0.11123
 |
Component:206; Jaccard:0.085377
 |
Component:206; Jaccard:0.046147
 |
Component:206; Jaccard:0.018807
 |
Component:187; Jaccard:0.008404
 |
Correlation:0.092111
  |
Correlation:0.092111
  |
Correlation:0.092111
  |
Correlation:0.20809
  |
Correlation:0.20809
  |
Correlation:0.20809
  |
Correlation:-0.0092071
  |
Component:153; Jaccard:0.087327
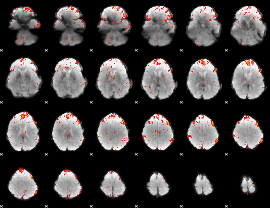 |
Component:153; Jaccard:0.093046
 |
Component:153; Jaccard:0.085473
 |
Component:153; Jaccard:0.06057
 |
Component:160; Jaccard:0.033116
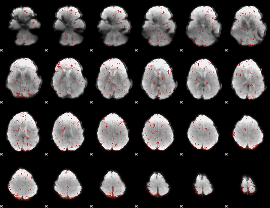 |
Component:160; Jaccard:0.016819
 |
Component:160; Jaccard:0.007206
 |
Correlation:0.10005
  |
Correlation:0.10005
  |
Correlation:0.10005
  |
Correlation:0.10005
  |
Correlation:0.12623
  |
Correlation:0.12623
  |
Correlation:0.12623
  |
Component:258; Jaccard:0.081511
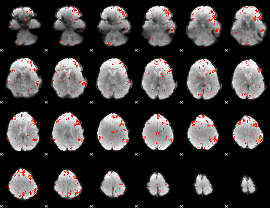 |
Component:258; Jaccard:0.086233
 |
Component:258; Jaccard:0.078663
 |
Component:303; Jaccard:0.057731
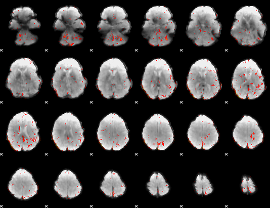 |
Component:303; Jaccard:0.032516
 |
Component:187; Jaccard:0.0162
 |
Component:206; Jaccard:0.006673
 |
Correlation:0.011315
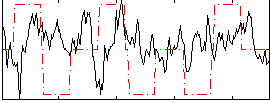 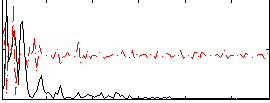 |
Correlation:0.011315
  |
Correlation:0.011315
  |
Correlation:0.029313
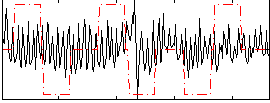 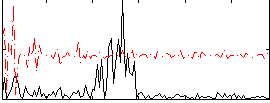 |
Correlation:0.029313
  |
Correlation:-0.0092071
  |
Correlation:0.20809
  |
Component:077; Jaccard:0.079082
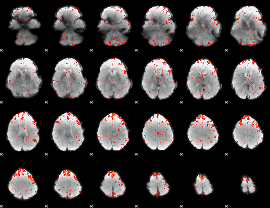 |
Component:078; Jaccard:0.077322
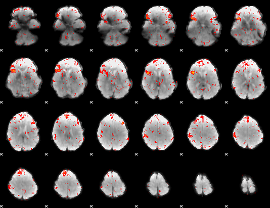 |
Component:303; Jaccard:0.075761
 |
Component:258; Jaccard:0.056444
 |
Component:153; Jaccard:0.031756
 |
Component:303; Jaccard:0.01612
 |
Component:313; Jaccard:0.00634
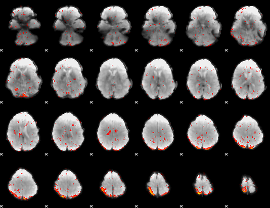 |
Correlation:-0.11838
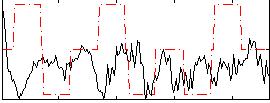 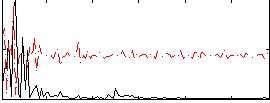 |
Correlation:0.074688
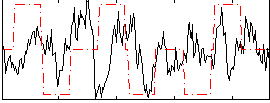 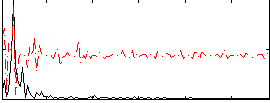 |
Correlation:0.029313
  |
Correlation:0.011315
  |
Correlation:0.10005
  |
Correlation:0.029313
  |
Correlation:-0.21198
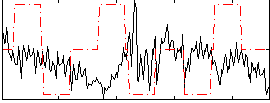 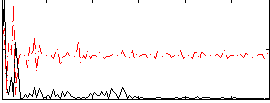 |
Component:218; Jaccard:0.076615
 |
Component:077; Jaccard:0.076096
 |
Component:160; Jaccard:0.06691
 |
Component:160; Jaccard:0.053713
 |
Component:258; Jaccard:0.031738
 |
Component:258; Jaccard:0.014802
 |
Component:065; Jaccard:0.006065
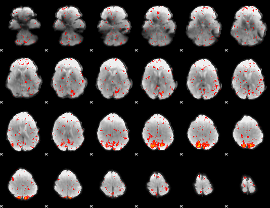 |
Correlation:-0.24384
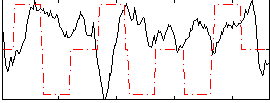 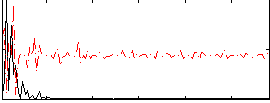 |
Correlation:-0.11838
  |
Correlation:0.12623
  |
Correlation:0.12623
  |
Correlation:0.011315
  |
Correlation:0.011315
  |
Correlation:0.010684
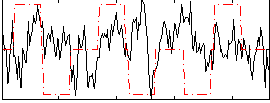 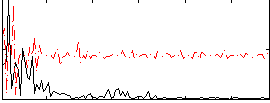 |
Component:078; Jaccard:0.07656
 |
Component:218; Jaccard:0.075311
 |
Component:080; Jaccard:0.066877
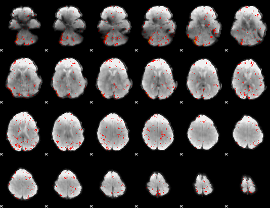 |
Component:148; Jaccard:0.048127
 |
Component:187; Jaccard:0.029887
 |
Component:153; Jaccard:0.013504
 |
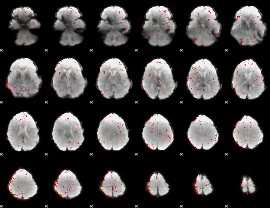
Component:163; Jaccard:0.005842
 |
Correlation:0.074688
  |
Correlation:-0.24384
  |
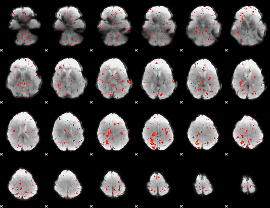
Correlation:0.071975
 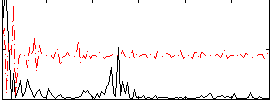 |
Correlation:0.15996
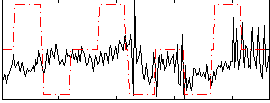 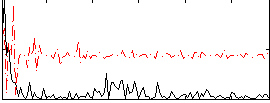 |
Correlation:-0.0092071
  |
Correlation:0.10005
  |
Correlation:-0.070951
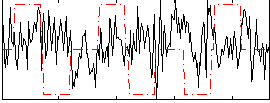 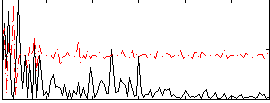 |
Component:065; Jaccard:0.07294
 |
Component:303; Jaccard:0.075121
 |
Component:174; Jaccard:0.065904
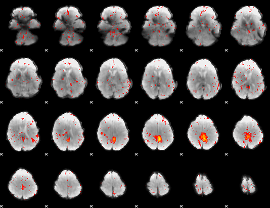 |
Component:174; Jaccard:0.04757
 |
Component:080; Jaccard:0.027807
 |
Component:148; Jaccard:0.013179
 |
Component:258; Jaccard:0.005486
 |
Correlation:0.010684
  |
Correlation:0.029313
  |
Correlation:0.19417
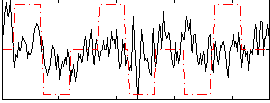 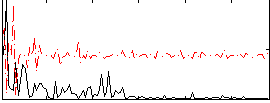 |
Correlation:0.19417
  |
Correlation:0.071975
  |
Correlation:0.15996
  |
Correlation:0.011315
  |
Component:034; Jaccard:0.072527
 |
Component:080; Jaccard:0.073061
 |
Component:078; Jaccard:0.064977
 |
Component:080; Jaccard:0.047528
 |
Component:148; Jaccard:0.027718
 |
Component:065; Jaccard:0.013066
 |
Component:080; Jaccard:0.005445
 |
Correlation:0.21626
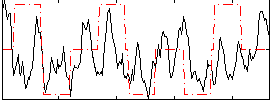  |
Correlation:0.071975
  |
Correlation:0.074688
  |
Correlation:0.071975
  |
Correlation:0.15996
  |
Correlation:0.010684
  |
Correlation:0.071975
  |
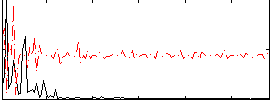
Component:059; Jaccard:0.07229
 |
Component:174; Jaccard:0.071605
 |
Component:218; Jaccard:0.064734
 |
Component:218; Jaccard:0.045757
 |
Component:174; Jaccard:0.026855
 |
Component:080; Jaccard:0.012498
 |
Component:218; Jaccard:0.005252
 |
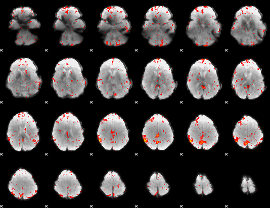
Correlation:-0.20368
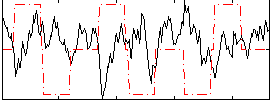  |
Correlation:0.19417
  |
Correlation:-0.24384
  |
Correlation:-0.24384
  |
Correlation:0.19417
  |
Correlation:0.071975
  |
Correlation:-0.24384
  |
Cope: 05:neg_MATCH BACK
|
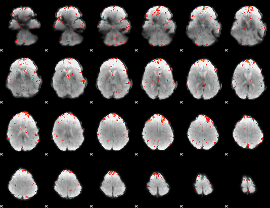
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
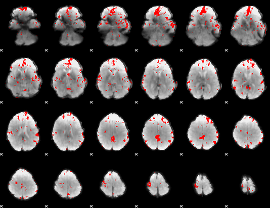 |
GLM binary result thresholded by 3
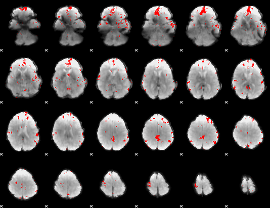 |
GLM binary result thresholded by 3.5
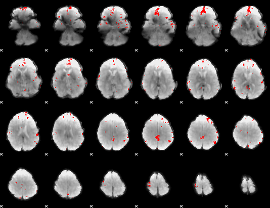 |
GLM binary result thresholded by 4
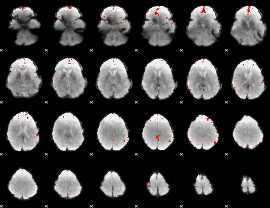 |
GLM Z-score result thresholded by 1
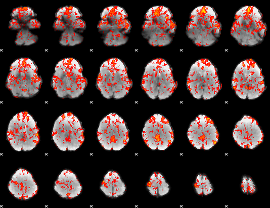 |
GLM Z-score result thresholded by 1.5
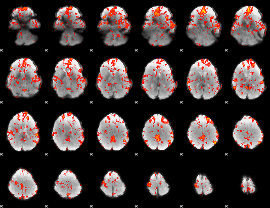 |
GLM Z-score result thresholded by 2
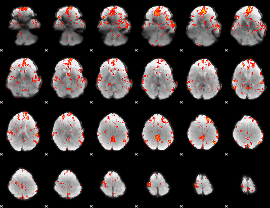 |
GLM Z-score result thresholded by 2.5
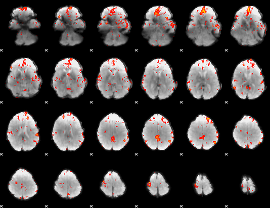 |
GLM Z-score result thresholded by 3
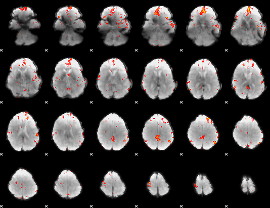 |
GLM Z-score result thresholded by 3.5
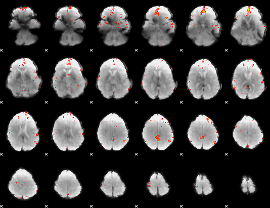 |
GLM Z-score result thresholded by 4
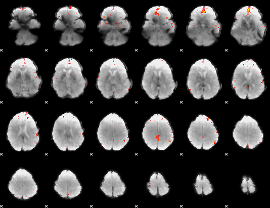 |
Component:034; Jaccard:0.12762
 |
Component:034; Jaccard:0.15563
 |
Component:034; Jaccard:0.17882
 |
Component:034; Jaccard:0.18199
 |
Component:034; Jaccard:0.16757
 |
Component:034; Jaccard:0.13212
 |
Component:034; Jaccard:0.09386
 |
Correlation:0.38071
 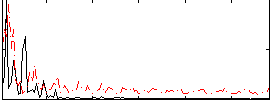 |
Correlation:0.38071
  |
Correlation:0.38071
  |
Correlation:0.38071
  |
Correlation:0.38071
  |
Correlation:0.38071
  |
Correlation:0.38071
  |
Component:259; Jaccard:0.12074
 |
Component:259; Jaccard:0.137
 |
Component:259; Jaccard:0.14455
 |
Component:043; Jaccard:0.14148
 |
Component:043; Jaccard:0.13546
 |
Component:043; Jaccard:0.11223
 |
Component:043; Jaccard:0.078399
 |
Correlation:0.26819
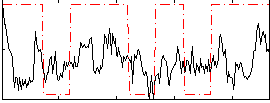 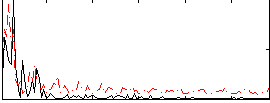 |
Correlation:0.26819
  |
Correlation:0.26819
  |
Correlation:0.28259
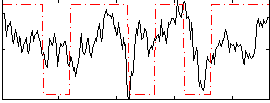  |
Correlation:0.28259
  |
Correlation:0.28259
  |
Correlation:0.28259
  |
Component:220; Jaccard:0.10323
 |
Component:387; Jaccard:0.11216
 |
Component:043; Jaccard:0.12958
 |
Component:259; Jaccard:0.13656
 |
Component:259; Jaccard:0.11683
 |
Component:259; Jaccard:0.082396
 |
Component:387; Jaccard:0.056098
 |
Correlation:0.13019
  |
Correlation:0.39601
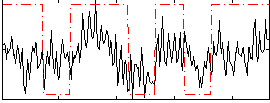  |
Correlation:0.28259
  |
Correlation:0.26819
  |
Correlation:0.26819
  |
Correlation:0.26819
  |
Correlation:0.39601
  |
Component:076; Jaccard:0.10163
 |
Component:220; Jaccard:0.11185
 |
Component:387; Jaccard:0.12523
 |
Component:387; Jaccard:0.12403
 |
Component:387; Jaccard:0.10686
 |
Component:387; Jaccard:0.081477
 |
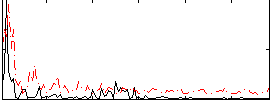
Component:392; Jaccard:0.055027
 |
Correlation:0.16531
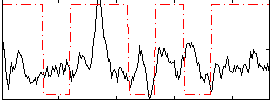 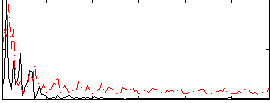 |
Correlation:0.13019
  |
Correlation:0.39601
  |
Correlation:0.39601
  |
Correlation:0.39601
  |
Correlation:0.39601
  |
Correlation:0.1805
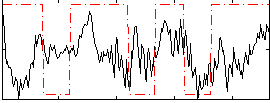 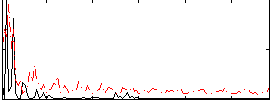 |
Component:016; Jaccard:0.10099
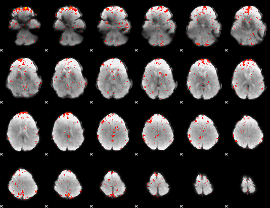 |
Component:076; Jaccard:0.11078
 |
Component:220; Jaccard:0.11415
 |
Component:220; Jaccard:0.10597
 |
Component:392; Jaccard:0.093296
 |
Component:392; Jaccard:0.073609
 |
Component:259; Jaccard:0.052064
 |
Correlation:0.15543
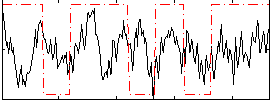  |
Correlation:0.16531
  |
Correlation:0.13019
  |
Correlation:0.13019
  |
Correlation:0.1805
  |
Correlation:0.1805
  |
Correlation:0.26819
  |
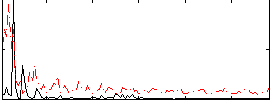
Component:333; Jaccard:0.097955
 |
Component:016; Jaccard:0.1107
 |
Component:016; Jaccard:0.11343
 |
Component:016; Jaccard:0.10533
 |
Component:016; Jaccard:0.089853
 |
Component:016; Jaccard:0.066172
 |
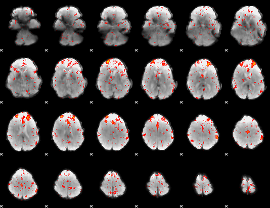
Component:167; Jaccard:0.049143
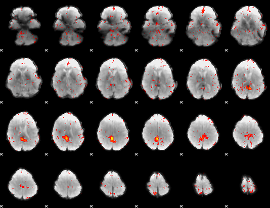 |
Correlation:0.16803
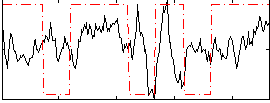 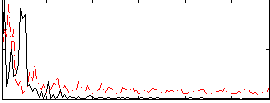 |
Correlation:0.15543
  |
Correlation:0.15543
  |
Correlation:0.15543
  |
Correlation:0.15543
  |
Correlation:0.15543
  |
Correlation:0.17287
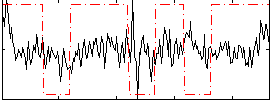 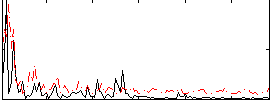 |
Component:387; Jaccard:0.094302
 |
Component:043; Jaccard:0.10931
 |
Component:076; Jaccard:0.11116
 |
Component:392; Jaccard:0.10232
 |
Component:220; Jaccard:0.086793
 |
Component:167; Jaccard:0.065877
 |
Component:322; Jaccard:0.045981
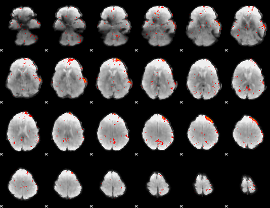 |
Correlation:0.39601
  |
Correlation:0.28259
  |
Correlation:0.16531
  |
Correlation:0.1805
  |
Correlation:0.13019
  |
Correlation:0.17287
  |
Correlation:0.18212
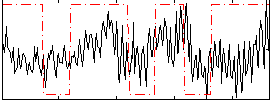 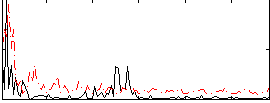 |
Component:149; Jaccard:0.094137
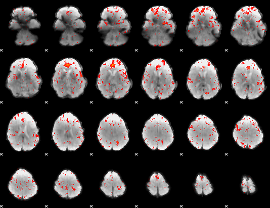 |
Component:149; Jaccard:0.10181
 |
Component:149; Jaccard:0.1036
 |
Component:076; Jaccard:0.10087
 |
Component:167; Jaccard:0.080471
 |
Component:174; Jaccard:0.063522
 |
Component:174; Jaccard:0.045495
 |
Correlation:0.064362
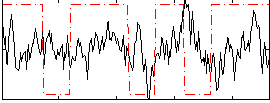 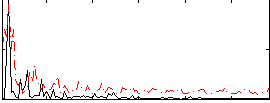 |
Correlation:0.064362
  |
Correlation:0.064362
  |
Correlation:0.16531
  |
Correlation:0.17287
  |
Correlation:0.24482
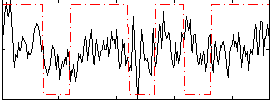 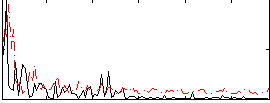 |
Correlation:0.24482
  |
Component:043; Jaccard:0.091214
 |
Component:333; Jaccard:0.099667
 |
Component:392; Jaccard:0.10156
 |
Component:149; Jaccard:0.095461
 |
Component:076; Jaccard:0.079685
 |
Component:220; Jaccard:0.062365
 |
Component:016; Jaccard:0.044008
 |
Correlation:0.28259
  |
Correlation:0.16803
  |
Correlation:0.1805
  |
Correlation:0.064362
  |
Correlation:0.16531
  |
Correlation:0.13019
  |
Correlation:0.15543
  |
Component:167; Jaccard:0.079946
 |
Component:392; Jaccard:0.091833
 |
Component:333; Jaccard:0.096847
 |
Component:167; Jaccard:0.08989
 |
Component:149; Jaccard:0.077565
 |
Component:322; Jaccard:0.060078
 |
Component:149; Jaccard:0.041578
 |
Correlation:0.17287
  |
Correlation:0.1805
  |
Correlation:0.16803
  |
Correlation:0.17287
  |
Correlation:0.064362
  |
Correlation:0.18212
  |
Correlation:0.064362
  |
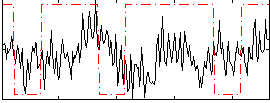
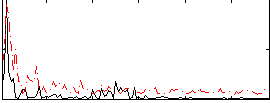
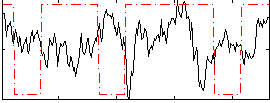
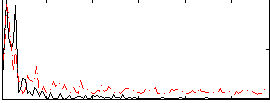
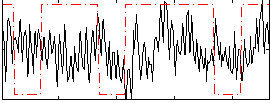
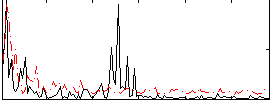
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
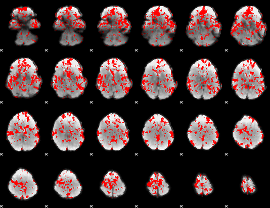 |
GLM binary result thresholded by 1.5
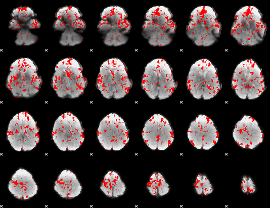 |
GLM binary result thresholded by 2
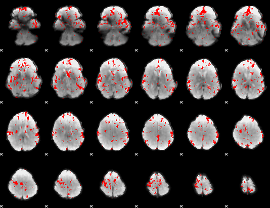 |
GLM binary result thresholded by 2.5
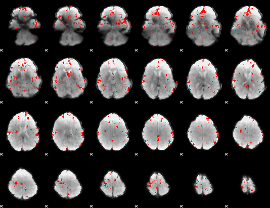 |
GLM binary result thresholded by 3
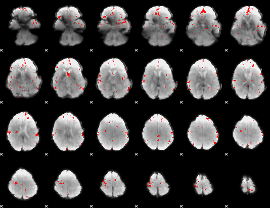 |
GLM binary result thresholded by 3.5
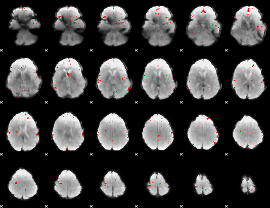 |
GLM binary result thresholded by 4
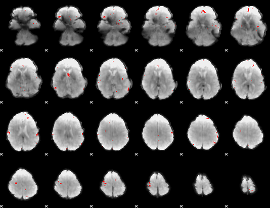 |
GLM Z-score result thresholded by 1
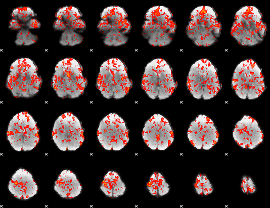 |
GLM Z-score result thresholded by 1.5
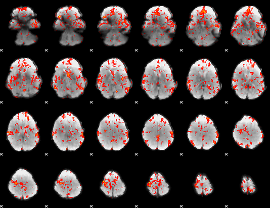 |
GLM Z-score result thresholded by 2
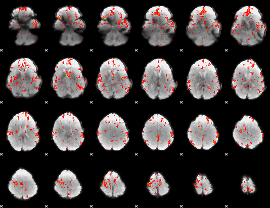 |
GLM Z-score result thresholded by 2.5
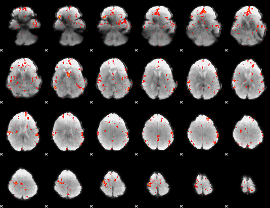 |
GLM Z-score result thresholded by 3
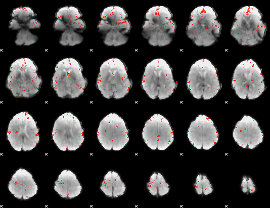 |
GLM Z-score result thresholded by 3.5
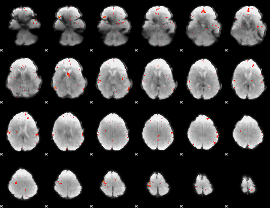 |
GLM Z-score result thresholded by 4
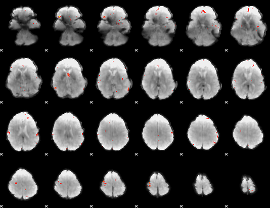 |
Component:076; Jaccard:0.11041
 |
Component:076; Jaccard:0.12449
 |
Component:076; Jaccard:0.12821
 |
Component:076; Jaccard:0.11875
 |
Component:124; Jaccard:0.099657
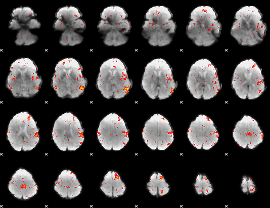 |
Component:124; Jaccard:0.0687
 |
Component:124; Jaccard:0.038912
 |
Correlation:-0.020224
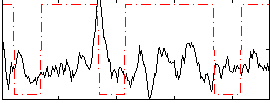 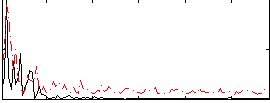 |
Correlation:-0.020224
  |
Correlation:-0.020224
  |
Correlation:-0.020224
  |
Correlation:0.096949
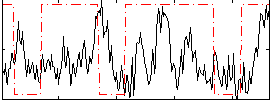 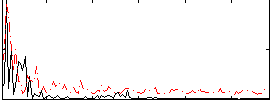 |
Correlation:0.096949
  |
Correlation:0.096949
  |
Component:259; Jaccard:0.099677
 |
Component:034; Jaccard:0.10705
 |
Component:124; Jaccard:0.11689
 |
Component:124; Jaccard:0.11812
 |
Component:076; Jaccard:0.092806
 |
Component:076; Jaccard:0.065904
 |
Component:076; Jaccard:0.037265
 |
Correlation:0.023328
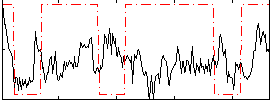  |
Correlation:0.015849
  |
Correlation:0.096949
  |
Correlation:0.096949
  |
Correlation:-0.020224
  |
Correlation:-0.020224
  |
Correlation:-0.020224
  |
Component:220; Jaccard:0.09663
 |
Component:124; Jaccard:0.10615
 |
Component:034; Jaccard:0.11148
 |
Component:034; Jaccard:0.10148
 |
Component:034; Jaccard:0.081612
 |
Component:387; Jaccard:0.052639
 |
Component:043; Jaccard:0.030352
 |
Correlation:-0.047873
  |
Correlation:0.096949
  |
Correlation:0.015849
  |
Correlation:0.015849
  |
Correlation:0.015849
  |
Correlation:-0.011418
  |
Correlation:-0.14948
  |
Component:034; Jaccard:0.096545
 |
Component:259; Jaccard:0.10611
 |
Component:259; Jaccard:0.10559
 |
Component:342; Jaccard:0.094337
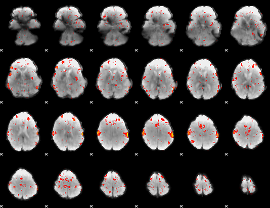 |
Component:387; Jaccard:0.076797
 |
Component:034; Jaccard:0.052345
 |
Component:387; Jaccard:0.029989
 |
Correlation:0.015849
  |
Correlation:0.023328
  |
Correlation:0.023328
  |
Correlation:-0.13588
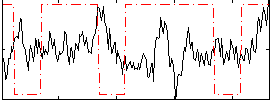 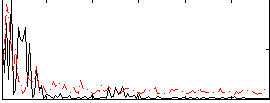 |
Correlation:-0.011418
  |
Correlation:0.015849
  |
Correlation:-0.011418
  |
Component:124; Jaccard:0.088965
 |
Component:220; Jaccard:0.099125
 |
Component:387; Jaccard:0.099763
 |
Component:387; Jaccard:0.09351
 |
Component:342; Jaccard:0.073904
 |
Component:274; Jaccard:0.049743
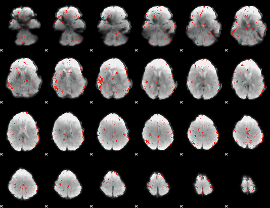 |
Component:034; Jaccard:0.028913
 |
Correlation:0.096949
  |
Correlation:-0.047873
  |
Correlation:-0.011418
  |
Correlation:-0.011418
  |
Correlation:-0.13588
  |
Correlation:0.049245
  |
Correlation:0.015849
  |
Component:342; Jaccard:0.082934
 |
Component:387; Jaccard:0.093386
 |
Component:342; Jaccard:0.098781
 |
Component:259; Jaccard:0.089252
 |
Component:043; Jaccard:0.072075
 |
Component:043; Jaccard:0.048798
 |
Component:342; Jaccard:0.027868
 |
Correlation:-0.13588
  |
Correlation:-0.011418
  |
Correlation:-0.13588
  |
Correlation:0.023328
  |
Correlation:-0.14948
  |
Correlation:-0.14948
  |
Correlation:-0.13588
  |
Component:387; Jaccard:0.082673
 |
Component:342; Jaccard:0.092837
 |
Component:220; Jaccard:0.093861
 |
Component:043; Jaccard:0.088459
 |
Component:274; Jaccard:0.06997
 |
Component:342; Jaccard:0.048559
 |
Component:274; Jaccard:0.027324
 |
Correlation:-0.011418
  |
Correlation:-0.13588
  |
Correlation:-0.047873
  |
Correlation:-0.14948
  |
Correlation:0.049245
  |
Correlation:-0.13588
  |
Correlation:0.049245
  |
Component:244; Jaccard:0.080707
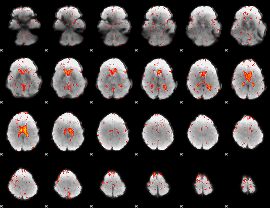 |
Component:263; Jaccard:0.085705
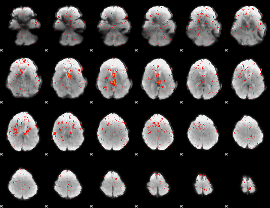 |
Component:043; Jaccard:0.091506
 |
Component:179; Jaccard:0.084625
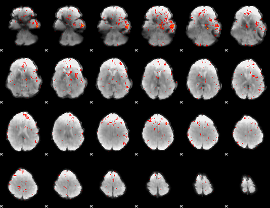 |
Component:179; Jaccard:0.067583
 |
Component:179; Jaccard:0.042457
 |
Component:392; Jaccard:0.023256
 |
Correlation:0.11517
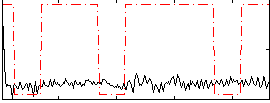 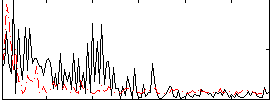 |
Correlation:0.10671
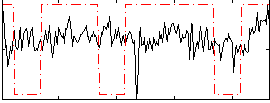 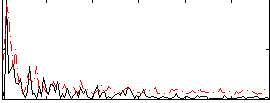 |
Correlation:-0.14948
  |
Correlation:0.066448
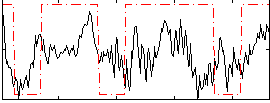
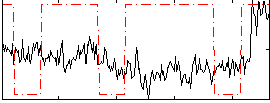  |
Correlation:0.066448
  |
Correlation:0.066448
  |
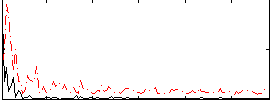
Correlation:0.29298
 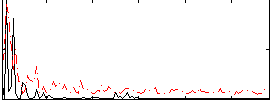 |
Component:263; Jaccard:0.079124
 |
Component:179; Jaccard:0.084708
 |
Component:179; Jaccard:0.089883
 |
Component:274; Jaccard:0.083771
 |
Component:259; Jaccard:0.064058
 |
Component:392; Jaccard:0.041221
 |
Component:179; Jaccard:0.022763
 |
Correlation:0.10671
  |
Correlation:0.066448
  |
Correlation:0.066448
  |
Correlation:0.049245
  |
Correlation:0.023328
  |
Correlation:0.29298
  |
Correlation:0.066448
  |
Component:016; Jaccard:0.078605
 |
Component:043; Jaccard:0.084483
 |
Component:274; Jaccard:0.08646
 |
Component:392; Jaccard:0.07887
 |
Component:392; Jaccard:0.063691
 |
Component:259; Jaccard:0.037461
 |
Component:357; Jaccard:0.020284
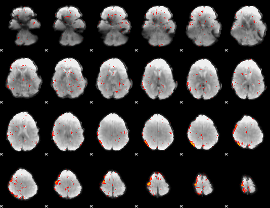 |
Correlation:0.25542
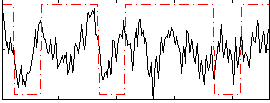 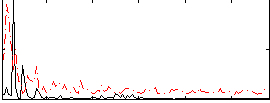 |
Correlation:-0.14948
  |
Correlation:0.049245
  |
Correlation:0.29298
  |
Correlation:0.29298
  |
Correlation:0.023328
  |
Correlation:0.10905
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































