Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
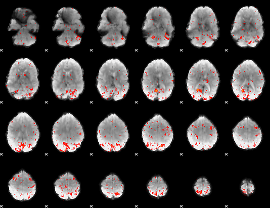
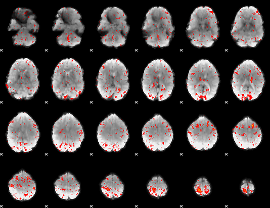
GLM result group-wise
 |
GLM binary result thresholded by 1
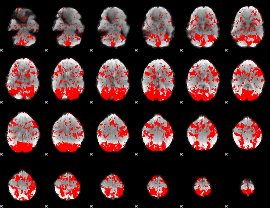 |
GLM binary result thresholded by 1.5
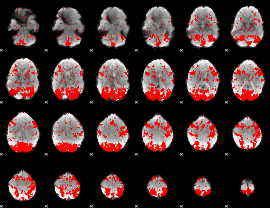 |
GLM binary result thresholded by 2
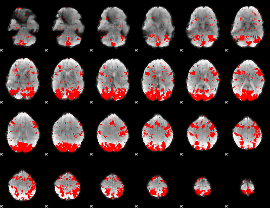 |
GLM binary result thresholded by 2.5
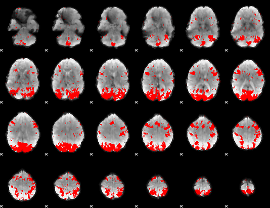 |
GLM binary result thresholded by 3
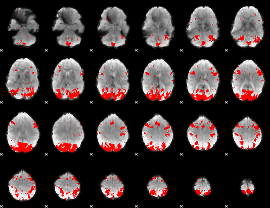 |
GLM binary result thresholded by 3.5
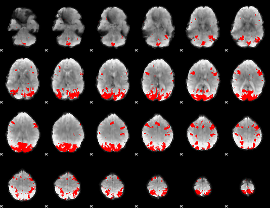 |
GLM binary result thresholded by 4
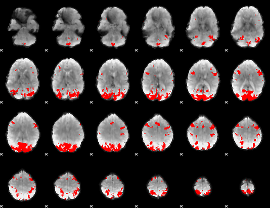 |
GLM Z-score result thresholded by 1
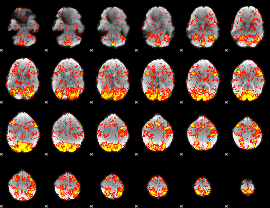 |
GLM Z-score result thresholded by 1.5
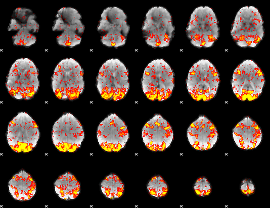 |
GLM Z-score result thresholded by 2
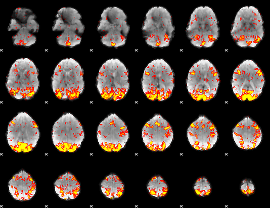 |
GLM Z-score result thresholded by 2.5
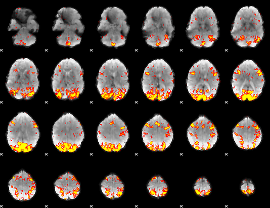 |
GLM Z-score result thresholded by 3
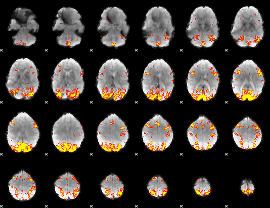 |
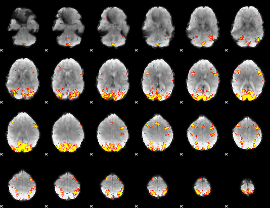
GLM Z-score result thresholded by 3.5
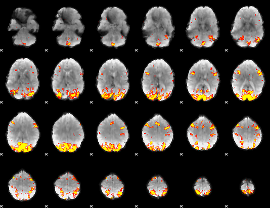 |
GLM Z-score result thresholded by 4
 |
Component:387; Jaccard:0.16897
 |
Component:387; Jaccard:0.22053
 |
Component:387; Jaccard:0.28176
 |
Component:387; Jaccard:0.34653
 |
Component:387; Jaccard:0.40529
 |
Component:387; Jaccard:0.45654
 |
Component:387; Jaccard:0.49258
 |
Correlation:0.25716
  |
Correlation:0.25716
  |
Correlation:0.25716
  |
Correlation:0.25716
  |
Correlation:0.25716
  |
Correlation:0.25716
  |
Correlation:0.25716
  |
Component:052; Jaccard:0.16252
 |
Component:052; Jaccard:0.20597
 |
Component:052; Jaccard:0.25058
 |
Component:052; Jaccard:0.29252
 |
Component:052; Jaccard:0.32125
 |
Component:052; Jaccard:0.33609
 |
Component:052; Jaccard:0.33629
 |
Correlation:0.23425
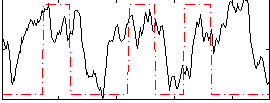 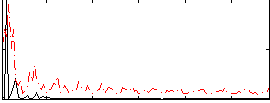 |
Correlation:0.23425
  |
Correlation:0.23425
  |
Correlation:0.23425
  |
Correlation:0.23425
  |
Correlation:0.23425
  |
Correlation:0.23425
  |
Component:142; Jaccard:0.15839
 |
Component:142; Jaccard:0.19437
 |
Component:142; Jaccard:0.23039
 |
Component:142; Jaccard:0.25899
 |
Component:142; Jaccard:0.27976
 |
Component:142; Jaccard:0.29018
 |
Component:142; Jaccard:0.29018
 |
Correlation:0.20501
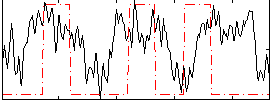 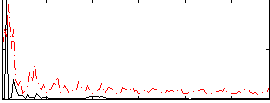 |
Correlation:0.20501
  |
Correlation:0.20501
  |
Correlation:0.20501
  |
Correlation:0.20501
  |
Correlation:0.20501
  |
Correlation:0.20501
  |
Component:031; Jaccard:0.15054
 |
Component:031; Jaccard:0.18291
 |
Component:031; Jaccard:0.2117
 |
Component:031; Jaccard:0.23517
 |
Component:031; Jaccard:0.24905
 |
Component:183; Jaccard:0.2553
 |
Component:183; Jaccard:0.2635
 |
Correlation:0.2653
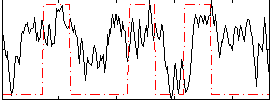 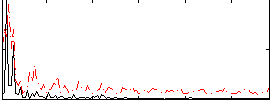 |
Correlation:0.2653
  |
Correlation:0.2653
  |
Correlation:0.2653
  |
Correlation:0.2653
  |
Correlation:0.3144
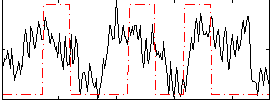 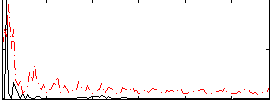 |
Correlation:0.3144
  |
Component:256; Jaccard:0.12745
 |
Component:183; Jaccard:0.15767
 |
Component:183; Jaccard:0.19093
 |
Component:183; Jaccard:0.22054
 |
Component:183; Jaccard:0.24213
 |
Component:031; Jaccard:0.25376
 |
Component:031; Jaccard:0.25246
 |
Correlation:0.13859
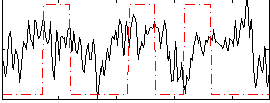 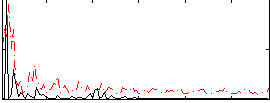 |
Correlation:0.3144
  |
Correlation:0.3144
  |
Correlation:0.3144
  |
Correlation:0.3144
  |
Correlation:0.2653
  |
Correlation:0.2653
  |
Component:183; Jaccard:0.12638
 |
Component:256; Jaccard:0.15003
 |
Component:256; Jaccard:0.17151
 |
Component:256; Jaccard:0.18595
 |
Component:256; Jaccard:0.19457
 |
Component:256; Jaccard:0.19299
 |
Component:256; Jaccard:0.18798
 |
Correlation:0.3144
  |
Correlation:0.13859
  |
Correlation:0.13859
  |
Correlation:0.13859
  |
Correlation:0.13859
  |
Correlation:0.13859
  |
Correlation:0.13859
  |
Component:286; Jaccard:0.11952
 |
Component:286; Jaccard:0.13605
 |
Component:182; Jaccard:0.15493
 |
Component:182; Jaccard:0.16787
 |
Component:182; Jaccard:0.1757
 |
Component:182; Jaccard:0.17738
 |
Component:182; Jaccard:0.17625
 |
Correlation:0.18794
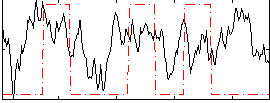 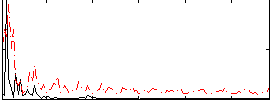 |
Correlation:0.18794
  |
Correlation:0.20615
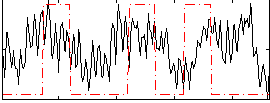  |
Correlation:0.20615
  |
Correlation:0.20615
  |
Correlation:0.20615
  |
Correlation:0.20615
  |
Component:182; Jaccard:0.11662
 |
Component:182; Jaccard:0.13587
 |
Component:286; Jaccard:0.14927
 |
Component:286; Jaccard:0.15479
 |
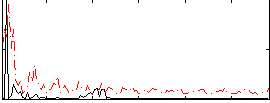
Component:172; Jaccard:0.15882
 |
Component:172; Jaccard:0.16246
 |
Component:172; Jaccard:0.1611
 |
Correlation:0.20615
  |
Correlation:0.20615
  |
Correlation:0.18794
  |
Correlation:0.18794
  |
Correlation:0.26322
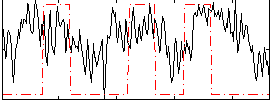 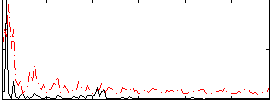 |
Correlation:0.26322
  |
Correlation:0.26322
  |
Component:316; Jaccard:0.11421
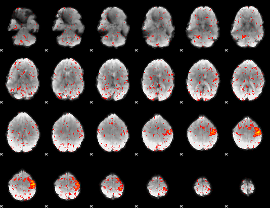 |
Component:155; Jaccard:0.12211
 |
Component:155; Jaccard:0.13715
 |
Component:172; Jaccard:0.14976
 |
Component:155; Jaccard:0.15176
 |
Component:155; Jaccard:0.14996
 |
Component:155; Jaccard:0.14406
 |
Correlation:0.22381
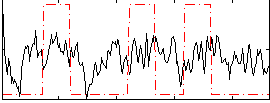 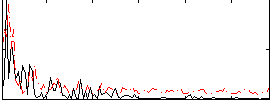 |
Correlation:-0.020108
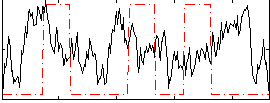  |
Correlation:-0.020108
  |
Correlation:0.26322
  |
Correlation:-0.020108
  |
Correlation:-0.020108
  |
Correlation:-0.020108
  |
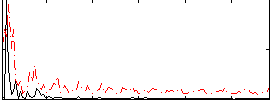
Component:128; Jaccard:0.10863
 |
Component:316; Jaccard:0.12082
 |
Component:172; Jaccard:0.13676
 |
Component:155; Jaccard:0.14716
 |
Component:286; Jaccard:0.15153
 |
Component:286; Jaccard:0.14379
 |
Component:286; Jaccard:0.1319
 |
Correlation:0.032168
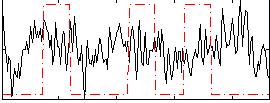 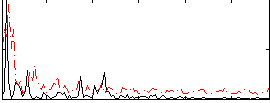 |
Correlation:0.22381
  |
Correlation:0.26322
  |
Correlation:-0.020108
  |
Correlation:0.18794
  |
Correlation:0.18794
  |
Correlation:0.18794
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
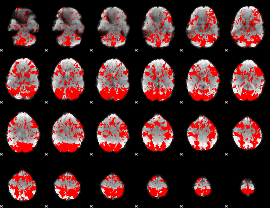 |
GLM binary result thresholded by 1.5
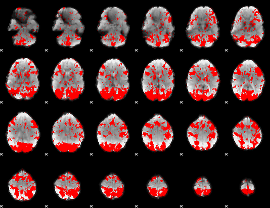 |
GLM binary result thresholded by 2
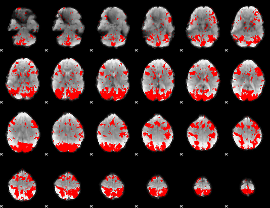 |
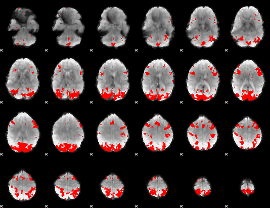
GLM binary result thresholded by 2.5
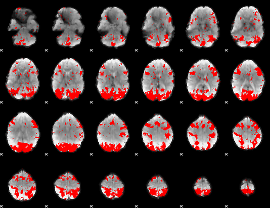 |
GLM binary result thresholded by 3
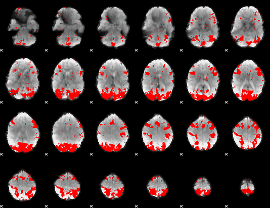 |
GLM binary result thresholded by 3.5
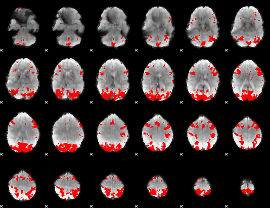 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
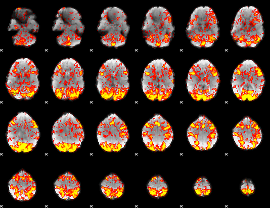 |
GLM Z-score result thresholded by 1.5
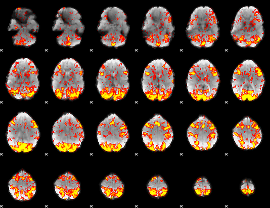 |
GLM Z-score result thresholded by 2
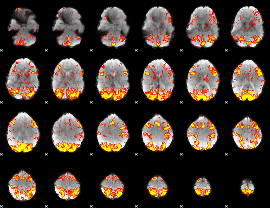 |
GLM Z-score result thresholded by 2.5
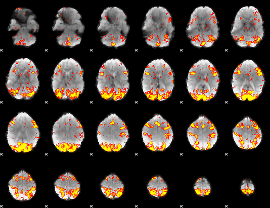 |
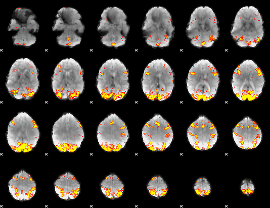
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
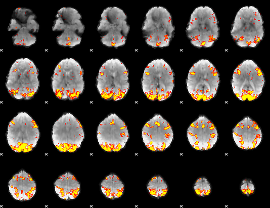 |
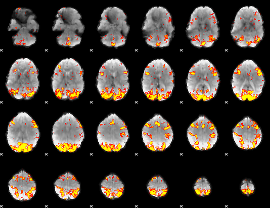
GLM Z-score result thresholded by 4
 |
Component:387; Jaccard:0.14483
 |
Component:387; Jaccard:0.18287
 |
Component:387; Jaccard:0.22754
 |
Component:387; Jaccard:0.27617
 |
Component:387; Jaccard:0.32377
 |
Component:387; Jaccard:0.36871
 |
Component:387; Jaccard:0.41064
 |
Correlation:0.22809
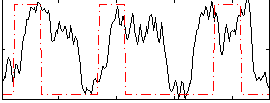 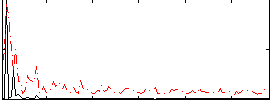 |
Correlation:0.22809
  |
Correlation:0.22809
  |
Correlation:0.22809
  |
Correlation:0.22809
  |
Correlation:0.22809
  |
Correlation:0.22809
  |
Component:052; Jaccard:0.14243
 |
Component:052; Jaccard:0.17732
 |
Component:052; Jaccard:0.21694
 |
Component:052; Jaccard:0.25726
 |
Component:052; Jaccard:0.29256
 |
Component:052; Jaccard:0.32036
 |
Component:052; Jaccard:0.34297
 |
Correlation:0.11615
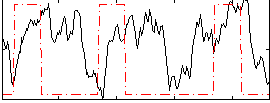 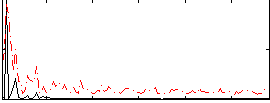 |
Correlation:0.11615
  |
Correlation:0.11615
  |
Correlation:0.11615
  |
Correlation:0.11615
  |
Correlation:0.11615
  |
Correlation:0.11615
  |
Component:142; Jaccard:0.13759
 |
Component:142; Jaccard:0.16608
 |
Component:142; Jaccard:0.19483
 |
Component:142; Jaccard:0.22115
 |
Component:142; Jaccard:0.24206
 |
Component:142; Jaccard:0.25608
 |
Component:142; Jaccard:0.26638
 |
Correlation:0.043697
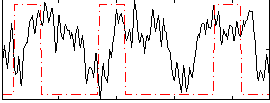 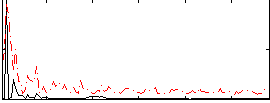 |
Correlation:0.043697
  |
Correlation:0.043697
  |
Correlation:0.043697
  |
Correlation:0.043697
  |
Correlation:0.043697
  |
Correlation:0.043697
  |
Component:286; Jaccard:0.12592
 |
Component:286; Jaccard:0.14869
 |
Component:286; Jaccard:0.1705
 |
Component:183; Jaccard:0.19023
 |
Component:183; Jaccard:0.21247
 |
Component:183; Jaccard:0.2322
 |
Component:183; Jaccard:0.246
 |
Correlation:0.070677
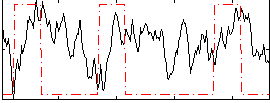 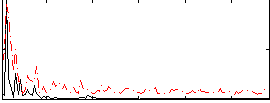 |
Correlation:0.070677
  |
Correlation:0.070677
  |
Correlation:0.17853
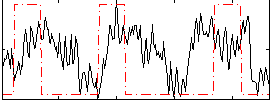 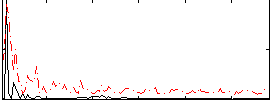 |
Correlation:0.17853
  |
Correlation:0.17853
  |
Correlation:0.17853
  |
Component:031; Jaccard:0.12275
 |
Component:031; Jaccard:0.14495
 |
Component:031; Jaccard:0.16523
 |
Component:286; Jaccard:0.18696
 |
Component:286; Jaccard:0.19721
 |
Component:155; Jaccard:0.20454
 |
Component:031; Jaccard:0.20884
 |
Correlation:0.13093
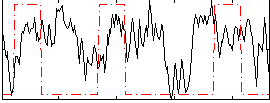 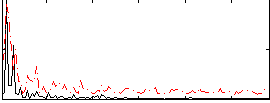 |
Correlation:0.13093
  |
Correlation:0.13093
  |
Correlation:0.070677
  |
Correlation:0.070677
  |
Correlation:0.27985
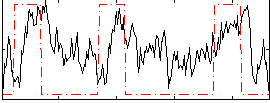 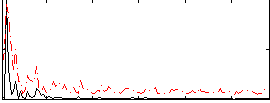 |
Correlation:0.13093
  |
Component:256; Jaccard:0.11871
 |
Component:256; Jaccard:0.14033
 |
Component:183; Jaccard:0.16335
 |
Component:031; Jaccard:0.18299
 |
Component:155; Jaccard:0.19637
 |
Component:031; Jaccard:0.20359
 |
Component:155; Jaccard:0.20803
 |
Correlation:0.044887
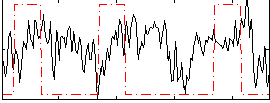 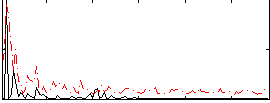 |
Correlation:0.044887
  |
Correlation:0.17853
  |
Correlation:0.13093
  |
Correlation:0.27985
  |
Correlation:0.13093
  |
Correlation:0.27985
  |
Component:155; Jaccard:0.11664
 |
Component:155; Jaccard:0.13944
 |
Component:155; Jaccard:0.16203
 |
Component:155; Jaccard:0.18183
 |
Component:031; Jaccard:0.19509
 |
Component:256; Jaccard:0.1992
 |
Component:256; Jaccard:0.20206
 |
Correlation:0.27985
  |
Correlation:0.27985
  |
Correlation:0.27985
  |
Correlation:0.27985
  |
Correlation:0.13093
  |
Correlation:0.044887
  |
Correlation:0.044887
  |
Component:128; Jaccard:0.11468
 |
Component:183; Jaccard:0.13625
 |
Component:256; Jaccard:0.16119
 |
Component:256; Jaccard:0.17857
 |
Component:256; Jaccard:0.19086
 |
Component:286; Jaccard:0.1991
 |
Component:286; Jaccard:0.19346
 |
Correlation:0.18503
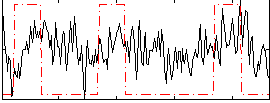 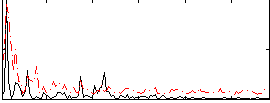 |
Correlation:0.17853
  |
Correlation:0.044887
  |
Correlation:0.044887
  |
Correlation:0.044887
  |
Correlation:0.070677
  |
Correlation:0.070677
  |
Component:183; Jaccard:0.11106
 |
Component:128; Jaccard:0.12766
 |
Component:182; Jaccard:0.14401
 |
Component:182; Jaccard:0.15814
 |
Component:182; Jaccard:0.16651
 |
Component:182; Jaccard:0.17104
 |
Component:182; Jaccard:0.17083
 |
Correlation:0.17853
  |
Correlation:0.18503
  |
Correlation:0.066798
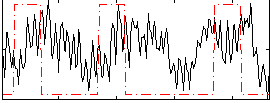 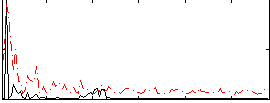 |
Correlation:0.066798
  |
Correlation:0.066798
  |
Correlation:0.066798
  |
Correlation:0.066798
  |
Component:182; Jaccard:0.10945
 |
Component:182; Jaccard:0.12718
 |
Component:128; Jaccard:0.13798
 |
Component:128; Jaccard:0.14435
 |
Component:128; Jaccard:0.14696
 |
Component:128; Jaccard:0.14422
 |
Component:172; Jaccard:0.14694
 |
Correlation:0.066798
  |
Correlation:0.066798
  |
Correlation:0.18503
  |
Correlation:0.18503
  |
Correlation:0.18503
  |
Correlation:0.18503
  |
Correlation:0.23428
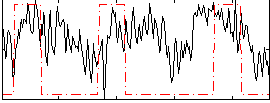 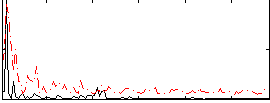 |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
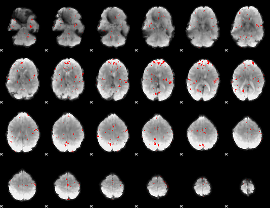 |
GLM binary result thresholded by 3
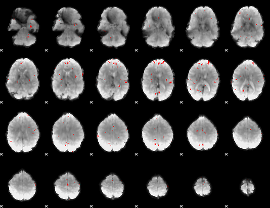 |
GLM binary result thresholded by 3.5
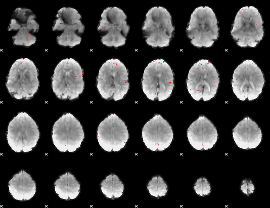 |
GLM binary result thresholded by 4
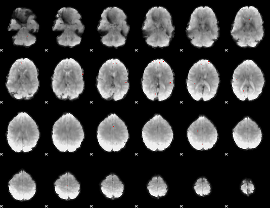 |
GLM Z-score result thresholded by 1
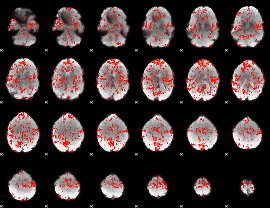 |
GLM Z-score result thresholded by 1.5
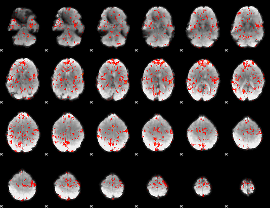 |
GLM Z-score result thresholded by 2
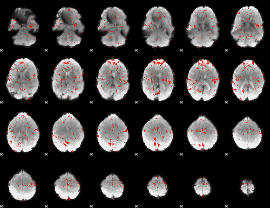 |
GLM Z-score result thresholded by 2.5
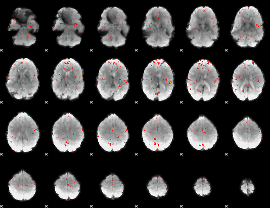 |
GLM Z-score result thresholded by 3
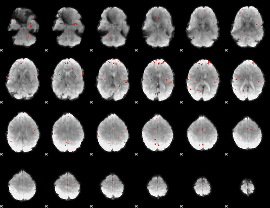 |
GLM Z-score result thresholded by 3.5
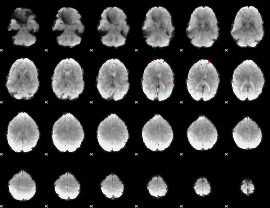 |
GLM Z-score result thresholded by 4
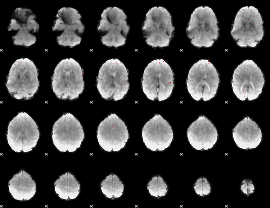 |
Component:347; Jaccard:0.097985
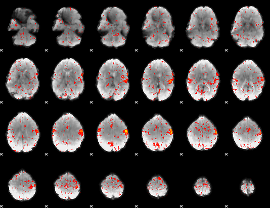 |
Component:347; Jaccard:0.099587
 |
Component:347; Jaccard:0.083344
 |
Component:347; Jaccard:0.053051
 |
Component:119; Jaccard:0.027588
 |
Component:347; Jaccard:0.012556
 |
Component:194; Jaccard:0.006049
 |
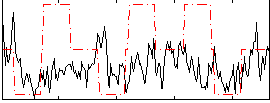
Correlation:0.19511
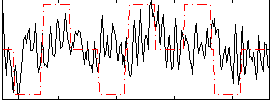  |
Correlation:0.19511
  |
Correlation:0.19511
  |
Correlation:0.19511
  |
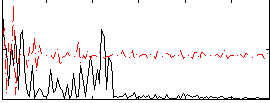
Correlation:0.064523
 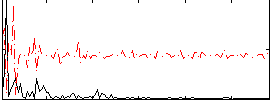 |
Correlation:0.19511
  |
Correlation:0.14497
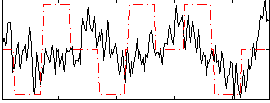 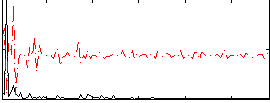 |
Component:200; Jaccard:0.090429
 |
Component:200; Jaccard:0.086418
 |
Component:119; Jaccard:0.069269
 |
Component:119; Jaccard:0.047893
 |
Component:347; Jaccard:0.027061
 |
Component:119; Jaccard:0.012292
 |
Component:119; Jaccard:0.005503
 |
Correlation:0.17658
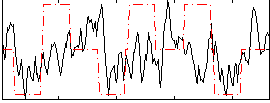 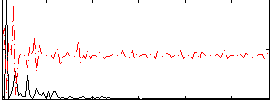 |
Correlation:0.17658
  |
Correlation:0.064523
  |
Correlation:0.064523
  |
Correlation:0.19511
  |
Correlation:0.064523
  |
Correlation:0.064523
  |
Component:119; Jaccard:0.083061
 |
Component:119; Jaccard:0.08396
 |
Component:200; Jaccard:0.066499
 |
Component:194; Jaccard:0.043294
 |
Component:194; Jaccard:0.023048
 |
Component:194; Jaccard:0.011013
 |
Component:347; Jaccard:0.005103
 |
Correlation:0.064523
  |
Correlation:0.064523
  |
Correlation:0.17658
  |
Correlation:0.14497
  |
Correlation:0.14497
  |
Correlation:0.14497
  |
Correlation:0.19511
  |
Component:118; Jaccard:0.080344
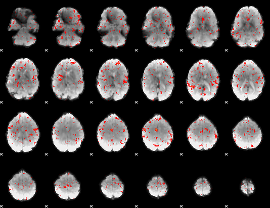 |
Component:118; Jaccard:0.080053
 |
Component:118; Jaccard:0.063467
 |
Component:200; Jaccard:0.042814
 |
Component:215; Jaccard:0.022672
 |
Component:118; Jaccard:0.009718
 |
Component:200; Jaccard:0.004321
 |
Correlation:0.11787
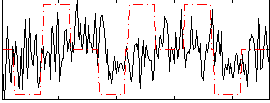 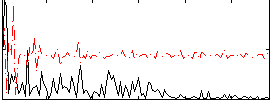 |
Correlation:0.11787
  |
Correlation:0.11787
  |
Correlation:0.17658
  |
Correlation:0.07514
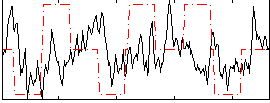  |
Correlation:0.11787
  |
Correlation:0.17658
  |
Component:215; Jaccard:0.078103
 |
Component:215; Jaccard:0.075879
 |
Component:215; Jaccard:0.0609
 |
Component:215; Jaccard:0.042497
 |
Component:200; Jaccard:0.022264
 |
Component:200; Jaccard:0.008992
 |
Component:118; Jaccard:0.004137
 |
Correlation:0.07514
  |
Correlation:0.07514
  |
Correlation:0.07514
  |
Correlation:0.07514
  |
Correlation:0.17658
  |
Correlation:0.17658
  |
Correlation:0.11787
  |
Component:284; Jaccard:0.070044
 |
Component:194; Jaccard:0.068458
 |
Component:194; Jaccard:0.059926
 |
Component:118; Jaccard:0.039469
 |
Component:118; Jaccard:0.021432
 |
Component:215; Jaccard:0.008817
 |
Component:215; Jaccard:0.004106
 |
Correlation:0.055641
  |
Correlation:0.14497
  |
Correlation:0.14497
  |
Correlation:0.11787
  |
Correlation:0.11787
  |
Correlation:0.07514
  |
Correlation:0.07514
  |
Component:041; Jaccard:0.069864
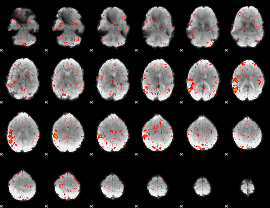 |
Component:041; Jaccard:0.065738
 |
Component:041; Jaccard:0.052592
 |
Component:344; Jaccard:0.03387
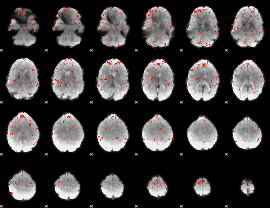 |
Component:344; Jaccard:0.018453
 |
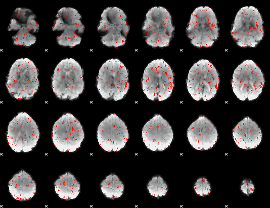
Component:126; Jaccard:0.007302
 |
Component:193; Jaccard:0.003941
 |
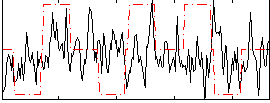
Correlation:0.063122
 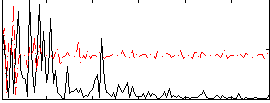 |
Correlation:0.063122
  |
Correlation:0.063122
  |
Correlation:0.06419
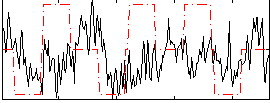 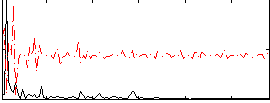 |
Correlation:0.06419
  |
Correlation:0.15598
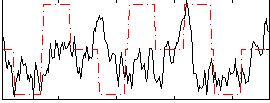 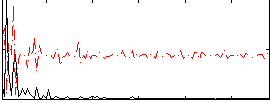 |
Correlation:0.13805
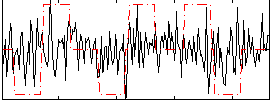 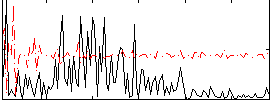 |
Component:121; Jaccard:0.067403
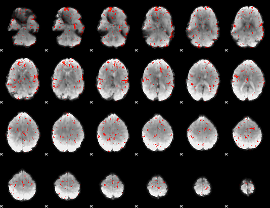 |
Component:121; Jaccard:0.06193
 |
Component:121; Jaccard:0.049189
 |
Component:126; Jaccard:0.032761
 |
Component:126; Jaccard:0.016309
 |
Component:121; Jaccard:0.006926
 |
Component:126; Jaccard:0.003747
 |
Correlation:0.0088334
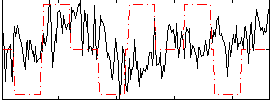 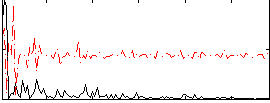 |
Correlation:0.0088334
  |
Correlation:0.0088334
  |
Correlation:0.15598
  |
Correlation:0.15598
  |
Correlation:0.0088334
  |
Correlation:0.15598
  |
Component:194; Jaccard:0.065839
 |
Component:284; Jaccard:0.06185
 |
Component:344; Jaccard:0.048709
 |
Component:041; Jaccard:0.031736
 |
Component:041; Jaccard:0.015875
 |
Component:124; Jaccard:0.00689
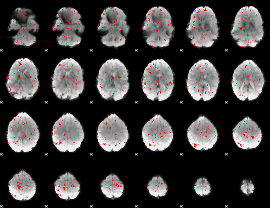 |
Component:124; Jaccard:0.003614
 |
Correlation:0.14497
  |
Correlation:0.055641
  |
Correlation:0.06419
  |
Correlation:0.063122
  |
Correlation:0.063122
  |
Correlation:0.19769
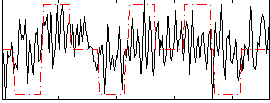 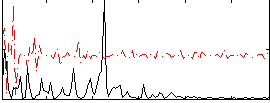 |
Correlation:0.19769
  |
Component:211; Jaccard:0.064557
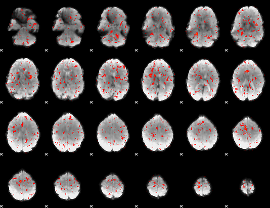 |
Component:211; Jaccard:0.059387
 |
Component:126; Jaccard:0.046139
 |
Component:121; Jaccard:0.031532
 |
Component:380; Jaccard:0.015673
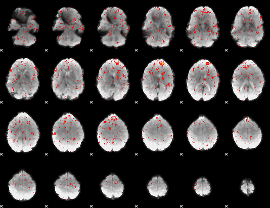 |
Component:380; Jaccard:0.006615
 |
Component:344; Jaccard:0.003186
 |
Correlation:0.12712
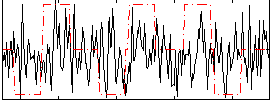 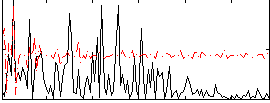 |
Correlation:0.12712
  |
Correlation:0.15598
  |
Correlation:0.0088334
  |
Correlation:0.17323
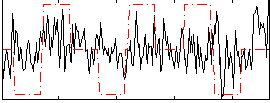 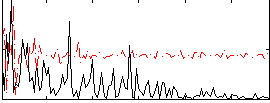 |
Correlation:0.17323
  |
Correlation:0.06419
  |
Cope: 04:REL-MATCH BACK
|
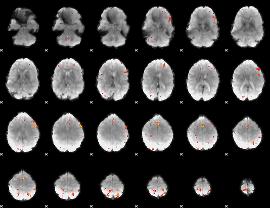
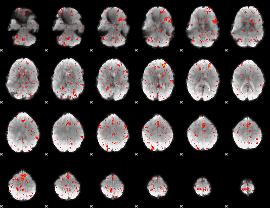
GLM result group-wise
 |
GLM binary result thresholded by 1
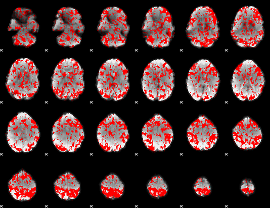 |
GLM binary result thresholded by 1.5
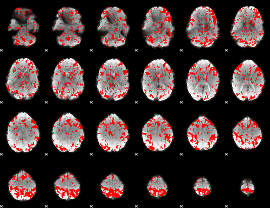 |
GLM binary result thresholded by 2
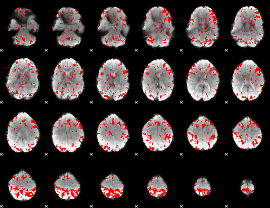 |
GLM binary result thresholded by 2.5
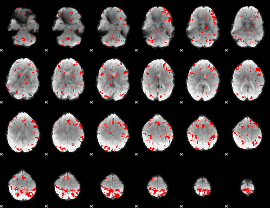 |
GLM binary result thresholded by 3
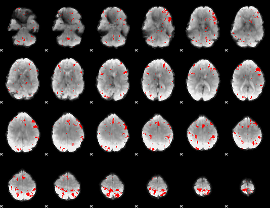 |
GLM binary result thresholded by 3.5
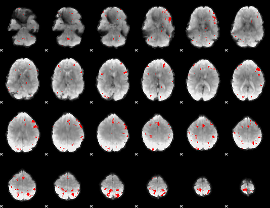 |
GLM binary result thresholded by 4
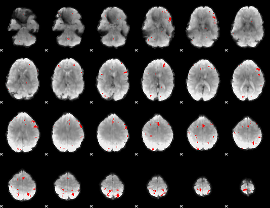 |
GLM Z-score result thresholded by 1
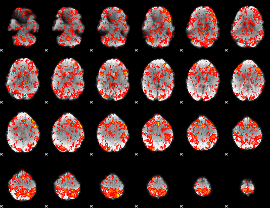 |
GLM Z-score result thresholded by 1.5
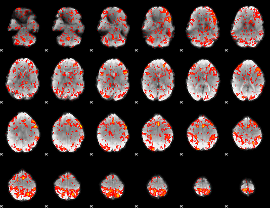 |
GLM Z-score result thresholded by 2
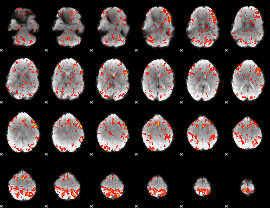 |
GLM Z-score result thresholded by 2.5
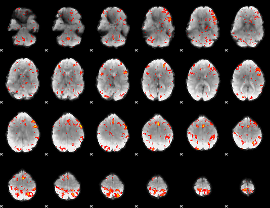 |
GLM Z-score result thresholded by 3
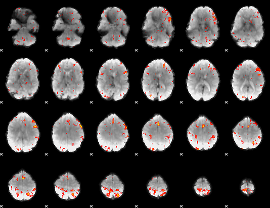 |
GLM Z-score result thresholded by 3.5
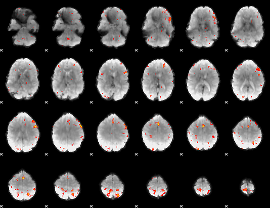 |
GLM Z-score result thresholded by 4
 |
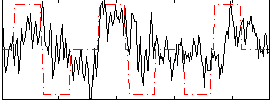
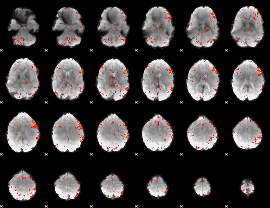
Component:212; Jaccard:0.12417
 |
Component:155; Jaccard:0.16067
 |
Component:155; Jaccard:0.20269
 |
Component:155; Jaccard:0.22037
 |
Component:155; Jaccard:0.20159
 |
Component:155; Jaccard:0.15244
 |
Component:155; Jaccard:0.10013
 |
Correlation:0.33338
 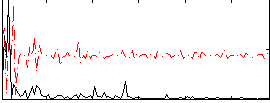 |
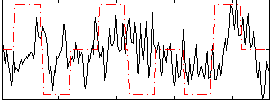
Correlation:0.17779
  |
Correlation:0.17779
  |
Correlation:0.17779
  |
Correlation:0.17779
  |
Correlation:0.17779
  |
Correlation:0.17779
  |
Component:155; Jaccard:0.12356
 |
Component:212; Jaccard:0.14927
 |
Component:212; Jaccard:0.16631
 |
Component:212; Jaccard:0.16655
 |
Component:212; Jaccard:0.1418
 |
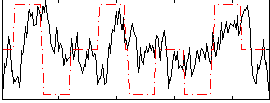
Component:348; Jaccard:0.10931
 |
Component:348; Jaccard:0.077957
 |
Correlation:0.17779
  |
Correlation:0.33338
  |
Correlation:0.33338
  |
Correlation:0.33338
  |
Correlation:0.33338
  |
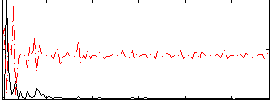
Correlation:0.16281
 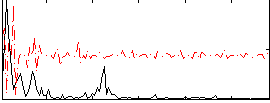 |
Correlation:0.16281
  |
Component:286; Jaccard:0.11361
 |
Component:286; Jaccard:0.13654
 |
Component:286; Jaccard:0.15517
 |
Component:348; Jaccard:0.15241
 |
Component:348; Jaccard:0.13992
 |
Component:212; Jaccard:0.10538
 |
Component:007; Jaccard:0.077945
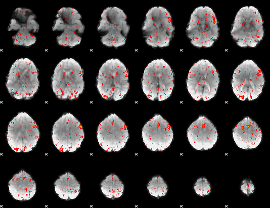 |
Correlation:-0.069501
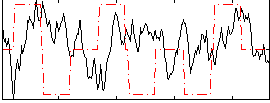 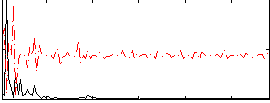 |
Correlation:-0.069501
  |
Correlation:-0.069501
  |
Correlation:0.16281
  |
Correlation:0.16281
  |
Correlation:0.33338
  |
Correlation:0.34935
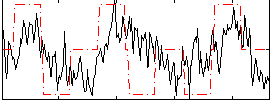 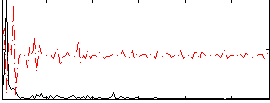 |
Component:188; Jaccard:0.10497
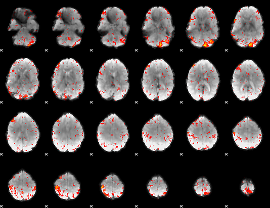 |
Component:348; Jaccard:0.12661
 |
Component:348; Jaccard:0.14776
 |
Component:286; Jaccard:0.14832
 |
Component:007; Jaccard:0.12808
 |
Component:007; Jaccard:0.10462
 |
Component:212; Jaccard:0.071338
 |
Correlation:-0.30897
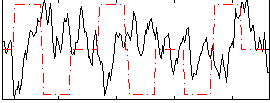 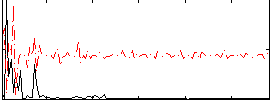 |
Correlation:0.16281
  |
Correlation:0.16281
  |
Correlation:-0.069501
  |
Correlation:0.34935
  |
Correlation:0.34935
  |
Correlation:0.33338
  |
Component:345; Jaccard:0.10351
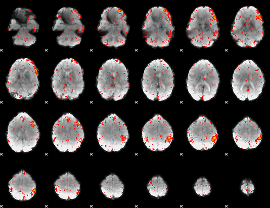 |
Component:007; Jaccard:0.11729
 |
Component:007; Jaccard:0.13515
 |
Component:007; Jaccard:0.14039
 |
Component:286; Jaccard:0.12126
 |
Component:286; Jaccard:0.08362
 |
Component:286; Jaccard:0.052597
 |
Correlation:-0.0096361
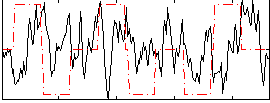 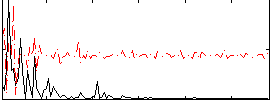 |
Correlation:0.34935
  |
Correlation:0.34935
  |
Correlation:0.34935
  |
Correlation:-0.069501
  |
Correlation:-0.069501
  |
Correlation:-0.069501
  |
Component:348; Jaccard:0.10303
 |
Component:345; Jaccard:0.11418
 |
Component:192; Jaccard:0.11752
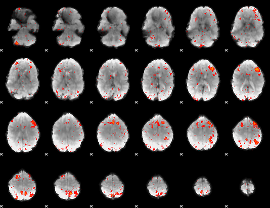 |
Component:192; Jaccard:0.11594
 |
Component:192; Jaccard:0.09842
 |
Component:192; Jaccard:0.07519
 |
Component:192; Jaccard:0.051579
 |
Correlation:0.16281
  |
Correlation:-0.0096361
  |
Correlation:0.063108
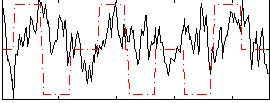 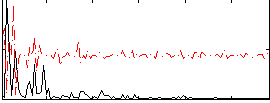 |
Correlation:0.063108
  |
Correlation:0.063108
  |
Correlation:0.063108
  |
Correlation:0.063108
  |
Component:007; Jaccard:0.095978
 |
Component:188; Jaccard:0.11172
 |
Component:052; Jaccard:0.11709
 |
Component:052; Jaccard:0.11207
 |
Component:052; Jaccard:0.092278
 |
Component:052; Jaccard:0.066114
 |
Component:052; Jaccard:0.045266
 |
Correlation:0.34935
  |
Correlation:-0.30897
  |
Correlation:-0.069997
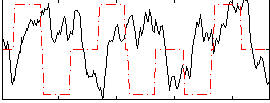 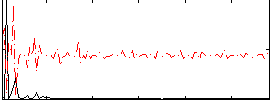 |
Correlation:-0.069997
  |
Correlation:-0.069997
  |
Correlation:-0.069997
  |
Correlation:-0.069997
  |
Component:134; Jaccard:0.095949
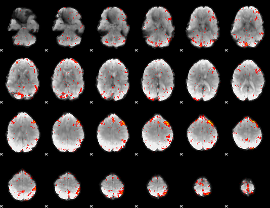 |
Component:052; Jaccard:0.10915
 |
Component:345; Jaccard:0.11536
 |
Component:345; Jaccard:0.10493
 |
Component:345; Jaccard:0.082984
 |
Component:134; Jaccard:0.059186
 |
Component:128; Jaccard:0.040397
 |
Correlation:-0.065743
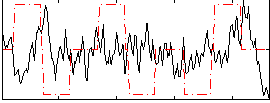 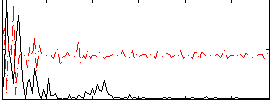 |
Correlation:-0.069997
  |
Correlation:-0.0096361
  |
Correlation:-0.0096361
  |
Correlation:-0.0096361
  |
Correlation:-0.065743
  |
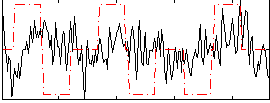
Correlation:0.090604
 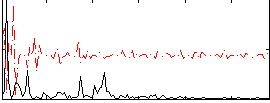 |
Component:266; Jaccard:0.09584
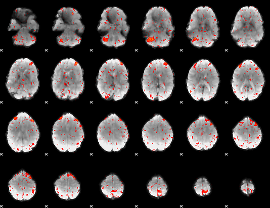 |
Component:266; Jaccard:0.10742
 |
Component:188; Jaccard:0.11093
 |
Component:134; Jaccard:0.10151
 |
Component:134; Jaccard:0.082836
 |
Component:128; Jaccard:0.058791
 |
Component:134; Jaccard:0.038789
 |
Correlation:0.17394
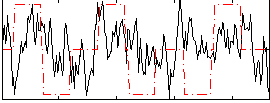 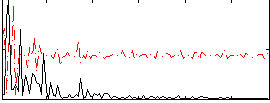 |
Correlation:0.17394
  |
Correlation:-0.30897
  |
Correlation:-0.065743
  |
Correlation:-0.065743
  |
Correlation:0.090604
  |
Correlation:-0.065743
  |
Component:052; Jaccard:0.095478
 |
Component:192; Jaccard:0.10642
 |
Component:134; Jaccard:0.10916
 |
Component:266; Jaccard:0.099044
 |
Component:128; Jaccard:0.082479
 |
Component:345; Jaccard:0.055948
 |
Component:387; Jaccard:0.035657
 |
Correlation:-0.069997
  |
Correlation:0.063108
  |
Correlation:-0.065743
  |
Correlation:0.17394
  |
Correlation:0.090604
  |
Correlation:-0.0096361
  |
Correlation:-0.017232
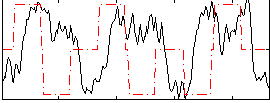 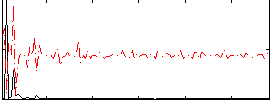 |
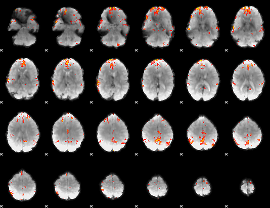
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
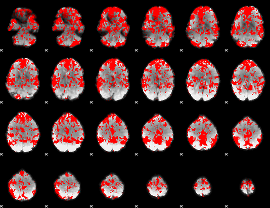 |
GLM binary result thresholded by 1.5
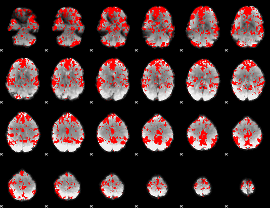 |
GLM binary result thresholded by 2
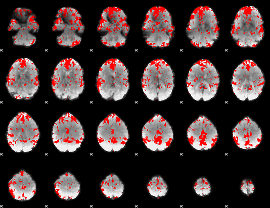 |
GLM binary result thresholded by 2.5
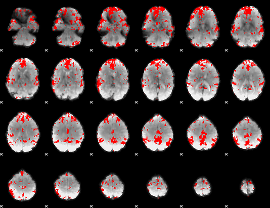 |
GLM binary result thresholded by 3
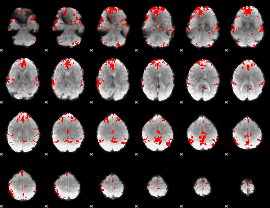 |
GLM binary result thresholded by 3.5
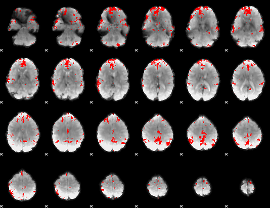 |
GLM binary result thresholded by 4
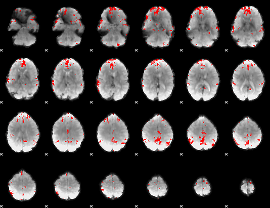 |
GLM Z-score result thresholded by 1
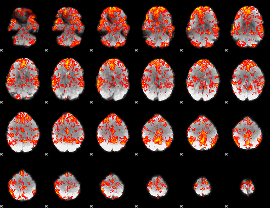 |
GLM Z-score result thresholded by 1.5
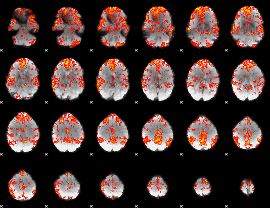 |
GLM Z-score result thresholded by 2
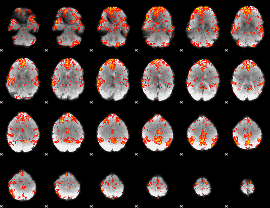 |
GLM Z-score result thresholded by 2.5
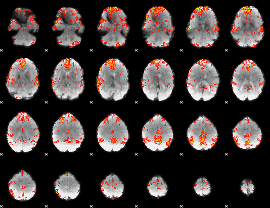 |
GLM Z-score result thresholded by 3
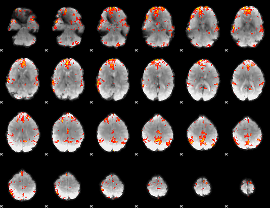 |
GLM Z-score result thresholded by 3.5
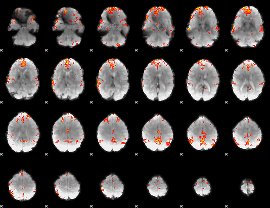 |
GLM Z-score result thresholded by 4
 |
Component:200; Jaccard:0.13196
 |
Component:200; Jaccard:0.16095
 |
Component:200; Jaccard:0.18973
 |
Component:075; Jaccard:0.22648
 |
Component:075; Jaccard:0.25297
 |
Component:075; Jaccard:0.26432
 |
Component:075; Jaccard:0.25274
 |
Correlation:0.13154
  |
Correlation:0.13154
  |
Correlation:0.13154
  |
Correlation:0.16444
 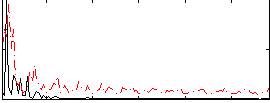 |
Correlation:0.16444
  |
Correlation:0.16444
  |
Correlation:0.16444
  |
Component:075; Jaccard:0.11757
 |
Component:075; Jaccard:0.15063
 |
Component:075; Jaccard:0.18774
 |
Component:200; Jaccard:0.21066
 |
Component:200; Jaccard:0.21087
 |
Component:063; Jaccard:0.21567
 |
Component:063; Jaccard:0.20008
 |
Correlation:0.16444
  |
Correlation:0.16444
  |
Correlation:0.16444
  |
Correlation:0.13154
  |
Correlation:0.13154
  |
Correlation:0.1333
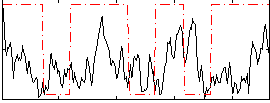 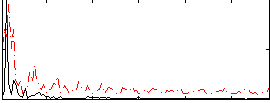 |
Correlation:0.1333
  |
Component:016; Jaccard:0.11621
 |
Component:063; Jaccard:0.13461
 |
Component:063; Jaccard:0.16304
 |
Component:063; Jaccard:0.18946
 |
Component:063; Jaccard:0.21024
 |
Component:200; Jaccard:0.19013
 |
Component:126; Jaccard:0.17258
 |
Correlation:0.058439
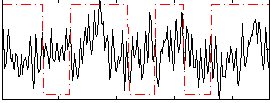 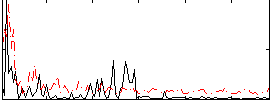 |
Correlation:0.1333
  |
Correlation:0.1333
  |
Correlation:0.1333
  |
Correlation:0.1333
  |
Correlation:0.13154
  |
Correlation:-0.025559
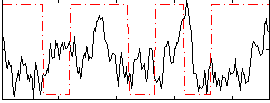 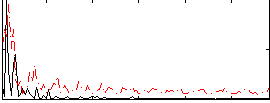 |
Component:063; Jaccard:0.10789
 |
Component:016; Jaccard:0.13142
 |
Component:126; Jaccard:0.15023
 |
Component:126; Jaccard:0.17281
 |
Component:126; Jaccard:0.18747
 |
Component:126; Jaccard:0.18666
 |
Component:215; Jaccard:0.16035
 |
Correlation:0.1333
  |
Correlation:0.058439
  |
Correlation:-0.025559
  |
Correlation:-0.025559
  |
Correlation:-0.025559
  |
Correlation:-0.025559
  |
Correlation:0.16164
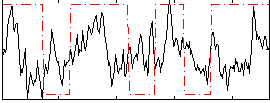  |
Component:153; Jaccard:0.10662
 |
Component:126; Jaccard:0.12544
 |
Component:016; Jaccard:0.142
 |
Component:215; Jaccard:0.15302
 |
Component:215; Jaccard:0.16821
 |
Component:215; Jaccard:0.17076
 |
Component:200; Jaccard:0.15808
 |
Correlation:0.16101
  |
Correlation:-0.025559
  |
Correlation:0.058439
  |
Correlation:0.16164
  |
Correlation:0.16164
  |
Correlation:0.16164
  |
Correlation:0.13154
  |
Component:126; Jaccard:0.10232
 |
Component:153; Jaccard:0.1127
 |
Component:215; Jaccard:0.13429
 |
Component:016; Jaccard:0.14366
 |
Component:144; Jaccard:0.15279
 |
Component:144; Jaccard:0.15858
 |
Component:144; Jaccard:0.14886
 |
Correlation:-0.025559
  |
Correlation:0.16101
  |
Correlation:0.16164
  |
Correlation:0.058439
  |
Correlation:0.13347
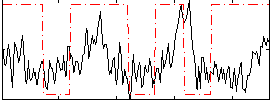 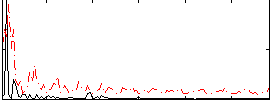 |
Correlation:0.13347
  |
Correlation:0.13347
  |
Component:167; Jaccard:0.098057
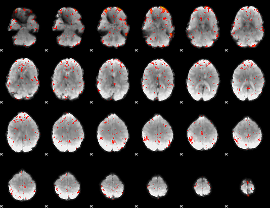 |
Component:215; Jaccard:0.11163
 |
Component:131; Jaccard:0.12334
 |
Component:144; Jaccard:0.13682
 |
Component:131; Jaccard:0.13863
 |
Component:131; Jaccard:0.13239
 |
Component:131; Jaccard:0.11417
 |
Correlation:0.0325
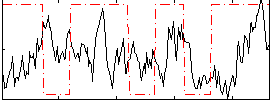 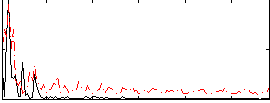 |
Correlation:0.16164
  |
Correlation:-0.14379
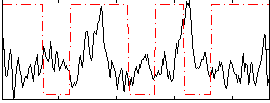 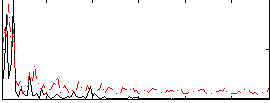 |
Correlation:0.13347
  |
Correlation:-0.14379
  |
Correlation:-0.14379
  |
Correlation:-0.14379
  |
Component:342; Jaccard:0.096387
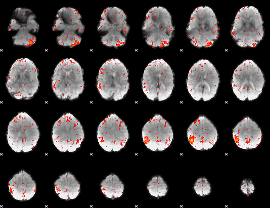 |
Component:167; Jaccard:0.111
 |
Component:167; Jaccard:0.122
 |
Component:131; Jaccard:0.13648
 |
Component:016; Jaccard:0.13488
 |
Component:016; Jaccard:0.11895
 |
Component:194; Jaccard:0.099298
 |
Correlation:0.037208
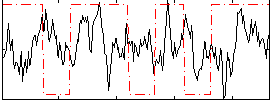 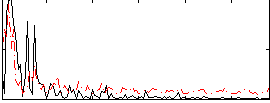 |
Correlation:0.0325
  |
Correlation:0.0325
  |
Correlation:-0.14379
  |
Correlation:0.058439
  |
Correlation:0.058439
  |
Correlation:0.065234
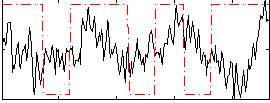 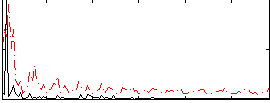 |
Component:131; Jaccard:0.093446
 |
Component:131; Jaccard:0.10904
 |
Component:144; Jaccard:0.11835
 |
Component:167; Jaccard:0.12678
 |
Component:167; Jaccard:0.12429
 |
Component:167; Jaccard:0.11369
 |
Component:016; Jaccard:0.096001
 |
Correlation:-0.14379
  |
Correlation:-0.14379
  |
Correlation:0.13347
  |
Correlation:0.0325
  |
Correlation:0.0325
  |
Correlation:0.0325
  |
Correlation:0.058439
  |
Component:218; Jaccard:0.093016
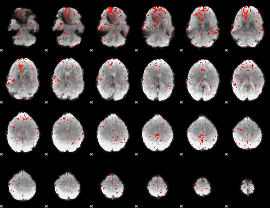 |
Component:342; Jaccard:0.105
 |
Component:153; Jaccard:0.11382
 |
Component:194; Jaccard:0.11449
 |
Component:194; Jaccard:0.11837
 |
Component:194; Jaccard:0.11175
 |
Component:167; Jaccard:0.092442
 |
Correlation:0.2204
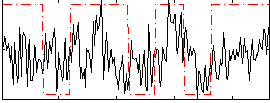 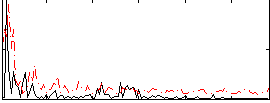 |
Correlation:0.037208
  |
Correlation:0.16101
  |
Correlation:0.065234
  |
Correlation:0.065234
  |
Correlation:0.065234
  |
Correlation:0.0325
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
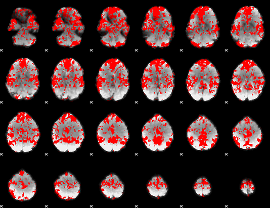 |
GLM binary result thresholded by 1.5
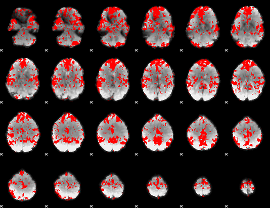 |
GLM binary result thresholded by 2
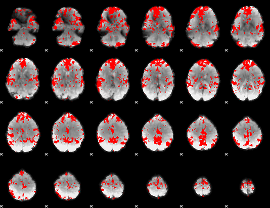 |
GLM binary result thresholded by 2.5
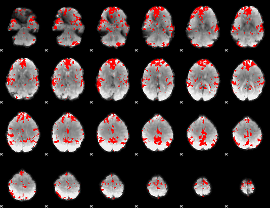 |
GLM binary result thresholded by 3
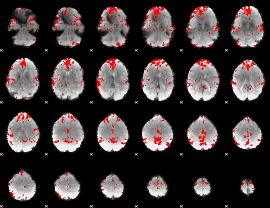 |
GLM binary result thresholded by 3.5
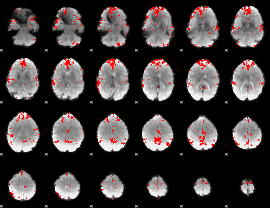 |
GLM binary result thresholded by 4
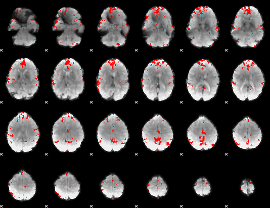 |
GLM Z-score result thresholded by 1
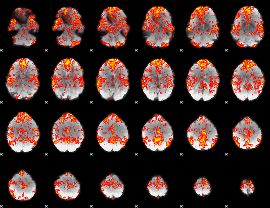 |
GLM Z-score result thresholded by 1.5
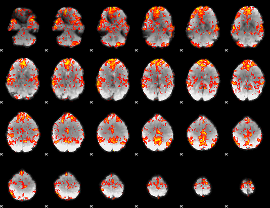 |
GLM Z-score result thresholded by 2
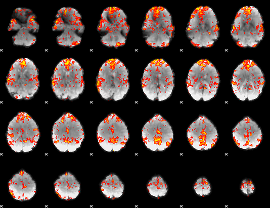 |
GLM Z-score result thresholded by 2.5
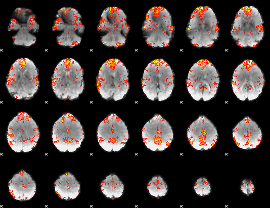 |
GLM Z-score result thresholded by 3
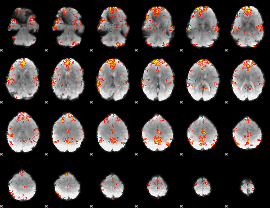 |
GLM Z-score result thresholded by 3.5
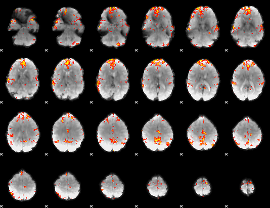 |
GLM Z-score result thresholded by 4
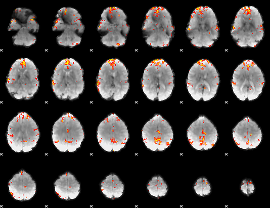 |
Component:200; Jaccard:0.14724
 |
Component:200; Jaccard:0.18157
 |
Component:200; Jaccard:0.21568
 |
Component:200; Jaccard:0.24274
 |
Component:200; Jaccard:0.25525
 |
Component:075; Jaccard:0.25036
 |
Component:075; Jaccard:0.24862
 |
Correlation:0.42947
  |
Correlation:0.42947
  |
Correlation:0.42947
  |
Correlation:0.42947
  |
Correlation:0.42947
  |
Correlation:0.14421
 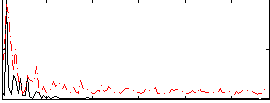 |
Correlation:0.14421
  |
Component:075; Jaccard:0.12396
 |
Component:075; Jaccard:0.15326
 |
Component:075; Jaccard:0.18649
 |
Component:075; Jaccard:0.21608
 |
Component:075; Jaccard:0.2389
 |
Component:200; Jaccard:0.24554
 |
Component:200; Jaccard:0.21817
 |
Correlation:0.14421
  |
Correlation:0.14421
  |
Correlation:0.14421
  |
Correlation:0.14421
  |
Correlation:0.14421
  |
Correlation:0.42947
  |
Correlation:0.42947
  |
Component:016; Jaccard:0.11696
 |
Component:126; Jaccard:0.13838
 |
Component:126; Jaccard:0.16267
 |
Component:126; Jaccard:0.18348
 |
Component:126; Jaccard:0.19815
 |
Component:215; Jaccard:0.20583
 |
Component:215; Jaccard:0.20214
 |
Correlation:0.21675
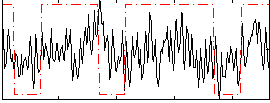 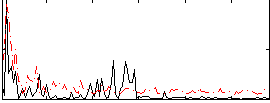 |
Correlation:0.23761
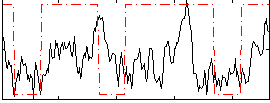 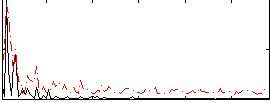 |
Correlation:0.23761
  |
Correlation:0.23761
  |
Correlation:0.23761
  |
Correlation:0.28842
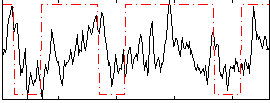  |
Correlation:0.28842
  |
Component:126; Jaccard:0.11424
 |
Component:063; Jaccard:0.13042
 |
Component:215; Jaccard:0.15578
 |
Component:215; Jaccard:0.18036
 |
Component:215; Jaccard:0.19772
 |
Component:126; Jaccard:0.20175
 |
Component:126; Jaccard:0.19439
 |
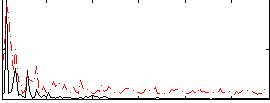
Correlation:0.23761
  |
Correlation:0.044119
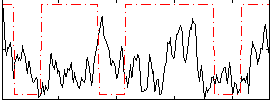 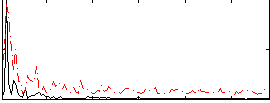 |
Correlation:0.28842
  |
Correlation:0.28842
  |
Correlation:0.28842
  |
Correlation:0.23761
  |
Correlation:0.23761
  |
Component:063; Jaccard:0.10926
 |
Component:215; Jaccard:0.13008
 |
Component:063; Jaccard:0.15325
 |
Component:063; Jaccard:0.17277
 |
Component:063; Jaccard:0.1863
 |
Component:063; Jaccard:0.18714
 |
Component:063; Jaccard:0.17854
 |
Correlation:0.044119
  |
Correlation:0.28842
  |
Correlation:0.044119
  |
Correlation:0.044119
  |
Correlation:0.044119
  |
Correlation:0.044119
  |
Correlation:0.044119
  |
Component:215; Jaccard:0.10674
 |
Component:016; Jaccard:0.12788
 |
Component:016; Jaccard:0.13486
 |
Component:144; Jaccard:0.14395
 |
Component:144; Jaccard:0.15559
 |
Component:144; Jaccard:0.15865
 |
Component:144; Jaccard:0.1544
 |
Correlation:0.28842
  |
Correlation:0.21675
  |
Correlation:0.21675
  |
Correlation:0.12808
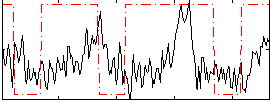 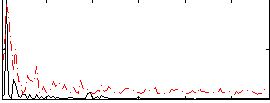 |
Correlation:0.12808
  |
Correlation:0.12808
  |
Correlation:0.12808
  |
Component:119; Jaccard:0.097798
 |
Component:119; Jaccard:0.11208
 |
Component:194; Jaccard:0.12734
 |
Component:194; Jaccard:0.14034
 |
Component:194; Jaccard:0.14655
 |
Component:194; Jaccard:0.14319
 |
Component:194; Jaccard:0.13628
 |
Correlation:0.215
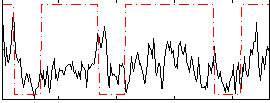 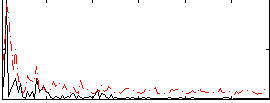 |
Correlation:0.215
  |
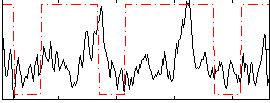
Correlation:0.30983
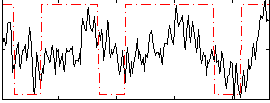  |
Correlation:0.30983
  |
Correlation:0.30983
  |
Correlation:0.30983
  |
Correlation:0.30983
  |
Component:131; Jaccard:0.09752
 |
Component:131; Jaccard:0.11027
 |
Component:119; Jaccard:0.12551
 |
Component:016; Jaccard:0.13604
 |
Component:119; Jaccard:0.1396
 |
Component:119; Jaccard:0.14087
 |
Component:119; Jaccard:0.1305
 |
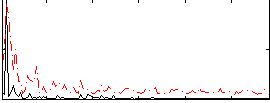
Correlation:0.18771
 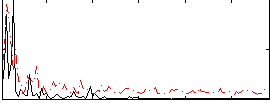 |
Correlation:0.18771
  |
Correlation:0.215
  |
Correlation:0.21675
  |
Correlation:0.215
  |
Correlation:0.215
  |
Correlation:0.215
  |
Component:153; Jaccard:0.095019
 |
Component:194; Jaccard:0.10903
 |
Component:144; Jaccard:0.12488
 |
Component:119; Jaccard:0.13487
 |
Component:131; Jaccard:0.13594
 |
Component:131; Jaccard:0.12966
 |
Component:131; Jaccard:0.12094
 |
Correlation:0.29489
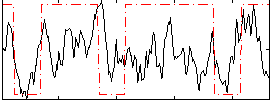 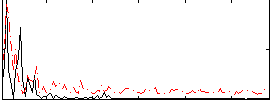 |
Correlation:0.30983
  |
Correlation:0.12808
  |
Correlation:0.215
  |
Correlation:0.18771
  |
Correlation:0.18771
  |
Correlation:0.18771
  |
Component:284; Jaccard:0.094932
 |
Component:144; Jaccard:0.10493
 |
Component:131; Jaccard:0.12441
 |
Component:131; Jaccard:0.13318
 |
Component:016; Jaccard:0.13074
 |
Component:016; Jaccard:0.11705
 |
Component:016; Jaccard:0.10008
 |
Correlation:0.2397
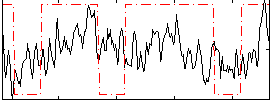  |
Correlation:0.12808
  |
Correlation:0.18771
  |
Correlation:0.18771
  |
Correlation:0.21675
  |
Correlation:0.21675
  |
Correlation:0.21675
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































