Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
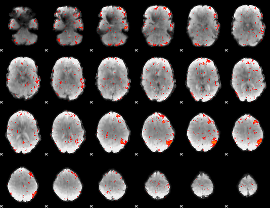
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
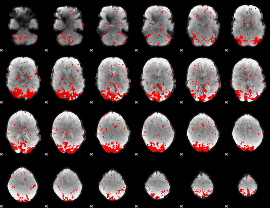 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
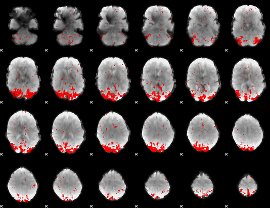
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
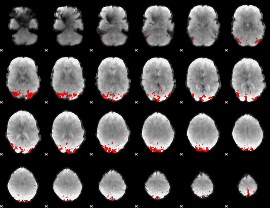
GLM Z-score result thresholded by 1.5
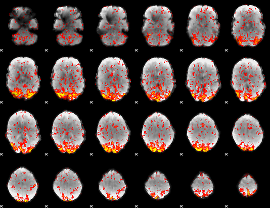 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
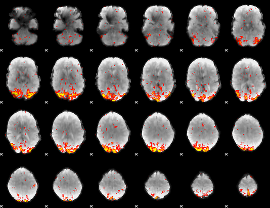 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:019; Jaccard:0.15761
 |
Component:019; Jaccard:0.21259
 |
Component:019; Jaccard:0.26445
 |
Component:019; Jaccard:0.28675
 |
Component:019; Jaccard:0.28403
 |
Component:019; Jaccard:0.25647
 |
Component:019; Jaccard:0.22463
 |
Correlation:0.086155
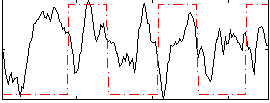 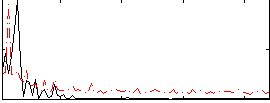 |
Correlation:0.086155
  |
Correlation:0.086155
  |
Correlation:0.086155
  |
Correlation:0.086155
  |
Correlation:0.086155
  |
Correlation:0.086155
  |
Component:399; Jaccard:0.1562
 |
Component:399; Jaccard:0.20173
 |
Component:399; Jaccard:0.23874
 |
Component:399; Jaccard:0.25327
 |
Component:006; Jaccard:0.2531
 |
Component:006; Jaccard:0.2373
 |
Component:006; Jaccard:0.2052
 |
Correlation:-0.087126
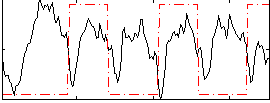  |
Correlation:-0.087126
  |
Correlation:-0.087126
  |
Correlation:-0.087126
  |
Correlation:0.44769
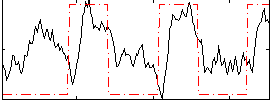 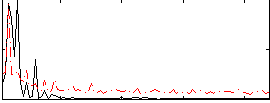 |
Correlation:0.44769
  |
Correlation:0.44769
  |
Component:006; Jaccard:0.15106
 |
Component:006; Jaccard:0.19327
 |
Component:006; Jaccard:0.23093
 |
Component:006; Jaccard:0.25271
 |
Component:399; Jaccard:0.24328
 |
Component:399; Jaccard:0.21721
 |
Component:399; Jaccard:0.18484
 |
Correlation:0.44769
  |
Correlation:0.44769
  |
Correlation:0.44769
  |
Correlation:0.44769
  |
Correlation:-0.087126
  |
Correlation:-0.087126
  |
Correlation:-0.087126
  |
Component:149; Jaccard:0.13057
 |
Component:149; Jaccard:0.15171
 |
Component:289; Jaccard:0.16354
 |
Component:234; Jaccard:0.16515
 |
Component:234; Jaccard:0.16459
 |
Component:234; Jaccard:0.15851
 |
Component:234; Jaccard:0.14458
 |
Correlation:-0.0058885
  |
Correlation:-0.0058885
  |
Correlation:0.17727
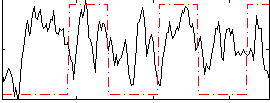 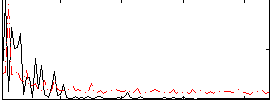 |
Correlation:0.49461
  |
Correlation:0.49461
  |
Correlation:0.49461
  |
Correlation:0.49461
  |
Component:289; Jaccard:0.12716
 |
Component:289; Jaccard:0.15138
 |
Component:149; Jaccard:0.1618
 |
Component:289; Jaccard:0.16312
 |
Component:289; Jaccard:0.15025
 |
Component:289; Jaccard:0.12594
 |
Component:041; Jaccard:0.10396
 |
Correlation:0.17727
  |
Correlation:0.17727
  |
Correlation:-0.0058885
  |
Correlation:0.17727
  |
Correlation:0.17727
  |
Correlation:0.17727
  |
Correlation:0.25883
  |
Component:120; Jaccard:0.11732
 |
Component:120; Jaccard:0.13466
 |
Component:234; Jaccard:0.1543
 |
Component:149; Jaccard:0.15748
 |
Component:149; Jaccard:0.14096
 |
Component:149; Jaccard:0.12014
 |
Component:289; Jaccard:0.10359
 |
Correlation:-0.20569
  |
Correlation:-0.20569
  |
Correlation:0.49461
  |
Correlation:-0.0058885
  |
Correlation:-0.0058885
  |
Correlation:-0.0058885
  |
Correlation:0.17727
  |
Component:281; Jaccard:0.10817
 |
Component:234; Jaccard:0.13243
 |
Component:120; Jaccard:0.14302
 |
Component:120; Jaccard:0.14075
 |
Component:120; Jaccard:0.12734
 |
Component:041; Jaccard:0.11774
 |
Component:149; Jaccard:0.10073
 |
Correlation:-0.22647
  |
Correlation:0.49461
  |
Correlation:-0.20569
  |
Correlation:-0.20569
  |
Correlation:-0.20569
  |
Correlation:0.25883
  |
Correlation:-0.0058885
  |
Component:234; Jaccard:0.10756
 |
Component:281; Jaccard:0.12509
 |
Component:281; Jaccard:0.13251
 |
Component:041; Jaccard:0.12934
 |
Component:041; Jaccard:0.12459
 |
Component:120; Jaccard:0.10863
 |
Component:120; Jaccard:0.092428
 |
Correlation:0.49461
  |
Correlation:-0.22647
  |
Correlation:-0.22647
  |
Correlation:0.25883
  |
Correlation:0.25883
  |
Correlation:-0.20569
  |
Correlation:-0.20569
  |
Component:348; Jaccard:0.10395
 |
Component:041; Jaccard:0.11653
 |
Component:041; Jaccard:0.12673
 |
Component:281; Jaccard:0.12639
 |
Component:281; Jaccard:0.11033
 |
Component:281; Jaccard:0.087795
 |
Component:348; Jaccard:0.070046
 |
Correlation:0.037543
  |
Correlation:0.25883
  |
Correlation:0.25883
  |
Correlation:-0.22647
  |
Correlation:-0.22647
  |
Correlation:-0.22647
  |
Correlation:0.037543
  |
Component:041; Jaccard:0.10117
 |
Component:348; Jaccard:0.11566
 |
Component:348; Jaccard:0.11878
 |
Component:348; Jaccard:0.11085
 |
Component:348; Jaccard:0.099012
 |
Component:348; Jaccard:0.083619
 |
Component:281; Jaccard:0.069542
 |
Correlation:0.25883
  |
Correlation:0.037543
  |
Correlation:0.037543
  |
Correlation:0.037543
  |
Correlation:0.037543
  |
Correlation:0.037543
  |
Correlation:-0.22647
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
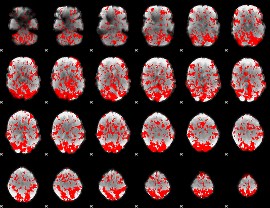 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
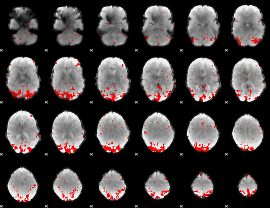 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
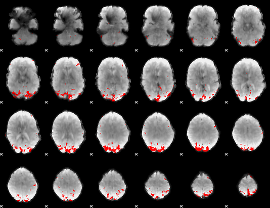 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
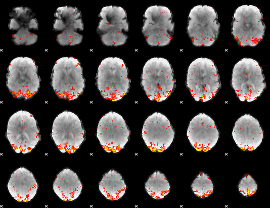 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:399; Jaccard:0.16157
 |
Component:399; Jaccard:0.21107
 |
Component:399; Jaccard:0.26099
 |
Component:399; Jaccard:0.29376
 |
Component:019; Jaccard:0.30574
 |
Component:019; Jaccard:0.29812
 |
Component:019; Jaccard:0.27024
 |
Correlation:0.36902
  |
Correlation:0.36902
  |
Correlation:0.36902
  |
Correlation:0.36902
  |
Correlation:0.16118
  |
Correlation:0.16118
  |
Correlation:0.16118
  |
Component:019; Jaccard:0.14683
 |
Component:019; Jaccard:0.19333
 |
Component:019; Jaccard:0.24322
 |
Component:019; Jaccard:0.28577
 |
Component:399; Jaccard:0.30137
 |
Component:399; Jaccard:0.28263
 |
Component:399; Jaccard:0.24642
 |
Correlation:0.16118
  |
Correlation:0.16118
  |
Correlation:0.16118
  |
Correlation:0.16118
  |
Correlation:0.36902
  |
Correlation:0.36902
  |
Correlation:0.36902
  |
Component:249; Jaccard:0.13062
 |
Component:249; Jaccard:0.15406
 |
Component:249; Jaccard:0.1696
 |
Component:120; Jaccard:0.17364
 |
Component:120; Jaccard:0.16903
 |
Component:120; Jaccard:0.15314
 |
Component:120; Jaccard:0.12929
 |
Correlation:0.52198
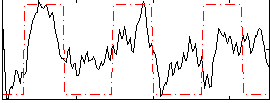  |
Correlation:0.52198
  |
Correlation:0.52198
  |
Correlation:0.33102
  |
Correlation:0.33102
  |
Correlation:0.33102
  |
Correlation:0.33102
  |
Component:120; Jaccard:0.12778
 |
Component:120; Jaccard:0.14811
 |
Component:120; Jaccard:0.16525
 |
Component:249; Jaccard:0.17071
 |
Component:281; Jaccard:0.16471
 |
Component:281; Jaccard:0.14233
 |
Component:281; Jaccard:0.11432
 |
Correlation:0.33102
  |
Correlation:0.33102
  |
Correlation:0.33102
  |
Correlation:0.52198
  |
Correlation:0.3625
 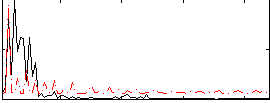 |
Correlation:0.3625
  |
Correlation:0.3625
  |
Component:214; Jaccard:0.12164
 |
Component:214; Jaccard:0.14432
 |
Component:214; Jaccard:0.16007
 |
Component:281; Jaccard:0.17006
 |
Component:249; Jaccard:0.15745
 |
Component:249; Jaccard:0.13484
 |
Component:214; Jaccard:0.1117
 |
Correlation:0.44452
  |
Correlation:0.44452
  |
Correlation:0.44452
  |
Correlation:0.3625
  |
Correlation:0.52198
  |
Correlation:0.52198
  |
Correlation:0.44452
  |
Component:149; Jaccard:0.12084
 |
Component:289; Jaccard:0.14034
 |
Component:281; Jaccard:0.15972
 |
Component:214; Jaccard:0.16193
 |
Component:214; Jaccard:0.15457
 |
Component:214; Jaccard:0.13434
 |
Component:006; Jaccard:0.11064
 |
Correlation:0.20169
  |
Correlation:0.14331
  |
Correlation:0.3625
  |
Correlation:0.44452
  |
Correlation:0.44452
  |
Correlation:0.44452
  |
Correlation:-0.24199
 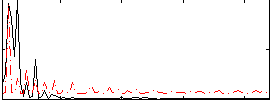 |
Component:289; Jaccard:0.11908
 |
Component:149; Jaccard:0.13925
 |
Component:289; Jaccard:0.15641
 |
Component:149; Jaccard:0.15848
 |
Component:149; Jaccard:0.14915
 |
Component:149; Jaccard:0.13044
 |
Component:249; Jaccard:0.10843
 |
Correlation:0.14331
  |
Correlation:0.20169
  |
Correlation:0.14331
  |
Correlation:0.20169
  |
Correlation:0.20169
  |
Correlation:0.20169
  |
Correlation:0.52198
  |
Component:281; Jaccard:0.11683
 |
Component:281; Jaccard:0.13914
 |
Component:149; Jaccard:0.15261
 |
Component:289; Jaccard:0.15787
 |
Component:289; Jaccard:0.14625
 |
Component:289; Jaccard:0.12771
 |
Component:289; Jaccard:0.10822
 |
Correlation:0.3625
  |
Correlation:0.3625
  |
Correlation:0.20169
  |
Correlation:0.14331
  |
Correlation:0.14331
  |
Correlation:0.14331
  |
Correlation:0.14331
  |
Component:132; Jaccard:0.10562
 |
Component:132; Jaccard:0.11528
 |
Component:006; Jaccard:0.12542
 |
Component:006; Jaccard:0.13238
 |
Component:006; Jaccard:0.13369
 |
Component:006; Jaccard:0.12752
 |
Component:149; Jaccard:0.10772
 |
Correlation:0.3592
  |
Correlation:0.3592
  |
Correlation:-0.24199
  |
Correlation:-0.24199
  |
Correlation:-0.24199
  |
Correlation:-0.24199
  |
Correlation:0.20169
  |
Component:346; Jaccard:0.098716
 |
Component:006; Jaccard:0.11243
 |
Component:132; Jaccard:0.1168
 |
Component:132; Jaccard:0.10662
 |
Component:132; Jaccard:0.090696
 |
Component:041; Jaccard:0.08389
 |
Component:041; Jaccard:0.076294
 |
Correlation:0.48913
  |
Correlation:-0.24199
  |
Correlation:0.3592
  |
Correlation:0.3592
  |
Correlation:0.3592
  |
Correlation:-0.19466
  |
Correlation:-0.19466
  |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
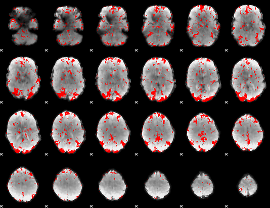 |
GLM binary result thresholded by 2.5
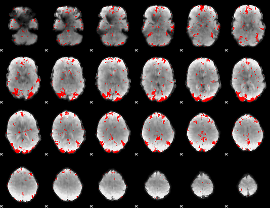 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
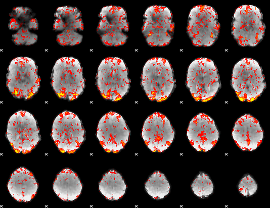 |
GLM Z-score result thresholded by 2
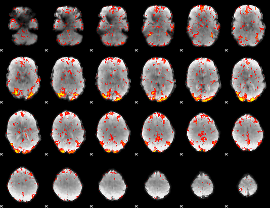 |
GLM Z-score result thresholded by 2.5
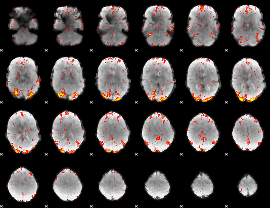 |
GLM Z-score result thresholded by 3
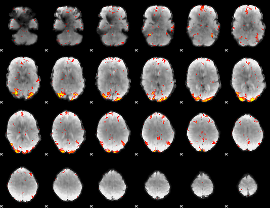 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:006; Jaccard:0.12969
 |
Component:006; Jaccard:0.15763
 |
Component:390; Jaccard:0.18823
 |
Component:390; Jaccard:0.21306
 |
Component:390; Jaccard:0.21615
 |
Component:390; Jaccard:0.20185
 |
Component:390; Jaccard:0.17679
 |
Correlation:0.37409
  |
Correlation:0.37409
  |
Correlation:0.65456
  |
Correlation:0.65456
  |
Correlation:0.65456
  |
Correlation:0.65456
  |
Correlation:0.65456
  |
Component:394; Jaccard:0.11991
 |
Component:390; Jaccard:0.15321
 |
Component:006; Jaccard:0.18378
 |
Component:006; Jaccard:0.19856
 |
Component:006; Jaccard:0.19427
 |
Component:006; Jaccard:0.17231
 |
Component:006; Jaccard:0.1439
 |
Correlation:0.20938
  |
Correlation:0.65456
  |
Correlation:0.37409
  |
Correlation:0.37409
  |
Correlation:0.37409
  |
Correlation:0.37409
  |
Correlation:0.37409
  |
Component:390; Jaccard:0.11782
 |
Component:394; Jaccard:0.1461
 |
Component:394; Jaccard:0.16122
 |
Component:394; Jaccard:0.16211
 |
Component:234; Jaccard:0.15527
 |
Component:234; Jaccard:0.14819
 |
Component:234; Jaccard:0.13484
 |
Correlation:0.65456
  |
Correlation:0.20938
  |
Correlation:0.20938
  |
Correlation:0.20938
  |
Correlation:0.43054
  |
Correlation:0.43054
  |
Correlation:0.43054
  |
Component:362; Jaccard:0.10135
 |
Component:234; Jaccard:0.11374
 |
Component:234; Jaccard:0.13248
 |
Component:234; Jaccard:0.14746
 |
Component:394; Jaccard:0.13521
 |
Component:394; Jaccard:0.094107
 |
Component:041; Jaccard:0.057064
 |
Correlation:-0.032756
  |
Correlation:0.43054
  |
Correlation:0.43054
  |
Correlation:0.43054
  |
Correlation:0.20938
  |
Correlation:0.20938
  |
Correlation:0.24598
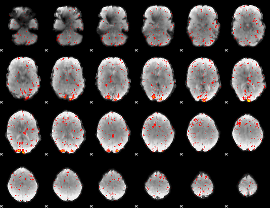
 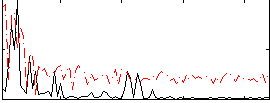 |
Component:234; Jaccard:0.094988
 |
Component:362; Jaccard:0.11116
 |
Component:362; Jaccard:0.11017
 |
Component:362; Jaccard:0.097722
 |
Component:041; Jaccard:0.077363
 |
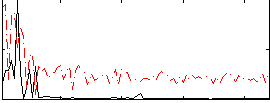
Component:095; Jaccard:0.068848
 |
Component:095; Jaccard:0.056
 |
Correlation:0.43054
  |
Correlation:-0.032756
  |
Correlation:-0.032756
  |
Correlation:-0.032756
  |
Correlation:0.24598
  |
Correlation:0.31234
  |
Correlation:0.31234
  |
Component:389; Jaccard:0.082767
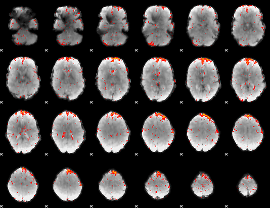 |
Component:071; Jaccard:0.08909
 |
Component:071; Jaccard:0.090876
 |
Component:071; Jaccard:0.086685
 |
Component:095; Jaccard:0.076774
 |
Component:041; Jaccard:0.067574
 |
Component:394; Jaccard:0.053306
 |
Correlation:0.12832
  |
Correlation:0.34034
  |
Correlation:0.34034
  |
Correlation:0.34034
  |
Correlation:0.31234
  |
Correlation:0.24598
  |
Correlation:0.20938
  |
Component:071; Jaccard:0.079926
 |
Component:389; Jaccard:0.087469
 |
Component:141; Jaccard:0.087599
 |
Component:306; Jaccard:0.081585
 |
Component:362; Jaccard:0.074521
 |
Component:121; Jaccard:0.06393
 |
Component:121; Jaccard:0.052662
 |
Correlation:0.34034
  |
Correlation:0.12832
  |
Correlation:0.13037
  |
Correlation:0.32683
  |
Correlation:-0.032756
  |
Correlation:0.39594
  |
Correlation:0.39594
  |
Component:171; Jaccard:0.078138
 |
Component:141; Jaccard:0.085159
 |
Component:389; Jaccard:0.085145
 |
Component:041; Jaccard:0.081569
 |
Component:071; Jaccard:0.07394
 |
Component:306; Jaccard:0.054204
 |
Component:306; Jaccard:0.035304
 |
Correlation:-0.18325
  |
Correlation:0.13037
  |
Correlation:0.12832
  |
Correlation:0.24598
  |
Correlation:0.34034
  |
Correlation:0.32683
  |
Correlation:0.32683
  |
Component:142; Jaccard:0.078088
 |
Component:142; Jaccard:0.08276
 |
Component:041; Jaccard:0.083761
 |
Component:095; Jaccard:0.08146
 |
Component:121; Jaccard:0.072078
 |
Component:071; Jaccard:0.052594
 |
Component:019; Jaccard:0.032762
 |
Correlation:0.12017
  |
Correlation:0.12017
  |
Correlation:0.24598
  |
Correlation:0.31234
  |
Correlation:0.39594
  |
Correlation:0.34034
  |
Correlation:-0.040694
  |
Component:141; Jaccard:0.076565
 |
Component:041; Jaccard:0.080324
 |
Component:306; Jaccard:0.082438
 |
Component:121; Jaccard:0.08137
 |
Component:306; Jaccard:0.071122
 |
Component:362; Jaccard:0.050588
 |
Component:071; Jaccard:0.032731
 |
Correlation:0.13037
  |
Correlation:0.24598
  |
Correlation:0.32683
  |
Correlation:0.39594
  |
Correlation:0.32683
  |
Correlation:-0.032756
  |
Correlation:0.34034
  |
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
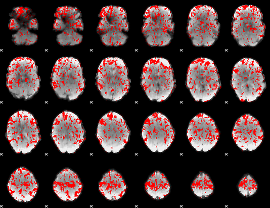 |
GLM binary result thresholded by 1.5
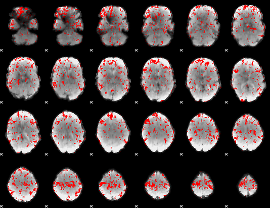 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
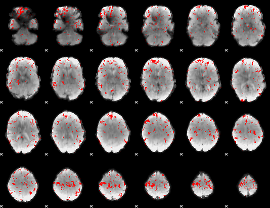
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
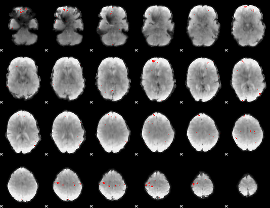 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
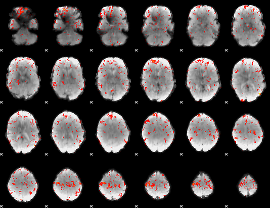 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
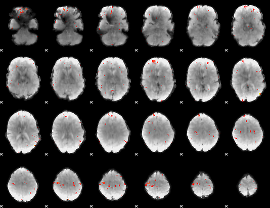 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:283; Jaccard:0.12089
 |
Component:283; Jaccard:0.12672
 |
Component:067; Jaccard:0.11493
 |
Component:039; Jaccard:0.093376
 |
Component:039; Jaccard:0.065381
 |
Component:039; Jaccard:0.038539
 |
Component:039; Jaccard:0.020802
 |
Correlation:0.19719
  |
Correlation:0.19719
  |
Correlation:0.23414
  |
Correlation:0.51209
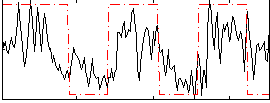  |
Correlation:0.51209
  |
Correlation:0.51209
  |
Correlation:0.51209
  |
Component:067; Jaccard:0.10636
 |
Component:067; Jaccard:0.11755
 |
Component:283; Jaccard:0.11196
 |
Component:067; Jaccard:0.090449
 |
Component:067; Jaccard:0.054306
 |
Component:067; Jaccard:0.027344
 |
Component:067; Jaccard:0.012637
 |
Correlation:0.23414
  |
Correlation:0.23414
  |
Correlation:0.19719
  |
Correlation:0.23414
  |
Correlation:0.23414
  |
Correlation:0.23414
  |
Correlation:0.23414
  |
Component:361; Jaccard:0.086368
 |
Component:039; Jaccard:0.097169
 |
Component:039; Jaccard:0.10644
 |
Component:283; Jaccard:0.075687
 |
Component:283; Jaccard:0.038903
 |
Component:283; Jaccard:0.01864
 |
Component:283; Jaccard:0.008954
 |
Correlation:0.29124
  |
Correlation:0.51209
  |
Correlation:0.51209
  |
Correlation:0.19719
  |
Correlation:0.19719
  |
Correlation:0.19719
  |
Correlation:0.19719
  |
Component:170; Jaccard:0.082029
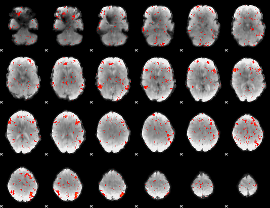 |
Component:170; Jaccard:0.089974
 |
Component:170; Jaccard:0.084273
 |
Component:170; Jaccard:0.063227
 |
Component:361; Jaccard:0.036089
 |
Component:361; Jaccard:0.017015
 |
Component:361; Jaccard:0.007921
 |
Correlation:0.15517
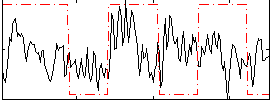  |
Correlation:0.15517
  |
Correlation:0.15517
  |
Correlation:0.15517
  |
Correlation:0.29124
  |
Correlation:0.29124
  |
Correlation:0.29124
  |
Component:039; Jaccard:0.081205
 |
Component:361; Jaccard:0.089329
 |
Component:361; Jaccard:0.083436
 |
Component:361; Jaccard:0.061395
 |
Component:170; Jaccard:0.032922
 |
Component:170; Jaccard:0.014347
 |
Component:091; Jaccard:0.006831
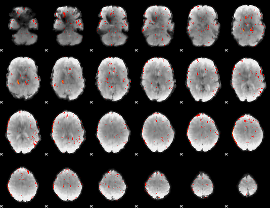 |
Correlation:0.51209
  |
Correlation:0.29124
  |
Correlation:0.29124
  |
Correlation:0.29124
  |
Correlation:0.15517
  |
Correlation:0.15517
  |
Correlation:0.3008
  |
Component:247; Jaccard:0.076949
 |
Component:080; Jaccard:0.07479
 |
Component:080; Jaccard:0.066859
 |
Component:080; Jaccard:0.044543
 |
Component:088; Jaccard:0.024965
 |
Component:088; Jaccard:0.014269
 |
Component:354; Jaccard:0.006566
 |
Correlation:0.26723
  |
Correlation:0.055812
  |
Correlation:0.055812
  |
Correlation:0.055812
  |
Correlation:0.18275
  |
Correlation:0.18275
  |
Correlation:0.22501
  |
Component:362; Jaccard:0.074535
 |
Component:247; Jaccard:0.074698
 |
Component:247; Jaccard:0.060347
 |
Component:362; Jaccard:0.041847
 |
Component:091; Jaccard:0.024496
 |
Component:091; Jaccard:0.013134
 |
Component:088; Jaccard:0.006387
 |
Correlation:0.13288
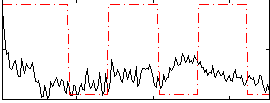 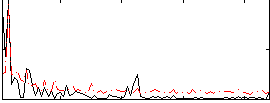 |
Correlation:0.26723
  |
Correlation:0.26723
  |
Correlation:0.13288
  |
Correlation:0.3008
  |
Correlation:0.3008
  |
Correlation:0.18275
  |
Component:080; Jaccard:0.073089
 |
Component:362; Jaccard:0.07163
 |
Component:362; Jaccard:0.059316
 |
Component:109; Jaccard:0.040787
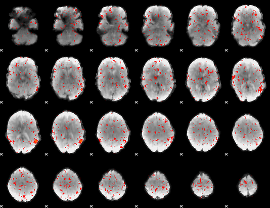 |
Component:080; Jaccard:0.024489
 |
Component:194; Jaccard:0.01087
 |
Component:170; Jaccard:0.005865
 |
Correlation:0.055812
  |
Correlation:0.13288
  |
Correlation:0.13288
  |
Correlation:0.16786
  |
Correlation:0.055812
  |
Correlation:0.1781
  |
Correlation:0.15517
  |
Component:235; Jaccard:0.072044
 |
Component:109; Jaccard:0.067167
 |
Component:109; Jaccard:0.058307
 |
Component:091; Jaccard:0.040724
 |
Component:193; Jaccard:0.023033
 |
Component:354; Jaccard:0.010699
 |
Component:261; Jaccard:0.005596
 |
Correlation:0.069662
  |
Correlation:0.16786
  |
Correlation:0.16786
  |
Correlation:0.3008
  |
Correlation:-0.019953
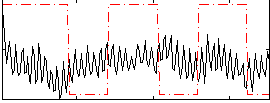  |
Correlation:0.22501
  |
Correlation:0.21877
  |
Component:254; Jaccard:0.068319
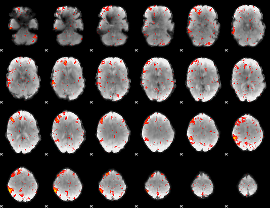 |
Component:235; Jaccard:0.065749
 |
Component:285; Jaccard:0.056679
 |
Component:088; Jaccard:0.040506
 |
Component:362; Jaccard:0.022759
 |
Component:080; Jaccard:0.010294
 |
Component:194; Jaccard:0.005251
 |
Correlation:0.019932
  |
Correlation:0.069662
  |
Correlation:0.21924
  |
Correlation:0.18275
  |
Correlation:0.13288
  |
Correlation:0.055812
  |
Correlation:0.1781
  |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
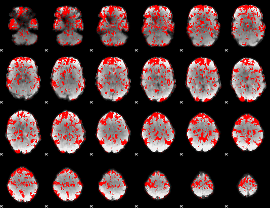 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
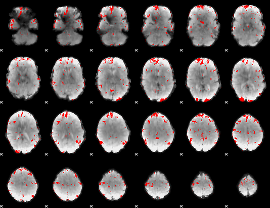 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
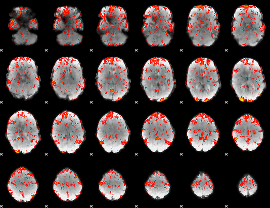 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:283; Jaccard:0.132
 |
Component:362; Jaccard:0.15827
 |
Component:362; Jaccard:0.17731
 |
Component:362; Jaccard:0.16554
 |
Component:362; Jaccard:0.12235
 |
Component:390; Jaccard:0.09218
 |
Component:390; Jaccard:0.065374
 |
Correlation:-0.056803
  |
Correlation:0.072492
  |
Correlation:0.072492
  |
Correlation:0.072492
  |
Correlation:0.072492
  |
Correlation:0.60721
  |
Correlation:0.60721
  |
Component:362; Jaccard:0.12971
 |
Component:283; Jaccard:0.14889
 |
Component:283; Jaccard:0.15322
 |
Component:283; Jaccard:0.13295
 |
Component:390; Jaccard:0.11672
 |
Component:362; Jaccard:0.072302
 |
Component:080; Jaccard:0.037122
 |
Correlation:0.072492
  |
Correlation:-0.056803
  |
Correlation:-0.056803
  |
Correlation:-0.056803
  |
Correlation:0.60721
  |
Correlation:0.072492
  |
Correlation:0.15421
  |
Component:080; Jaccard:0.10546
 |
Component:080; Jaccard:0.12429
 |
Component:080; Jaccard:0.13467
 |
Component:390; Jaccard:0.1281
 |
Component:080; Jaccard:0.10031
 |
Component:080; Jaccard:0.068285
 |
Component:362; Jaccard:0.036304
 |
Correlation:0.15421
  |
Correlation:0.15421
  |
Correlation:0.15421
  |
Correlation:0.60721
  |
Correlation:0.15421
  |
Correlation:0.15421
  |
Correlation:0.072492
  |
Component:394; Jaccard:0.097119
 |
Component:390; Jaccard:0.1115
 |
Component:390; Jaccard:0.12588
 |
Component:080; Jaccard:0.12755
 |
Component:283; Jaccard:0.098432
 |
Component:283; Jaccard:0.059679
 |
Component:394; Jaccard:0.029283
 |
Correlation:0.28412
  |
Correlation:0.60721
  |
Correlation:0.60721
  |
Correlation:0.15421
  |
Correlation:-0.056803
  |
Correlation:-0.056803
  |
Correlation:0.28412
  |
Component:235; Jaccard:0.095472
 |
Component:394; Jaccard:0.10815
 |
Component:394; Jaccard:0.11287
 |
Component:394; Jaccard:0.10419
 |
Component:394; Jaccard:0.079852
 |
Component:394; Jaccard:0.052329
 |
Component:283; Jaccard:0.028701
 |
Correlation:0.088351
  |
Correlation:0.28412
  |
Correlation:0.28412
  |
Correlation:0.28412
  |
Correlation:0.28412
  |
Correlation:0.28412
  |
Correlation:-0.056803
  |
Component:390; Jaccard:0.094674
 |
Component:235; Jaccard:0.10335
 |
Component:235; Jaccard:0.10359
 |
Component:235; Jaccard:0.083225
 |
Component:141; Jaccard:0.060343
 |
Component:024; Jaccard:0.043964
 |
Component:024; Jaccard:0.022277
 |
Correlation:0.60721
  |
Correlation:0.088351
  |
Correlation:0.088351
  |
Correlation:0.088351
  |
Correlation:0.16915
  |
Correlation:0.10585
  |
Correlation:0.10585
  |
Component:067; Jaccard:0.089594
 |
Component:067; Jaccard:0.094928
 |
Component:067; Jaccard:0.09177
 |
Component:067; Jaccard:0.08133
 |
Component:024; Jaccard:0.060279
 |
Component:067; Jaccard:0.037125
 |
Component:022; Jaccard:0.021103
 |
Correlation:-0.26116
  |
Correlation:-0.26116
  |
Correlation:-0.26116
  |
Correlation:-0.26116
  |
Correlation:0.10585
  |
Correlation:-0.26116
  |
Correlation:0.00096651
  |
Component:171; Jaccard:0.087933
 |
Component:022; Jaccard:0.092071
 |
Component:022; Jaccard:0.090015
 |
Component:022; Jaccard:0.077071
 |
Component:067; Jaccard:0.059548
 |
Component:141; Jaccard:0.036997
 |
Component:141; Jaccard:0.01955
 |
Correlation:-0.16305
  |
Correlation:0.00096651
  |
Correlation:0.00096651
  |
Correlation:0.00096651
  |
Correlation:-0.26116
  |
Correlation:0.16915
  |
Correlation:0.16915
  |
Component:022; Jaccard:0.08435
 |
Component:171; Jaccard:0.090959
 |
Component:171; Jaccard:0.087397
 |
Component:141; Jaccard:0.074769
 |
Component:022; Jaccard:0.055771
 |
Component:022; Jaccard:0.034995
 |
Component:215; Jaccard:0.019406
 |
Correlation:0.00096651
  |
Correlation:-0.16305
  |
Correlation:-0.16305
  |
Correlation:0.16915
  |
Correlation:0.00096651
  |
Correlation:0.00096651
  |
Correlation:0.032824
  |
Component:361; Jaccard:0.080782
 |
Component:361; Jaccard:0.082668
 |
Component:384; Jaccard:0.083412
 |
Component:171; Jaccard:0.073599
 |
Component:235; Jaccard:0.055331
 |
Component:215; Jaccard:0.034144
 |
Component:091; Jaccard:0.018761
 |
Correlation:-0.096251
  |
Correlation:-0.096251
  |
Correlation:0.10046
  |
Correlation:-0.16305
  |
Correlation:0.088351
  |
Correlation:0.032824
  |
Correlation:-0.10936
  |
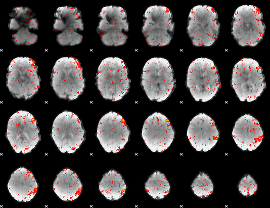
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
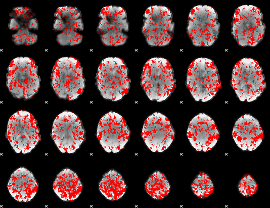 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
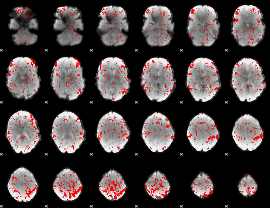 |
GLM binary result thresholded by 2.5
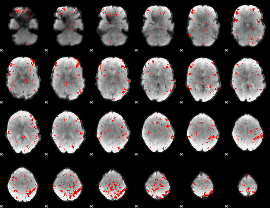 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
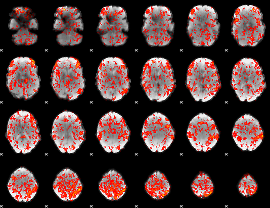 |
GLM Z-score result thresholded by 1.5
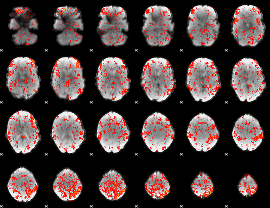 |
GLM Z-score result thresholded by 2
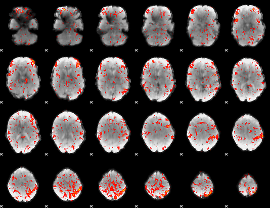 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:249; Jaccard:0.12065
 |
Component:249; Jaccard:0.14246
 |
Component:249; Jaccard:0.15592
 |
Component:249; Jaccard:0.14869
 |
Component:249; Jaccard:0.11429
 |
Component:249; Jaccard:0.077077
 |
Component:249; Jaccard:0.044562
 |
Correlation:0.45102
 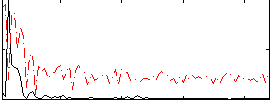 |
Correlation:0.45102
  |
Correlation:0.45102
  |
Correlation:0.45102
  |
Correlation:0.45102
  |
Correlation:0.45102
  |
Correlation:0.45102
  |
Component:275; Jaccard:0.10745
 |
Component:275; Jaccard:0.13027
 |
Component:275; Jaccard:0.14313
 |
Component:275; Jaccard:0.13711
 |
Component:275; Jaccard:0.11037
 |
Component:275; Jaccard:0.073556
 |
Component:275; Jaccard:0.041499
 |
Correlation:0.33414
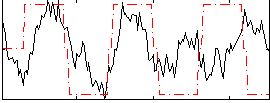  |
Correlation:0.33414
  |
Correlation:0.33414
  |
Correlation:0.33414
  |
Correlation:0.33414
  |
Correlation:0.33414
  |
Correlation:0.33414
  |
Component:268; Jaccard:0.10674
 |
Component:179; Jaccard:0.12484
 |
Component:179; Jaccard:0.13646
 |
Component:179; Jaccard:0.13257
 |
Component:179; Jaccard:0.10426
 |
Component:179; Jaccard:0.069807
 |
Component:262; Jaccard:0.039757
 |
Correlation:0.33088
 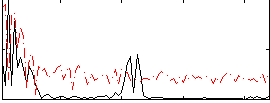 |
Correlation:0.29219
  |
Correlation:0.29219
  |
Correlation:0.29219
  |
Correlation:0.29219
  |
Correlation:0.29219
  |
Correlation:0.28791
  |
Component:179; Jaccard:0.10595
 |
Component:268; Jaccard:0.11378
 |
Component:262; Jaccard:0.11651
 |
Component:262; Jaccard:0.11123
 |
Component:262; Jaccard:0.089938
 |
Component:262; Jaccard:0.063958
 |
Component:179; Jaccard:0.038741
 |
Correlation:0.29219
  |
Correlation:0.33088
  |
Correlation:0.28791
  |
Correlation:0.28791
  |
Correlation:0.28791
  |
Correlation:0.28791
  |
Correlation:0.29219
  |
Component:262; Jaccard:0.097611
 |
Component:262; Jaccard:0.10917
 |
Component:268; Jaccard:0.11233
 |
Component:214; Jaccard:0.10272
 |
Component:325; Jaccard:0.081477
 |
Component:325; Jaccard:0.054662
 |
Component:039; Jaccard:0.033505
 |
Correlation:0.28791
  |
Correlation:0.28791
  |
Correlation:0.33088
  |
Correlation:0.36338
  |
Correlation:0.39147
  |
Correlation:0.39147
  |
Correlation:0.52606
  |
Component:214; Jaccard:0.096717
 |
Component:214; Jaccard:0.10845
 |
Component:214; Jaccard:0.11179
 |
Component:093; Jaccard:0.10215
 |
Component:093; Jaccard:0.079922
 |
Component:039; Jaccard:0.053399
 |
Component:325; Jaccard:0.032333
 |
Correlation:0.36338
  |
Correlation:0.36338
  |
Correlation:0.36338
  |
Correlation:0.21927
  |
Correlation:0.21927
  |
Correlation:0.52606
  |
Correlation:0.39147
  |
Component:346; Jaccard:0.092425
 |
Component:093; Jaccard:0.10422
 |
Component:093; Jaccard:0.11048
 |
Component:325; Jaccard:0.10186
 |
Component:214; Jaccard:0.079094
 |
Component:093; Jaccard:0.051009
 |
Component:271; Jaccard:0.030949
 |
Correlation:0.37953
  |
Correlation:0.21927
  |
Correlation:0.21927
  |
Correlation:0.39147
  |
Correlation:0.36338
  |
Correlation:0.21927
  |
Correlation:0.31272
  |
Component:093; Jaccard:0.090164
 |
Component:346; Jaccard:0.10022
 |
Component:325; Jaccard:0.10533
 |
Component:268; Jaccard:0.094791
 |
Component:039; Jaccard:0.071325
 |
Component:271; Jaccard:0.049876
 |
Component:214; Jaccard:0.030261
 |
Correlation:0.21927
  |
Correlation:0.37953
  |
Correlation:0.39147
  |
Correlation:0.33088
  |
Correlation:0.52606
  |
Correlation:0.31272
  |
Correlation:0.36338
  |
Component:153; Jaccard:0.089859
 |
Component:325; Jaccard:0.099288
 |
Component:014; Jaccard:0.096843
 |
Component:039; Jaccard:0.084812
 |
Component:271; Jaccard:0.067849
 |
Component:214; Jaccard:0.048592
 |
Component:093; Jaccard:0.026759
 |
Correlation:0.34451
  |
Correlation:0.39147
  |
Correlation:0.41089
  |
Correlation:0.52606
  |
Correlation:0.31272
  |
Correlation:0.36338
  |
Correlation:0.21927
  |
Component:014; Jaccard:0.089146
 |
Component:014; Jaccard:0.096638
 |
Component:346; Jaccard:0.096731
 |
Component:346; Jaccard:0.082345
 |
Component:268; Jaccard:0.064958
 |
Component:392; Jaccard:0.042187
 |
Component:392; Jaccard:0.02522
 |
Correlation:0.41089
  |
Correlation:0.41089
  |
Correlation:0.37953
  |
Correlation:0.37953
  |
Correlation:0.33088
  |
Correlation:0.36191
  |
Correlation:0.36191
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































