Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
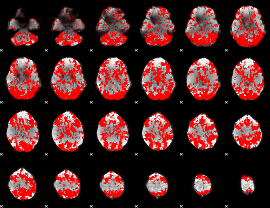 |
GLM binary result thresholded by 1.5
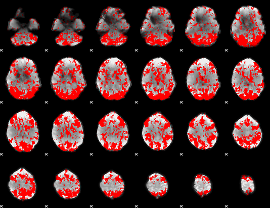 |
GLM binary result thresholded by 2
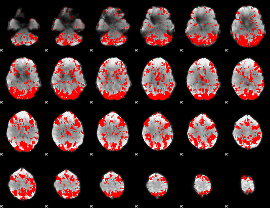 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
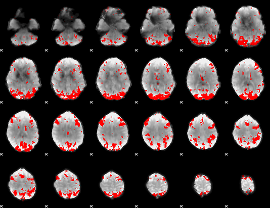 |
GLM binary result thresholded by 4
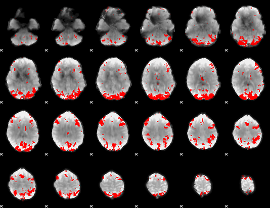 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
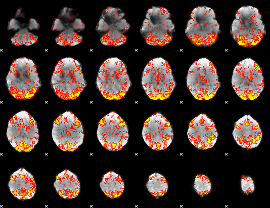 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
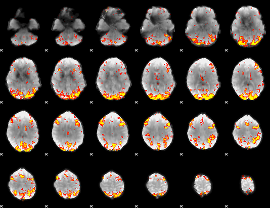 |
GLM Z-score result thresholded by 4
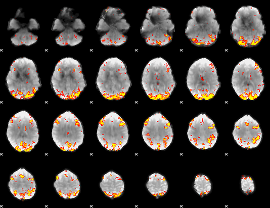 |
Component:308; Jaccard:0.11728
 |
Component:308; Jaccard:0.14673
 |
Component:308; Jaccard:0.18256
 |
Component:308; Jaccard:0.22283
 |
Component:308; Jaccard:0.26175
 |
Component:288; Jaccard:0.29472
 |
Component:288; Jaccard:0.32216
 |
Correlation:0.26953
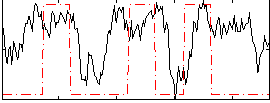 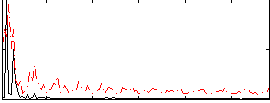 |
Correlation:0.26953
  |
Correlation:0.26953
  |
Correlation:0.26953
  |
Correlation:0.26953
  |
Correlation:0.16589
  |
Correlation:0.16589
  |
Component:072; Jaccard:0.11613
 |
Component:288; Jaccard:0.13867
 |
Component:288; Jaccard:0.17375
 |
Component:288; Jaccard:0.21538
 |
Component:288; Jaccard:0.25773
 |
Component:308; Jaccard:0.28919
 |
Component:308; Jaccard:0.30184
 |
Correlation:0.034124
  |
Correlation:0.16589
  |
Correlation:0.16589
  |
Correlation:0.16589
  |
Correlation:0.16589
  |
Correlation:0.26953
  |
Correlation:0.26953
  |
Component:288; Jaccard:0.11102
 |
Component:101; Jaccard:0.13533
 |
Component:101; Jaccard:0.16654
 |
Component:101; Jaccard:0.20055
 |
Component:101; Jaccard:0.23209
 |
Component:382; Jaccard:0.25548
 |
Component:382; Jaccard:0.27982
 |
Correlation:0.16589
  |
Correlation:0.20927
  |
Correlation:0.20927
  |
Correlation:0.20927
  |
Correlation:0.20927
  |
Correlation:0.24732
  |
Correlation:0.24732
  |
Component:101; Jaccard:0.10963
 |
Component:072; Jaccard:0.13232
 |
Component:188; Jaccard:0.15725
 |
Component:188; Jaccard:0.18881
 |
Component:382; Jaccard:0.2225
 |
Component:101; Jaccard:0.25194
 |
Component:101; Jaccard:0.26371
 |
Correlation:0.20927
  |
Correlation:0.034124
  |
Correlation:0.30314
  |
Correlation:0.30314
  |
Correlation:0.24732
  |
Correlation:0.20927
  |
Correlation:0.20927
  |
Component:188; Jaccard:0.10446
 |
Component:188; Jaccard:0.12856
 |
Component:081; Jaccard:0.15153
 |
Component:382; Jaccard:0.1852
 |
Component:188; Jaccard:0.21745
 |
Component:081; Jaccard:0.24116
 |
Component:081; Jaccard:0.2579
 |
Correlation:0.30314
  |
Correlation:0.30314
  |
Correlation:0.21062
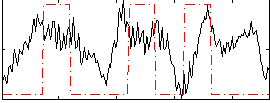 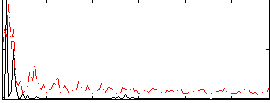 |
Correlation:0.24732
  |
Correlation:0.30314
  |
Correlation:0.21062
  |
Correlation:0.21062
  |
Component:299; Jaccard:0.10384
 |
Component:299; Jaccard:0.12498
 |
Component:382; Jaccard:0.15007
 |
Component:081; Jaccard:0.18433
 |
Component:081; Jaccard:0.21488
 |
Component:188; Jaccard:0.23995
 |
Component:188; Jaccard:0.25366
 |
Correlation:0.2751
  |
Correlation:0.2751
  |
Correlation:0.24732
  |
Correlation:0.21062
  |
Correlation:0.21062
  |
Correlation:0.30314
  |
Correlation:0.30314
  |
Component:082; Jaccard:0.10232
 |
Component:081; Jaccard:0.12152
 |
Component:299; Jaccard:0.14764
 |
Component:332; Jaccard:0.17318
 |
Component:332; Jaccard:0.2007
 |
Component:332; Jaccard:0.21973
 |
Component:332; Jaccard:0.23177
 |
Correlation:-0.17988
  |
Correlation:0.21062
  |
Correlation:0.2751
  |
Correlation:0.21509
  |
Correlation:0.21509
  |
Correlation:0.21509
  |
Correlation:0.21509
  |
Component:374; Jaccard:0.10079
 |
Component:382; Jaccard:0.11962
 |
Component:072; Jaccard:0.14619
 |
Component:299; Jaccard:0.16962
 |
Component:299; Jaccard:0.18391
 |
Component:299; Jaccard:0.19132
 |
Component:299; Jaccard:0.19
 |
Correlation:0.061989
  |
Correlation:0.24732
  |
Correlation:0.034124
  |
Correlation:0.2751
  |
Correlation:0.2751
  |
Correlation:0.2751
  |
Correlation:0.2751
  |
Component:217; Jaccard:0.098169
 |
Component:374; Jaccard:0.11785
 |
Component:332; Jaccard:0.1441
 |
Component:072; Jaccard:0.15616
 |
Component:324; Jaccard:0.16162
 |
Component:324; Jaccard:0.17116
 |
Component:324; Jaccard:0.1733
 |
Correlation:0.088999
  |
Correlation:0.061989
  |
Correlation:0.21509
  |
Correlation:0.034124
  |
Correlation:0.23942
  |
Correlation:0.23942
  |
Correlation:0.23942
  |
Component:081; Jaccard:0.097613
 |
Component:332; Jaccard:0.11734
 |
Component:374; Jaccard:0.13522
 |
Component:374; Jaccard:0.14971
 |
Component:072; Jaccard:0.16138
 |
Component:374; Jaccard:0.16462
 |
Component:374; Jaccard:0.15889
 |
Correlation:0.21062
  |
Correlation:0.21509
  |
Correlation:0.061989
  |
Correlation:0.061989
  |
Correlation:0.034124
  |
Correlation:0.061989
  |
Correlation:0.061989
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
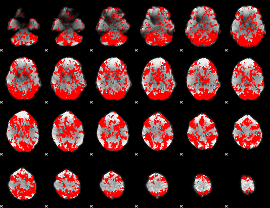 |
GLM binary result thresholded by 1.5
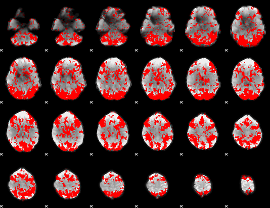 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
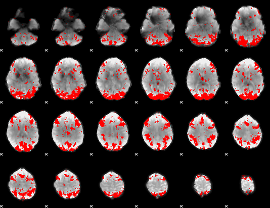 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
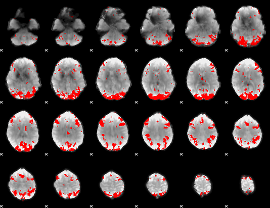 |
GLM Z-score result thresholded by 1
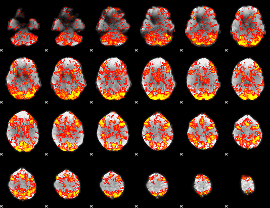 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
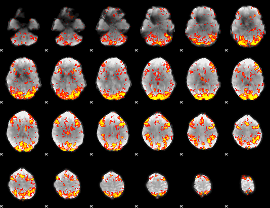 |
GLM Z-score result thresholded by 3
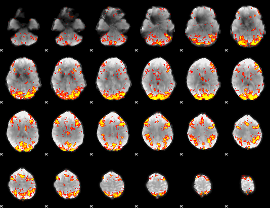 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
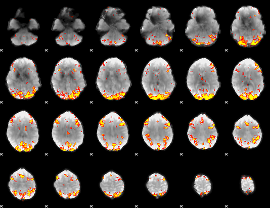 |
Component:308; Jaccard:0.12121
 |
Component:308; Jaccard:0.14704
 |
Component:308; Jaccard:0.17869
 |
Component:308; Jaccard:0.21176
 |
Component:288; Jaccard:0.24697
 |
Component:288; Jaccard:0.28005
 |
Component:288; Jaccard:0.3049
 |
Correlation:0.15084
 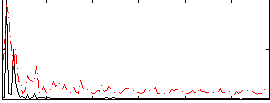 |
Correlation:0.15084
  |
Correlation:0.15084
  |
Correlation:0.15084
  |
Correlation:-0.032812
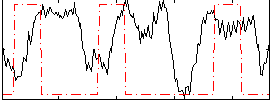  |
Correlation:-0.032812
  |
Correlation:-0.032812
  |
Component:101; Jaccard:0.11567
 |
Component:288; Jaccard:0.14162
 |
Component:288; Jaccard:0.17334
 |
Component:288; Jaccard:0.20959
 |
Component:308; Jaccard:0.24252
 |
Component:188; Jaccard:0.26724
 |
Component:188; Jaccard:0.28535
 |
Correlation:0.09302
 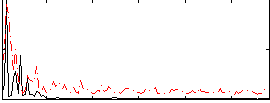 |
Correlation:-0.032812
  |
Correlation:-0.032812
  |
Correlation:-0.032812
  |
Correlation:0.15084
  |
Correlation:0.20545
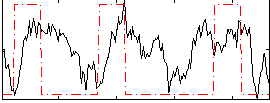 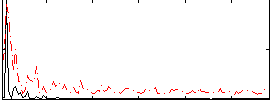 |
Correlation:0.20545
  |
Component:288; Jaccard:0.11565
 |
Component:101; Jaccard:0.14123
 |
Component:188; Jaccard:0.17274
 |
Component:188; Jaccard:0.20791
 |
Component:188; Jaccard:0.24144
 |
Component:308; Jaccard:0.26459
 |
Component:308; Jaccard:0.27669
 |
Correlation:-0.032812
  |
Correlation:0.09302
  |
Correlation:0.20545
  |
Correlation:0.20545
  |
Correlation:0.20545
  |
Correlation:0.15084
  |
Correlation:0.15084
  |
Component:188; Jaccard:0.1141
 |
Component:188; Jaccard:0.14097
 |
Component:101; Jaccard:0.17123
 |
Component:101; Jaccard:0.20366
 |
Component:101; Jaccard:0.23231
 |
Component:101; Jaccard:0.25658
 |
Component:382; Jaccard:0.27185
 |
Correlation:0.20545
  |
Correlation:0.20545
  |
Correlation:0.09302
  |
Correlation:0.09302
  |
Correlation:0.09302
  |
Correlation:0.09302
  |
Correlation:0.079148
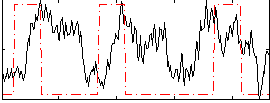  |
Component:072; Jaccard:0.11302
 |
Component:299; Jaccard:0.12826
 |
Component:081; Jaccard:0.15162
 |
Component:382; Jaccard:0.18428
 |
Component:382; Jaccard:0.21687
 |
Component:382; Jaccard:0.24626
 |
Component:101; Jaccard:0.26921
 |
Correlation:-0.029881
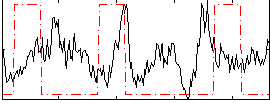  |
Correlation:0.11299
  |
Correlation:0.044841
  |
Correlation:0.079148
  |
Correlation:0.079148
  |
Correlation:0.079148
  |
Correlation:0.09302
  |
Component:082; Jaccard:0.11028
 |
Component:072; Jaccard:0.12548
 |
Component:382; Jaccard:0.1515
 |
Component:081; Jaccard:0.17928
 |
Component:081; Jaccard:0.20713
 |
Component:081; Jaccard:0.23182
 |
Component:081; Jaccard:0.24766
 |
Correlation:0.18587
  |
Correlation:-0.029881
  |
Correlation:0.079148
  |
Correlation:0.044841
  |
Correlation:0.044841
  |
Correlation:0.044841
  |
Correlation:0.044841
  |
Component:299; Jaccard:0.10818
 |
Component:081; Jaccard:0.1246
 |
Component:299; Jaccard:0.15016
 |
Component:332; Jaccard:0.1775
 |
Component:332; Jaccard:0.20349
 |
Component:332; Jaccard:0.22215
 |
Component:332; Jaccard:0.23164
 |
Correlation:0.11299
  |
Correlation:0.044841
  |
Correlation:0.11299
  |
Correlation:0.10475
 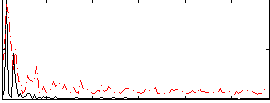 |
Correlation:0.10475
  |
Correlation:0.10475
  |
Correlation:0.10475
  |
Component:374; Jaccard:0.1066
 |
Component:382; Jaccard:0.12392
 |
Component:332; Jaccard:0.14853
 |
Component:299; Jaccard:0.17048
 |
Component:299; Jaccard:0.18517
 |
Component:299; Jaccard:0.19196
 |
Component:299; Jaccard:0.19253
 |
Correlation:0.13416
  |
Correlation:0.079148
  |
Correlation:0.10475
  |
Correlation:0.11299
  |
Correlation:0.11299
  |
Correlation:0.11299
  |
Correlation:0.11299
  |
Component:217; Jaccard:0.10294
 |
Component:374; Jaccard:0.12324
 |
Component:374; Jaccard:0.14094
 |
Component:374; Jaccard:0.1571
 |
Component:324; Jaccard:0.16841
 |
Component:324; Jaccard:0.17982
 |
Component:324; Jaccard:0.18357
 |
Correlation:0.12375
  |
Correlation:0.13416
  |
Correlation:0.13416
  |
Correlation:0.13416
  |
Correlation:0.033675
  |
Correlation:0.033675
  |
Correlation:0.033675
  |
Component:081; Jaccard:0.10205
 |
Component:332; Jaccard:0.12222
 |
Component:072; Jaccard:0.13567
 |
Component:324; Jaccard:0.15181
 |
Component:374; Jaccard:0.16766
 |
Component:374; Jaccard:0.17133
 |
Component:374; Jaccard:0.16853
 |
Correlation:0.044841
  |
Correlation:0.10475
  |
Correlation:-0.029881
  |
Correlation:0.033675
  |
Correlation:0.13416
  |
Correlation:0.13416
  |
Correlation:0.13416
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
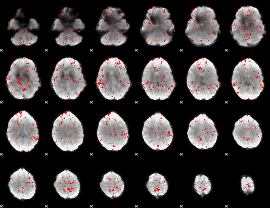 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
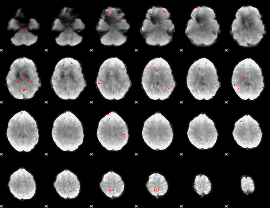 |
GLM binary result thresholded by 3.5
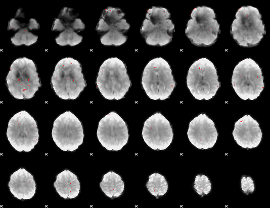 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
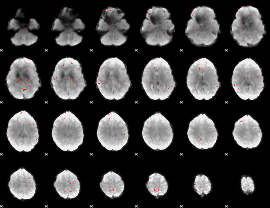 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
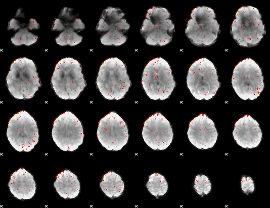
Component:105; Jaccard:0.082988
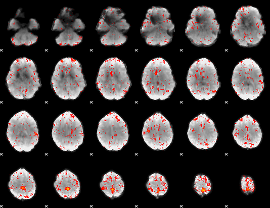 |
Component:312; Jaccard:0.083586
 |
Component:197; Jaccard:0.072984
 |
Component:252; Jaccard:0.061059
 |
Component:252; Jaccard:0.035191
 |
Component:252; Jaccard:0.016769
 |
Component:252; Jaccard:0.00681
 |
Correlation:-0.00062899
 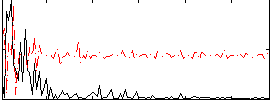 |
Correlation:0.22342
  |
Correlation:0.21578
  |
Correlation:0.098868
  |
Correlation:0.098868
  |
Correlation:0.098868
  |
Correlation:0.098868
  |
Component:312; Jaccard:0.080116
 |
Component:197; Jaccard:0.08081
 |
Component:252; Jaccard:0.072757
 |
Component:197; Jaccard:0.054809
 |
Component:093; Jaccard:0.031072
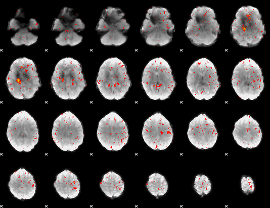 |
Component:093; Jaccard:0.014892
 |
Component:093; Jaccard:0.00646
 |
Correlation:0.22342
  |
Correlation:0.21578
  |
Correlation:0.098868
  |
Correlation:0.21578
  |
Correlation:0.020431
  |
Correlation:0.020431
  |
Correlation:0.020431
  |
Component:118; Jaccard:0.077784
 |
Component:238; Jaccard:0.075321
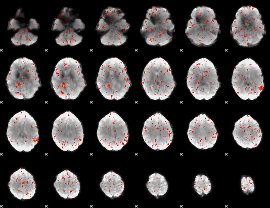 |
Component:312; Jaccard:0.072264
 |
Component:093; Jaccard:0.05415
 |
Component:197; Jaccard:0.030945
 |
Component:289; Jaccard:0.012976
 |
Component:289; Jaccard:0.004808
 |
Correlation:0.14392
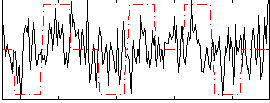  |
Correlation:0.20363
 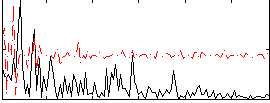 |
Correlation:0.22342
  |
Correlation:0.020431
  |
Correlation:0.21578
  |
Correlation:0.049376
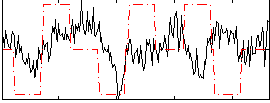 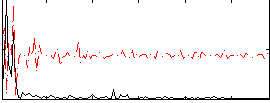 |
Correlation:0.049376
  |
Component:352; Jaccard:0.075677
 |
Component:032; Jaccard:0.073562
 |
Component:093; Jaccard:0.067542
 |
Component:312; Jaccard:0.049975
 |
Component:289; Jaccard:0.028802
 |
Component:197; Jaccard:0.012675
 |
Component:001; Jaccard:0.004528
 |
Correlation:0.30566
  |
Correlation:0.044205
  |
Correlation:0.020431
  |
Correlation:0.22342
  |
Correlation:0.049376
  |
Correlation:0.21578
  |
Correlation:0.15461
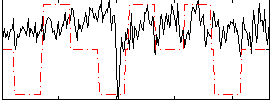  |
Component:197; Jaccard:0.074413
 |
Component:352; Jaccard:0.073013
 |
Component:238; Jaccard:0.064275
 |
Component:001; Jaccard:0.04772
 |
Component:001; Jaccard:0.027926
 |
Component:001; Jaccard:0.012444
 |
Component:197; Jaccard:0.003622
 |
Correlation:0.21578
  |
Correlation:0.30566
  |
Correlation:0.20363
  |
Correlation:0.15461
  |
Correlation:0.15461
  |
Correlation:0.15461
  |
Correlation:0.21578
  |
Component:238; Jaccard:0.072038
 |
Component:105; Jaccard:0.072718
 |
Component:032; Jaccard:0.063907
 |
Component:112; Jaccard:0.046276
 |
Component:112; Jaccard:0.024593
 |
Component:389; Jaccard:0.010453
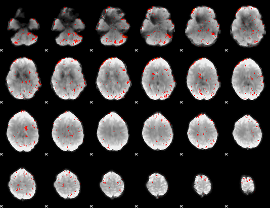 |
Component:389; Jaccard:0.002917
 |
Correlation:0.20363
  |
Correlation:-0.00062899
  |
Correlation:0.044205
  |
Correlation:0.1626
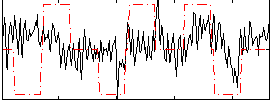  |
Correlation:0.1626
  |
Correlation:0.25008
 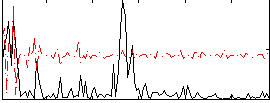 |
Correlation:0.25008
  |
Component:216; Jaccard:0.071211
 |
Component:118; Jaccard:0.071273
 |
Component:001; Jaccard:0.063856
 |
Component:289; Jaccard:0.046124
 |
Component:312; Jaccard:0.023655
 |
Component:112; Jaccard:0.008943
 |
Component:355; Jaccard:0.002691
 |
Correlation:0.029111
 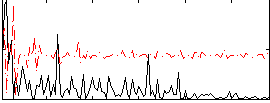 |
Correlation:0.14392
  |
Correlation:0.15461
  |
Correlation:0.049376
  |
Correlation:0.22342
  |
Correlation:0.1626
  |
Correlation:0.076175
 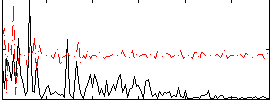 |
Component:376; Jaccard:0.070413
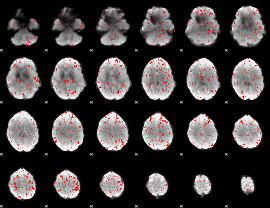 |
Component:216; Jaccard:0.07097
 |
Component:112; Jaccard:0.061312
 |
Component:238; Jaccard:0.043173
 |
Component:389; Jaccard:0.023537
 |
Component:312; Jaccard:0.008827
 |
Component:238; Jaccard:0.002621
 |
Correlation:-0.03563
  |
Correlation:0.029111
  |
Correlation:0.1626
  |
Correlation:0.20363
  |
Correlation:0.25008
  |
Correlation:0.22342
  |
Correlation:0.20363
  |
Component:032; Jaccard:0.069903
 |
Component:203; Jaccard:0.067927
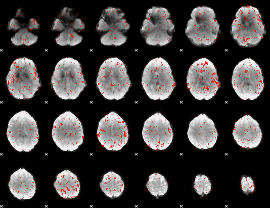 |
Component:216; Jaccard:0.060027
 |
Component:389; Jaccard:0.040986
 |
Component:238; Jaccard:0.020781
 |
Component:355; Jaccard:0.007957
 |
Component:107; Jaccard:0.002579
 |
Correlation:0.044205
  |
Correlation:-0.099629
  |
Correlation:0.029111
  |
Correlation:0.25008
  |
Correlation:0.20363
  |
Correlation:0.076175
  |
Correlation:-0.022634
  |
Component:203; Jaccard:0.069282
 |
Component:252; Jaccard:0.067671
 |
Component:352; Jaccard:0.05827
 |
Component:032; Jaccard:0.040361
 |
Component:191; Jaccard:0.019535
 |
Component:376; Jaccard:0.007801
 |
Component:376; Jaccard:0.002503
 |
Correlation:-0.099629
  |
Correlation:0.098868
  |
Correlation:0.30566
  |
Correlation:0.044205
  |
Correlation:0.038307
  |
Correlation:-0.03563
  |
Correlation:-0.03563
  |
Cope: 04:REL-MATCH BACK
|
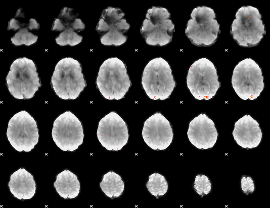
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
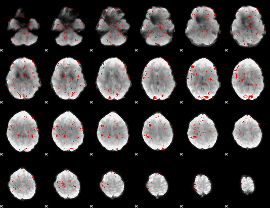 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
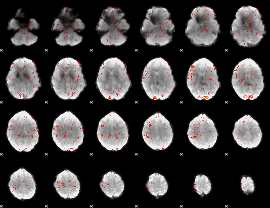 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
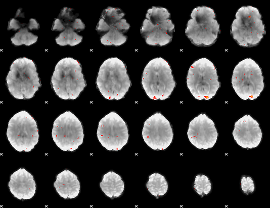 |
GLM Z-score result thresholded by 3.5
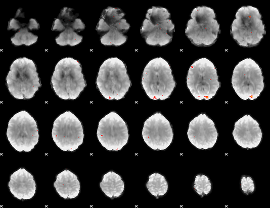 |
GLM Z-score result thresholded by 4
 |
Component:022; Jaccard:0.11832
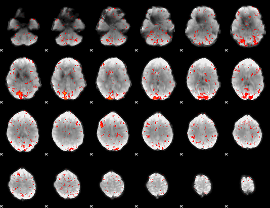 |
Component:022; Jaccard:0.12322
 |
Component:022; Jaccard:0.10436
 |
Component:022; Jaccard:0.073901
 |
Component:022; Jaccard:0.043929
 |
Component:022; Jaccard:0.023493
 |
Component:022; Jaccard:0.013424
 |
Correlation:0.02126
 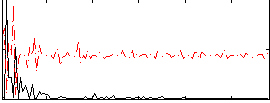 |
Correlation:0.02126
  |
Correlation:0.02126
  |
Correlation:0.02126
  |
Correlation:0.02126
  |
Correlation:0.02126
  |
Correlation:0.02126
  |
Component:076; Jaccard:0.085128
 |
Component:076; Jaccard:0.087667
 |
Component:076; Jaccard:0.075438
 |
Component:076; Jaccard:0.047636
 |
Component:188; Jaccard:0.026318
 |
Component:188; Jaccard:0.016758
 |
Component:188; Jaccard:0.010888
 |
Correlation:0.11705
  |
Correlation:0.11705
  |
Correlation:0.11705
  |
Correlation:0.11705
  |
Correlation:-0.0579
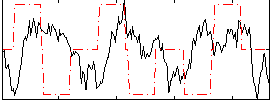  |
Correlation:-0.0579
  |
Correlation:-0.0579
  |
Component:347; Jaccard:0.076897
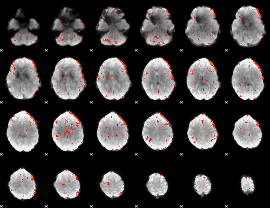 |
Component:347; Jaccard:0.077021
 |
Component:347; Jaccard:0.062474
 |
Component:347; Jaccard:0.041202
 |
Component:347; Jaccard:0.020954
 |
Component:382; Jaccard:0.012083
 |
Component:382; Jaccard:0.008032
 |
Correlation:0.032397
 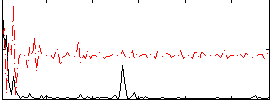 |
Correlation:0.032397
  |
Correlation:0.032397
  |
Correlation:0.032397
  |
Correlation:0.032397
  |
Correlation:-0.099675
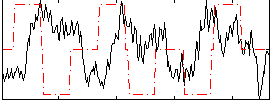  |
Correlation:-0.099675
  |
Component:188; Jaccard:0.071476
 |
Component:325; Jaccard:0.069165
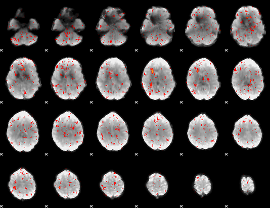 |
Component:188; Jaccard:0.055544
 |
Component:188; Jaccard:0.040838
 |
Component:076; Jaccard:0.019459
 |
Component:076; Jaccard:0.007905
 |
Component:332; Jaccard:0.00497
 |
Correlation:-0.0579
  |
Correlation:0.05448
  |
Correlation:-0.0579
  |
Correlation:-0.0579
  |
Correlation:0.11705
  |
Correlation:0.11705
  |
Correlation:-0.065396
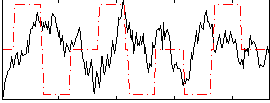  |
Component:325; Jaccard:0.07113
 |
Component:188; Jaccard:0.067808
 |
Component:325; Jaccard:0.055114
 |
Component:008; Jaccard:0.031681
 |
Component:382; Jaccard:0.016163
 |
Component:347; Jaccard:0.007462
 |
Component:081; Jaccard:0.004298
 |
Correlation:0.05448
  |
Correlation:-0.0579
  |
Correlation:0.05448
  |
Correlation:0.1072
  |
Correlation:-0.099675
  |
Correlation:0.032397
  |
Correlation:-0.098258
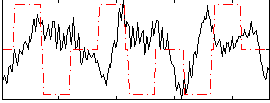  |
Component:008; Jaccard:0.070013
 |
Component:008; Jaccard:0.064814
 |
Component:008; Jaccard:0.05053
 |
Component:325; Jaccard:0.029216
 |
Component:008; Jaccard:0.01602
 |
Component:008; Jaccard:0.007238
 |
Component:008; Jaccard:0.004076
 |
Correlation:0.1072
  |
Correlation:0.1072
  |
Correlation:0.1072
  |
Correlation:0.05448
  |
Correlation:0.1072
  |
Correlation:0.1072
  |
Correlation:0.1072
  |
Component:124; Jaccard:0.068804
 |
Component:124; Jaccard:0.058414
 |
Component:083; Jaccard:0.045025
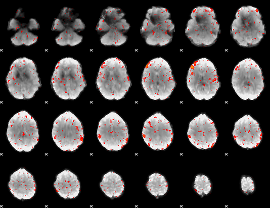 |
Component:379; Jaccard:0.025602
 |
Component:293; Jaccard:0.011749
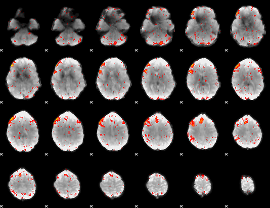 |
Component:133; Jaccard:0.006635
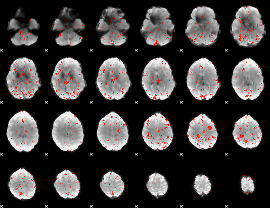 |
Component:133; Jaccard:0.00374
 |
Correlation:0.1592
  |
Correlation:0.1592
  |
Correlation:0.061109
  |
Correlation:0.23271
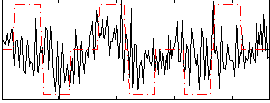  |
Correlation:-0.093838
  |
Correlation:-0.082707
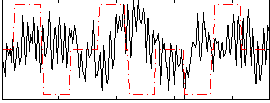  |
Correlation:-0.082707
  |
Component:397; Jaccard:0.066509
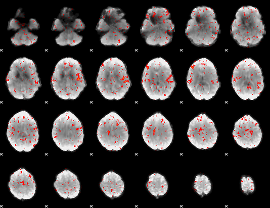 |
Component:397; Jaccard:0.058091
 |
Component:379; Jaccard:0.043379
 |
Component:083; Jaccard:0.024978
 |
Component:083; Jaccard:0.011461
 |
Component:332; Jaccard:0.006618
 |
Component:293; Jaccard:0.003336
 |
Correlation:0.079408
  |
Correlation:0.079408
  |
Correlation:0.23271
  |
Correlation:0.061109
  |
Correlation:0.061109
  |
Correlation:-0.065396
  |
Correlation:-0.093838
  |
Component:360; Jaccard:0.065898
 |
Component:083; Jaccard:0.057013
 |
Component:397; Jaccard:0.043131
 |
Component:033; Jaccard:0.024978
 |
Component:325; Jaccard:0.010896
 |
Component:293; Jaccard:0.006124
 |
Component:101; Jaccard:0.002642
 |
Correlation:0.071878
  |
Correlation:0.061109
  |
Correlation:0.079408
  |
Correlation:0.019615
  |
Correlation:0.05448
  |
Correlation:-0.093838
  |
Correlation:-0.068903
  |
Component:048; Jaccard:0.062006
 |
Component:360; Jaccard:0.056844
 |
Component:360; Jaccard:0.042278
 |
Component:293; Jaccard:0.024925
 |
Component:332; Jaccard:0.010655
 |
Component:101; Jaccard:0.005808
 |
Component:076; Jaccard:0.00264
 |
Correlation:0.17767
  |
Correlation:0.071878
  |
Correlation:0.071878
  |
Correlation:-0.093838
  |
Correlation:-0.065396
  |
Correlation:-0.068903
  |
Correlation:0.11705
  |
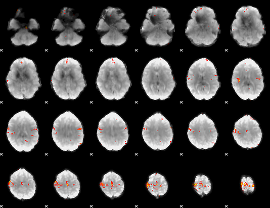
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
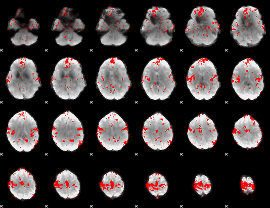 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
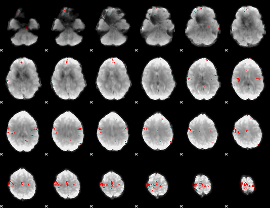 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
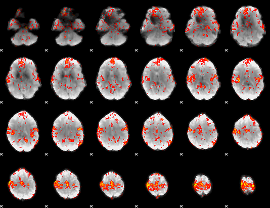 |
GLM Z-score result thresholded by 2
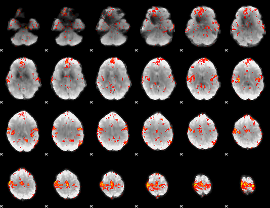 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
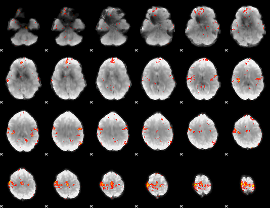 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:311; Jaccard:0.14819
 |
Component:311; Jaccard:0.17831
 |
Component:311; Jaccard:0.20405
 |
Component:311; Jaccard:0.21672
 |
Component:311; Jaccard:0.21602
 |
Component:214; Jaccard:0.19639
 |
Component:311; Jaccard:0.16899
 |
Correlation:0.36329
  |
Correlation:0.36329
  |
Correlation:0.36329
  |
Correlation:0.36329
  |
Correlation:0.36329
  |
Correlation:0.34105
  |
Correlation:0.36329
  |
Component:214; Jaccard:0.13696
 |
Component:214; Jaccard:0.16517
 |
Component:214; Jaccard:0.19066
 |
Component:214; Jaccard:0.20857
 |
Component:214; Jaccard:0.21109
 |
Component:311; Jaccard:0.19615
 |
Component:214; Jaccard:0.16448
 |
Correlation:0.34105
  |
Correlation:0.34105
  |
Correlation:0.34105
  |
Correlation:0.34105
  |
Correlation:0.34105
  |
Correlation:0.36329
  |
Correlation:0.34105
  |
Component:174; Jaccard:0.11943
 |
Component:174; Jaccard:0.13813
 |
Component:174; Jaccard:0.15069
 |
Component:174; Jaccard:0.14799
 |
Component:066; Jaccard:0.13672
 |
Component:066; Jaccard:0.12312
 |
Component:066; Jaccard:0.10293
 |
Correlation:0.23157
  |
Correlation:0.23157
  |
Correlation:0.23157
  |
Correlation:0.23157
  |
Correlation:0.29687
  |
Correlation:0.29687
  |
Correlation:0.29687
  |
Component:048; Jaccard:0.11561
 |
Component:048; Jaccard:0.12354
 |
Component:066; Jaccard:0.13279
 |
Component:066; Jaccard:0.14213
 |
Component:042; Jaccard:0.13341
 |
Component:042; Jaccard:0.11879
 |
Component:042; Jaccard:0.10174
 |
Correlation:0.19219
  |
Correlation:0.19219
  |
Correlation:0.29687
  |
Correlation:0.29687
  |
Correlation:0.22091
  |
Correlation:0.22091
  |
Correlation:0.22091
  |
Component:265; Jaccard:0.11199
 |
Component:265; Jaccard:0.12284
 |
Component:042; Jaccard:0.13002
 |
Component:042; Jaccard:0.13931
 |
Component:174; Jaccard:0.1313
 |
Component:174; Jaccard:0.10709
 |
Component:174; Jaccard:0.079357
 |
Correlation:0.1089
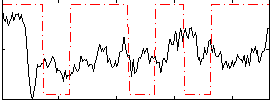 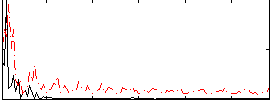 |
Correlation:0.1089
  |
Correlation:0.22091
  |
Correlation:0.22091
  |
Correlation:0.23157
  |
Correlation:0.23157
  |
Correlation:0.23157
  |
Component:291; Jaccard:0.10346
 |
Component:066; Jaccard:0.11708
 |
Component:048; Jaccard:0.12624
 |
Component:048; Jaccard:0.12037
 |
Component:048; Jaccard:0.1055
 |
Component:048; Jaccard:0.084925
 |
Component:048; Jaccard:0.067955
 |
Correlation:0.25021
  |
Correlation:0.29687
  |
Correlation:0.19219
  |
Correlation:0.19219
  |
Correlation:0.19219
  |
Correlation:0.19219
  |
Correlation:0.19219
  |
Component:066; Jaccard:0.099707
 |
Component:042; Jaccard:0.11516
 |
Component:265; Jaccard:0.12455
 |
Component:265; Jaccard:0.11138
 |
Component:090; Jaccard:0.090701
 |
Component:090; Jaccard:0.076257
 |
Component:090; Jaccard:0.0611
 |
Correlation:0.29687
  |
Correlation:0.22091
  |
Correlation:0.1089
  |
Correlation:0.1089
  |
Correlation:0.17518
  |
Correlation:0.17518
  |
Correlation:0.17518
  |
Component:025; Jaccard:0.099627
 |
Component:291; Jaccard:0.10804
 |
Component:291; Jaccard:0.10723
 |
Component:291; Jaccard:0.10285
 |
Component:291; Jaccard:0.089254
 |
Component:291; Jaccard:0.075483
 |
Component:291; Jaccard:0.056648
 |
Correlation:0.037439
  |
Correlation:0.25021
  |
Correlation:0.25021
  |
Correlation:0.25021
  |
Correlation:0.25021
  |
Correlation:0.25021
  |
Correlation:0.25021
  |
Component:042; Jaccard:0.097531
 |
Component:025; Jaccard:0.10587
 |
Component:090; Jaccard:0.10683
 |
Component:090; Jaccard:0.10191
 |
Component:265; Jaccard:0.088166
 |
Component:292; Jaccard:0.068646
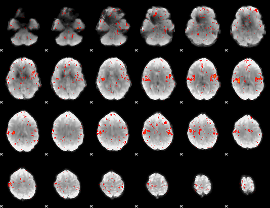 |
Component:292; Jaccard:0.052676
 |
Correlation:0.22091
  |
Correlation:0.037439
  |
Correlation:0.17518
  |
Correlation:0.17518
  |
Correlation:0.1089
  |
Correlation:0.23666
 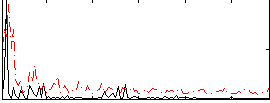 |
Correlation:0.23666
  |
Component:292; Jaccard:0.096958
 |
Component:292; Jaccard:0.10437
 |
Component:292; Jaccard:0.10471
 |
Component:292; Jaccard:0.0977
 |
Component:292; Jaccard:0.083697
 |
Component:220; Jaccard:0.065062
 |
Component:220; Jaccard:0.050975
 |
Correlation:0.23666
  |
Correlation:0.23666
  |
Correlation:0.23666
  |
Correlation:0.23666
  |
Correlation:0.23666
  |
Correlation:0.29104
  |
Correlation:0.29104
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
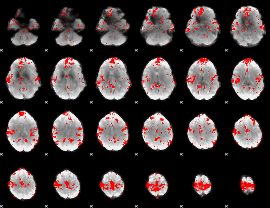
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
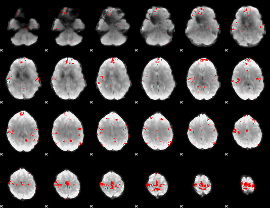
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
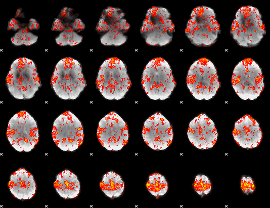
GLM Z-score result thresholded by 1
 |
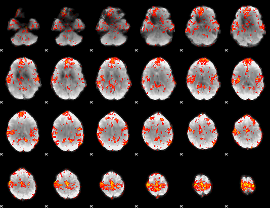
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
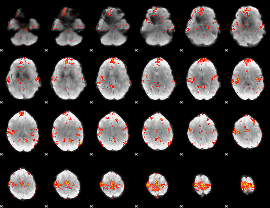 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:214; Jaccard:0.12274
 |
Component:214; Jaccard:0.14682
 |
Component:214; Jaccard:0.17096
 |
Component:214; Jaccard:0.18938
 |
Component:214; Jaccard:0.19911
 |
Component:214; Jaccard:0.19354
 |
Component:214; Jaccard:0.17535
 |
Correlation:0.18824
  |
Correlation:0.18824
  |
Correlation:0.18824
  |
Correlation:0.18824
  |
Correlation:0.18824
  |
Correlation:0.18824
  |
Correlation:0.18824
  |
Component:311; Jaccard:0.11851
 |
Component:311; Jaccard:0.139
 |
Component:311; Jaccard:0.15943
 |
Component:311; Jaccard:0.17564
 |
Component:311; Jaccard:0.17968
 |
Component:066; Jaccard:0.17178
 |
Component:066; Jaccard:0.15954
 |
Correlation:-0.094774
  |
Correlation:-0.094774
  |
Correlation:-0.094774
  |
Correlation:-0.094774
  |
Correlation:-0.094774
  |
Correlation:0.21507
  |
Correlation:0.21507
  |
Component:265; Jaccard:0.1138
 |
Component:174; Jaccard:0.12796
 |
Component:066; Jaccard:0.1451
 |
Component:066; Jaccard:0.16403
 |
Component:066; Jaccard:0.17612
 |
Component:311; Jaccard:0.16921
 |
Component:311; Jaccard:0.14954
 |
Correlation:0.049417
  |
Correlation:0.063736
  |
Correlation:0.21507
  |
Correlation:0.21507
  |
Correlation:0.21507
  |
Correlation:-0.094774
  |
Correlation:-0.094774
  |
Component:312; Jaccard:0.11215
 |
Component:265; Jaccard:0.12747
 |
Component:174; Jaccard:0.14225
 |
Component:174; Jaccard:0.15405
 |
Component:174; Jaccard:0.14935
 |
Component:174; Jaccard:0.13142
 |
Component:174; Jaccard:0.10733
 |
Correlation:0.38168
  |
Correlation:0.049417
  |
Correlation:0.063736
  |
Correlation:0.063736
  |
Correlation:0.063736
  |
Correlation:0.063736
  |
Correlation:0.063736
  |
Component:174; Jaccard:0.10947
 |
Component:066; Jaccard:0.12215
 |
Component:265; Jaccard:0.13878
 |
Component:265; Jaccard:0.1416
 |
Component:265; Jaccard:0.13012
 |
Component:042; Jaccard:0.11751
 |
Component:042; Jaccard:0.10633
 |
Correlation:0.063736
  |
Correlation:0.21507
  |
Correlation:0.049417
  |
Correlation:0.049417
  |
Correlation:0.049417
  |
Correlation:0.094881
  |
Correlation:0.094881
  |
Component:291; Jaccard:0.1091
 |
Component:312; Jaccard:0.12116
 |
Component:025; Jaccard:0.12842
 |
Component:025; Jaccard:0.12754
 |
Component:042; Jaccard:0.12076
 |
Component:265; Jaccard:0.10171
 |
Component:090; Jaccard:0.085046
 |
Correlation:0.24069
  |
Correlation:0.38168
  |
Correlation:0.076784
  |
Correlation:0.076784
  |
Correlation:0.094881
  |
Correlation:0.049417
  |
Correlation:0.13218
  |
Component:025; Jaccard:0.10775
 |
Component:025; Jaccard:0.1199
 |
Component:312; Jaccard:0.12422
 |
Component:291; Jaccard:0.12369
 |
Component:025; Jaccard:0.11564
 |
Component:090; Jaccard:0.10007
 |
Component:291; Jaccard:0.082772
 |
Correlation:0.076784
  |
Correlation:0.076784
  |
Correlation:0.38168
  |
Correlation:0.24069
  |
Correlation:0.076784
  |
Correlation:0.13218
  |
Correlation:0.24069
  |
Component:066; Jaccard:0.10006
 |
Component:291; Jaccard:0.11772
 |
Component:291; Jaccard:0.12331
 |
Component:312; Jaccard:0.12295
 |
Component:291; Jaccard:0.11311
 |
Component:291; Jaccard:0.098155
 |
Component:265; Jaccard:0.071281
 |
Correlation:0.21507
  |
Correlation:0.24069
  |
Correlation:0.24069
  |
Correlation:0.38168
  |
Correlation:0.24069
  |
Correlation:0.24069
  |
Correlation:0.049417
  |
Component:090; Jaccard:0.095748
 |
Component:090; Jaccard:0.10761
 |
Component:090; Jaccard:0.1166
 |
Component:090; Jaccard:0.12143
 |
Component:090; Jaccard:0.1126
 |
Component:025; Jaccard:0.095174
 |
Component:025; Jaccard:0.071
 |
Correlation:0.13218
  |
Correlation:0.13218
  |
Correlation:0.13218
  |
Correlation:0.13218
  |
Correlation:0.13218
  |
Correlation:0.076784
  |
Correlation:0.076784
  |
Component:085; Jaccard:0.0943
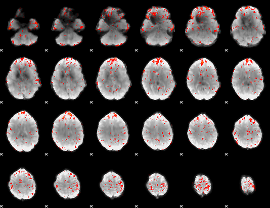 |
Component:048; Jaccard:0.09859
 |
Component:042; Jaccard:0.10814
 |
Component:042; Jaccard:0.11808
 |
Component:312; Jaccard:0.1102
 |
Component:312; Jaccard:0.088775
 |
Component:312; Jaccard:0.065654
 |
Correlation:0.070037
  |
Correlation:-0.10757
  |
Correlation:0.094881
  |
Correlation:0.094881
  |
Correlation:0.38168
  |
Correlation:0.38168
  |
Correlation:0.38168
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































