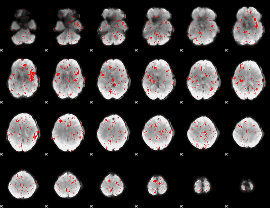
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
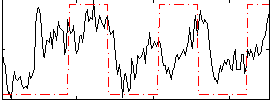
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
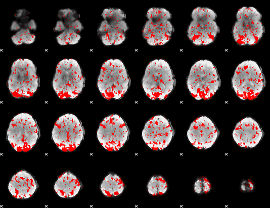 |
GLM binary result thresholded by 2
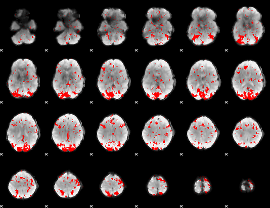 |
GLM binary result thresholded by 2.5
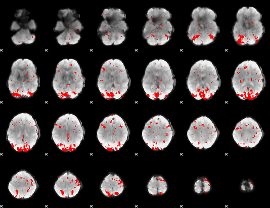 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
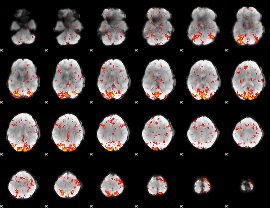 |
GLM Z-score result thresholded by 2.5
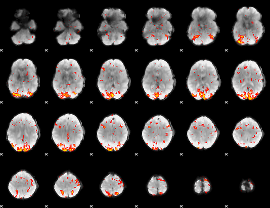 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:373; Jaccard:0.16257
 |
Component:373; Jaccard:0.22622
 |
Component:373; Jaccard:0.28832
 |
Component:373; Jaccard:0.32184
 |
Component:373; Jaccard:0.32027
 |
Component:373; Jaccard:0.28492
 |
Component:373; Jaccard:0.235
 |
Correlation:0.14397
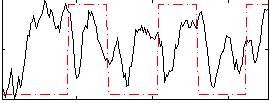 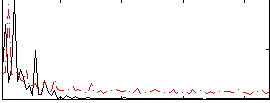 |
Correlation:0.14397
  |
Correlation:0.14397
  |
Correlation:0.14397
  |
Correlation:0.14397
  |
Correlation:0.14397
  |
Correlation:0.14397
  |
Component:323; Jaccard:0.13124
 |
Component:363; Jaccard:0.16272
 |
Component:363; Jaccard:0.19646
 |
Component:363; Jaccard:0.2085
 |
Component:363; Jaccard:0.19483
 |
Component:363; Jaccard:0.16154
 |
Component:363; Jaccard:0.1259
 |
Correlation:0.37692
  |
Correlation:0.10055
 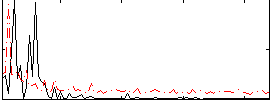 |
Correlation:0.10055
  |
Correlation:0.10055
  |
Correlation:0.10055
  |
Correlation:0.10055
  |
Correlation:0.10055
  |
Component:363; Jaccard:0.12819
 |
Component:323; Jaccard:0.16075
 |
Component:323; Jaccard:0.17632
 |
Component:323; Jaccard:0.17393
 |
Component:061; Jaccard:0.16181
 |
Component:061; Jaccard:0.1471
 |
Component:061; Jaccard:0.12478
 |
Correlation:0.10055
  |
Correlation:0.37692
  |
Correlation:0.37692
  |
Correlation:0.37692
  |
Correlation:-0.15243
  |
Correlation:-0.15243
  |
Correlation:-0.15243
  |
Component:291; Jaccard:0.11399
 |
Component:061; Jaccard:0.13426
 |
Component:061; Jaccard:0.15861
 |
Component:061; Jaccard:0.168
 |
Component:323; Jaccard:0.15317
 |
Component:321; Jaccard:0.12645
 |
Component:321; Jaccard:0.10752
 |
Correlation:-0.32439
  |
Correlation:-0.15243
  |
Correlation:-0.15243
  |
Correlation:-0.15243
  |
Correlation:0.37692
  |
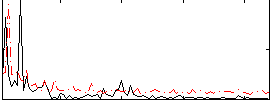
Correlation:0.34549
  |
Correlation:0.34549
  |
Component:061; Jaccard:0.10664
 |
Component:291; Jaccard:0.13355
 |
Component:321; Jaccard:0.14913
 |
Component:321; Jaccard:0.15233
 |
Component:321; Jaccard:0.14351
 |
Component:323; Jaccard:0.12611
 |
Component:323; Jaccard:0.10082
 |
Correlation:-0.15243
  |
Correlation:-0.32439
  |
Correlation:0.34549
  |
Correlation:0.34549
  |
Correlation:0.34549
  |
Correlation:0.37692
  |
Correlation:0.37692
  |
Component:321; Jaccard:0.10525
 |
Component:321; Jaccard:0.13111
 |
Component:291; Jaccard:0.14413
 |
Component:291; Jaccard:0.13985
 |
Component:291; Jaccard:0.12288
 |
Component:291; Jaccard:0.099812
 |
Component:291; Jaccard:0.07876
 |
Correlation:0.34549
  |
Correlation:0.34549
  |
Correlation:-0.32439
  |
Correlation:-0.32439
  |
Correlation:-0.32439
  |
Correlation:-0.32439
  |
Correlation:-0.32439
  |
Component:201; Jaccard:0.1032
 |
Component:201; Jaccard:0.12189
 |
Component:201; Jaccard:0.13401
 |
Component:201; Jaccard:0.13298
 |
Component:201; Jaccard:0.11975
 |
Component:201; Jaccard:0.099742
 |
Component:201; Jaccard:0.076995
 |
Correlation:-0.10861
  |
Correlation:-0.10861
  |
Correlation:-0.10861
  |
Correlation:-0.10861
  |
Correlation:-0.10861
  |
Correlation:-0.10861
  |
Correlation:-0.10861
  |
Component:335; Jaccard:0.09841
 |
Component:335; Jaccard:0.10437
 |
Component:082; Jaccard:0.10591
 |
Component:082; Jaccard:0.098734
 |
Component:190; Jaccard:0.084095
 |
Component:190; Jaccard:0.070124
 |
Component:335; Jaccard:0.05414
 |
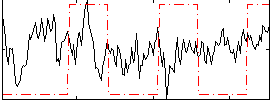
Correlation:-0.081272
  |
Correlation:-0.081272
  |
Correlation:-0.050352
  |
Correlation:-0.050352
  |
Correlation:-0.21853
  |
Correlation:-0.21853
  |
Correlation:-0.081272
  |
Component:067; Jaccard:0.092833
 |
Component:067; Jaccard:0.104
 |
Component:067; Jaccard:0.10534
 |
Component:335; Jaccard:0.096378
 |
Component:082; Jaccard:0.083368
 |
Component:335; Jaccard:0.067792
 |
Component:190; Jaccard:0.051117
 |
Correlation:0.022432
 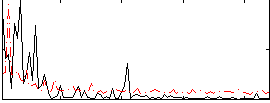 |
Correlation:0.022432
  |
Correlation:0.022432
  |
Correlation:-0.081272
  |
Correlation:-0.050352
  |
Correlation:-0.081272
  |
Correlation:-0.21853
  |
Component:213; Jaccard:0.091489
 |
Component:082; Jaccard:0.10385
 |
Component:335; Jaccard:0.10318
 |
Component:190; Jaccard:0.09521
 |
Component:335; Jaccard:0.082956
 |
Component:082; Jaccard:0.065068
 |
Component:082; Jaccard:0.04732
 |
Correlation:0.0023849
  |
Correlation:-0.050352
  |
Correlation:-0.081272
  |
Correlation:-0.21853
  |
Correlation:-0.081272
  |
Correlation:-0.050352
  |
Correlation:-0.050352
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
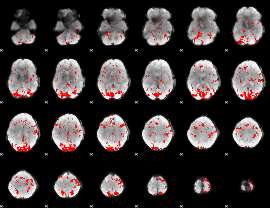 |
GLM binary result thresholded by 2.5
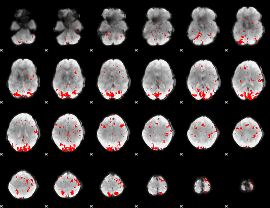 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
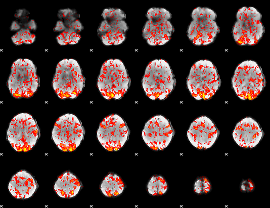 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
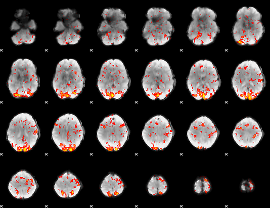 |
GLM Z-score result thresholded by 2.5
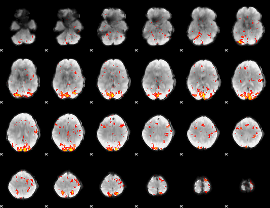 |
GLM Z-score result thresholded by 3
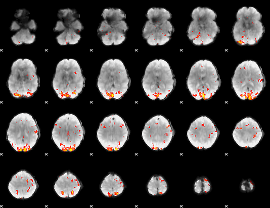 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:373; Jaccard:0.16085
 |
Component:373; Jaccard:0.21822
 |
Component:373; Jaccard:0.26517
 |
Component:373; Jaccard:0.28906
 |
Component:373; Jaccard:0.27275
 |
Component:373; Jaccard:0.23269
 |
Component:373; Jaccard:0.19055
 |
Correlation:0.070902
  |
Correlation:0.070902
  |
Correlation:0.070902
  |
Correlation:0.070902
  |
Correlation:0.070902
  |
Correlation:0.070902
  |
Correlation:0.070902
  |
Component:291; Jaccard:0.13897
 |
Component:061; Jaccard:0.19242
 |
Component:061; Jaccard:0.24722
 |
Component:061; Jaccard:0.27179
 |
Component:061; Jaccard:0.26208
 |
Component:061; Jaccard:0.22696
 |
Component:061; Jaccard:0.18901
 |
Correlation:0.43728
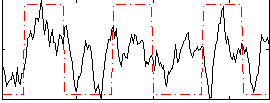 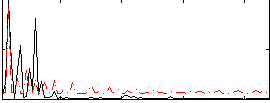 |
Correlation:0.37468
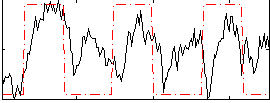 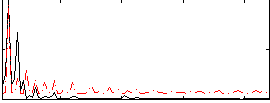 |
Correlation:0.37468
  |
Correlation:0.37468
  |
Correlation:0.37468
  |
Correlation:0.37468
  |
Correlation:0.37468
  |
Component:061; Jaccard:0.1354
 |
Component:291; Jaccard:0.1715
 |
Component:291; Jaccard:0.19081
 |
Component:363; Jaccard:0.19572
 |
Component:363; Jaccard:0.17971
 |
Component:363; Jaccard:0.1551
 |
Component:363; Jaccard:0.11797
 |
Correlation:0.37468
  |
Correlation:0.43728
  |
Correlation:0.43728
  |
Correlation:0.061178
  |
Correlation:0.061178
  |
Correlation:0.061178
  |
Correlation:0.061178
  |
Component:363; Jaccard:0.12401
 |
Component:363; Jaccard:0.15581
 |
Component:363; Jaccard:0.18328
 |
Component:291; Jaccard:0.18851
 |
Component:291; Jaccard:0.16511
 |
Component:291; Jaccard:0.13586
 |
Component:291; Jaccard:0.10437
 |
Correlation:0.061178
  |
Correlation:0.061178
  |
Correlation:0.061178
  |
Correlation:0.43728
  |
Correlation:0.43728
  |
Correlation:0.43728
  |
Correlation:0.43728
  |
Component:366; Jaccard:0.10595
 |
Component:201; Jaccard:0.11824
 |
Component:201; Jaccard:0.12781
 |
Component:201; Jaccard:0.12265
 |
Component:201; Jaccard:0.10746
 |
Component:321; Jaccard:0.088996
 |
Component:321; Jaccard:0.073325
 |
Correlation:0.2405
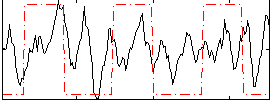  |
Correlation:0.14951
  |
Correlation:0.14951
  |
Correlation:0.14951
  |
Correlation:0.14951
  |
Correlation:-0.21236
  |
Correlation:-0.21236
  |
Component:201; Jaccard:0.10212
 |
Component:366; Jaccard:0.11819
 |
Component:366; Jaccard:0.11686
 |
Component:321; Jaccard:0.11624
 |
Component:321; Jaccard:0.10587
 |
Component:201; Jaccard:0.086454
 |
Component:201; Jaccard:0.06538
 |
Correlation:0.14951
  |
Correlation:0.2405
  |
Correlation:0.2405
  |
Correlation:-0.21236
  |
Correlation:-0.21236
  |
Correlation:0.14951
  |
Correlation:0.14951
  |
Component:067; Jaccard:0.098393
 |
Component:067; Jaccard:0.1125
 |
Component:067; Jaccard:0.11574
 |
Component:067; Jaccard:0.10411
 |
Component:067; Jaccard:0.084235
 |
Component:366; Jaccard:0.065575
 |
Component:366; Jaccard:0.050242
 |
Correlation:0.016411
  |
Correlation:0.016411
  |
Correlation:0.016411
  |
Correlation:0.016411
  |
Correlation:0.016411
  |
Correlation:0.2405
  |
Correlation:0.2405
  |
Component:190; Jaccard:0.096119
 |
Component:190; Jaccard:0.10585
 |
Component:321; Jaccard:0.11065
 |
Component:366; Jaccard:0.10325
 |
Component:366; Jaccard:0.082206
 |
Component:067; Jaccard:0.064333
 |
Component:335; Jaccard:0.046861
 |
Correlation:0.33544
  |
Correlation:0.33544
  |
Correlation:-0.21236
  |
Correlation:0.2405
  |
Correlation:0.2405
  |
Correlation:0.016411
  |
Correlation:0.25351
  |
Component:335; Jaccard:0.095779
 |
Component:265; Jaccard:0.099876
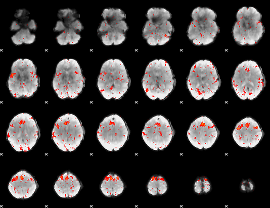 |
Component:190; Jaccard:0.10496
 |
Component:190; Jaccard:0.093927
 |
Component:190; Jaccard:0.078732
 |
Component:190; Jaccard:0.062293
 |
Component:190; Jaccard:0.045966
 |
Correlation:0.25351
  |
Correlation:0.1463
  |
Correlation:0.33544
  |
Correlation:0.33544
  |
Correlation:0.33544
  |
Correlation:0.33544
  |
Correlation:0.33544
  |
Component:265; Jaccard:0.092651
 |
Component:321; Jaccard:0.099458
 |
Component:265; Jaccard:0.099543
 |
Component:265; Jaccard:0.088192
 |
Component:335; Jaccard:0.069799
 |
Component:335; Jaccard:0.057484
 |
Component:067; Jaccard:0.044196
 |
Correlation:0.1463
  |
Correlation:-0.21236
  |
Correlation:0.1463
  |
Correlation:0.1463
  |
Correlation:0.25351
  |
Correlation:0.25351
  |
Correlation:0.016411
  |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
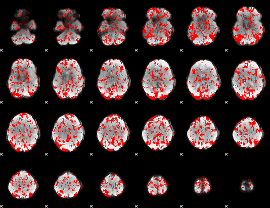 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
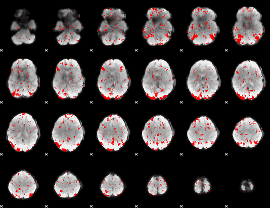 |
GLM binary result thresholded by 2.5
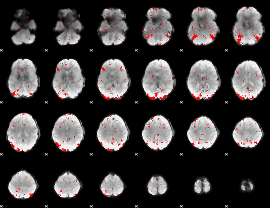 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
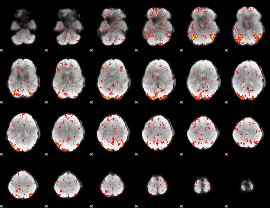 |
GLM Z-score result thresholded by 2.5
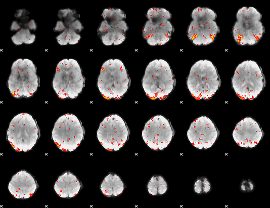 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:323; Jaccard:0.16594
 |
Component:323; Jaccard:0.23439
 |
Component:323; Jaccard:0.29413
 |
Component:323; Jaccard:0.29953
 |
Component:323; Jaccard:0.25097
 |
Component:323; Jaccard:0.18879
 |
Component:323; Jaccard:0.13458
 |
Correlation:0.37765
  |
Correlation:0.37765
  |
Correlation:0.37765
  |
Correlation:0.37765
  |
Correlation:0.37765
  |
Correlation:0.37765
  |
Correlation:0.37765
  |
Component:078; Jaccard:0.12258
 |
Component:078; Jaccard:0.14237
 |
Component:078; Jaccard:0.14408
 |
Component:304; Jaccard:0.14063
 |
Component:304; Jaccard:0.12141
 |
Component:304; Jaccard:0.098393
 |
Component:304; Jaccard:0.074362
 |
Correlation:0.36525
  |
Correlation:0.36525
  |
Correlation:0.36525
  |
Correlation:0.30452
  |
Correlation:0.30452
  |
Correlation:0.30452
  |
Correlation:0.30452
  |
Component:304; Jaccard:0.09746
 |
Component:304; Jaccard:0.11836
 |
Component:304; Jaccard:0.13607
 |
Component:078; Jaccard:0.12605
 |
Component:078; Jaccard:0.096707
 |
Component:078; Jaccard:0.074022
 |
Component:078; Jaccard:0.054545
 |
Correlation:0.30452
  |
Correlation:0.30452
  |
Correlation:0.30452
  |
Correlation:0.36525
  |
Correlation:0.36525
  |
Correlation:0.36525
  |
Correlation:0.36525
  |
Component:218; Jaccard:0.09379
 |
Component:218; Jaccard:0.10075
 |
Component:321; Jaccard:0.10391
 |
Component:321; Jaccard:0.096494
 |
Component:321; Jaccard:0.079018
 |
Component:373; Jaccard:0.055349
 |
Component:373; Jaccard:0.034891
 |
Correlation:0.079161
  |
Correlation:0.079161
  |
Correlation:0.30258
  |
Correlation:0.30258
  |
Correlation:0.30258
  |
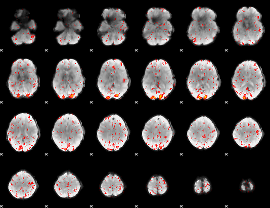
Correlation:0.039633
  |
Correlation:0.039633
  |
Component:082; Jaccard:0.081657
 |
Component:321; Jaccard:0.091939
 |
Component:218; Jaccard:0.10019
 |
Component:373; Jaccard:0.090604
 |
Component:373; Jaccard:0.076302
 |
Component:321; Jaccard:0.055323
 |
Component:321; Jaccard:0.034218
 |
Correlation:-0.031561
  |
Correlation:0.30258
  |
Correlation:0.079161
  |
Correlation:0.039633
  |
Correlation:0.039633
  |
Correlation:0.30258
  |
Correlation:0.30258
  |
Component:373; Jaccard:0.080615
 |
Component:082; Jaccard:0.089869
 |
Component:373; Jaccard:0.096044
 |
Component:218; Jaccard:0.079197
 |
Component:082; Jaccard:0.06178
 |
Component:082; Jaccard:0.045613
 |
Component:082; Jaccard:0.030814
 |
Correlation:0.039633
  |
Correlation:-0.031561
  |
Correlation:0.039633
  |
Correlation:0.079161
  |
Correlation:-0.031561
  |
Correlation:-0.031561
  |
Correlation:-0.031561
  |
Component:321; Jaccard:0.077132
 |
Component:373; Jaccard:0.089474
 |
Component:082; Jaccard:0.090417
 |
Component:082; Jaccard:0.07911
 |
Component:218; Jaccard:0.057308
 |
Component:218; Jaccard:0.037849
 |
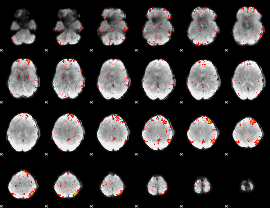
Component:021; Jaccard:0.025393
 |
Correlation:0.30258
  |
Correlation:0.039633
  |
Correlation:-0.031561
  |
Correlation:-0.031561
  |
Correlation:0.079161
  |
Correlation:0.079161
  |
Correlation:0.20928
  |
Component:216; Jaccard:0.074675
 |
Component:216; Jaccard:0.082242
 |
Component:216; Jaccard:0.083353
 |
Component:216; Jaccard:0.070112
 |
Component:216; Jaccard:0.051285
 |
Component:021; Jaccard:0.03651
 |
Component:218; Jaccard:0.023598
 |
Correlation:0.30343
  |
Correlation:0.30343
  |
Correlation:0.30343
  |
Correlation:0.30343
  |
Correlation:0.30343
  |
Correlation:0.20928
  |
Correlation:0.079161
  |
Component:021; Jaccard:0.074184
 |
Component:129; Jaccard:0.079216
 |
Component:129; Jaccard:0.078218
 |
Component:021; Jaccard:0.05998
 |
Component:021; Jaccard:0.047806
 |
Component:216; Jaccard:0.03529
 |
Component:216; Jaccard:0.021127
 |
Correlation:0.20928
  |
Correlation:0.22005
  |
Correlation:0.22005
  |
Correlation:0.20928
  |
Correlation:0.20928
  |
Correlation:0.30343
  |
Correlation:0.30343
  |
Component:391; Jaccard:0.071994
 |
Component:021; Jaccard:0.074684
 |
Component:021; Jaccard:0.068806
 |
Component:129; Jaccard:0.059841
 |
Component:363; Jaccard:0.040443
 |
Component:335; Jaccard:0.028491
 |
Component:349; Jaccard:0.021092
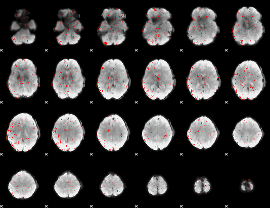 |
Correlation:0.096362
 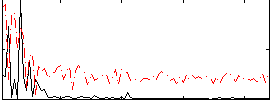 |
Correlation:0.20928
  |
Correlation:0.20928
  |
Correlation:0.22005
  |
Correlation:0.021354
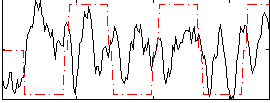  |
Correlation:-0.18159
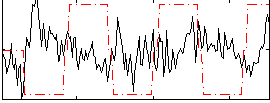  |
Correlation:0.11647
  |
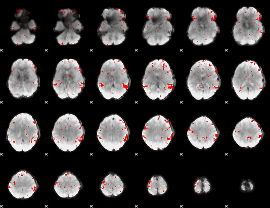
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
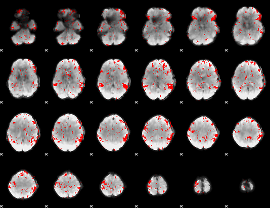 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:345; Jaccard:0.09531
 |
Component:107; Jaccard:0.1221
 |
Component:107; Jaccard:0.14509
 |
Component:107; Jaccard:0.14254
 |
Component:107; Jaccard:0.11463
 |
Component:054; Jaccard:0.081495
 |
Component:054; Jaccard:0.053641
 |
Correlation:-0.077826
  |
Correlation:0.10182
  |
Correlation:0.10182
  |
Correlation:0.10182
  |
Correlation:0.10182
  |
Correlation:0.08517
  |
Correlation:0.08517
  |
Component:107; Jaccard:0.094351
 |
Component:013; Jaccard:0.10686
 |
Component:013; Jaccard:0.11797
 |
Component:054; Jaccard:0.12634
 |
Component:054; Jaccard:0.11388
 |
Component:107; Jaccard:0.072459
 |
Component:107; Jaccard:0.042015
 |
Correlation:0.10182
  |
Correlation:-0.11576
  |
Correlation:-0.11576
  |
Correlation:0.08517
  |
Correlation:0.08517
  |
Correlation:0.10182
  |
Correlation:0.10182
  |
Component:315; Jaccard:0.09174
 |
Component:345; Jaccard:0.10443
 |
Component:054; Jaccard:0.11516
 |
Component:013; Jaccard:0.10626
 |
Component:013; Jaccard:0.082483
 |
Component:013; Jaccard:0.051993
 |
Component:013; Jaccard:0.030702
 |
Correlation:0.01306
  |
Correlation:-0.077826
  |
Correlation:0.08517
  |
Correlation:-0.11576
  |
Correlation:-0.11576
  |
Correlation:-0.11576
  |
Correlation:-0.11576
  |
Component:227; Jaccard:0.090625
 |
Component:227; Jaccard:0.10376
 |
Component:227; Jaccard:0.10709
 |
Component:040; Jaccard:0.093254
 |
Component:040; Jaccard:0.077868
 |
Component:040; Jaccard:0.050962
 |
Component:040; Jaccard:0.03049
 |
Correlation:0.07927
  |
Correlation:0.07927
  |
Correlation:0.07927
  |
Correlation:0.041222
  |
Correlation:0.041222
  |
Correlation:0.041222
  |
Correlation:0.041222
  |
Component:013; Jaccard:0.089177
 |
Component:315; Jaccard:0.10052
 |
Component:345; Jaccard:0.099509
 |
Component:227; Jaccard:0.090847
 |
Component:099; Jaccard:0.064982
 |
Component:099; Jaccard:0.037176
 |
Component:149; Jaccard:0.024201
 |
Correlation:-0.11576
  |
Correlation:0.01306
  |
Correlation:-0.077826
  |
Correlation:0.07927
  |
Correlation:-0.12594
  |
Correlation:-0.12594
  |
Correlation:0.2061
  |
Component:209; Jaccard:0.081797
 |
Component:054; Jaccard:0.096873
 |
Component:040; Jaccard:0.097188
 |
Component:099; Jaccard:0.087747
 |
Component:227; Jaccard:0.059244
 |
Component:149; Jaccard:0.03712
 |
Component:227; Jaccard:0.020609
 |
Correlation:0.24733
  |
Correlation:0.08517
  |
Correlation:0.041222
  |
Correlation:-0.12594
  |
Correlation:0.07927
  |
Correlation:0.2061
  |
Correlation:0.07927
  |
Component:266; Jaccard:0.080164
 |
Component:099; Jaccard:0.086448
 |
Component:315; Jaccard:0.096736
 |
Component:345; Jaccard:0.080617
 |
Component:149; Jaccard:0.052586
 |
Component:227; Jaccard:0.033626
 |
Component:206; Jaccard:0.016626
 |
Correlation:0.15345
  |
Correlation:-0.12594
  |
Correlation:0.01306
  |
Correlation:-0.077826
  |
Correlation:0.2061
  |
Correlation:0.07927
  |
Correlation:0.033816
  |
Component:011; Jaccard:0.077502
 |
Component:040; Jaccard:0.086402
 |
Component:099; Jaccard:0.094556
 |
Component:315; Jaccard:0.078479
 |
Component:345; Jaccard:0.051453
 |
Component:345; Jaccard:0.029657
 |
Component:315; Jaccard:0.016552
 |
Correlation:0.052554
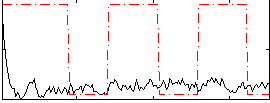  |
Correlation:0.041222
  |
Correlation:-0.12594
  |
Correlation:0.01306
  |
Correlation:-0.077826
  |
Correlation:-0.077826
  |
Correlation:0.01306
  |
Component:054; Jaccard:0.07687
 |
Component:209; Jaccard:0.084894
 |
Component:288; Jaccard:0.080891
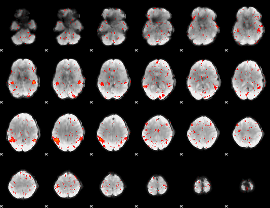 |
Component:149; Jaccard:0.068726
 |
Component:315; Jaccard:0.051415
 |
Component:315; Jaccard:0.028882
 |
Component:345; Jaccard:0.015909
 |
Correlation:0.08517
  |
Correlation:0.24733
  |
Correlation:0.0038612
 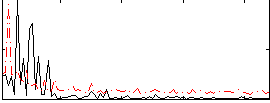 |
Correlation:0.2061
  |
Correlation:0.01306
  |
Correlation:0.01306
  |
Correlation:-0.077826
  |
Component:189; Jaccard:0.075134
 |
Component:266; Jaccard:0.082069
 |
Component:149; Jaccard:0.07758
 |
Component:288; Jaccard:0.065929
 |
Component:288; Jaccard:0.045944
 |
Component:288; Jaccard:0.028026
 |
Component:099; Jaccard:0.015614
 |
Correlation:-0.0078583
  |
Correlation:0.15345
  |
Correlation:0.2061
  |
Correlation:0.0038612
  |
Correlation:0.0038612
  |
Correlation:0.0038612
  |
Correlation:-0.12594
  |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
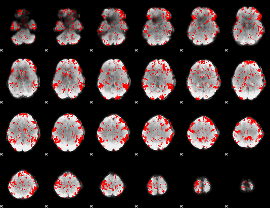 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:345; Jaccard:0.11397
 |
Component:345; Jaccard:0.13017
 |
Component:345; Jaccard:0.13653
 |
Component:107; Jaccard:0.13251
 |
Component:107; Jaccard:0.1068
 |
Component:107; Jaccard:0.069405
 |
Component:054; Jaccard:0.039031
 |
Correlation:0.080287
  |
Correlation:0.080287
  |
Correlation:0.080287
  |
Correlation:-0.056582
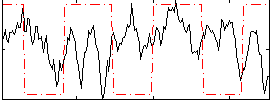  |
Correlation:-0.056582
  |
Correlation:-0.056582
  |
Correlation:-0.12604
  |
Component:078; Jaccard:0.10024
 |
Component:078; Jaccard:0.11108
 |
Component:107; Jaccard:0.12933
 |
Component:345; Jaccard:0.1274
 |
Component:345; Jaccard:0.097316
 |
Component:054; Jaccard:0.066266
 |
Component:107; Jaccard:0.035714
 |
Correlation:0.28246
 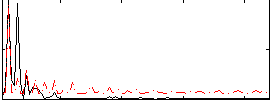 |
Correlation:0.28246
  |
Correlation:-0.056582
  |
Correlation:0.080287
  |
Correlation:0.080287
  |
Correlation:-0.12604
  |
Correlation:-0.056582
  |
Component:209; Jaccard:0.092351
 |
Component:107; Jaccard:0.10984
 |
Component:013; Jaccard:0.1127
 |
Component:013; Jaccard:0.10871
 |
Component:054; Jaccard:0.090574
 |
Component:345; Jaccard:0.062925
 |
Component:345; Jaccard:0.034396
 |
Correlation:-0.19314
  |
Correlation:-0.056582
  |
Correlation:0.17011
  |
Correlation:0.17011
  |
Correlation:-0.12604
  |
Correlation:0.080287
  |
Correlation:0.080287
  |
Component:011; Jaccard:0.088586
 |
Component:209; Jaccard:0.10171
 |
Component:078; Jaccard:0.10852
 |
Component:054; Jaccard:0.10594
 |
Component:040; Jaccard:0.086106
 |
Component:040; Jaccard:0.061763
 |
Component:040; Jaccard:0.03421
 |
Correlation:0.090444
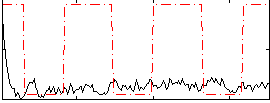 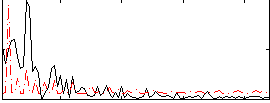 |
Correlation:-0.19314
  |
Correlation:0.28246
  |
Correlation:-0.12604
  |
Correlation:-0.13494
  |
Correlation:-0.13494
  |
Correlation:-0.13494
  |
Component:107; Jaccard:0.086919
 |
Component:013; Jaccard:0.10168
 |
Component:040; Jaccard:0.10278
 |
Component:040; Jaccard:0.10357
 |
Component:013; Jaccard:0.083435
 |
Component:013; Jaccard:0.055842
 |
Component:013; Jaccard:0.03082
 |
Correlation:-0.056582
  |
Correlation:0.17011
  |
Correlation:-0.13494
  |
Correlation:-0.13494
  |
Correlation:0.17011
  |
Correlation:0.17011
  |
Correlation:0.17011
  |
Component:013; Jaccard:0.085334
 |
Component:040; Jaccard:0.0911
 |
Component:209; Jaccard:0.10256
 |
Component:099; Jaccard:0.093004
 |
Component:099; Jaccard:0.074258
 |
Component:099; Jaccard:0.044062
 |
Component:273; Jaccard:0.019635
 |
Correlation:0.17011
  |
Correlation:-0.13494
  |
Correlation:-0.19314
  |
Correlation:0.097305
  |
Correlation:0.097305
  |
Correlation:0.097305
  |
Correlation:0.037839
  |
Component:391; Jaccard:0.08236
 |
Component:391; Jaccard:0.088255
 |
Component:054; Jaccard:0.10108
 |
Component:078; Jaccard:0.091359
 |
Component:078; Jaccard:0.06513
 |
Component:273; Jaccard:0.037462
 |
Component:149; Jaccard:0.019534
 |
Correlation:0.07986
  |
Correlation:0.07986
  |
Correlation:-0.12604
  |
Correlation:0.28246
  |
Correlation:0.28246
  |
Correlation:0.037839
  |
Correlation:-0.12074
  |
Component:315; Jaccard:0.080823
 |
Component:315; Jaccard:0.088131
 |
Component:099; Jaccard:0.097752
 |
Component:209; Jaccard:0.087725
 |
Component:209; Jaccard:0.060902
 |
Component:078; Jaccard:0.03666
 |
Component:078; Jaccard:0.018872
 |
Correlation:-0.039756
  |
Correlation:-0.039756
  |
Correlation:0.097305
  |
Correlation:-0.19314
  |
Correlation:-0.19314
  |
Correlation:0.28246
  |
Correlation:0.28246
  |
Component:148; Jaccard:0.078348
 |
Component:288; Jaccard:0.086855
 |
Component:288; Jaccard:0.089324
 |
Component:288; Jaccard:0.082844
 |
Component:288; Jaccard:0.058322
 |
Component:288; Jaccard:0.036368
 |
Component:206; Jaccard:0.018699
 |
Correlation:-0.12396
  |
Correlation:-0.10442
  |
Correlation:-0.10442
  |
Correlation:-0.10442
  |
Correlation:-0.10442
  |
Correlation:-0.10442
  |
Correlation:-0.1212
 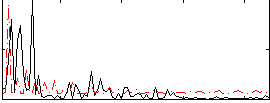 |
Component:288; Jaccard:0.077717
 |
Component:099; Jaccard:0.086571
 |
Component:315; Jaccard:0.086904
 |
Component:148; Jaccard:0.074484
 |
Component:273; Jaccard:0.052831
 |
Component:209; Jaccard:0.03514
 |
Component:209; Jaccard:0.017868
 |
Correlation:-0.10442
  |
Correlation:0.097305
  |
Correlation:-0.039756
  |
Correlation:-0.12396
  |
Correlation:0.037839
  |
Correlation:-0.19314
  |
Correlation:-0.19314
  |
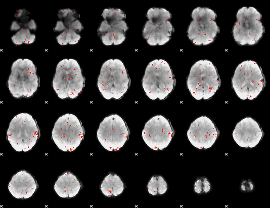
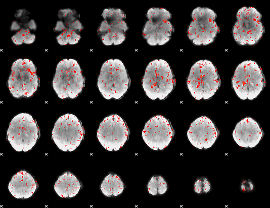
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:346; Jaccard:0.1003
 |
Component:346; Jaccard:0.10626
 |
Component:346; Jaccard:0.094171
 |
Component:346; Jaccard:0.060408
 |
Component:346; Jaccard:0.029045
 |
Component:061; Jaccard:0.014398
 |
Component:061; Jaccard:0.008607
 |
Correlation:0.38335
  |
Correlation:0.38335
  |
Correlation:0.38335
  |
Correlation:0.38335
  |
Correlation:0.38335
  |
Correlation:0.28591
  |
Correlation:0.28591
  |
Component:227; Jaccard:0.075397
 |
Component:227; Jaccard:0.076964
 |
Component:227; Jaccard:0.061738
 |
Component:134; Jaccard:0.04255
 |
Component:061; Jaccard:0.026353
 |
Component:346; Jaccard:0.012638
 |
Component:346; Jaccard:0.005862
 |
Correlation:0.15848
  |
Correlation:0.15848
  |
Correlation:0.15848
  |
Correlation:0.13854
  |
Correlation:0.28591
  |
Correlation:0.38335
  |
Correlation:0.38335
  |
Component:366; Jaccard:0.074608
 |
Component:366; Jaccard:0.071993
 |
Component:134; Jaccard:0.058965
 |
Component:061; Jaccard:0.042523
 |
Component:134; Jaccard:0.020166
 |
Component:134; Jaccard:0.009539
 |
Component:134; Jaccard:0.004506
 |
Correlation:0.25928
  |
Correlation:0.25928
  |
Correlation:0.13854
  |
Correlation:0.28591
  |
Correlation:0.13854
  |
Correlation:0.13854
  |
Correlation:0.13854
  |
Component:292; Jaccard:0.07371
 |
Component:292; Jaccard:0.071506
 |
Component:061; Jaccard:0.056993
 |
Component:227; Jaccard:0.037106
 |
Component:366; Jaccard:0.01735
 |
Component:054; Jaccard:0.007796
 |
Component:291; Jaccard:0.003727
 |
Correlation:0.2956
  |
Correlation:0.2956
  |
Correlation:0.28591
  |
Correlation:0.15848
  |
Correlation:0.25928
  |
Correlation:0.11456
  |
Correlation:0.41314
  |
Component:266; Jaccard:0.072417
 |
Component:009; Jaccard:0.067586
 |
Component:366; Jaccard:0.056408
 |
Component:366; Jaccard:0.03507
 |
Component:227; Jaccard:0.015637
 |
Component:291; Jaccard:0.007443
 |
Component:054; Jaccard:0.003415
 |
Correlation:0.20848
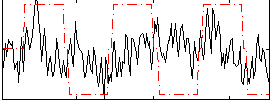  |
Correlation:0.10311
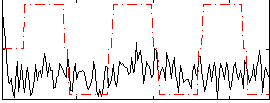  |
Correlation:0.25928
  |
Correlation:0.25928
  |
Correlation:0.15848
  |
Correlation:0.41314
  |
Correlation:0.11456
  |
Component:180; Jaccard:0.071009
 |
Component:180; Jaccard:0.06725
 |
Component:292; Jaccard:0.053607
 |
Component:098; Jaccard:0.034148
 |
Component:291; Jaccard:0.015038
 |
Component:366; Jaccard:0.006712
 |
Component:025; Jaccard:0.003153
 |
Correlation:0.18214
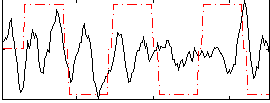  |
Correlation:0.18214
  |
Correlation:0.2956
  |
Correlation:0.21019
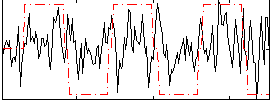  |
Correlation:0.41314
  |
Correlation:0.25928
  |
Correlation:0.15163
  |
Component:009; Jaccard:0.070588
 |
Component:183; Jaccard:0.066436
 |
Component:098; Jaccard:0.053321
 |
Component:292; Jaccard:0.032839
 |
Component:054; Jaccard:0.014449
 |
Component:227; Jaccard:0.005721
 |
Component:171; Jaccard:0.002916
 |
Correlation:0.10311
  |
Correlation:0.045435
  |
Correlation:0.21019
  |
Correlation:0.2956
  |
Correlation:0.11456
  |
Correlation:0.15848
  |
Correlation:-0.01663
  |
Component:186; Jaccard:0.070238
 |
Component:098; Jaccard:0.066433
 |
Component:186; Jaccard:0.051687
 |
Component:186; Jaccard:0.030244
 |
Component:094; Jaccard:0.01418
 |
Component:332; Jaccard:0.005321
 |
Component:366; Jaccard:0.002894
 |
Correlation:0.155
  |
Correlation:0.21019
  |
Correlation:0.155
  |
Correlation:0.155
  |
Correlation:0.095763
  |
Correlation:0.21333
  |
Correlation:0.25928
  |
Component:183; Jaccard:0.068474
 |
Component:186; Jaccard:0.065269
 |
Component:009; Jaccard:0.049486
 |
Component:183; Jaccard:0.02902
 |
Component:098; Jaccard:0.013231
 |
Component:171; Jaccard:0.005237
 |
Component:204; Jaccard:0.002012
 |
Correlation:0.045435
  |
Correlation:0.155
  |
Correlation:0.10311
  |
Correlation:0.045435
  |
Correlation:0.21019
  |
Correlation:-0.01663
  |
Correlation:0.0707
  |
Component:098; Jaccard:0.066994
 |
Component:266; Jaccard:0.063454
 |
Component:183; Jaccard:0.048773
 |
Component:332; Jaccard:0.027784
 |
Component:292; Jaccard:0.012872
 |
Component:149; Jaccard:0.00496
 |
Component:168; Jaccard:0.001902
 |
Correlation:0.21019
  |
Correlation:0.20848
  |
Correlation:0.045435
  |
Correlation:0.21333
  |
Correlation:0.2956
  |
Correlation:0.17729
  |
Correlation:0.075927
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































