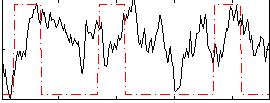
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
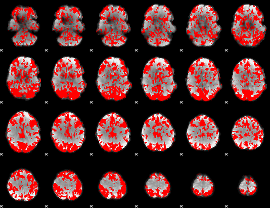 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
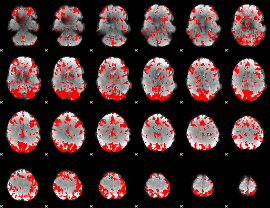 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
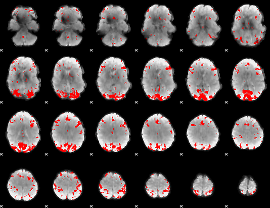 |
GLM Z-score result thresholded by 1
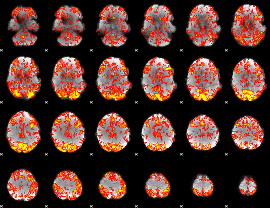 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
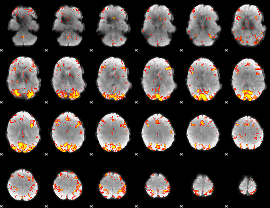 |
GLM Z-score result thresholded by 4
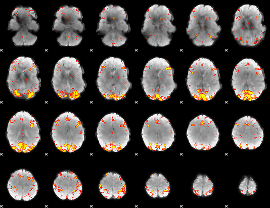 |
Component:038; Jaccard:0.13383
 |
Component:038; Jaccard:0.16641
 |
Component:038; Jaccard:0.20574
 |
Component:038; Jaccard:0.24636
 |
Component:272; Jaccard:0.28831
 |
Component:272; Jaccard:0.32505
 |
Component:272; Jaccard:0.34641
 |
Correlation:0.29108
  |
Correlation:0.29108
  |
Correlation:0.29108
  |
Correlation:0.29108
  |
Correlation:0.18965
  |
Correlation:0.18965
  |
Correlation:0.18965
  |
Component:255; Jaccard:0.12867
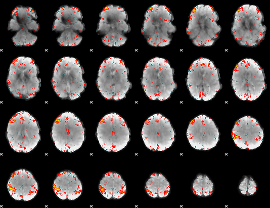 |
Component:272; Jaccard:0.16071
 |
Component:272; Jaccard:0.20138
 |
Component:272; Jaccard:0.2451
 |
Component:038; Jaccard:0.28108
 |
Component:038; Jaccard:0.30539
 |
Component:114; Jaccard:0.31711
 |
Correlation:0.18256
  |
Correlation:0.18965
  |
Correlation:0.18965
  |
Correlation:0.18965
  |
Correlation:0.29108
  |
Correlation:0.29108
  |
Correlation:0.32299
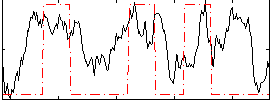 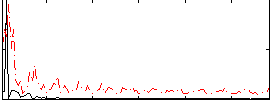 |
Component:272; Jaccard:0.12846
 |
Component:114; Jaccard:0.15004
 |
Component:114; Jaccard:0.18965
 |
Component:114; Jaccard:0.23264
 |
Component:114; Jaccard:0.27382
 |
Component:114; Jaccard:0.30436
 |
Component:038; Jaccard:0.31229
 |
Correlation:0.18965
  |
Correlation:0.32299
  |
Correlation:0.32299
  |
Correlation:0.32299
  |
Correlation:0.32299
  |
Correlation:0.32299
  |
Correlation:0.29108
  |
Component:331; Jaccard:0.12166
 |
Component:255; Jaccard:0.14768
 |
Component:331; Jaccard:0.16659
 |
Component:331; Jaccard:0.18618
 |
Component:331; Jaccard:0.19847
 |
Component:331; Jaccard:0.20028
 |
Component:331; Jaccard:0.19271
 |
Correlation:0.34479
  |
Correlation:0.18256
  |
Correlation:0.34479
  |
Correlation:0.34479
  |
Correlation:0.34479
  |
Correlation:0.34479
  |
Correlation:0.34479
  |
Component:387; Jaccard:0.12101
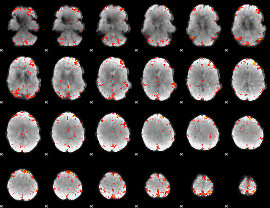 |
Component:331; Jaccard:0.14317
 |
Component:255; Jaccard:0.16369
 |
Component:255; Jaccard:0.17272
 |
Component:179; Jaccard:0.17507
 |
Component:179; Jaccard:0.17863
 |
Component:373; Jaccard:0.1837
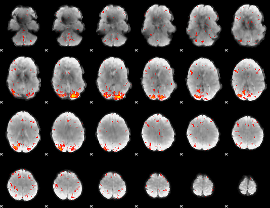 |
Correlation:0.28064
  |
Correlation:0.34479
  |
Correlation:0.18256
  |
Correlation:0.18256
  |
Correlation:0.28261
  |
Correlation:0.28261
  |
Correlation:0.2549
  |
Component:114; Jaccard:0.119
 |
Component:387; Jaccard:0.1345
 |
Component:387; Jaccard:0.14466
 |
Component:179; Jaccard:0.16079
 |
Component:255; Jaccard:0.17027
 |
Component:373; Jaccard:0.16866
 |
Component:179; Jaccard:0.17317
 |
Correlation:0.32299
  |
Correlation:0.28064
  |
Correlation:0.28064
  |
Correlation:0.28261
  |
Correlation:0.18256
  |
Correlation:0.2549
  |
Correlation:0.28261
  |
Component:163; Jaccard:0.10625
 |
Component:163; Jaccard:0.12137
 |
Component:179; Jaccard:0.14135
 |
Component:387; Jaccard:0.1482
 |
Component:373; Jaccard:0.14941
 |
Component:255; Jaccard:0.15763
 |
Component:006; Jaccard:0.13971
 |
Correlation:0.1551
  |
Correlation:0.1551
  |
Correlation:0.28261
  |
Correlation:0.28064
  |
Correlation:0.2549
  |
Correlation:0.18256
  |
Correlation:0.19224
  |
Component:306; Jaccard:0.10534
 |
Component:179; Jaccard:0.11967
 |
Component:163; Jaccard:0.13498
 |
Component:163; Jaccard:0.14322
 |
Component:163; Jaccard:0.14483
 |
Component:006; Jaccard:0.1412
 |
Component:255; Jaccard:0.13817
 |
Correlation:0.31713
  |
Correlation:0.28261
  |
Correlation:0.1551
  |
Correlation:0.1551
  |
Correlation:0.1551
  |
Correlation:0.19224
  |
Correlation:0.18256
  |
Component:179; Jaccard:0.10015
 |
Component:306; Jaccard:0.11311
 |
Component:006; Jaccard:0.11851
 |
Component:006; Jaccard:0.13043
 |
Component:387; Jaccard:0.14317
 |
Component:163; Jaccard:0.1382
 |
Component:163; Jaccard:0.12407
 |
Correlation:0.28261
  |
Correlation:0.31713
  |
Correlation:0.19224
  |
Correlation:0.19224
  |
Correlation:0.28064
  |
Correlation:0.1551
  |
Correlation:0.1551
  |
Component:103; Jaccard:0.094895
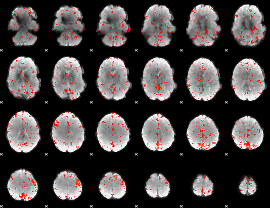 |
Component:006; Jaccard:0.10523
 |
Component:306; Jaccard:0.11794
 |
Component:373; Jaccard:0.12829
 |
Component:006; Jaccard:0.13822
 |
Component:387; Jaccard:0.1298
 |
Component:387; Jaccard:0.11021
 |
Correlation:0.31752
  |
Correlation:0.19224
  |
Correlation:0.31713
  |
Correlation:0.2549
  |
Correlation:0.19224
  |
Correlation:0.28064
  |
Correlation:0.28064
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
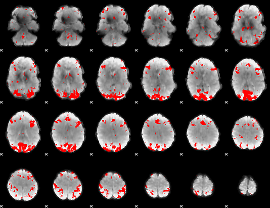 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:038; Jaccard:0.16691
 |
Component:272; Jaccard:0.20258
 |
Component:272; Jaccard:0.24565
 |
Component:272; Jaccard:0.28911
 |
Component:272; Jaccard:0.3251
 |
Component:272; Jaccard:0.34585
 |
Component:272; Jaccard:0.35263
 |
Correlation:0.097494
  |
Correlation:0.093272
  |
Correlation:0.093272
  |
Correlation:0.093272
  |
Correlation:0.093272
  |
Correlation:0.093272
  |
Correlation:0.093272
  |
Component:272; Jaccard:0.16502
 |
Component:038; Jaccard:0.20141
 |
Component:038; Jaccard:0.23568
 |
Component:114; Jaccard:0.27589
 |
Component:114; Jaccard:0.31039
 |
Component:114; Jaccard:0.32993
 |
Component:114; Jaccard:0.32865
 |
Correlation:0.093272
  |
Correlation:0.097494
  |
Correlation:0.097494
  |
Correlation:0.088094
  |
Correlation:0.088094
  |
Correlation:0.088094
  |
Correlation:0.088094
  |
Component:114; Jaccard:0.15241
 |
Component:114; Jaccard:0.18921
 |
Component:114; Jaccard:0.23222
 |
Component:038; Jaccard:0.26752
 |
Component:038; Jaccard:0.28796
 |
Component:038; Jaccard:0.2965
 |
Component:038; Jaccard:0.28862
 |
Correlation:0.088094
  |
Correlation:0.088094
  |
Correlation:0.088094
  |
Correlation:0.097494
  |
Correlation:0.097494
  |
Correlation:0.097494
  |
Correlation:0.097494
  |
Component:163; Jaccard:0.1294
 |
Component:163; Jaccard:0.14493
 |
Component:163; Jaccard:0.1589
 |
Component:163; Jaccard:0.16529
 |
Component:373; Jaccard:0.18594
 |
Component:373; Jaccard:0.2043
 |
Component:373; Jaccard:0.21188
 |
Correlation:0.015387
  |
Correlation:0.015387
  |
Correlation:0.015387
  |
Correlation:0.015387
  |
Correlation:0.25191
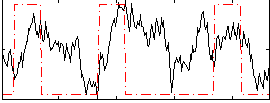 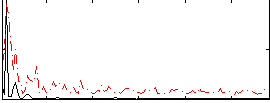 |
Correlation:0.25191
  |
Correlation:0.25191
  |
Component:255; Jaccard:0.12446
 |
Component:331; Jaccard:0.13777
 |
Component:331; Jaccard:0.15136
 |
Component:373; Jaccard:0.16469
 |
Component:179; Jaccard:0.16604
 |
Component:179; Jaccard:0.1653
 |
Component:179; Jaccard:0.16128
 |
Correlation:-0.11824
  |
Correlation:-0.20365
  |
Correlation:-0.20365
  |
Correlation:0.25191
  |
Correlation:0.073024
  |
Correlation:0.073024
  |
Correlation:0.073024
  |
Component:331; Jaccard:0.12324
 |
Component:255; Jaccard:0.13431
 |
Component:179; Jaccard:0.14785
 |
Component:331; Jaccard:0.16125
 |
Component:331; Jaccard:0.16585
 |
Component:331; Jaccard:0.16509
 |
Component:331; Jaccard:0.15746
 |
Correlation:-0.20365
  |
Correlation:-0.11824
  |
Correlation:0.073024
  |
Correlation:-0.20365
  |
Correlation:-0.20365
  |
Correlation:-0.20365
  |
Correlation:-0.20365
  |
Component:193; Jaccard:0.11841
 |
Component:193; Jaccard:0.13346
 |
Component:205; Jaccard:0.14758
 |
Component:179; Jaccard:0.16005
 |
Component:205; Jaccard:0.1642
 |
Component:205; Jaccard:0.15779
 |
Component:205; Jaccard:0.14444
 |
Correlation:0.099967
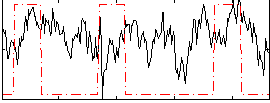 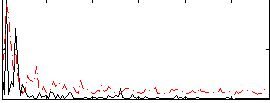 |
Correlation:0.099967
  |
Correlation:0.10763
  |
Correlation:0.073024
  |
Correlation:0.10763
  |
Correlation:0.10763
  |
Correlation:0.10763
  |
Component:179; Jaccard:0.11478
 |
Component:179; Jaccard:0.13193
 |
Component:193; Jaccard:0.14596
 |
Component:205; Jaccard:0.1578
 |
Component:163; Jaccard:0.16187
 |
Component:163; Jaccard:0.14987
 |
Component:006; Jaccard:0.14004
 |
Correlation:0.073024
  |
Correlation:0.073024
  |
Correlation:0.099967
  |
Correlation:0.10763
  |
Correlation:0.015387
  |
Correlation:0.015387
  |
Correlation:0.106
  |
Component:205; Jaccard:0.11425
 |
Component:205; Jaccard:0.13147
 |
Component:255; Jaccard:0.14061
 |
Component:193; Jaccard:0.15269
 |
Component:193; Jaccard:0.15346
 |
Component:193; Jaccard:0.14964
 |
Component:193; Jaccard:0.13924
 |
Correlation:0.10763
  |
Correlation:0.10763
  |
Correlation:-0.11824
  |
Correlation:0.099967
  |
Correlation:0.099967
  |
Correlation:0.099967
  |
Correlation:0.099967
  |
Component:006; Jaccard:0.10799
 |
Component:006; Jaccard:0.12142
 |
Component:373; Jaccard:0.14017
 |
Component:006; Jaccard:0.14079
 |
Component:006; Jaccard:0.14527
 |
Component:006; Jaccard:0.14606
 |
Component:163; Jaccard:0.13313
 |
Correlation:0.106
  |
Correlation:0.106
  |
Correlation:0.25191
  |
Correlation:0.106
  |
Correlation:0.106
  |
Correlation:0.106
  |
Correlation:0.015387
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
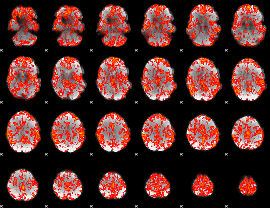
GLM binary result thresholded by 2
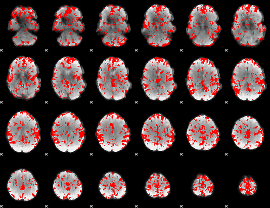 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
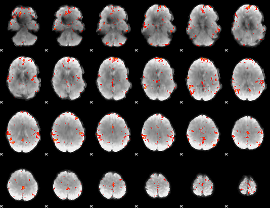
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:018; Jaccard:0.10838
 |
Component:018; Jaccard:0.12789
 |
Component:018; Jaccard:0.1483
 |
Component:018; Jaccard:0.16015
 |
Component:018; Jaccard:0.15694
 |
Component:018; Jaccard:0.13512
 |
Component:018; Jaccard:0.096946
 |
Correlation:0.45795
  |
Correlation:0.45795
  |
Correlation:0.45795
  |
Correlation:0.45795
  |
Correlation:0.45795
  |
Correlation:0.45795
  |
Correlation:0.45795
  |
Component:327; Jaccard:0.099993
 |
Component:327; Jaccard:0.11381
 |
Component:327; Jaccard:0.12459
 |
Component:314; Jaccard:0.13417
 |
Component:314; Jaccard:0.13725
 |
Component:314; Jaccard:0.12357
 |
Component:314; Jaccard:0.093683
 |
Correlation:0.39719
  |
Correlation:0.39719
  |
Correlation:0.39719
  |
Correlation:0.35518
  |
Correlation:0.35518
  |
Correlation:0.35518
  |
Correlation:0.35518
  |
Component:370; Jaccard:0.095517
 |
Component:053; Jaccard:0.10914
 |
Component:053; Jaccard:0.12442
 |
Component:053; Jaccard:0.1338
 |
Component:053; Jaccard:0.13201
 |
Component:024; Jaccard:0.11718
 |
Component:024; Jaccard:0.085975
 |
Correlation:0.28147
  |
Correlation:0.30832
  |
Correlation:0.30832
  |
Correlation:0.30832
  |
Correlation:0.30832
  |
Correlation:0.05193
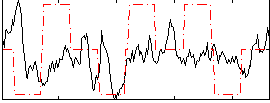  |
Correlation:0.05193
  |
Component:387; Jaccard:0.095221
 |
Component:370; Jaccard:0.10852
 |
Component:370; Jaccard:0.11868
 |
Component:024; Jaccard:0.12781
 |
Component:024; Jaccard:0.13031
 |
Component:053; Jaccard:0.10856
 |
Component:053; Jaccard:0.075219
 |
Correlation:0.37685
  |
Correlation:0.28147
  |
Correlation:0.28147
  |
Correlation:0.05193
  |
Correlation:0.05193
  |
Correlation:0.30832
  |
Correlation:0.30832
  |
Component:059; Jaccard:0.09313
 |
Component:266; Jaccard:0.10666
 |
Component:266; Jaccard:0.11867
 |
Component:327; Jaccard:0.12717
 |
Component:370; Jaccard:0.1193
 |
Component:050; Jaccard:0.098405
 |
Component:050; Jaccard:0.070731
 |
Correlation:0.13619
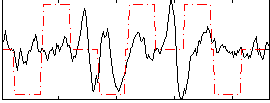 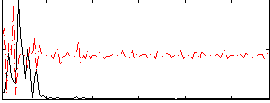 |
Correlation:0.32199
  |
Correlation:0.32199
  |
Correlation:0.39719
  |
Correlation:0.28147
  |
Correlation:0.048636
  |
Correlation:0.048636
  |
Component:266; Jaccard:0.092599
 |
Component:387; Jaccard:0.10401
 |
Component:314; Jaccard:0.11776
 |
Component:370; Jaccard:0.12435
 |
Component:266; Jaccard:0.11515
 |
Component:370; Jaccard:0.09696
 |
Component:370; Jaccard:0.067042
 |
Correlation:0.32199
  |
Correlation:0.37685
  |
Correlation:0.35518
  |
Correlation:0.28147
  |
Correlation:0.32199
  |
Correlation:0.28147
  |
Correlation:0.28147
  |
Component:053; Jaccard:0.092506
 |
Component:059; Jaccard:0.10326
 |
Component:024; Jaccard:0.11454
 |
Component:266; Jaccard:0.12374
 |
Component:327; Jaccard:0.1146
 |
Component:266; Jaccard:0.092369
 |
Component:032; Jaccard:0.064842
 |
Correlation:0.30832
  |
Correlation:0.13619
  |
Correlation:0.05193
  |
Correlation:0.32199
  |
Correlation:0.39719
  |
Correlation:0.32199
  |
Correlation:0.1342
  |
Component:308; Jaccard:0.090221
 |
Component:382; Jaccard:0.10008
 |
Component:382; Jaccard:0.11139
 |
Component:382; Jaccard:0.11734
 |
Component:050; Jaccard:0.11395
 |
Component:327; Jaccard:0.09028
 |
Component:266; Jaccard:0.062461
 |
Correlation:0.29126
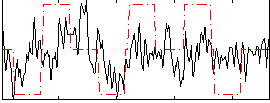  |
Correlation:0.2473
  |
Correlation:0.2473
  |
Correlation:0.2473
  |
Correlation:0.048636
  |
Correlation:0.39719
  |
Correlation:0.32199
  |
Component:381; Jaccard:0.088806
 |
Component:024; Jaccard:0.10002
 |
Component:059; Jaccard:0.10959
 |
Component:050; Jaccard:0.11423
 |
Component:382; Jaccard:0.11127
 |
Component:382; Jaccard:0.088553
 |
Component:327; Jaccard:0.060799
 |
Correlation:0.25042
  |
Correlation:0.05193
  |
Correlation:0.13619
  |
Correlation:0.048636
  |
Correlation:0.2473
  |
Correlation:0.2473
  |
Correlation:0.39719
  |
Component:371; Jaccard:0.087483
 |
Component:381; Jaccard:0.099634
 |
Component:387; Jaccard:0.10803
 |
Component:059; Jaccard:0.10958
 |
Component:059; Jaccard:0.099076
 |
Component:032; Jaccard:0.080457
 |
Component:382; Jaccard:0.056275
 |
Correlation:0.15335
  |
Correlation:0.25042
  |
Correlation:0.37685
  |
Correlation:0.13619
  |
Correlation:0.13619
  |
Correlation:0.1342
  |
Correlation:0.2473
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
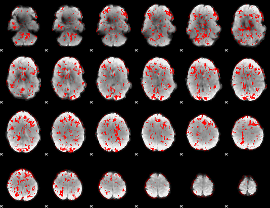 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
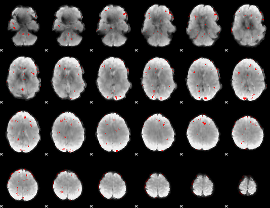 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
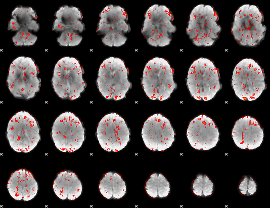 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
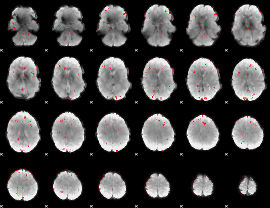 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:390; Jaccard:0.11791
 |
Component:390; Jaccard:0.13354
 |
Component:390; Jaccard:0.13213
 |
Component:390; Jaccard:0.11382
 |
Component:390; Jaccard:0.075464
 |
Component:390; Jaccard:0.035857
 |
Component:390; Jaccard:0.012926
 |
Correlation:0.34912
  |
Correlation:0.34912
  |
Correlation:0.34912
  |
Correlation:0.34912
  |
Correlation:0.34912
  |
Correlation:0.34912
  |
Correlation:0.34912
  |
Component:169; Jaccard:0.082877
 |
Component:149; Jaccard:0.095561
 |
Component:149; Jaccard:0.10042
 |
Component:189; Jaccard:0.091067
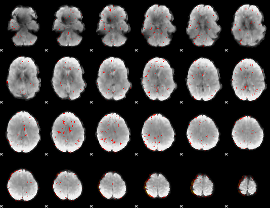 |
Component:189; Jaccard:0.063494
 |
Component:189; Jaccard:0.031397
 |
Component:149; Jaccard:0.012876
 |
Correlation:0.30228
  |
Correlation:0.17932
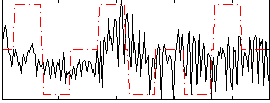  |
Correlation:0.17932
  |
Correlation:0.37181
  |
Correlation:0.37181
  |
Correlation:0.37181
  |
Correlation:0.17932
  |
Component:149; Jaccard:0.082847
 |
Component:169; Jaccard:0.089854
 |
Component:189; Jaccard:0.098608
 |
Component:149; Jaccard:0.088195
 |
Component:149; Jaccard:0.061117
 |
Component:149; Jaccard:0.031324
 |
Component:189; Jaccard:0.011192
 |
Correlation:0.17932
  |
Correlation:0.30228
  |
Correlation:0.37181
  |
Correlation:0.17932
  |
Correlation:0.17932
  |
Correlation:0.17932
  |
Correlation:0.37181
  |
Component:205; Jaccard:0.082316
 |
Component:189; Jaccard:0.088348
 |
Component:169; Jaccard:0.084396
 |
Component:169; Jaccard:0.06383
 |
Component:322; Jaccard:0.039051
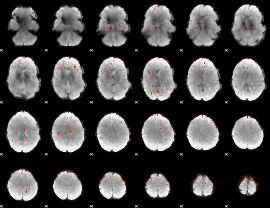 |
Component:079; Jaccard:0.019381
 |
Component:079; Jaccard:0.008278
 |
Correlation:-0.087641
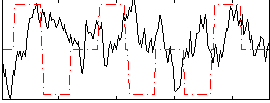  |
Correlation:0.37181
  |
Correlation:0.30228
  |
Correlation:0.30228
  |
Correlation:0.30173
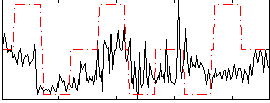  |
Correlation:0.2299
  |
Correlation:0.2299
  |
Component:200; Jaccard:0.080955
 |
Component:200; Jaccard:0.082149
 |
Component:200; Jaccard:0.075923
 |
Component:200; Jaccard:0.058348
 |
Component:169; Jaccard:0.037091
 |
Component:322; Jaccard:0.019099
 |
Component:322; Jaccard:0.007388
 |
Correlation:0.24431
  |
Correlation:0.24431
  |
Correlation:0.24431
  |
Correlation:0.24431
  |
Correlation:0.30228
  |
Correlation:0.30173
  |
Correlation:0.30173
  |
Component:193; Jaccard:0.078232
 |
Component:131; Jaccard:0.079797
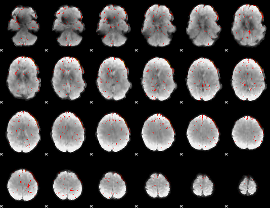 |
Component:131; Jaccard:0.069166
 |
Component:322; Jaccard:0.057734
 |
Component:200; Jaccard:0.036354
 |
Component:394; Jaccard:0.017177
 |
Component:394; Jaccard:0.006766
 |
Correlation:0.010017
 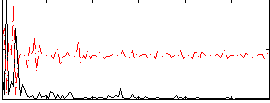 |
Correlation:0.14797
  |
Correlation:0.14797
  |
Correlation:0.30173
  |
Correlation:0.24431
  |
Correlation:0.19913
  |
Correlation:0.19913
  |
Component:131; Jaccard:0.077973
 |
Component:205; Jaccard:0.074639
 |
Component:151; Jaccard:0.068249
 |
Component:151; Jaccard:0.052853
 |
Component:079; Jaccard:0.035432
 |
Component:200; Jaccard:0.016963
 |
Component:294; Jaccard:0.006606
 |
Correlation:0.14797
  |
Correlation:-0.087641
  |
Correlation:0.36385
 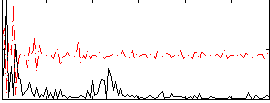 |
Correlation:0.36385
  |
Correlation:0.2299
  |
Correlation:0.24431
  |
Correlation:0.1548
  |
Component:189; Jaccard:0.071749
 |
Component:193; Jaccard:0.069914
 |
Component:322; Jaccard:0.066656
 |
Component:131; Jaccard:0.049879
 |
Component:394; Jaccard:0.034179
 |
Component:294; Jaccard:0.015799
 |
Component:200; Jaccard:0.006573
 |
Correlation:0.37181
  |
Correlation:0.010017
  |
Correlation:0.30173
  |
Correlation:0.14797
  |
Correlation:0.19913
  |
Correlation:0.1548
  |
Correlation:0.24431
  |
Component:026; Jaccard:0.071304
 |
Component:335; Jaccard:0.068975
 |
Component:335; Jaccard:0.058999
 |
Component:079; Jaccard:0.049766
 |
Component:151; Jaccard:0.033111
 |
Component:169; Jaccard:0.015397
 |
Component:373; Jaccard:0.005695
 |
Correlation:0.12667
  |
Correlation:0.30582
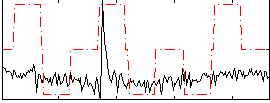  |
Correlation:0.30582
  |
Correlation:0.2299
  |
Correlation:0.36385
  |
Correlation:0.30228
  |
Correlation:-0.0017768
 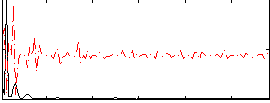 |
Component:211; Jaccard:0.070289
 |
Component:151; Jaccard:0.066956
 |
Component:236; Jaccard:0.058188
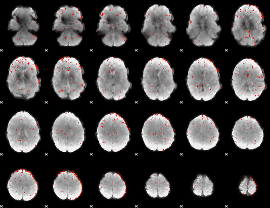 |
Component:394; Jaccard:0.04531
 |
Component:294; Jaccard:0.028441
 |
Component:151; Jaccard:0.014708
 |
Component:335; Jaccard:0.005605
 |
Correlation:-0.041995
 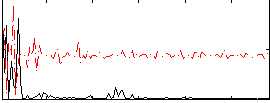 |
Correlation:0.36385
  |
Correlation:0.15493
  |
Correlation:0.19913
  |
Correlation:0.1548
  |
Correlation:0.36385
  |
Correlation:0.30582
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
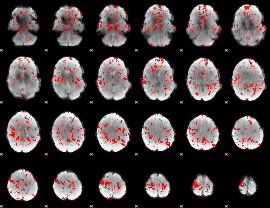 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
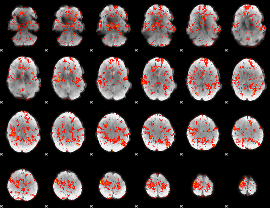 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
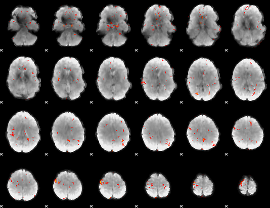 |
GLM Z-score result thresholded by 4
 |
Component:326; Jaccard:0.1226
 |
Component:326; Jaccard:0.13861
 |
Component:326; Jaccard:0.14535
 |
Component:326; Jaccard:0.13438
 |
Component:326; Jaccard:0.10743
 |
Component:326; Jaccard:0.075415
 |
Component:249; Jaccard:0.05475
 |
Correlation:0.10401
  |
Correlation:0.10401
  |
Correlation:0.10401
  |
Correlation:0.10401
  |
Correlation:0.10401
  |
Correlation:0.10401
  |
Correlation:0.30915
  |
Component:186; Jaccard:0.096182
 |
Component:186; Jaccard:0.095185
 |
Component:032; Jaccard:0.096939
 |
Component:195; Jaccard:0.095714
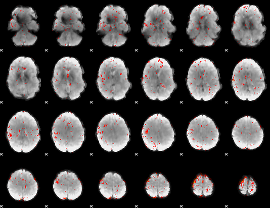 |
Component:195; Jaccard:0.08334
 |
Component:249; Jaccard:0.068653
 |
Component:195; Jaccard:0.047751
 |
Correlation:0.0085299
  |
Correlation:0.0085299
  |
Correlation:0.086028
  |
Correlation:0.2509
  |
Correlation:0.2509
  |
Correlation:0.30915
  |
Correlation:0.2509
  |
Component:209; Jaccard:0.088303
 |
Component:032; Jaccard:0.094046
 |
Component:204; Jaccard:0.094668
 |
Component:032; Jaccard:0.090086
 |
Component:032; Jaccard:0.074982
 |
Component:195; Jaccard:0.068304
 |
Component:326; Jaccard:0.047009
 |
Correlation:-0.028022
  |
Correlation:0.086028
  |
Correlation:0.21575
  |
Correlation:0.086028
  |
Correlation:0.086028
  |
Correlation:0.2509
  |
Correlation:0.10401
  |
Component:234; Jaccard:0.086281
 |
Component:234; Jaccard:0.093843
 |
Component:234; Jaccard:0.094407
 |
Component:204; Jaccard:0.086511
 |
Component:249; Jaccard:0.074888
 |
Component:032; Jaccard:0.05103
 |
Component:079; Jaccard:0.035305
 |
Correlation:0.18395
  |
Correlation:0.18395
  |
Correlation:0.18395
  |
Correlation:0.21575
  |
Correlation:0.30915
  |
Correlation:0.086028
  |
Correlation:0.30027
  |
Component:180; Jaccard:0.085269
 |
Component:204; Jaccard:0.091712
 |
Component:195; Jaccard:0.093899
 |
Component:234; Jaccard:0.086509
 |
Component:204; Jaccard:0.070727
 |
Component:079; Jaccard:0.050234
 |
Component:384; Jaccard:0.034401
 |
Correlation:0.34557
  |
Correlation:0.21575
  |
Correlation:0.2509
  |
Correlation:0.18395
  |
Correlation:0.21575
  |
Correlation:0.30027
  |
Correlation:0.089818
  |
Component:380; Jaccard:0.085102
 |
Component:209; Jaccard:0.090916
 |
Component:384; Jaccard:0.091615
 |
Component:384; Jaccard:0.08385
 |
Component:234; Jaccard:0.070663
 |
Component:234; Jaccard:0.048656
 |
Component:032; Jaccard:0.027366
 |
Correlation:0.096409
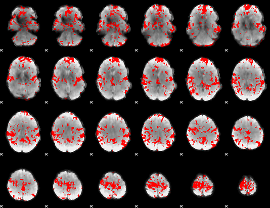
  |
Correlation:-0.028022
  |
Correlation:0.089818
  |
Correlation:0.089818
  |
Correlation:0.18395
  |
Correlation:0.18395
  |
Correlation:0.086028
  |
Component:032; Jaccard:0.083547
 |
Component:180; Jaccard:0.089315
 |
Component:209; Jaccard:0.089877
 |
Component:375; Jaccard:0.078454
 |
Component:384; Jaccard:0.06625
 |
Component:384; Jaccard:0.048526
 |
Component:234; Jaccard:0.02718
 |
Correlation:0.086028
  |
Correlation:0.34557
  |
Correlation:-0.028022
  |
Correlation:0.24457
  |
Correlation:0.089818
  |
Correlation:0.089818
  |
Correlation:0.18395
  |
Component:384; Jaccard:0.083333
 |
Component:384; Jaccard:0.089274
 |
Component:186; Jaccard:0.086332
 |
Component:209; Jaccard:0.077329
 |
Component:375; Jaccard:0.063931
 |
Component:204; Jaccard:0.047101
 |
Component:204; Jaccard:0.026593
 |
Correlation:0.089818
  |
Correlation:0.089818
  |
Correlation:0.0085299
  |
Correlation:-0.028022
  |
Correlation:0.24457
  |
Correlation:0.21575
  |
Correlation:0.21575
  |
Component:204; Jaccard:0.082997
 |
Component:195; Jaccard:0.087051
 |
Component:180; Jaccard:0.086004
 |
Component:180; Jaccard:0.073505
 |
Component:079; Jaccard:0.06154
 |
Component:375; Jaccard:0.043868
 |
Component:375; Jaccard:0.026459
 |
Correlation:0.21575
  |
Correlation:0.2509
  |
Correlation:0.34557
  |
Correlation:0.34557
  |
Correlation:0.30027
  |
Correlation:0.24457
  |
Correlation:0.24457
  |
Component:024; Jaccard:0.080595
 |
Component:380; Jaccard:0.086999
 |
Component:380; Jaccard:0.08447
 |
Component:249; Jaccard:0.073016
 |
Component:209; Jaccard:0.059135
 |
Component:160; Jaccard:0.040261
 |
Component:160; Jaccard:0.025034
 |
Correlation:0.057435
  |
Correlation:0.096409
  |
Correlation:0.096409
  |
Correlation:0.30915
  |
Correlation:-0.028022
  |
Correlation:0.26412
  |
Correlation:0.26412
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
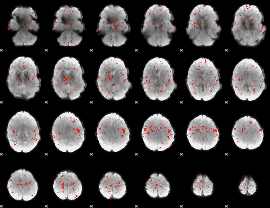
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
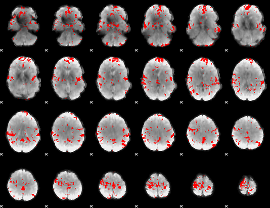 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
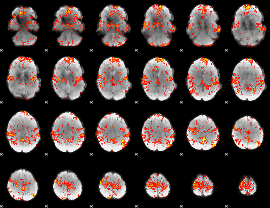 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:186; Jaccard:0.10747
 |
Component:326; Jaccard:0.12333
 |
Component:326; Jaccard:0.14115
 |
Component:326; Jaccard:0.1577
 |
Component:326; Jaccard:0.16832
 |
Component:326; Jaccard:0.16965
 |
Component:314; Jaccard:0.15287
 |
Correlation:0.45402
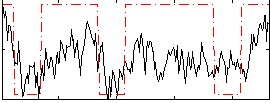  |
Correlation:0.22568
 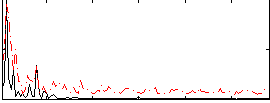 |
Correlation:0.22568
  |
Correlation:0.22568
  |
Correlation:0.22568
  |
Correlation:0.22568
  |
Correlation:0.5202
  |
Component:326; Jaccard:0.10656
 |
Component:186; Jaccard:0.12185
 |
Component:186; Jaccard:0.13652
 |
Component:024; Jaccard:0.14981
 |
Component:024; Jaccard:0.16233
 |
Component:314; Jaccard:0.1645
 |
Component:024; Jaccard:0.15276
 |
Correlation:0.22568
  |
Correlation:0.45402
  |
Correlation:0.45402
  |
Correlation:0.14505
  |
Correlation:0.14505
  |
Correlation:0.5202
  |
Correlation:0.14505
  |
Component:024; Jaccard:0.10145
 |
Component:024; Jaccard:0.1169
 |
Component:024; Jaccard:0.13376
 |
Component:186; Jaccard:0.14901
 |
Component:314; Jaccard:0.16109
 |
Component:024; Jaccard:0.16341
 |
Component:326; Jaccard:0.15211
 |
Correlation:0.14505
  |
Correlation:0.14505
  |
Correlation:0.14505
  |
Correlation:0.45402
  |
Correlation:0.5202
  |
Correlation:0.14505
  |
Correlation:0.22568
  |
Component:050; Jaccard:0.099383
 |
Component:314; Jaccard:0.11332
 |
Component:314; Jaccard:0.13053
 |
Component:314; Jaccard:0.14884
 |
Component:186; Jaccard:0.153
 |
Component:186; Jaccard:0.14963
 |
Component:032; Jaccard:0.14753
 |
Correlation:0.26605
  |
Correlation:0.5202
  |
Correlation:0.5202
  |
Correlation:0.5202
  |
Correlation:0.45402
  |
Correlation:0.45402
  |
Correlation:0.31246
  |
Component:314; Jaccard:0.096988
 |
Component:050; Jaccard:0.11024
 |
Component:050; Jaccard:0.12072
 |
Component:050; Jaccard:0.12956
 |
Component:050; Jaccard:0.13404
 |
Component:032; Jaccard:0.14334
 |
Component:186; Jaccard:0.13402
 |
Correlation:0.5202
  |
Correlation:0.26605
  |
Correlation:0.26605
  |
Correlation:0.26605
  |
Correlation:0.26605
  |
Correlation:0.31246
  |
Correlation:0.45402
  |
Component:266; Jaccard:0.096064
 |
Component:382; Jaccard:0.10525
 |
Component:382; Jaccard:0.11466
 |
Component:032; Jaccard:0.12022
 |
Component:032; Jaccard:0.13365
 |
Component:050; Jaccard:0.13064
 |
Component:050; Jaccard:0.12353
 |
Correlation:0.43355
  |
Correlation:0.37016
  |
Correlation:0.37016
  |
Correlation:0.31246
  |
Correlation:0.31246
  |
Correlation:0.26605
  |
Correlation:0.26605
  |
Component:382; Jaccard:0.095275
 |
Component:053; Jaccard:0.10463
 |
Component:053; Jaccard:0.11367
 |
Component:382; Jaccard:0.1182
 |
Component:053; Jaccard:0.11909
 |
Component:053; Jaccard:0.11385
 |
Component:209; Jaccard:0.098933
 |
Correlation:0.37016
  |
Correlation:0.29536
  |
Correlation:0.29536
  |
Correlation:0.37016
  |
Correlation:0.29536
  |
Correlation:0.29536
  |
Correlation:0.30066
  |
Component:053; Jaccard:0.094306
 |
Component:266; Jaccard:0.10393
 |
Component:266; Jaccard:0.11132
 |
Component:053; Jaccard:0.11782
 |
Component:209; Jaccard:0.11754
 |
Component:209; Jaccard:0.11305
 |
Component:053; Jaccard:0.098576
 |
Correlation:0.29536
  |
Correlation:0.43355
  |
Correlation:0.43355
  |
Correlation:0.29536
  |
Correlation:0.30066
  |
Correlation:0.30066
  |
Correlation:0.29536
  |
Component:370; Jaccard:0.092396
 |
Component:370; Jaccard:0.098807
 |
Component:209; Jaccard:0.10773
 |
Component:266; Jaccard:0.11624
 |
Component:266; Jaccard:0.11552
 |
Component:266; Jaccard:0.1096
 |
Component:266; Jaccard:0.098069
 |
Correlation:0.38472
  |
Correlation:0.38472
  |
Correlation:0.30066
  |
Correlation:0.43355
  |
Correlation:0.43355
  |
Correlation:0.43355
  |
Correlation:0.43355
  |
Component:282; Jaccard:0.089563
 |
Component:209; Jaccard:0.098598
 |
Component:032; Jaccard:0.10554
 |
Component:209; Jaccard:0.11554
 |
Component:382; Jaccard:0.11429
 |
Component:382; Jaccard:0.1038
 |
Component:234; Jaccard:0.088207
 |
Correlation:0.29696
  |
Correlation:0.30066
  |
Correlation:0.31246
  |
Correlation:0.30066
  |
Correlation:0.37016
  |
Correlation:0.37016
  |
Correlation:0.19648
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































