Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
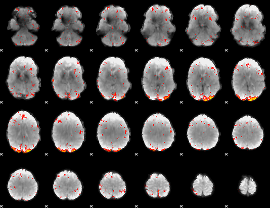
GLM result group-wise
 |
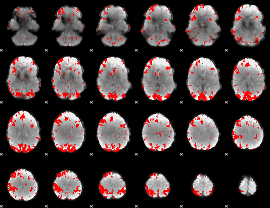
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
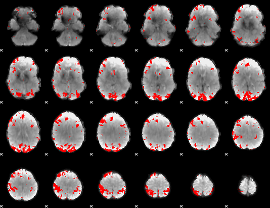
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
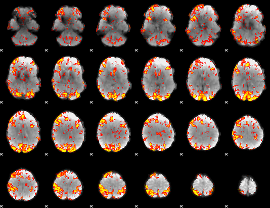
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
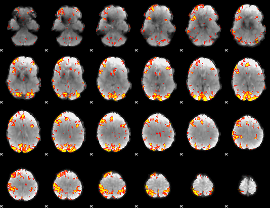
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
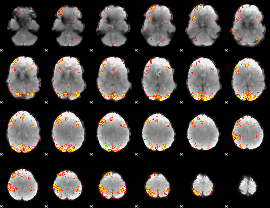 |
Component:220; Jaccard:0.14974
 |
Component:220; Jaccard:0.17728
 |
Component:220; Jaccard:0.20639
 |
Component:220; Jaccard:0.23038
 |
Component:220; Jaccard:0.24821
 |
Component:220; Jaccard:0.25244
 |
Component:027; Jaccard:0.25134
 |
Correlation:0.080725
  |
Correlation:0.080725
  |
Correlation:0.080725
  |
Correlation:0.080725
  |
Correlation:0.080725
  |
Correlation:0.080725
  |
Correlation:0.27268
  |
Component:168; Jaccard:0.14192
 |
Component:168; Jaccard:0.16994
 |
Component:168; Jaccard:0.20024
 |
Component:168; Jaccard:0.22514
 |
Component:168; Jaccard:0.24234
 |
Component:168; Jaccard:0.25052
 |
Component:168; Jaccard:0.25084
 |
Correlation:0.13942
  |
Correlation:0.13942
  |
Correlation:0.13942
  |
Correlation:0.13942
  |
Correlation:0.13942
  |
Correlation:0.13942
  |
Correlation:0.13942
  |
Component:043; Jaccard:0.13444
 |
Component:027; Jaccard:0.15185
 |
Component:027; Jaccard:0.18213
 |
Component:027; Jaccard:0.20788
 |
Component:027; Jaccard:0.23141
 |
Component:027; Jaccard:0.2472
 |
Component:220; Jaccard:0.25055
 |
Correlation:0.13521
  |
Correlation:0.27268
  |
Correlation:0.27268
  |
Correlation:0.27268
  |
Correlation:0.27268
  |
Correlation:0.27268
  |
Correlation:0.080725
  |
Component:398; Jaccard:0.12637
 |
Component:043; Jaccard:0.15054
 |
Component:009; Jaccard:0.17797
 |
Component:009; Jaccard:0.20364
 |
Component:009; Jaccard:0.22433
 |
Component:009; Jaccard:0.23847
 |
Component:009; Jaccard:0.2458
 |
Correlation:0.071326
  |
Correlation:0.13521
  |
Correlation:0.12795
  |
Correlation:0.12795
  |
Correlation:0.12795
  |
Correlation:0.12795
  |
Correlation:0.12795
  |
Component:027; Jaccard:0.12539
 |
Component:009; Jaccard:0.15023
 |
Component:043; Jaccard:0.16329
 |
Component:043; Jaccard:0.1738
 |
Component:096; Jaccard:0.18716
 |
Component:159; Jaccard:0.19724
 |
Component:159; Jaccard:0.21063
 |
Correlation:0.27268
  |
Correlation:0.12795
  |
Correlation:0.13521
  |
Correlation:0.13521
  |
Correlation:0.1641
  |
Correlation:0.35952
  |
Correlation:0.35952
  |
Component:009; Jaccard:0.12534
 |
Component:398; Jaccard:0.14072
 |
Component:096; Jaccard:0.15498
 |
Component:096; Jaccard:0.17362
 |
Component:159; Jaccard:0.18092
 |
Component:096; Jaccard:0.19417
 |
Component:096; Jaccard:0.19564
 |
Correlation:0.12795
  |
Correlation:0.071326
  |
Correlation:0.1641
  |
Correlation:0.1641
  |
Correlation:0.35952
  |
Correlation:0.1641
  |
Correlation:0.1641
  |
Component:021; Jaccard:0.11767
 |
Component:021; Jaccard:0.13625
 |
Component:021; Jaccard:0.15346
 |
Component:021; Jaccard:0.16563
 |
Component:043; Jaccard:0.17662
 |
Component:021; Jaccard:0.17913
 |
Component:038; Jaccard:0.18405
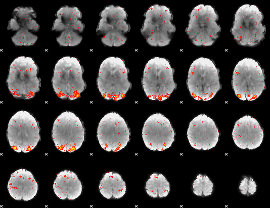 |
Correlation:0.25875
  |
Correlation:0.25875
  |
Correlation:0.25875
  |
Correlation:0.25875
  |
Correlation:0.13521
  |
Correlation:0.25875
  |
Correlation:0.15301
  |
Component:096; Jaccard:0.11214
 |
Component:096; Jaccard:0.13254
 |
Component:398; Jaccard:0.15319
 |
Component:159; Jaccard:0.15901
 |
Component:021; Jaccard:0.17597
 |
Component:043; Jaccard:0.17387
 |
Component:021; Jaccard:0.17605
 |
Correlation:0.1641
  |
Correlation:0.1641
  |
Correlation:0.071326
  |
Correlation:0.35952
  |
Correlation:0.25875
  |
Correlation:0.13521
  |
Correlation:0.25875
  |
Component:090; Jaccard:0.10959
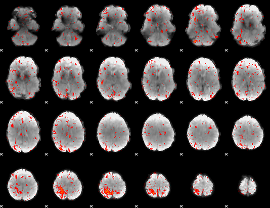 |
Component:295; Jaccard:0.11921
 |
Component:159; Jaccard:0.13798
 |
Component:398; Jaccard:0.15861
 |
Component:398; Jaccard:0.15965
 |
Component:038; Jaccard:0.17167
 |
Component:043; Jaccard:0.16558
 |
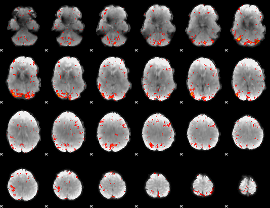
Correlation:0.20057
  |
Correlation:0.074316
  |
Correlation:0.35952
  |
Correlation:0.071326
  |
Correlation:0.071326
  |
Correlation:0.15301
  |
Correlation:0.13521
  |
Component:304; Jaccard:0.10908
 |
Component:304; Jaccard:0.11842
 |
Component:295; Jaccard:0.1319
 |
Component:295; Jaccard:0.14093
 |
Component:038; Jaccard:0.15654
 |
Component:398; Jaccard:0.15384
 |
Component:398; Jaccard:0.14567
 |
Correlation:-0.0067406
  |
Correlation:-0.0067406
  |
Correlation:0.074316
  |
Correlation:0.074316
  |
Correlation:0.15301
  |
Correlation:0.071326
  |
Correlation:0.071326
  |
Cope: 02:REWARD BACK
|
GLM result group-wise
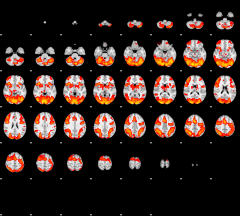 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:220; Jaccard:0.14013
 |
Component:220; Jaccard:0.16922
 |
Component:220; Jaccard:0.20041
 |
Component:220; Jaccard:0.2311
 |
Component:220; Jaccard:0.25509
 |
Component:220; Jaccard:0.27252
 |
Component:220; Jaccard:0.27922
 |
Correlation:0.12419
  |
Correlation:0.12419
  |
Correlation:0.12419
  |
Correlation:0.12419
  |
Correlation:0.12419
  |
Correlation:0.12419
  |
Correlation:0.12419
  |
Component:168; Jaccard:0.12586
 |
Component:168; Jaccard:0.15095
 |
Component:168; Jaccard:0.17813
 |
Component:168; Jaccard:0.20453
 |
Component:168; Jaccard:0.22471
 |
Component:168; Jaccard:0.23831
 |
Component:009; Jaccard:0.24603
 |
Correlation:0.19417
  |
Correlation:0.19417
  |
Correlation:0.19417
  |
Correlation:0.19417
  |
Correlation:0.19417
  |
Correlation:0.19417
  |
Correlation:0.32528
  |
Component:398; Jaccard:0.11647
 |
Component:009; Jaccard:0.13425
 |
Component:009; Jaccard:0.15949
 |
Component:009; Jaccard:0.18567
 |
Component:009; Jaccard:0.21093
 |
Component:009; Jaccard:0.23175
 |
Component:168; Jaccard:0.24306
 |
Correlation:0.030754
  |
Correlation:0.32528
  |
Correlation:0.32528
  |
Correlation:0.32528
  |
Correlation:0.32528
  |
Correlation:0.32528
  |
Correlation:0.19417
  |
Component:043; Jaccard:0.11349
 |
Component:027; Jaccard:0.13023
 |
Component:027; Jaccard:0.15387
 |
Component:027; Jaccard:0.17822
 |
Component:027; Jaccard:0.1996
 |
Component:027; Jaccard:0.21723
 |
Component:027; Jaccard:0.22819
 |
Correlation:-0.14782
  |
Correlation:0.1171
  |
Correlation:0.1171
  |
Correlation:0.1171
  |
Correlation:0.1171
  |
Correlation:0.1171
  |
Correlation:0.1171
  |
Component:009; Jaccard:0.11148
 |
Component:398; Jaccard:0.12968
 |
Component:398; Jaccard:0.14059
 |
Component:096; Jaccard:0.15124
 |
Component:096; Jaccard:0.16581
 |
Component:159; Jaccard:0.18554
 |
Component:159; Jaccard:0.20494
 |
Correlation:0.32528
  |
Correlation:0.030754
  |
Correlation:0.030754
  |
Correlation:0.12135
  |
Correlation:0.12135
  |
Correlation:0.18054
  |
Correlation:0.18054
  |
Component:027; Jaccard:0.10796
 |
Component:043; Jaccard:0.12631
 |
Component:043; Jaccard:0.13744
 |
Component:221; Jaccard:0.14966
 |
Component:159; Jaccard:0.16505
 |
Component:038; Jaccard:0.18002
 |
Component:038; Jaccard:0.19802
 |
Correlation:0.1171
  |
Correlation:-0.14782
  |
Correlation:-0.14782
  |
Correlation:0.36051
  |
Correlation:0.18054
  |
Correlation:0.50041
 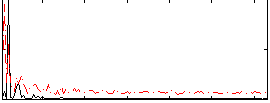 |
Correlation:0.50041
  |
Component:304; Jaccard:0.10772
 |
Component:221; Jaccard:0.11966
 |
Component:221; Jaccard:0.13553
 |
Component:398; Jaccard:0.14891
 |
Component:038; Jaccard:0.15968
 |
Component:096; Jaccard:0.17413
 |
Component:096; Jaccard:0.17426
 |
Correlation:0.016664
 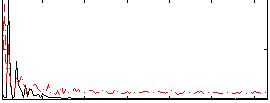 |
Correlation:0.36051
  |
Correlation:0.36051
  |
Correlation:0.030754
  |
Correlation:0.50041
  |
Correlation:0.12135
  |
Correlation:0.12135
  |
Component:021; Jaccard:0.10422
 |
Component:304; Jaccard:0.11896
 |
Component:096; Jaccard:0.13323
 |
Component:021; Jaccard:0.14664
 |
Component:221; Jaccard:0.15953
 |
Component:221; Jaccard:0.16758
 |
Component:221; Jaccard:0.17014
 |
Correlation:0.16101
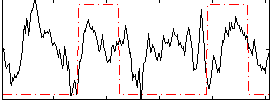  |
Correlation:0.016664
  |
Correlation:0.12135
  |
Correlation:0.16101
  |
Correlation:0.36051
  |
Correlation:0.36051
  |
Correlation:0.36051
  |
Component:221; Jaccard:0.10294
 |
Component:021; Jaccard:0.11868
 |
Component:021; Jaccard:0.13301
 |
Component:043; Jaccard:0.14655
 |
Component:021; Jaccard:0.15767
 |
Component:021; Jaccard:0.16253
 |
Component:021; Jaccard:0.16319
 |
Correlation:0.36051
  |
Correlation:0.16101
  |
Correlation:0.16101
  |
Correlation:-0.14782
  |
Correlation:0.16101
  |
Correlation:0.16101
  |
Correlation:0.16101
  |
Component:089; Jaccard:0.099136
 |
Component:096; Jaccard:0.11468
 |
Component:304; Jaccard:0.12908
 |
Component:159; Jaccard:0.14341
 |
Component:043; Jaccard:0.15322
 |
Component:043; Jaccard:0.1528
 |
Component:043; Jaccard:0.14748
 |
Correlation:0.15211
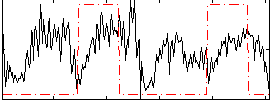  |
Correlation:0.12135
  |
Correlation:0.016664
  |
Correlation:0.18054
  |
Correlation:-0.14782
  |
Correlation:-0.14782
  |
Correlation:-0.14782
  |
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
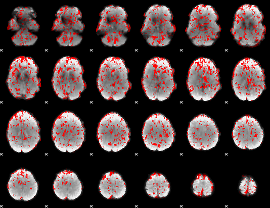
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
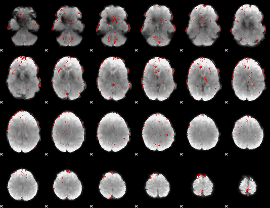
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
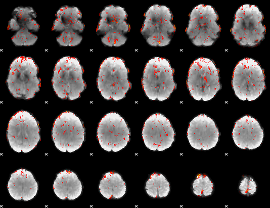
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
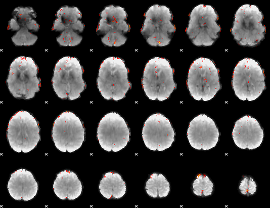
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
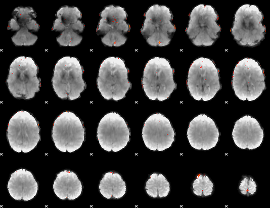 |
Component:147; Jaccard:0.10746
 |
Component:157; Jaccard:0.12523
 |
Component:157; Jaccard:0.15001
 |
Component:157; Jaccard:0.15711
 |
Component:157; Jaccard:0.14014
 |
Component:157; Jaccard:0.10943
 |
Component:157; Jaccard:0.076987
 |
Correlation:0.20063
 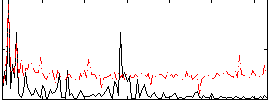 |
Correlation:0.43673
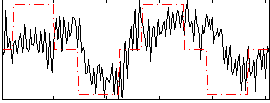  |
Correlation:0.43673
  |
Correlation:0.43673
  |
Correlation:0.43673
  |
Correlation:0.43673
  |
Correlation:0.43673
  |
Component:157; Jaccard:0.098476
 |
Component:342; Jaccard:0.11656
 |
Component:281; Jaccard:0.12613
 |
Component:281; Jaccard:0.12548
 |
Component:281; Jaccard:0.11229
 |
Component:281; Jaccard:0.082696
 |
Component:281; Jaccard:0.057544
 |
Correlation:0.43673
  |
Correlation:0.15908
  |
Correlation:0.35121
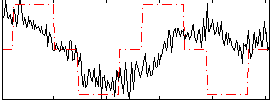 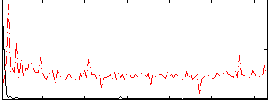 |
Correlation:0.35121
  |
Correlation:0.35121
  |
Correlation:0.35121
  |
Correlation:0.35121
  |
Component:342; Jaccard:0.098138
 |
Component:281; Jaccard:0.10954
 |
Component:342; Jaccard:0.12563
 |
Component:342; Jaccard:0.11585
 |
Component:342; Jaccard:0.089252
 |
Component:342; Jaccard:0.063886
 |
Component:342; Jaccard:0.044396
 |
Correlation:0.15908
  |
Correlation:0.35121
  |
Correlation:0.15908
  |
Correlation:0.15908
  |
Correlation:0.15908
  |
Correlation:0.15908
  |
Correlation:0.15908
  |
Component:314; Jaccard:0.097837
 |
Component:147; Jaccard:0.10674
 |
Component:237; Jaccard:0.1006
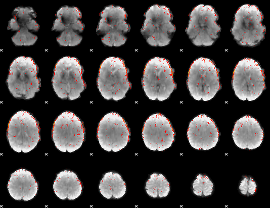 |
Component:237; Jaccard:0.093097
 |
Component:237; Jaccard:0.073917
 |
Component:237; Jaccard:0.052401
 |
Component:237; Jaccard:0.035451
 |
Correlation:0.28435
  |
Correlation:0.20063
  |
Correlation:0.42337
  |
Correlation:0.42337
  |
Correlation:0.42337
  |
Correlation:0.42337
  |
Correlation:0.42337
  |
Component:281; Jaccard:0.091164
 |
Component:314; Jaccard:0.096367
 |
Component:147; Jaccard:0.090649
 |
Component:374; Jaccard:0.074447
 |
Component:374; Jaccard:0.053049
 |
Component:374; Jaccard:0.034594
 |
Component:374; Jaccard:0.01982
 |
Correlation:0.35121
  |
Correlation:0.28435
  |
Correlation:0.20063
  |
Correlation:0.20215
  |
Correlation:0.20215
  |
Correlation:0.20215
  |
Correlation:0.20215
  |
Component:053; Jaccard:0.086348
 |
Component:237; Jaccard:0.095782
 |
Component:374; Jaccard:0.08727
 |
Component:082; Jaccard:0.066303
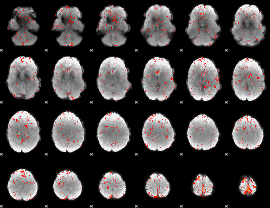 |
Component:082; Jaccard:0.046405
 |
Component:082; Jaccard:0.029093
 |
Component:240; Jaccard:0.018176
 |
Correlation:0.28702
  |
Correlation:0.42337
  |
Correlation:0.20215
  |
Correlation:0.10491
 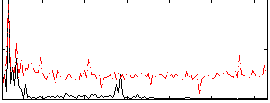 |
Correlation:0.10491
  |
Correlation:0.10491
  |
Correlation:0.046382
 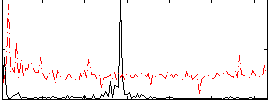 |
Component:237; Jaccard:0.085754
 |
Component:053; Jaccard:0.092844
 |
Component:053; Jaccard:0.0865
 |
Component:053; Jaccard:0.065478
 |
Component:053; Jaccard:0.043987
 |
Component:053; Jaccard:0.027784
 |
Component:082; Jaccard:0.017109
 |
Correlation:0.42337
  |
Correlation:0.28702
  |
Correlation:0.28702
  |
Correlation:0.28702
  |
Correlation:0.28702
  |
Correlation:0.28702
  |
Correlation:0.10491
  |
Component:082; Jaccard:0.083265
 |
Component:374; Jaccard:0.0891
 |
Component:314; Jaccard:0.082736
 |
Component:147; Jaccard:0.065067
 |
Component:147; Jaccard:0.040009
 |
Component:240; Jaccard:0.026179
 |
Component:217; Jaccard:0.016659
 |
Correlation:0.10491
  |
Correlation:0.20215
  |
Correlation:0.28435
  |
Correlation:0.20063
  |
Correlation:0.20063
  |
Correlation:0.046382
  |
Correlation:0.40381
  |
Component:139; Jaccard:0.08259
 |
Component:082; Jaccard:0.086018
 |
Component:082; Jaccard:0.079262
 |
Component:314; Jaccard:0.05779
 |
Component:240; Jaccard:0.038392
 |
Component:217; Jaccard:0.026176
 |
Component:053; Jaccard:0.016241
 |
Correlation:0.26571
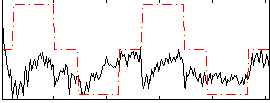  |
Correlation:0.10491
  |
Correlation:0.10491
  |
Correlation:0.28435
  |
Correlation:0.046382
  |
Correlation:0.40381
  |
Correlation:0.28702
  |
Component:374; Jaccard:0.08172
 |
Component:139; Jaccard:0.080834
 |
Component:139; Jaccard:0.070015
 |
Component:139; Jaccard:0.054219
 |
Component:314; Jaccard:0.035828
 |
Component:370; Jaccard:0.024059
 |
Component:370; Jaccard:0.016173
 |
Correlation:0.20215
  |
Correlation:0.26571
  |
Correlation:0.26571
  |
Correlation:0.26571
  |
Correlation:0.28435
  |
Correlation:0.3452
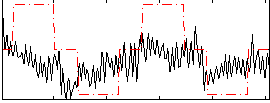  |
Correlation:0.3452
  |
Cope: 04:neg_PUNISH BACK
|
GLM result group-wise
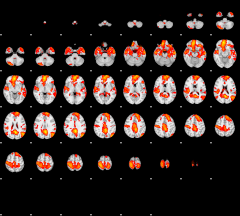 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
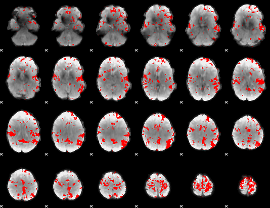 |
GLM binary result thresholded by 3.5
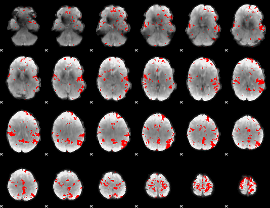 |
GLM binary result thresholded by 4
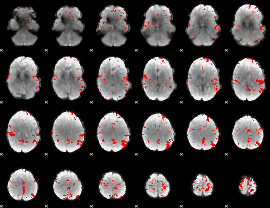 |
GLM Z-score result thresholded by 1
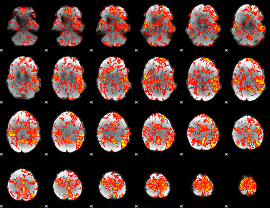 |
GLM Z-score result thresholded by 1.5
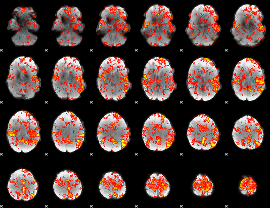 |
GLM Z-score result thresholded by 2
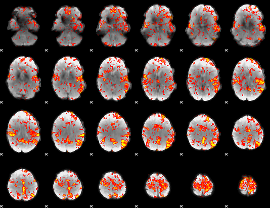 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
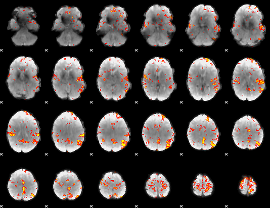 |
GLM Z-score result thresholded by 4
 |
Component:254; Jaccard:0.11069
 |
Component:232; Jaccard:0.1312
 |
Component:232; Jaccard:0.16215
 |
Component:232; Jaccard:0.19535
 |
Component:232; Jaccard:0.22582
 |
Component:232; Jaccard:0.24463
 |
Component:232; Jaccard:0.24997
 |
Correlation:0.46671
  |
Correlation:0.34371
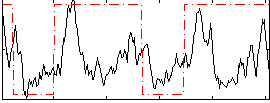  |
Correlation:0.34371
  |
Correlation:0.34371
  |
Correlation:0.34371
  |
Correlation:0.34371
  |
Correlation:0.34371
  |
Component:257; Jaccard:0.10845
 |
Component:253; Jaccard:0.12738
 |
Component:253; Jaccard:0.15131
 |
Component:253; Jaccard:0.17019
 |
Component:253; Jaccard:0.18328
 |
Component:253; Jaccard:0.18446
 |
Component:253; Jaccard:0.17377
 |
Correlation:0.24484
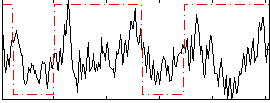  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Component:111; Jaccard:0.10815
 |
Component:257; Jaccard:0.12581
 |
Component:257; Jaccard:0.14511
 |
Component:257; Jaccard:0.1607
 |
Component:257; Jaccard:0.16513
 |
Component:257; Jaccard:0.16113
 |
Component:257; Jaccard:0.14746
 |
Correlation:0.3317
  |
Correlation:0.24484
  |
Correlation:0.24484
  |
Correlation:0.24484
  |
Correlation:0.24484
  |
Correlation:0.24484
  |
Correlation:0.24484
  |
Component:012; Jaccard:0.10795
 |
Component:254; Jaccard:0.12316
 |
Component:111; Jaccard:0.13398
 |
Component:111; Jaccard:0.14104
 |
Component:111; Jaccard:0.1413
 |
Component:111; Jaccard:0.13841
 |
Component:116; Jaccard:0.13444
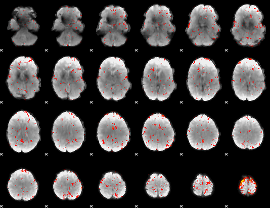 |
Correlation:0.2686
  |
Correlation:0.46671
  |
Correlation:0.3317
  |
Correlation:0.3317
  |
Correlation:0.3317
  |
Correlation:0.3317
  |
Correlation:0.36197
  |
Component:253; Jaccard:0.1062
 |
Component:111; Jaccard:0.12239
 |
Component:012; Jaccard:0.13382
 |
Component:254; Jaccard:0.13906
 |
Component:012; Jaccard:0.13771
 |
Component:116; Jaccard:0.13816
 |
Component:111; Jaccard:0.12641
 |
Correlation:0.28494
  |
Correlation:0.3317
  |
Correlation:0.2686
  |
Correlation:0.46671
  |
Correlation:0.2686
  |
Correlation:0.36197
  |
Correlation:0.3317
  |
Component:232; Jaccard:0.10467
 |
Component:012; Jaccard:0.12181
 |
Component:254; Jaccard:0.13359
 |
Component:012; Jaccard:0.1389
 |
Component:254; Jaccard:0.13629
 |
Component:012; Jaccard:0.12687
 |
Component:026; Jaccard:0.11291
 |
Correlation:0.34371
  |
Correlation:0.2686
  |
Correlation:0.46671
  |
Correlation:0.2686
  |
Correlation:0.46671
  |
Correlation:0.2686
  |
Correlation:0.25162
  |
Component:392; Jaccard:0.097671
 |
Component:392; Jaccard:0.1034
 |
Component:116; Jaccard:0.11349
 |
Component:116; Jaccard:0.12629
 |
Component:116; Jaccard:0.13366
 |
Component:254; Jaccard:0.12636
 |
Component:012; Jaccard:0.10988
 |
Correlation:0.26095
  |
Correlation:0.26095
  |
Correlation:0.36197
  |
Correlation:0.36197
  |
Correlation:0.36197
  |
Correlation:0.46671
  |
Correlation:0.2686
  |
Component:301; Jaccard:0.094849
 |
Component:301; Jaccard:0.10251
 |
Component:371; Jaccard:0.1096
 |
Component:371; Jaccard:0.11338
 |
Component:026; Jaccard:0.11211
 |
Component:026; Jaccard:0.11662
 |
Component:254; Jaccard:0.10934
 |
Correlation:0.25231
  |
Correlation:0.25231
  |
Correlation:0.30056
  |
Correlation:0.30056
  |
Correlation:0.25162
  |
Correlation:0.25162
  |
Correlation:0.46671
  |
Component:083; Jaccard:0.092575
 |
Component:371; Jaccard:0.10105
 |
Component:301; Jaccard:0.109
 |
Component:132; Jaccard:0.11141
 |
Component:132; Jaccard:0.11024
 |
Component:344; Jaccard:0.10843
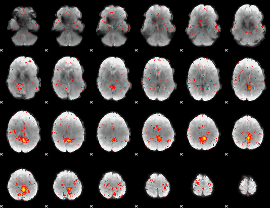 |
Component:344; Jaccard:0.10489
 |
Correlation:0.33976
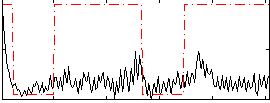  |
Correlation:0.30056
  |
Correlation:0.25231
  |
Correlation:0.24576
  |
Correlation:0.24576
  |
Correlation:0.085066
  |
Correlation:0.085066
  |
Component:371; Jaccard:0.091959
 |
Component:132; Jaccard:0.10069
 |
Component:132; Jaccard:0.10854
 |
Component:301; Jaccard:0.10978
 |
Component:371; Jaccard:0.10955
 |
Component:132; Jaccard:0.10316
 |
Component:132; Jaccard:0.091673
 |
Correlation:0.30056
  |
Correlation:0.24576
  |
Correlation:0.24576
  |
Correlation:0.25231
  |
Correlation:0.30056
  |
Correlation:0.24576
  |
Correlation:0.24576
  |
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
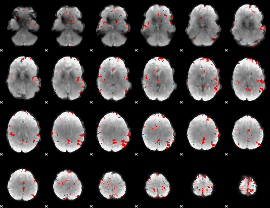 |
GLM Z-score result thresholded by 1
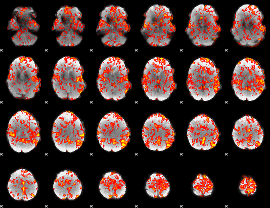 |
GLM Z-score result thresholded by 1.5
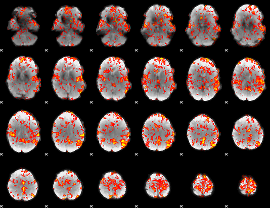 |
GLM Z-score result thresholded by 2
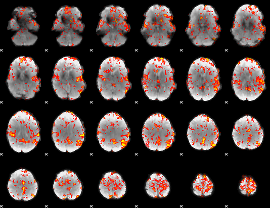 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
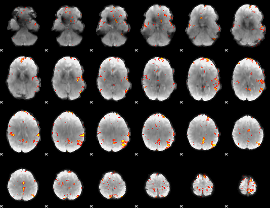 |
Component:253; Jaccard:0.12381
 |
Component:253; Jaccard:0.14766
 |
Component:253; Jaccard:0.17312
 |
Component:253; Jaccard:0.19447
 |
Component:232; Jaccard:0.2145
 |
Component:232; Jaccard:0.22124
 |
Component:232; Jaccard:0.20745
 |
Correlation:0.16865
  |
Correlation:0.16865
  |
Correlation:0.16865
  |
Correlation:0.16865
  |
Correlation:0.30003
  |
Correlation:0.30003
  |
Correlation:0.30003
  |
Component:301; Jaccard:0.11911
 |
Component:232; Jaccard:0.13721
 |
Component:232; Jaccard:0.1676
 |
Component:232; Jaccard:0.19398
 |
Component:253; Jaccard:0.19985
 |
Component:253; Jaccard:0.18882
 |
Component:253; Jaccard:0.1646
 |
Correlation:0.28382
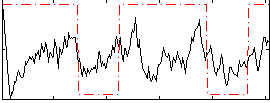  |
Correlation:0.30003
  |
Correlation:0.30003
  |
Correlation:0.30003
  |
Correlation:0.16865
  |
Correlation:0.16865
  |
Correlation:0.16865
  |
Component:257; Jaccard:0.11343
 |
Component:301; Jaccard:0.13311
 |
Component:301; Jaccard:0.14309
 |
Component:257; Jaccard:0.14902
 |
Component:257; Jaccard:0.14758
 |
Component:257; Jaccard:0.13817
 |
Component:257; Jaccard:0.11789
 |
Correlation:0.30535
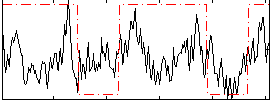  |
Correlation:0.28382
  |
Correlation:0.28382
  |
Correlation:0.30535
  |
Correlation:0.30535
  |
Correlation:0.30535
  |
Correlation:0.30535
  |
Component:232; Jaccard:0.11258
 |
Component:257; Jaccard:0.1281
 |
Component:257; Jaccard:0.14101
 |
Component:301; Jaccard:0.14603
 |
Component:132; Jaccard:0.14266
 |
Component:132; Jaccard:0.13369
 |
Component:132; Jaccard:0.11448
 |
Correlation:0.30003
  |
Correlation:0.30535
  |
Correlation:0.30535
  |
Correlation:0.28382
  |
Correlation:0.34394
  |
Correlation:0.34394
  |
Correlation:0.34394
  |
Component:132; Jaccard:0.10921
 |
Component:132; Jaccard:0.12301
 |
Component:132; Jaccard:0.13426
 |
Component:132; Jaccard:0.14241
 |
Component:301; Jaccard:0.13958
 |
Component:301; Jaccard:0.12645
 |
Component:301; Jaccard:0.10869
 |
Correlation:0.34394
  |
Correlation:0.34394
  |
Correlation:0.34394
  |
Correlation:0.34394
  |
Correlation:0.28382
  |
Correlation:0.28382
  |
Correlation:0.28382
  |
Component:012; Jaccard:0.10096
 |
Component:012; Jaccard:0.1071
 |
Component:218; Jaccard:0.11382
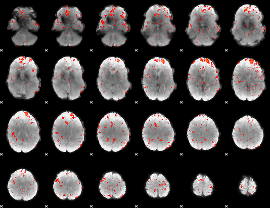 |
Component:218; Jaccard:0.12134
 |
Component:218; Jaccard:0.1209
 |
Component:342; Jaccard:0.11775
 |
Component:342; Jaccard:0.10796
 |
Correlation:0.19209
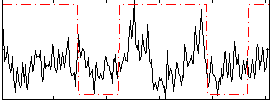  |
Correlation:0.19209
  |
Correlation:0.42473
  |
Correlation:0.42473
  |
Correlation:0.42473
  |
Correlation:0.31701
  |
Correlation:0.31701
  |
Component:111; Jaccard:0.098765
 |
Component:111; Jaccard:0.10638
 |
Component:111; Jaccard:0.11185
 |
Component:111; Jaccard:0.11379
 |
Component:342; Jaccard:0.11872
 |
Component:218; Jaccard:0.11394
 |
Component:218; Jaccard:0.098225
 |
Correlation:0.16198
  |
Correlation:0.16198
  |
Correlation:0.16198
  |
Correlation:0.16198
  |
Correlation:0.31701
  |
Correlation:0.42473
  |
Correlation:0.42473
  |
Component:371; Jaccard:0.096343
 |
Component:371; Jaccard:0.10412
 |
Component:012; Jaccard:0.10802
 |
Component:342; Jaccard:0.11179
 |
Component:111; Jaccard:0.10959
 |
Component:111; Jaccard:0.101
 |
Component:111; Jaccard:0.088228
 |
Correlation:0.15829
  |
Correlation:0.15829
  |
Correlation:0.19209
  |
Correlation:0.31701
  |
Correlation:0.16198
  |
Correlation:0.16198
  |
Correlation:0.16198
  |
Component:149; Jaccard:0.092192
 |
Component:218; Jaccard:0.10319
 |
Component:371; Jaccard:0.10795
 |
Component:371; Jaccard:0.10592
 |
Component:371; Jaccard:0.10158
 |
Component:116; Jaccard:0.093196
 |
Component:116; Jaccard:0.082414
 |
Correlation:0.28757
  |
Correlation:0.42473
  |
Correlation:0.15829
  |
Correlation:0.15829
  |
Correlation:0.15829
  |
Correlation:0.045206
  |
Correlation:0.045206
  |
Component:218; Jaccard:0.091888
 |
Component:149; Jaccard:0.0946
 |
Component:342; Jaccard:0.10095
 |
Component:012; Jaccard:0.1021
 |
Component:116; Jaccard:0.095921
 |
Component:371; Jaccard:0.090534
 |
Component:026; Jaccard:0.080855
 |
Correlation:0.42473
  |
Correlation:0.28757
  |
Correlation:0.31701
  |
Correlation:0.19209
  |
Correlation:0.045206
  |
Correlation:0.15829
  |
Correlation:0.062465
  |
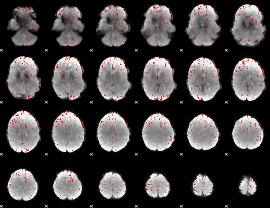
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
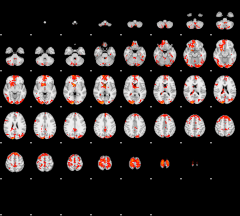 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
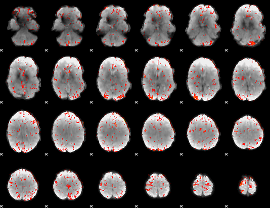 |
GLM Z-score result thresholded by 3
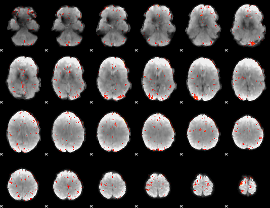 |
GLM Z-score result thresholded by 3.5
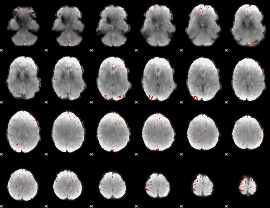 |
GLM Z-score result thresholded by 4
 |
Component:089; Jaccard:0.097281
 |
Component:089; Jaccard:0.10865
 |
Component:219; Jaccard:0.11207
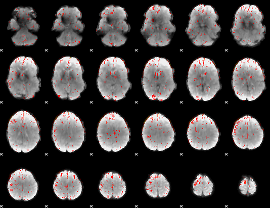 |
Component:219; Jaccard:0.11486
 |
Component:057; Jaccard:0.10302
 |
Component:057; Jaccard:0.084932
 |
Component:057; Jaccard:0.061861
 |
Correlation:0.18657
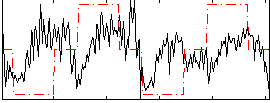  |
Correlation:0.18657
  |
Correlation:0.24107
 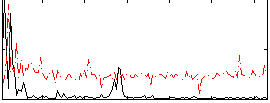 |
Correlation:0.24107
  |
Correlation:0.27662
  |
Correlation:0.27662
  |
Correlation:0.27662
  |
Component:103; Jaccard:0.089414
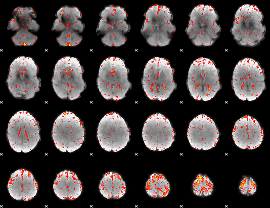 |
Component:219; Jaccard:0.10037
 |
Component:089; Jaccard:0.11048
 |
Component:283; Jaccard:0.11173
 |
Component:283; Jaccard:0.10213
 |
Component:034; Jaccard:0.07817
 |
Component:144; Jaccard:0.055351
 |
Correlation:0.10528
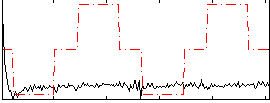 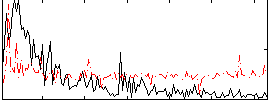 |
Correlation:0.24107
  |
Correlation:0.18657
  |
Correlation:0.17745
  |
Correlation:0.17745
  |
Correlation:0.20563
  |
Correlation:0.48485
  |
Component:219; Jaccard:0.085501
 |
Component:283; Jaccard:0.092416
 |
Component:283; Jaccard:0.10464
 |
Component:034; Jaccard:0.10718
 |
Component:034; Jaccard:0.10111
 |
Component:283; Jaccard:0.077976
 |
Component:283; Jaccard:0.052436
 |
Correlation:0.24107
  |
Correlation:0.17745
  |
Correlation:0.17745
  |
Correlation:0.20563
  |
Correlation:0.20563
  |
Correlation:0.17745
  |
Correlation:0.17745
  |
Component:392; Jaccard:0.083099
 |
Component:144; Jaccard:0.090499
 |
Component:144; Jaccard:0.10235
 |
Component:144; Jaccard:0.10555
 |
Component:219; Jaccard:0.10015
 |
Component:219; Jaccard:0.077493
 |
Component:034; Jaccard:0.051449
 |
Correlation:0.083301
 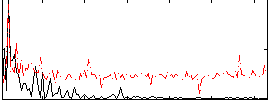 |
Correlation:0.48485
  |
Correlation:0.48485
  |
Correlation:0.48485
  |
Correlation:0.24107
  |
Correlation:0.24107
  |
Correlation:0.20563
  |
Component:052; Jaccard:0.081803
 |
Component:025; Jaccard:0.088817
 |
Component:034; Jaccard:0.098177
 |
Component:057; Jaccard:0.1009
 |
Component:144; Jaccard:0.095808
 |
Component:144; Jaccard:0.076964
 |
Component:219; Jaccard:0.050112
 |
Correlation:-0.0015529
 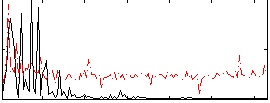 |
Correlation:0.1637
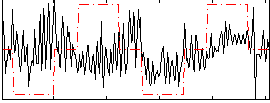 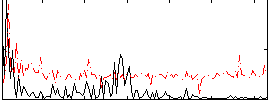 |
Correlation:0.20563
  |
Correlation:0.27662
  |
Correlation:0.48485
  |
Correlation:0.48485
  |
Correlation:0.24107
  |
Component:025; Jaccard:0.079331
 |
Component:103; Jaccard:0.086112
 |
Component:025; Jaccard:0.094397
 |
Component:089; Jaccard:0.095585
 |
Component:025; Jaccard:0.073289
 |
Component:025; Jaccard:0.053067
 |
Component:364; Jaccard:0.035792
 |
Correlation:0.1637
  |
Correlation:0.10528
  |
Correlation:0.1637
  |
Correlation:0.18657
  |
Correlation:0.1637
  |
Correlation:0.1637
  |
Correlation:0.11453
  |
Component:254; Jaccard:0.078789
 |
Component:034; Jaccard:0.083542
 |
Component:057; Jaccard:0.087718
 |
Component:025; Jaccard:0.087699
 |
Component:089; Jaccard:0.07084
 |
Component:364; Jaccard:0.051797
 |
Component:282; Jaccard:0.03259
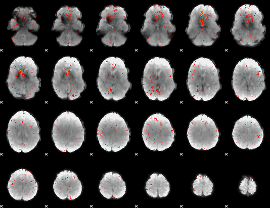 |
Correlation:0.28866
  |
Correlation:0.20563
  |
Correlation:0.27662
  |
Correlation:0.1637
  |
Correlation:0.18657
  |
Correlation:0.11453
  |
Correlation:0.31454
  |
Component:283; Jaccard:0.078223
 |
Component:052; Jaccard:0.082944
 |
Component:060; Jaccard:0.085987
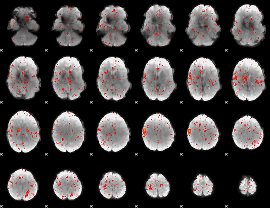 |
Component:060; Jaccard:0.079799
 |
Component:364; Jaccard:0.063928
 |
Component:282; Jaccard:0.046077
 |
Component:025; Jaccard:0.032059
 |
Correlation:0.17745
  |
Correlation:-0.0015529
  |
Correlation:0.3909
  |
Correlation:0.3909
  |
Correlation:0.11453
  |
Correlation:0.31454
  |
Correlation:0.1637
  |
Component:134; Jaccard:0.076549
 |
Component:060; Jaccard:0.0829
 |
Component:269; Jaccard:0.085887
 |
Component:269; Jaccard:0.079788
 |
Component:269; Jaccard:0.062778
 |
Component:089; Jaccard:0.046065
 |
Component:376; Jaccard:0.026942
 |
Correlation:0.17568
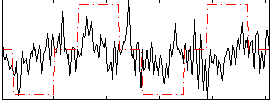  |
Correlation:0.3909
  |
Correlation:0.16077
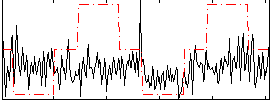 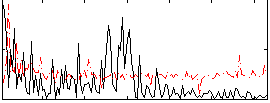 |
Correlation:0.16077
  |
Correlation:0.16077
  |
Correlation:0.18657
  |
Correlation:0.22848
  |
Component:144; Jaccard:0.076053
 |
Component:392; Jaccard:0.081267
 |
Component:376; Jaccard:0.082804
 |
Component:120; Jaccard:0.076923
 |
Component:376; Jaccard:0.062126
 |
Component:269; Jaccard:0.044807
 |
Component:269; Jaccard:0.025562
 |
Correlation:0.48485
  |
Correlation:0.083301
  |
Correlation:0.22848
  |
Correlation:0.38804
  |
Correlation:0.22848
  |
Correlation:0.16077
  |
Correlation:0.16077
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































