Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
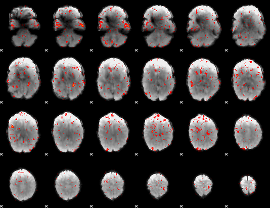
GLM result group-wise
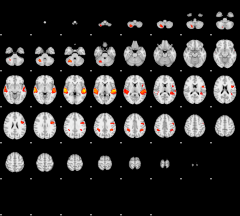 |
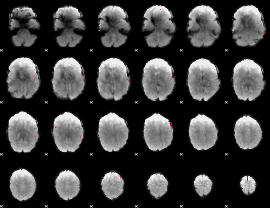
GLM binary result thresholded by 1
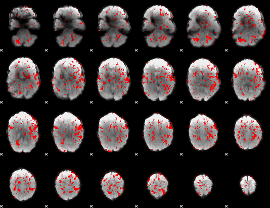 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
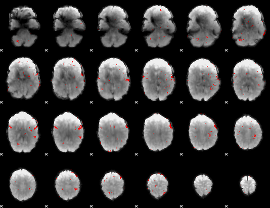 |
GLM binary result thresholded by 3
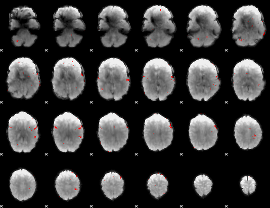 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
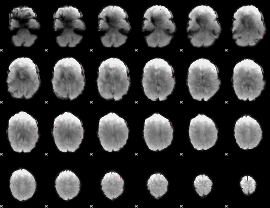 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
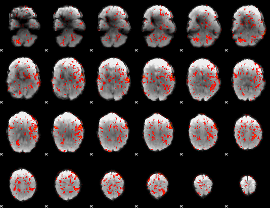
GLM Z-score result thresholded by 4
 |
Component:324; Jaccard:0.12218
 |
Component:324; Jaccard:0.12303
 |
Component:324; Jaccard:0.10297
 |
Component:324; Jaccard:0.070202
 |
Component:324; Jaccard:0.044535
 |
Component:324; Jaccard:0.024499
 |
Component:324; Jaccard:0.010232
 |
Correlation:0.57511
  |
Correlation:0.57511
  |
Correlation:0.57511
  |
Correlation:0.57511
  |
Correlation:0.57511
  |
Correlation:0.57511
  |
Correlation:0.57511
  |
Component:064; Jaccard:0.09707
 |
Component:064; Jaccard:0.091115
 |
Component:064; Jaccard:0.074002
 |
Component:064; Jaccard:0.050472
 |
Component:064; Jaccard:0.030701
 |
Component:064; Jaccard:0.017419
 |
Component:064; Jaccard:0.008337
 |
Correlation:-0.23057
  |
Correlation:-0.23057
  |
Correlation:-0.23057
  |
Correlation:-0.23057
  |
Correlation:-0.23057
  |
Correlation:-0.23057
  |
Correlation:-0.23057
  |
Component:081; Jaccard:0.090044
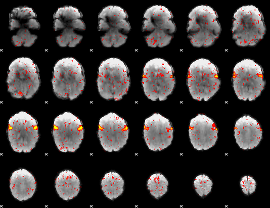 |
Component:081; Jaccard:0.082203
 |
Component:190; Jaccard:0.066058
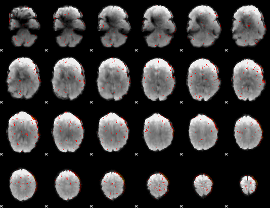 |
Component:190; Jaccard:0.045048
 |
Component:081; Jaccard:0.026097
 |
Component:081; Jaccard:0.014841
 |
Component:397; Jaccard:0.007588
 |
Correlation:0.35684
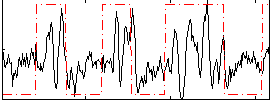 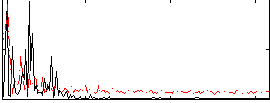 |
Correlation:0.35684
  |
Correlation:0.11376
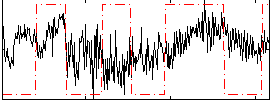  |
Correlation:0.11376
  |
Correlation:0.35684
  |
Correlation:0.35684
  |
Correlation:-0.25107
  |
Component:041; Jaccard:0.085378
 |
Component:190; Jaccard:0.077512
 |
Component:081; Jaccard:0.064238
 |
Component:081; Jaccard:0.04365
 |
Component:190; Jaccard:0.02592
 |
Component:060; Jaccard:0.014118
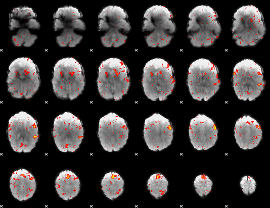 |
Component:081; Jaccard:0.006346
 |
Correlation:0.71199
  |
Correlation:0.11376
  |
Correlation:0.35684
  |
Correlation:0.35684
  |
Correlation:0.11376
  |
Correlation:0.521
  |
Correlation:0.35684
  |
Component:190; Jaccard:0.07773
 |
Component:041; Jaccard:0.073196
 |
Component:060; Jaccard:0.057182
 |
Component:060; Jaccard:0.038648
 |
Component:060; Jaccard:0.024234
 |
Component:397; Jaccard:0.013788
 |
Component:190; Jaccard:0.005843
 |
Correlation:0.11376
  |
Correlation:0.71199
  |
Correlation:0.521
  |
Correlation:0.521
  |
Correlation:0.521
  |
Correlation:-0.25107
  |
Correlation:0.11376
  |
Component:281; Jaccard:0.076622
 |
Component:060; Jaccard:0.069902
 |
Component:041; Jaccard:0.051058
 |
Component:397; Jaccard:0.03192
 |
Component:397; Jaccard:0.021099
 |
Component:190; Jaccard:0.013695
 |
Component:060; Jaccard:0.005782
 |
Correlation:0.32981
  |
Correlation:0.521
  |
Correlation:0.71199
  |
Correlation:-0.25107
  |
Correlation:-0.25107
  |
Correlation:0.11376
  |
Correlation:0.521
  |
Component:060; Jaccard:0.072788
 |
Component:281; Jaccard:0.064991
 |
Component:281; Jaccard:0.044648
 |
Component:041; Jaccard:0.030862
 |
Component:313; Jaccard:0.018396
 |
Component:045; Jaccard:0.010721
 |
Component:045; Jaccard:0.005082
 |
Correlation:0.521
  |
Correlation:0.32981
  |
Correlation:0.32981
  |
Correlation:0.71199
  |
Correlation:-0.25475
  |
Correlation:0.18514
  |
Correlation:0.18514
  |
Component:400; Jaccard:0.070153
 |
Component:400; Jaccard:0.057465
 |
Component:397; Jaccard:0.042713
 |
Component:313; Jaccard:0.028725
 |
Component:045; Jaccard:0.017463
 |
Component:313; Jaccard:0.010168
 |
Component:132; Jaccard:0.004705
 |
Correlation:0.46738
  |
Correlation:0.46738
  |
Correlation:-0.25107
  |
Correlation:-0.25475
  |
Correlation:0.18514
  |
Correlation:-0.25475
  |
Correlation:-0.28756
  |
Component:354; Jaccard:0.062117
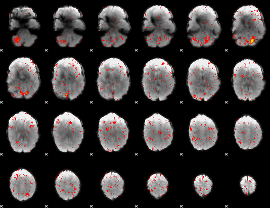 |
Component:189; Jaccard:0.055097
 |
Component:313; Jaccard:0.042161
 |
Component:045; Jaccard:0.028344
 |
Component:041; Jaccard:0.017075
 |
Component:041; Jaccard:0.008917
 |
Component:313; Jaccard:0.004254
 |
Correlation:0.41689
  |
Correlation:-0.0084224
  |
Correlation:-0.25475
  |
Correlation:0.18514
  |
Correlation:0.71199
  |
Correlation:0.71199
  |
Correlation:-0.25475
  |
Component:157; Jaccard:0.060952
 |
Component:313; Jaccard:0.053966
 |
Component:107; Jaccard:0.040259
 |
Component:107; Jaccard:0.026275
 |
Component:321; Jaccard:0.014404
 |
Component:321; Jaccard:0.008787
 |
Component:321; Jaccard:0.003875
 |
Correlation:0.24256
  |
Correlation:-0.25475
  |
Correlation:-0.017082
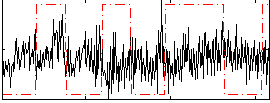 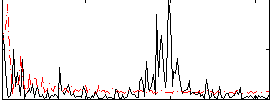 |
Correlation:-0.017082
  |
Correlation:-0.1342
  |
Correlation:-0.1342
  |
Correlation:-0.1342
  |
Cope: 02:STORY BACK
|
GLM result group-wise
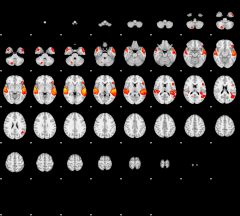 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
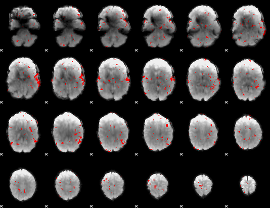 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
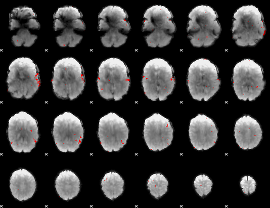 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
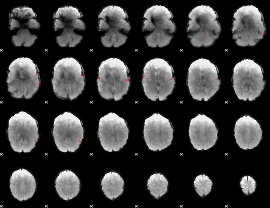 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
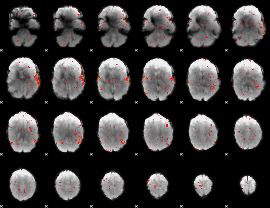 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:064; Jaccard:0.13752
 |
Component:064; Jaccard:0.14133
 |
Component:064; Jaccard:0.12399
 |
Component:064; Jaccard:0.090276
 |
Component:064; Jaccard:0.061111
 |
Component:064; Jaccard:0.040445
 |
Component:064; Jaccard:0.025325
 |
Correlation:0.16928
  |
Correlation:0.16928
  |
Correlation:0.16928
  |
Correlation:0.16928
  |
Correlation:0.16928
  |
Correlation:0.16928
  |
Correlation:0.16928
  |
Component:146; Jaccard:0.11079
 |
Component:146; Jaccard:0.11931
 |
Component:146; Jaccard:0.10188
 |
Component:146; Jaccard:0.076409
 |
Component:146; Jaccard:0.048294
 |
Component:397; Jaccard:0.031115
 |
Component:397; Jaccard:0.020624
 |
Correlation:0.48664
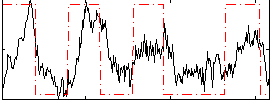  |
Correlation:0.48664
  |
Correlation:0.48664
  |
Correlation:0.48664
  |
Correlation:0.48664
  |
Correlation:0.25216
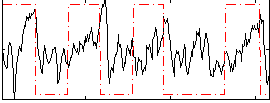
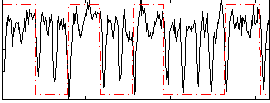  |
Correlation:0.25216
  |
Component:313; Jaccard:0.089877
 |
Component:313; Jaccard:0.094951
 |
Component:313; Jaccard:0.088581
 |
Component:313; Jaccard:0.071236
 |
Component:313; Jaccard:0.046399
 |
Component:313; Jaccard:0.029212
 |
Component:313; Jaccard:0.018238
 |
Correlation:0.22634
  |
Correlation:0.22634
  |
Correlation:0.22634
  |
Correlation:0.22634
  |
Correlation:0.22634
  |
Correlation:0.22634
  |
Correlation:0.22634
  |
Component:247; Jaccard:0.08874
 |
Component:247; Jaccard:0.086941
 |
Component:397; Jaccard:0.074351
 |
Component:397; Jaccard:0.061093
 |
Component:397; Jaccard:0.045222
 |
Component:146; Jaccard:0.026414
 |
Component:132; Jaccard:0.016774
 |
Correlation:0.38324
  |
Correlation:0.38324
  |
Correlation:0.25216
  |
Correlation:0.25216
  |
Correlation:0.25216
  |
Correlation:0.48664
  |
Correlation:0.3098
  |
Component:167; Jaccard:0.081794
 |
Component:167; Jaccard:0.08204
 |
Component:167; Jaccard:0.070932
 |
Component:167; Jaccard:0.052772
 |
Component:167; Jaccard:0.034193
 |
Component:132; Jaccard:0.02214
 |
Component:146; Jaccard:0.015737
 |
Correlation:0.50895
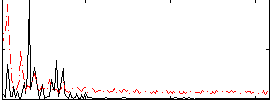
  |
Correlation:0.50895
  |
Correlation:0.50895
  |
Correlation:0.50895
  |
Correlation:0.50895
  |
Correlation:0.3098
  |
Correlation:0.48664
  |
Component:399; Jaccard:0.079446
 |
Component:397; Jaccard:0.077862
 |
Component:247; Jaccard:0.068511
 |
Component:247; Jaccard:0.04764
 |
Component:132; Jaccard:0.028054
 |
Component:167; Jaccard:0.017805
 |
Component:321; Jaccard:0.010417
 |
Correlation:0.15111
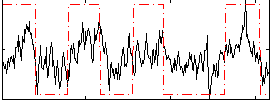
  |
Correlation:0.25216
  |
Correlation:0.38324
  |
Correlation:0.38324
  |
Correlation:0.3098
  |
Correlation:0.50895
  |
Correlation:0.10481
  |
Component:397; Jaccard:0.073125
 |
Component:399; Jaccard:0.07237
 |
Component:399; Jaccard:0.054667
 |
Component:399; Jaccard:0.036119
 |
Component:247; Jaccard:0.027063
 |
Component:321; Jaccard:0.016801
 |
Component:167; Jaccard:0.009584
 |
Correlation:0.25216
  |
Correlation:0.15111
  |
Correlation:0.15111
  |
Correlation:0.15111
  |
Correlation:0.38324
  |
Correlation:0.10481
  |
Correlation:0.50895
  |
Component:366; Jaccard:0.064102
 |
Component:366; Jaccard:0.061
 |
Component:185; Jaccard:0.049116
 |
Component:132; Jaccard:0.035264
 |
Component:399; Jaccard:0.02198
 |
Component:399; Jaccard:0.014105
 |
Component:399; Jaccard:0.009274
 |
Correlation:0.43855
  |
Correlation:0.43855
  |
Correlation:0.1841
  |
Correlation:0.3098
  |
Correlation:0.15111
  |
Correlation:0.15111
  |
Correlation:0.15111
  |
Component:144; Jaccard:0.061256
 |
Component:185; Jaccard:0.057679
 |
Component:366; Jaccard:0.048503
 |
Component:366; Jaccard:0.034783
 |
Component:321; Jaccard:0.021901
 |
Component:247; Jaccard:0.013215
 |
Component:247; Jaccard:0.007417
 |
Correlation:0.16294
  |
Correlation:0.1841
  |
Correlation:0.43855
  |
Correlation:0.43855
  |
Correlation:0.10481
  |
Correlation:0.38324
  |
Correlation:0.38324
  |
Component:123; Jaccard:0.06106
 |
Component:315; Jaccard:0.057186
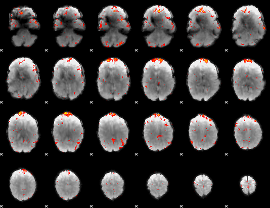 |
Component:315; Jaccard:0.045435
 |
Component:185; Jaccard:0.032021
 |
Component:366; Jaccard:0.019349
 |
Component:366; Jaccard:0.009923
 |
Component:366; Jaccard:0.006228
 |
Correlation:0.1094
  |
Correlation:0.67436
  |
Correlation:0.67436
  |
Correlation:0.1841
  |
Correlation:0.43855
  |
Correlation:0.43855
  |
Correlation:0.43855
  |
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
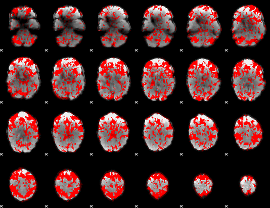 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
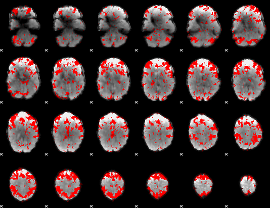 |
GLM binary result thresholded by 2.5
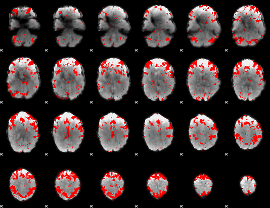 |
GLM binary result thresholded by 3
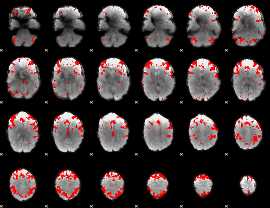 |
GLM binary result thresholded by 3.5
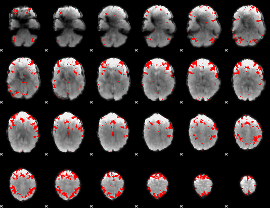 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
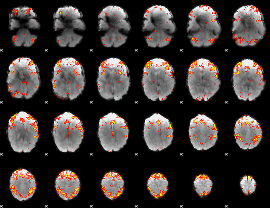 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
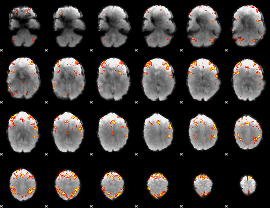 |
Component:383; Jaccard:0.13953
 |
Component:383; Jaccard:0.17146
 |
Component:383; Jaccard:0.20646
 |
Component:383; Jaccard:0.23778
 |
Component:383; Jaccard:0.26265
 |
Component:383; Jaccard:0.27227
 |
Component:041; Jaccard:0.27312
 |
Correlation:0.591
  |
Correlation:0.591
  |
Correlation:0.591
  |
Correlation:0.591
  |
Correlation:0.591
  |
Correlation:0.591
  |
Correlation:0.74109
  |
Component:020; Jaccard:0.13428
 |
Component:020; Jaccard:0.15437
 |
Component:041; Jaccard:0.17937
 |
Component:041; Jaccard:0.21438
 |
Component:041; Jaccard:0.24441
 |
Component:041; Jaccard:0.26626
 |
Component:383; Jaccard:0.26379
 |
Correlation:0.5466
  |
Correlation:0.5466
  |
Correlation:0.74109
  |
Correlation:0.74109
  |
Correlation:0.74109
  |
Correlation:0.74109
  |
Correlation:0.591
  |
Component:268; Jaccard:0.11943
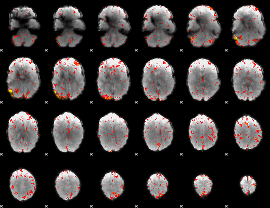 |
Component:041; Jaccard:0.14708
 |
Component:020; Jaccard:0.17169
 |
Component:324; Jaccard:0.18724
 |
Component:324; Jaccard:0.20484
 |
Component:324; Jaccard:0.21533
 |
Component:324; Jaccard:0.21695
 |
Correlation:0.44365
  |
Correlation:0.74109
  |
Correlation:0.5466
  |
Correlation:0.60318
  |
Correlation:0.60318
  |
Correlation:0.60318
  |
Correlation:0.60318
  |
Component:283; Jaccard:0.11894
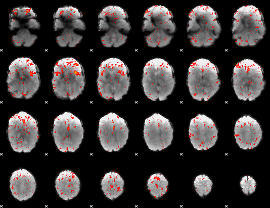 |
Component:324; Jaccard:0.14225
 |
Component:324; Jaccard:0.16612
 |
Component:020; Jaccard:0.18079
 |
Component:020; Jaccard:0.17977
 |
Component:020; Jaccard:0.17157
 |
Component:128; Jaccard:0.15799
 |
Correlation:0.54877
  |
Correlation:0.60318
  |
Correlation:0.60318
  |
Correlation:0.5466
  |
Correlation:0.5466
  |
Correlation:0.5466
  |
Correlation:0.37537
  |
Component:324; Jaccard:0.11882
 |
Component:283; Jaccard:0.1365
 |
Component:128; Jaccard:0.15083
 |
Component:128; Jaccard:0.16337
 |
Component:128; Jaccard:0.16931
 |
Component:128; Jaccard:0.16611
 |
Component:020; Jaccard:0.15644
 |
Correlation:0.60318
  |
Correlation:0.54877
  |
Correlation:0.37537
  |
Correlation:0.37537
  |
Correlation:0.37537
  |
Correlation:0.37537
  |
Correlation:0.5466
  |
Component:041; Jaccard:0.11841
 |
Component:128; Jaccard:0.13518
 |
Component:283; Jaccard:0.14898
 |
Component:278; Jaccard:0.15511
 |
Component:278; Jaccard:0.16089
 |
Component:278; Jaccard:0.15756
 |
Component:278; Jaccard:0.14975
 |
Correlation:0.74109
  |
Correlation:0.37537
  |
Correlation:0.54877
  |
Correlation:0.55679
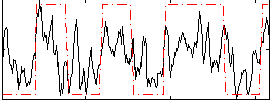  |
Correlation:0.55679
  |
Correlation:0.55679
  |
Correlation:0.55679
  |
Component:128; Jaccard:0.11692
 |
Component:268; Jaccard:0.13118
 |
Component:278; Jaccard:0.1448
 |
Component:283; Jaccard:0.15405
 |
Component:283; Jaccard:0.15114
 |
Component:282; Jaccard:0.14926
 |
Component:282; Jaccard:0.14558
 |
Correlation:0.37537
  |
Correlation:0.44365
  |
Correlation:0.55679
  |
Correlation:0.54877
  |
Correlation:0.54877
  |
Correlation:0.40418
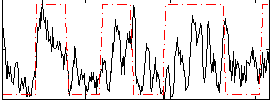  |
Correlation:0.40418
  |
Component:278; Jaccard:0.11642
 |
Component:278; Jaccard:0.13114
 |
Component:268; Jaccard:0.14056
 |
Component:268; Jaccard:0.14354
 |
Component:282; Jaccard:0.14977
 |
Component:400; Jaccard:0.14323
 |
Component:400; Jaccard:0.13628
 |
Correlation:0.55679
  |
Correlation:0.55679
  |
Correlation:0.44365
  |
Correlation:0.44365
  |
Correlation:0.40418
  |
Correlation:0.49268
 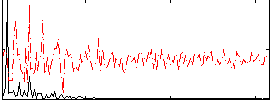 |
Correlation:0.49268
  |
Component:181; Jaccard:0.11259
 |
Component:181; Jaccard:0.12667
 |
Component:181; Jaccard:0.13805
 |
Component:181; Jaccard:0.14269
 |
Component:400; Jaccard:0.14555
 |
Component:283; Jaccard:0.14297
 |
Component:238; Jaccard:0.12898
 |
Correlation:0.44538
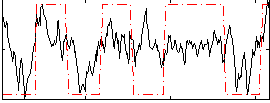  |
Correlation:0.44538
  |
Correlation:0.44538
  |
Correlation:0.44538
  |
Correlation:0.49268
  |
Correlation:0.54877
  |
Correlation:0.42934
  |
Component:001; Jaccard:0.11232
 |
Component:282; Jaccard:0.12125
 |
Component:282; Jaccard:0.13345
 |
Component:282; Jaccard:0.14223
 |
Component:181; Jaccard:0.14116
 |
Component:238; Jaccard:0.13757
 |
Component:283; Jaccard:0.12768
 |
Correlation:0.36091
  |
Correlation:0.40418
  |
Correlation:0.40418
  |
Correlation:0.40418
  |
Correlation:0.44538
  |
Correlation:0.42934
  |
Correlation:0.54877
  |
Cope: 04:STORY-MATH BACK
|
GLM result group-wise
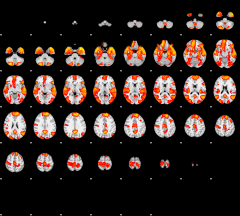 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
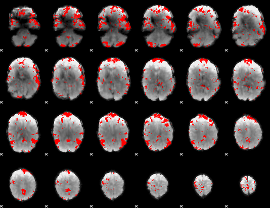 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
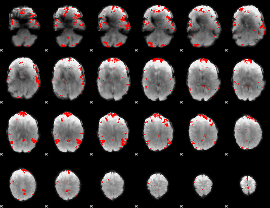 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
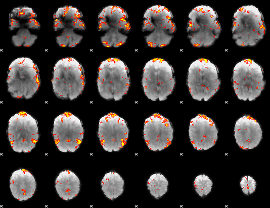 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:139; Jaccard:0.12586
 |
Component:146; Jaccard:0.15273
 |
Component:146; Jaccard:0.19312
 |
Component:146; Jaccard:0.23533
 |
Component:146; Jaccard:0.27026
 |
Component:146; Jaccard:0.29001
 |
Component:315; Jaccard:0.29222
 |
Correlation:0.40592
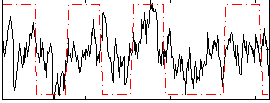  |
Correlation:0.49728
  |
Correlation:0.49728
  |
Correlation:0.49728
  |
Correlation:0.49728
  |
Correlation:0.49728
  |
Correlation:0.69335
  |
Component:285; Jaccard:0.12441
 |
Component:315; Jaccard:0.15257
 |
Component:315; Jaccard:0.19264
 |
Component:315; Jaccard:0.23339
 |
Component:315; Jaccard:0.26575
 |
Component:315; Jaccard:0.28655
 |
Component:146; Jaccard:0.29211
 |
Correlation:0.34973
  |
Correlation:0.69335
  |
Correlation:0.69335
  |
Correlation:0.69335
  |
Correlation:0.69335
  |
Correlation:0.69335
  |
Correlation:0.49728
  |
Component:336; Jaccard:0.12006
 |
Component:285; Jaccard:0.14721
 |
Component:285; Jaccard:0.1683
 |
Component:285; Jaccard:0.1847
 |
Component:065; Jaccard:0.19626
 |
Component:065; Jaccard:0.19839
 |
Component:065; Jaccard:0.18919
 |
Correlation:0.29771
  |
Correlation:0.34973
  |
Correlation:0.34973
  |
Correlation:0.34973
  |
Correlation:0.56769
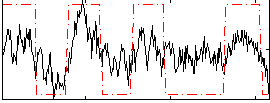  |
Correlation:0.56769
  |
Correlation:0.56769
  |
Component:315; Jaccard:0.1179
 |
Component:139; Jaccard:0.14304
 |
Component:065; Jaccard:0.16406
 |
Component:065; Jaccard:0.18402
 |
Component:285; Jaccard:0.19143
 |
Component:285; Jaccard:0.1889
 |
Component:285; Jaccard:0.17559
 |
Correlation:0.69335
  |
Correlation:0.40592
  |
Correlation:0.56769
  |
Correlation:0.56769
  |
Correlation:0.34973
  |
Correlation:0.34973
  |
Correlation:0.34973
  |
Component:146; Jaccard:0.11763
 |
Component:065; Jaccard:0.13926
 |
Component:139; Jaccard:0.15535
 |
Component:139; Jaccard:0.16182
 |
Component:399; Jaccard:0.16059
 |
Component:399; Jaccard:0.15553
 |
Component:171; Jaccard:0.1459
 |
Correlation:0.49728
  |
Correlation:0.56769
  |
Correlation:0.40592
  |
Correlation:0.40592
  |
Correlation:0.19223
  |
Correlation:0.19223
  |
Correlation:0.4842
 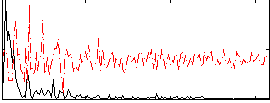 |
Component:399; Jaccard:0.11428
 |
Component:336; Jaccard:0.13379
 |
Component:399; Jaccard:0.14684
 |
Component:399; Jaccard:0.15659
 |
Component:139; Jaccard:0.16005
 |
Component:171; Jaccard:0.15382
 |
Component:399; Jaccard:0.14383
 |
Correlation:0.19223
  |
Correlation:0.29771
  |
Correlation:0.19223
  |
Correlation:0.19223
  |
Correlation:0.40592
  |
Correlation:0.4842
  |
Correlation:0.19223
  |
Component:065; Jaccard:0.11306
 |
Component:399; Jaccard:0.13198
 |
Component:336; Jaccard:0.14208
 |
Component:171; Jaccard:0.14986
 |
Component:171; Jaccard:0.15628
 |
Component:139; Jaccard:0.15049
 |
Component:139; Jaccard:0.13585
 |
Correlation:0.56769
  |
Correlation:0.19223
  |
Correlation:0.29771
  |
Correlation:0.4842
  |
Correlation:0.4842
  |
Correlation:0.40592
  |
Correlation:0.40592
  |
Component:211; Jaccard:0.10503
 |
Component:171; Jaccard:0.12082
 |
Component:171; Jaccard:0.13939
 |
Component:247; Jaccard:0.1434
 |
Component:247; Jaccard:0.14685
 |
Component:247; Jaccard:0.14356
 |
Component:247; Jaccard:0.13583
 |
Correlation:0.39714
  |
Correlation:0.4842
  |
Correlation:0.4842
  |
Correlation:0.39005
  |
Correlation:0.39005
  |
Correlation:0.39005
  |
Correlation:0.39005
  |
Component:171; Jaccard:0.10333
 |
Component:211; Jaccard:0.11669
 |
Component:247; Jaccard:0.13102
 |
Component:336; Jaccard:0.14219
 |
Component:336; Jaccard:0.13904
 |
Component:336; Jaccard:0.12935
 |
Component:167; Jaccard:0.1181
 |
Correlation:0.4842
  |
Correlation:0.39714
  |
Correlation:0.39005
  |
Correlation:0.29771
  |
Correlation:0.29771
  |
Correlation:0.29771
  |
Correlation:0.51838
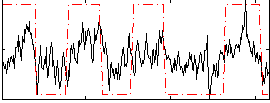  |
Component:210; Jaccard:0.10054
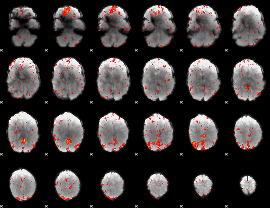 |
Component:247; Jaccard:0.11359
 |
Component:211; Jaccard:0.12587
 |
Component:211; Jaccard:0.12955
 |
Component:211; Jaccard:0.12746
 |
Component:167; Jaccard:0.12144
 |
Component:336; Jaccard:0.11624
 |
Correlation:0.21187
  |
Correlation:0.39005
  |
Correlation:0.39714
  |
Correlation:0.39714
  |
Correlation:0.39714
  |
Correlation:0.51838
  |
Correlation:0.29771
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
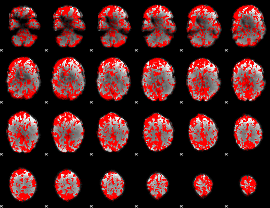 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
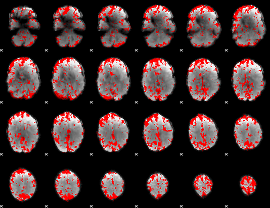 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
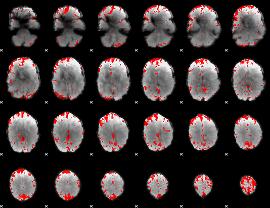 |
GLM binary result thresholded by 3.5
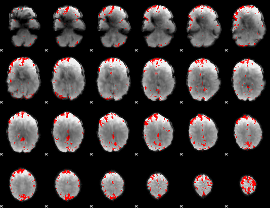 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
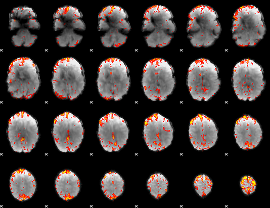
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
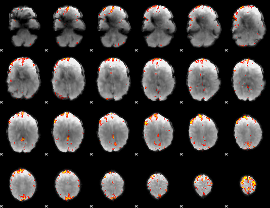
GLM Z-score result thresholded by 4
 |
Component:267; Jaccard:0.14156
 |
Component:267; Jaccard:0.16864
 |
Component:267; Jaccard:0.19473
 |
Component:267; Jaccard:0.2147
 |
Component:267; Jaccard:0.22382
 |
Component:267; Jaccard:0.21755
 |
Component:267; Jaccard:0.19816
 |
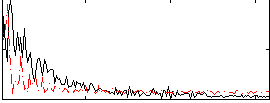
Correlation:0.01872
  |
Correlation:0.01872
  |
Correlation:0.01872
  |
Correlation:0.01872
  |
Correlation:0.01872
  |
Correlation:0.01872
  |
Correlation:0.01872
  |
Component:260; Jaccard:0.10916
 |
Component:260; Jaccard:0.1329
 |
Component:260; Jaccard:0.1573
 |
Component:260; Jaccard:0.17739
 |
Component:260; Jaccard:0.1879
 |
Component:260; Jaccard:0.18089
 |
Component:260; Jaccard:0.15902
 |
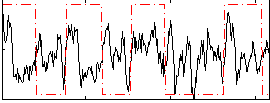
Correlation:0.053413
  |
Correlation:0.053413
  |
Correlation:0.053413
  |
Correlation:0.053413
  |
Correlation:0.053413
  |
Correlation:0.053413
  |
Correlation:0.053413
  |
Component:007; Jaccard:0.10057
 |
Component:007; Jaccard:0.11764
 |
Component:007; Jaccard:0.1344
 |
Component:007; Jaccard:0.14464
 |
Component:007; Jaccard:0.14651
 |
Component:007; Jaccard:0.13627
 |
Component:129; Jaccard:0.12076
 |
Correlation:-0.16878
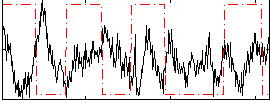  |
Correlation:-0.16878
  |
Correlation:-0.16878
  |
Correlation:-0.16878
  |
Correlation:-0.16878
  |
Correlation:-0.16878
  |
Correlation:0.19993
  |
Component:272; Jaccard:0.097769
 |
Component:272; Jaccard:0.11155
 |
Component:272; Jaccard:0.12477
 |
Component:272; Jaccard:0.13241
 |
Component:272; Jaccard:0.13202
 |
Component:129; Jaccard:0.12946
 |
Component:007; Jaccard:0.11877
 |
Correlation:-0.12868
  |
Correlation:-0.12868
  |
Correlation:-0.12868
  |
Correlation:-0.12868
  |
Correlation:-0.12868
  |
Correlation:0.19993
  |
Correlation:-0.16878
  |
Component:243; Jaccard:0.09627
 |
Component:243; Jaccard:0.10733
 |
Component:243; Jaccard:0.11712
 |
Component:181; Jaccard:0.123
 |
Component:129; Jaccard:0.12843
 |
Component:272; Jaccard:0.12442
 |
Component:389; Jaccard:0.11464
 |
Correlation:-0.16373
  |
Correlation:-0.16373
  |
Correlation:-0.16373
  |
Correlation:-0.42965
  |
Correlation:0.19993
  |
Correlation:-0.12868
  |
Correlation:0.2403
 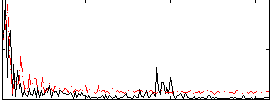 |
Component:048; Jaccard:0.091638
 |
Component:048; Jaccard:0.10383
 |
Component:181; Jaccard:0.11472
 |
Component:243; Jaccard:0.12245
 |
Component:181; Jaccard:0.12293
 |
Component:389; Jaccard:0.1158
 |
Component:272; Jaccard:0.10692
 |
Correlation:-0.3935
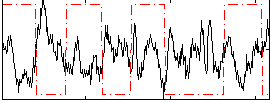  |
Correlation:-0.3935
  |
Correlation:-0.42965
  |
Correlation:-0.16373
  |
Correlation:-0.42965
  |
Correlation:0.2403
  |
Correlation:-0.12868
  |
Component:181; Jaccard:0.090609
 |
Component:181; Jaccard:0.10267
 |
Component:048; Jaccard:0.11406
 |
Component:129; Jaccard:0.12141
 |
Component:243; Jaccard:0.12062
 |
Component:223; Jaccard:0.11321
 |
Component:223; Jaccard:0.10594
 |
Correlation:-0.42965
  |
Correlation:-0.42965
  |
Correlation:-0.3935
  |
Correlation:0.19993
  |
Correlation:-0.16373
  |
Correlation:-0.027916
 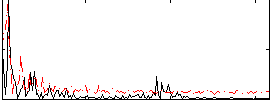 |
Correlation:-0.027916
  |
Component:053; Jaccard:0.088622
 |
Component:053; Jaccard:0.1014
 |
Component:053; Jaccard:0.11254
 |
Component:048; Jaccard:0.12029
 |
Component:053; Jaccard:0.1192
 |
Component:243; Jaccard:0.11273
 |
Component:243; Jaccard:0.10001
 |
Correlation:-0.27185
  |
Correlation:-0.27185
  |
Correlation:-0.27185
  |
Correlation:-0.3935
  |
Correlation:-0.27185
  |
Correlation:-0.16373
  |
Correlation:-0.16373
  |
Component:210; Jaccard:0.088178
 |
Component:210; Jaccard:0.099688
 |
Component:129; Jaccard:0.10952
 |
Component:053; Jaccard:0.11977
 |
Component:048; Jaccard:0.11741
 |
Component:181; Jaccard:0.11158
 |
Component:181; Jaccard:0.09337
 |
Correlation:0.20551
  |
Correlation:0.20551
  |
Correlation:0.19993
  |
Correlation:-0.27185
  |
Correlation:-0.3935
  |
Correlation:-0.42965
  |
Correlation:-0.42965
  |
Component:129; Jaccard:0.087399
 |
Component:129; Jaccard:0.098475
 |
Component:210; Jaccard:0.10795
 |
Component:210; Jaccard:0.11224
 |
Component:223; Jaccard:0.11654
 |
Component:053; Jaccard:0.10844
 |
Component:053; Jaccard:0.089597
 |
Correlation:0.19993
  |
Correlation:0.19993
  |
Correlation:0.20551
  |
Correlation:0.20551
  |
Correlation:-0.027916
  |
Correlation:-0.27185
  |
Correlation:-0.27185
  |
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
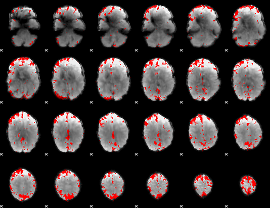 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
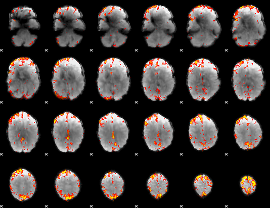 |
GLM Z-score result thresholded by 4
 |
Component:267; Jaccard:0.1364
 |
Component:267; Jaccard:0.15984
 |
Component:267; Jaccard:0.18443
 |
Component:267; Jaccard:0.20281
 |
Component:267; Jaccard:0.21462
 |
Component:267; Jaccard:0.21286
 |
Component:267; Jaccard:0.19901
 |
Correlation:-0.010389
  |
Correlation:-0.010389
  |
Correlation:-0.010389
  |
Correlation:-0.010389
  |
Correlation:-0.010389
  |
Correlation:-0.010389
  |
Correlation:-0.010389
  |
Component:181; Jaccard:0.1037
 |
Component:181; Jaccard:0.12545
 |
Component:181; Jaccard:0.14935
 |
Component:181; Jaccard:0.17072
 |
Component:181; Jaccard:0.18569
 |
Component:181; Jaccard:0.18986
 |
Component:181; Jaccard:0.18241
 |
Correlation:0.44332
  |
Correlation:0.44332
  |
Correlation:0.44332
  |
Correlation:0.44332
  |
Correlation:0.44332
  |
Correlation:0.44332
  |
Correlation:0.44332
  |
Component:268; Jaccard:0.10333
 |
Component:260; Jaccard:0.1222
 |
Component:260; Jaccard:0.14309
 |
Component:260; Jaccard:0.16069
 |
Component:260; Jaccard:0.1711
 |
Component:260; Jaccard:0.17492
 |
Component:260; Jaccard:0.1695
 |
Correlation:0.45237
  |
Correlation:-0.063956
  |
Correlation:-0.063956
  |
Correlation:-0.063956
  |
Correlation:-0.063956
  |
Correlation:-0.063956
  |
Correlation:-0.063956
  |
Component:260; Jaccard:0.10316
 |
Component:048; Jaccard:0.12038
 |
Component:048; Jaccard:0.13853
 |
Component:048; Jaccard:0.15555
 |
Component:048; Jaccard:0.16428
 |
Component:048; Jaccard:0.16337
 |
Component:048; Jaccard:0.15062
 |
Correlation:-0.063956
  |
Correlation:0.34554
  |
Correlation:0.34554
  |
Correlation:0.34554
  |
Correlation:0.34554
  |
Correlation:0.34554
  |
Correlation:0.34554
  |
Component:048; Jaccard:0.10259
 |
Component:268; Jaccard:0.11669
 |
Component:383; Jaccard:0.13376
 |
Component:053; Jaccard:0.14704
 |
Component:053; Jaccard:0.15675
 |
Component:053; Jaccard:0.15778
 |
Component:053; Jaccard:0.14987
 |
Correlation:0.34554
  |
Correlation:0.45237
  |
Correlation:0.59388
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Component:272; Jaccard:0.10049
 |
Component:272; Jaccard:0.11577
 |
Component:272; Jaccard:0.13079
 |
Component:383; Jaccard:0.1466
 |
Component:272; Jaccard:0.15059
 |
Component:272; Jaccard:0.14869
 |
Component:272; Jaccard:0.13805
 |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.59388
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
Correlation:0.15665
  |
Component:243; Jaccard:0.09807
 |
Component:383; Jaccard:0.11556
 |
Component:053; Jaccard:0.13053
 |
Component:272; Jaccard:0.14328
 |
Component:383; Jaccard:0.14845
 |
Component:007; Jaccard:0.14276
 |
Component:007; Jaccard:0.131
 |
Correlation:0.14021
  |
Correlation:0.59388
  |
Correlation:0.27895
  |
Correlation:0.15665
  |
Correlation:0.59388
  |
Correlation:0.14709
  |
Correlation:0.14709
  |
Component:383; Jaccard:0.097429
 |
Component:053; Jaccard:0.11343
 |
Component:268; Jaccard:0.12896
 |
Component:007; Jaccard:0.13561
 |
Component:007; Jaccard:0.14314
 |
Component:383; Jaccard:0.1407
 |
Component:383; Jaccard:0.13028
 |
Correlation:0.59388
  |
Correlation:0.27895
  |
Correlation:0.45237
  |
Correlation:0.14709
  |
Correlation:0.14709
  |
Correlation:0.59388
  |
Correlation:0.59388
  |
Component:053; Jaccard:0.097295
 |
Component:007; Jaccard:0.11106
 |
Component:007; Jaccard:0.12458
 |
Component:268; Jaccard:0.13486
 |
Component:268; Jaccard:0.13464
 |
Component:208; Jaccard:0.13395
 |
Component:208; Jaccard:0.12923
 |
Correlation:0.27895
  |
Correlation:0.14709
  |
Correlation:0.14709
  |
Correlation:0.45237
  |
Correlation:0.45237
  |
Correlation:0.29207
  |
Correlation:0.29207
  |
Component:007; Jaccard:0.096924
 |
Component:243; Jaccard:0.11075
 |
Component:243; Jaccard:0.12243
 |
Component:243; Jaccard:0.12989
 |
Component:208; Jaccard:0.1334
 |
Component:243; Jaccard:0.12754
 |
Component:243; Jaccard:0.11818
 |
Correlation:0.14709
  |
Correlation:0.14021
  |
Correlation:0.14021
  |
Correlation:0.14021
  |
Correlation:0.29207
  |
Correlation:0.14021
  |
Correlation:0.14021
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































