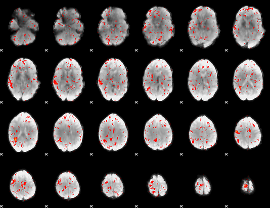
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
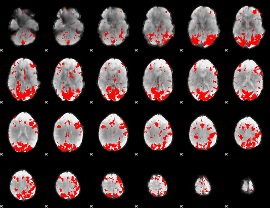 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
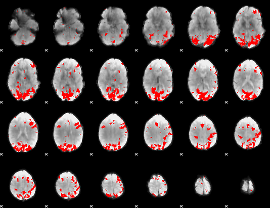 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
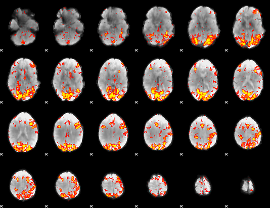 |
GLM Z-score result thresholded by 3
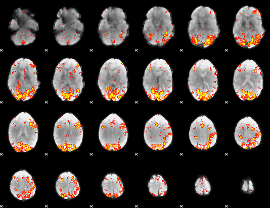 |
GLM Z-score result thresholded by 3.5
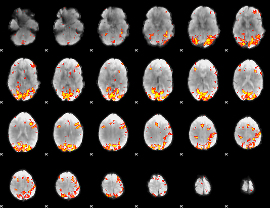 |
GLM Z-score result thresholded by 4
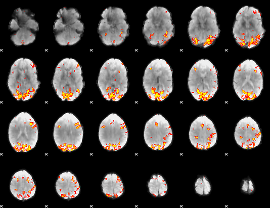 |
Component:223; Jaccard:0.14983
 |
Component:223; Jaccard:0.17933
 |
Component:342; Jaccard:0.21698
 |
Component:342; Jaccard:0.25648
 |
Component:072; Jaccard:0.28903
 |
Component:072; Jaccard:0.31466
 |
Component:072; Jaccard:0.32515
 |
Correlation:0.18446
  |
Correlation:0.18446
  |
Correlation:0.25262
  |
Correlation:0.25262
  |
Correlation:0.34976
  |
Correlation:0.34976
  |
Correlation:0.34976
  |
Component:186; Jaccard:0.14549
 |
Component:342; Jaccard:0.17802
 |
Component:072; Jaccard:0.21216
 |
Component:072; Jaccard:0.25393
 |
Component:342; Jaccard:0.28694
 |
Component:342; Jaccard:0.31114
 |
Component:342; Jaccard:0.3216
 |
Correlation:0.059451
  |
Correlation:0.25262
  |
Correlation:0.34976
  |
Correlation:0.34976
  |
Correlation:0.25262
  |
Correlation:0.25262
  |
Correlation:0.25262
  |
Component:342; Jaccard:0.14324
 |
Component:072; Jaccard:0.17154
 |
Component:223; Jaccard:0.20736
 |
Component:223; Jaccard:0.22518
 |
Component:368; Jaccard:0.23486
 |
Component:368; Jaccard:0.24392
 |
Component:368; Jaccard:0.24277
 |
Correlation:0.25262
  |
Correlation:0.34976
  |
Correlation:0.18446
  |
Correlation:0.18446
  |
Correlation:0.17179
  |
Correlation:0.17179
  |
Correlation:0.17179
  |
Component:088; Jaccard:0.14103
 |
Component:088; Jaccard:0.16555
 |
Component:368; Jaccard:0.1918
 |
Component:368; Jaccard:0.21768
 |
Component:223; Jaccard:0.23359
 |
Component:088; Jaccard:0.23381
 |
Component:036; Jaccard:0.23195
 |
Correlation:0.33474
  |
Correlation:0.33474
  |
Correlation:0.17179
  |
Correlation:0.17179
  |
Correlation:0.18446
  |
Correlation:0.33474
  |
Correlation:0.19106
  |
Component:263; Jaccard:0.13813
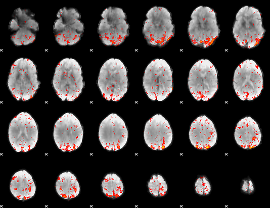 |
Component:368; Jaccard:0.16481
 |
Component:088; Jaccard:0.191
 |
Component:088; Jaccard:0.2126
 |
Component:088; Jaccard:0.22867
 |
Component:223; Jaccard:0.23094
 |
Component:088; Jaccard:0.23079
 |
Correlation:0.2137
  |
Correlation:0.17179
  |
Correlation:0.33474
  |
Correlation:0.33474
  |
Correlation:0.33474
  |
Correlation:0.18446
  |
Correlation:0.33474
  |
Component:368; Jaccard:0.13792
 |
Component:186; Jaccard:0.16314
 |
Component:186; Jaccard:0.17704
 |
Component:036; Jaccard:0.19681
 |
Component:036; Jaccard:0.21639
 |
Component:036; Jaccard:0.22753
 |
Component:223; Jaccard:0.2118
 |
Correlation:0.17179
  |
Correlation:0.059451
  |
Correlation:0.059451
  |
Correlation:0.19106
  |
Correlation:0.19106
  |
Correlation:0.19106
  |
Correlation:0.18446
  |
Component:072; Jaccard:0.13694
 |
Component:263; Jaccard:0.15815
 |
Component:263; Jaccard:0.17543
 |
Component:263; Jaccard:0.18742
 |
Component:263; Jaccard:0.18869
 |
Component:263; Jaccard:0.18513
 |
Component:108; Jaccard:0.17276
 |
Correlation:0.34976
  |
Correlation:0.2137
  |
Correlation:0.2137
  |
Correlation:0.2137
  |
Correlation:0.2137
  |
Correlation:0.2137
  |
Correlation:0.29294
  |
Component:381; Jaccard:0.12998
 |
Component:036; Jaccard:0.1476
 |
Component:036; Jaccard:0.17213
 |
Component:186; Jaccard:0.18195
 |
Component:031; Jaccard:0.18199
 |
Component:031; Jaccard:0.17914
 |
Component:263; Jaccard:0.17232
 |
Correlation:0.055803
  |
Correlation:0.19106
  |
Correlation:0.19106
  |
Correlation:0.059451
  |
Correlation:0.3762
  |
Correlation:0.3762
  |
Correlation:0.2137
  |
Component:280; Jaccard:0.12863
 |
Component:031; Jaccard:0.14614
 |
Component:031; Jaccard:0.16376
 |
Component:031; Jaccard:0.17755
 |
Component:186; Jaccard:0.1794
 |
Component:280; Jaccard:0.17456
 |
Component:031; Jaccard:0.168
 |
Correlation:0.080199
  |
Correlation:0.3762
  |
Correlation:0.3762
  |
Correlation:0.3762
  |
Correlation:0.059451
  |
Correlation:0.080199
  |
Correlation:0.3762
  |
Component:031; Jaccard:0.12699
 |
Component:280; Jaccard:0.14599
 |
Component:280; Jaccard:0.1617
 |
Component:280; Jaccard:0.17343
 |
Component:280; Jaccard:0.17749
 |
Component:108; Jaccard:0.17411
 |
Component:280; Jaccard:0.16305
 |
Correlation:0.3762
  |
Correlation:0.080199
  |
Correlation:0.080199
  |
Correlation:0.080199
  |
Correlation:0.080199
  |
Correlation:0.29294
  |
Correlation:0.080199
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
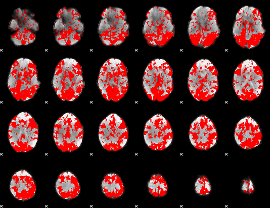 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
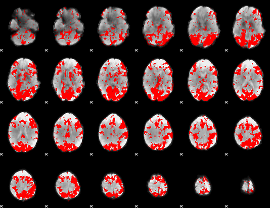 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
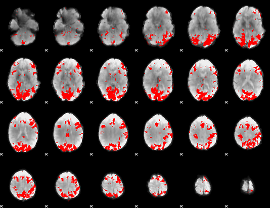 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
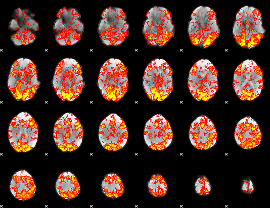 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
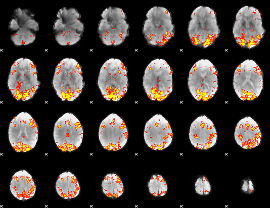 |
GLM Z-score result thresholded by 4
 |
Component:186; Jaccard:0.13362
 |
Component:186; Jaccard:0.15548
 |
Component:186; Jaccard:0.17677
 |
Component:342; Jaccard:0.21084
 |
Component:342; Jaccard:0.24951
 |
Component:342; Jaccard:0.28569
 |
Component:342; Jaccard:0.31652
 |
Correlation:0.20806
  |
Correlation:0.20806
  |
Correlation:0.20806
  |
Correlation:0.22381
  |
Correlation:0.22381
  |
Correlation:0.22381
  |
Correlation:0.22381
  |
Component:088; Jaccard:0.12345
 |
Component:223; Jaccard:0.14789
 |
Component:342; Jaccard:0.17505
 |
Component:223; Jaccard:0.20132
 |
Component:072; Jaccard:0.2245
 |
Component:072; Jaccard:0.2507
 |
Component:072; Jaccard:0.2721
 |
Correlation:0.14761
  |
Correlation:0.10347
  |
Correlation:0.22381
  |
Correlation:0.10347
  |
Correlation:0.18901
  |
Correlation:0.18901
  |
Correlation:0.18901
  |
Component:223; Jaccard:0.12341
 |
Component:088; Jaccard:0.1459
 |
Component:223; Jaccard:0.17467
 |
Component:088; Jaccard:0.19439
 |
Component:223; Jaccard:0.22293
 |
Component:036; Jaccard:0.24347
 |
Component:036; Jaccard:0.26547
 |
Correlation:0.10347
  |
Correlation:0.14761
  |
Correlation:0.10347
  |
Correlation:0.14761
  |
Correlation:0.10347
  |
Correlation:0.41803
  |
Correlation:0.41803
  |
Component:280; Jaccard:0.12289
 |
Component:280; Jaccard:0.14397
 |
Component:088; Jaccard:0.17032
 |
Component:186; Jaccard:0.19279
 |
Component:036; Jaccard:0.21771
 |
Component:223; Jaccard:0.23473
 |
Component:088; Jaccard:0.23828
 |
Correlation:0.18185
  |
Correlation:0.18185
  |
Correlation:0.14761
  |
Correlation:0.20806
  |
Correlation:0.41803
  |
Correlation:0.10347
  |
Correlation:0.14761
  |
Component:381; Jaccard:0.12146
 |
Component:342; Jaccard:0.14276
 |
Component:280; Jaccard:0.16685
 |
Component:072; Jaccard:0.19232
 |
Component:088; Jaccard:0.21556
 |
Component:088; Jaccard:0.22932
 |
Component:223; Jaccard:0.23548
 |
Correlation:0.24869
  |
Correlation:0.22381
  |
Correlation:0.18185
  |
Correlation:0.18901
  |
Correlation:0.14761
  |
Correlation:0.14761
  |
Correlation:0.10347
  |
Component:342; Jaccard:0.11648
 |
Component:368; Jaccard:0.13728
 |
Component:368; Jaccard:0.16126
 |
Component:036; Jaccard:0.18759
 |
Component:368; Jaccard:0.20742
 |
Component:368; Jaccard:0.22253
 |
Component:368; Jaccard:0.23183
 |
Correlation:0.22381
  |
Correlation:0.19461
  |
Correlation:0.19461
  |
Correlation:0.41803
  |
Correlation:0.19461
  |
Correlation:0.19461
  |
Correlation:0.19461
  |
Component:368; Jaccard:0.11547
 |
Component:381; Jaccard:0.13624
 |
Component:072; Jaccard:0.16019
 |
Component:280; Jaccard:0.18722
 |
Component:280; Jaccard:0.20533
 |
Component:280; Jaccard:0.21726
 |
Component:280; Jaccard:0.21957
 |
Correlation:0.19461
  |
Correlation:0.24869
  |
Correlation:0.18901
  |
Correlation:0.18185
  |
Correlation:0.18185
  |
Correlation:0.18185
  |
Correlation:0.18185
  |
Component:275; Jaccard:0.11291
 |
Component:072; Jaccard:0.13216
 |
Component:036; Jaccard:0.15744
 |
Component:368; Jaccard:0.18582
 |
Component:186; Jaccard:0.20076
 |
Component:186; Jaccard:0.20113
 |
Component:186; Jaccard:0.19499
 |
Correlation:-0.10751
  |
Correlation:0.18901
  |
Correlation:0.41803
  |
Correlation:0.19461
  |
Correlation:0.20806
  |
Correlation:0.20806
  |
Correlation:0.20806
  |
Component:263; Jaccard:0.11102
 |
Component:036; Jaccard:0.13124
 |
Component:381; Jaccard:0.14894
 |
Component:381; Jaccard:0.15946
 |
Component:381; Jaccard:0.16407
 |
Component:031; Jaccard:0.16809
 |
Component:031; Jaccard:0.16726
 |
Correlation:0.12276
  |
Correlation:0.41803
  |
Correlation:0.24869
  |
Correlation:0.24869
  |
Correlation:0.24869
  |
Correlation:0.13169
  |
Correlation:0.13169
  |
Component:031; Jaccard:0.10906
 |
Component:263; Jaccard:0.12496
 |
Component:031; Jaccard:0.13992
 |
Component:031; Jaccard:0.15354
 |
Component:031; Jaccard:0.16387
 |
Component:381; Jaccard:0.16179
 |
Component:263; Jaccard:0.15389
 |
Correlation:0.13169
  |
Correlation:0.12276
  |
Correlation:0.13169
  |
Correlation:0.13169
  |
Correlation:0.13169
  |
Correlation:0.24869
  |
Correlation:0.12276
  |
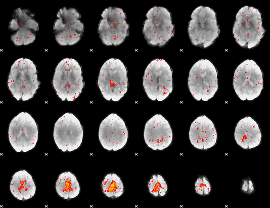
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
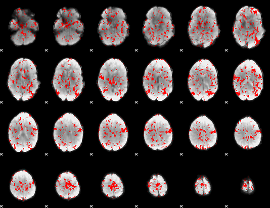 |
GLM binary result thresholded by 1.5
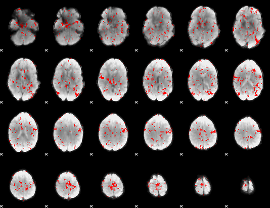 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
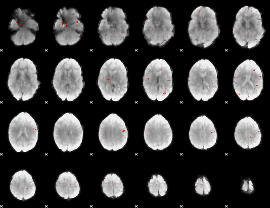 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
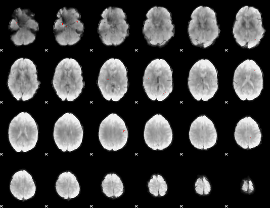 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
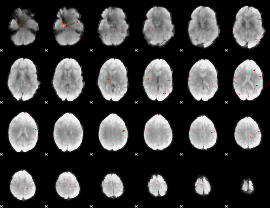 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
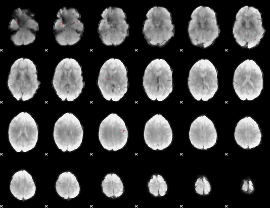 |
Component:317; Jaccard:0.093
 |
Component:145; Jaccard:0.092705
 |
Component:145; Jaccard:0.079816
 |
Component:157; Jaccard:0.053647
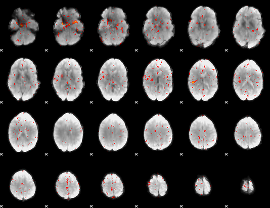 |
Component:157; Jaccard:0.033078
 |
Component:157; Jaccard:0.020875
 |
Component:157; Jaccard:0.008559
 |
Correlation:0.13573
  |
Correlation:0.13157
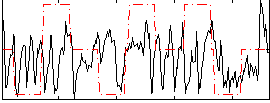  |
Correlation:0.13157
  |
Correlation:0.0030072
  |
Correlation:0.0030072
  |
Correlation:0.0030072
  |
Correlation:0.0030072
  |
Component:145; Jaccard:0.087818
 |
Component:317; Jaccard:0.087699
 |
Component:157; Jaccard:0.073086
 |
Component:145; Jaccard:0.053078
 |
Component:145; Jaccard:0.030681
 |
Component:145; Jaccard:0.015878
 |
Component:145; Jaccard:0.004235
 |
Correlation:0.13157
  |
Correlation:0.13573
  |
Correlation:0.0030072
  |
Correlation:0.13157
  |
Correlation:0.13157
  |
Correlation:0.13157
  |
Correlation:0.13157
  |
Component:007; Jaccard:0.076134
 |
Component:157; Jaccard:0.078032
 |
Component:317; Jaccard:0.066024
 |
Component:097; Jaccard:0.041163
 |
Component:097; Jaccard:0.021809
 |
Component:097; Jaccard:0.011549
 |
Component:097; Jaccard:0.003815
 |
Correlation:0.12404
  |
Correlation:0.0030072
  |
Correlation:0.13573
  |
Correlation:0.23689
  |
Correlation:0.23689
  |
Correlation:0.23689
  |
Correlation:0.23689
  |
Component:120; Jaccard:0.072315
 |
Component:007; Jaccard:0.074127
 |
Component:097; Jaccard:0.059216
 |
Component:317; Jaccard:0.039128
 |
Component:120; Jaccard:0.019965
 |
Component:120; Jaccard:0.009646
 |
Component:120; Jaccard:0.003534
 |
Correlation:0.16464
  |
Correlation:0.12404
  |
Correlation:0.23689
  |
Correlation:0.13573
  |
Correlation:0.16464
  |
Correlation:0.16464
  |
Correlation:0.16464
  |
Component:157; Jaccard:0.069216
 |
Component:120; Jaccard:0.070254
 |
Component:120; Jaccard:0.05595
 |
Component:120; Jaccard:0.035178
 |
Component:317; Jaccard:0.018406
 |
Component:317; Jaccard:0.008908
 |
Component:001; Jaccard:0.002813
 |
Correlation:0.0030072
  |
Correlation:0.16464
  |
Correlation:0.16464
  |
Correlation:0.16464
  |
Correlation:0.13573
  |
Correlation:0.13573
  |
Correlation:0.092515
  |
Component:034; Jaccard:0.065853
 |
Component:097; Jaccard:0.06729
 |
Component:007; Jaccard:0.055485
 |
Component:068; Jaccard:0.032072
 |
Component:068; Jaccard:0.017095
 |
Component:068; Jaccard:0.007887
 |
Component:355; Jaccard:0.00252
 |
Correlation:0.23582
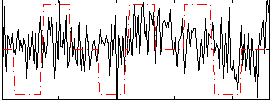  |
Correlation:0.23689
  |
Correlation:0.12404
  |
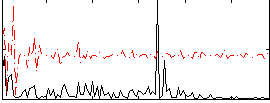
Correlation:0.10783
 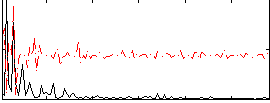 |
Correlation:0.10783
  |
Correlation:0.10783
  |
Correlation:0.076624
  |
Component:097; Jaccard:0.064506
 |
Component:034; Jaccard:0.062167
 |
Component:068; Jaccard:0.051452
 |
Component:007; Jaccard:0.030426
 |
Component:034; Jaccard:0.017035
 |
Component:134; Jaccard:0.007021
 |
Component:068; Jaccard:0.002352
 |
Correlation:0.23689
  |
Correlation:0.23582
  |
Correlation:0.10783
  |
Correlation:0.12404
  |
Correlation:0.23582
  |
Correlation:0.020574
 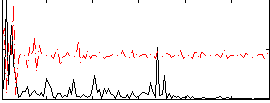 |
Correlation:0.10783
  |
Component:068; Jaccard:0.063363
 |
Component:068; Jaccard:0.061512
 |
Component:034; Jaccard:0.048321
 |
Component:034; Jaccard:0.030171
 |
Component:355; Jaccard:0.01423
 |
Component:034; Jaccard:0.006863
 |
Component:317; Jaccard:0.002197
 |
Correlation:0.10783
  |
Correlation:0.10783
  |
Correlation:0.23582
  |
Correlation:0.23582
  |
Correlation:0.076624
  |
Correlation:0.23582
  |
Correlation:0.13573
  |
Component:373; Jaccard:0.061839
 |
Component:355; Jaccard:0.0562
 |
Component:355; Jaccard:0.043438
 |
Component:355; Jaccard:0.028105
 |
Component:134; Jaccard:0.011817
 |
Component:355; Jaccard:0.006398
 |
Component:034; Jaccard:0.002072
 |
Correlation:0.092059
  |
Correlation:0.076624
  |
Correlation:0.076624
  |
Correlation:0.076624
  |
Correlation:0.020574
  |
Correlation:0.076624
  |
Correlation:0.23582
  |
Component:273; Jaccard:0.059832
 |
Component:373; Jaccard:0.055905
 |
Component:024; Jaccard:0.039579
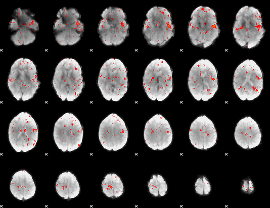 |
Component:024; Jaccard:0.024009
 |
Component:001; Jaccard:0.011381
 |
Component:024; Jaccard:0.005613
 |
Component:324; Jaccard:0.002033
 |
Correlation:0.10283
  |
Correlation:0.092059
  |
Correlation:0.096671
  |
Correlation:0.096671
  |
Correlation:0.092515
  |
Correlation:0.096671
  |
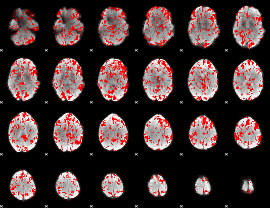
Correlation:0.073382
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
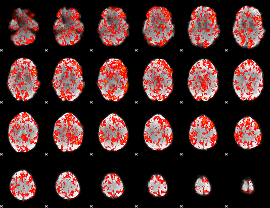
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
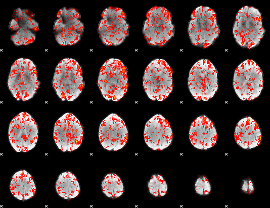
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
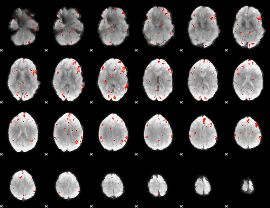 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:059; Jaccard:0.11978
 |
Component:059; Jaccard:0.14337
 |
Component:059; Jaccard:0.16122
 |
Component:059; Jaccard:0.15448
 |
Component:059; Jaccard:0.11717
 |
Component:117; Jaccard:0.075944
 |
Component:117; Jaccard:0.042959
 |
Correlation:0.0099493
  |
Correlation:0.0099493
  |
Correlation:0.0099493
  |
Correlation:0.0099493
  |
Correlation:0.0099493
  |
Correlation:0.15773
  |
Correlation:0.15773
  |
Component:130; Jaccard:0.10806
 |
Component:130; Jaccard:0.1195
 |
Component:117; Jaccard:0.136
 |
Component:117; Jaccard:0.13849
 |
Component:117; Jaccard:0.11261
 |
Component:059; Jaccard:0.070773
 |
Component:059; Jaccard:0.036239
 |
Correlation:0.17156
  |
Correlation:0.17156
  |
Correlation:0.15773
  |
Correlation:0.15773
  |
Correlation:0.15773
  |
Correlation:0.0099493
  |
Correlation:0.0099493
  |
Component:117; Jaccard:0.096814
 |
Component:117; Jaccard:0.11828
 |
Component:130; Jaccard:0.12134
 |
Component:130; Jaccard:0.10363
 |
Component:118; Jaccard:0.078782
 |
Component:118; Jaccard:0.050701
 |
Component:118; Jaccard:0.0292
 |
Correlation:0.15773
  |
Correlation:0.15773
  |
Correlation:0.17156
  |
Correlation:0.17156
  |
Correlation:0.12663
  |
Correlation:0.12663
  |
Correlation:0.12663
  |
Component:312; Jaccard:0.083597
 |
Component:118; Jaccard:0.093989
 |
Component:118; Jaccard:0.10221
 |
Component:118; Jaccard:0.09771
 |
Component:334; Jaccard:0.072637
 |
Component:334; Jaccard:0.045594
 |
Component:334; Jaccard:0.025418
 |
Correlation:0.093349
  |
Correlation:0.12663
  |
Correlation:0.12663
  |
Correlation:0.12663
  |
Correlation:0.17942
  |
Correlation:0.17942
  |
Correlation:0.17942
  |
Component:287; Jaccard:0.083417
 |
Component:287; Jaccard:0.091735
 |
Component:287; Jaccard:0.093912
 |
Component:334; Jaccard:0.089578
 |
Component:130; Jaccard:0.072335
 |
Component:130; Jaccard:0.041507
 |
Component:044; Jaccard:0.023115
 |
Correlation:-0.0060951
  |
Correlation:-0.0060951
  |
Correlation:-0.0060951
  |
Correlation:0.17942
  |
Correlation:0.17156
  |
Correlation:0.17156
  |
Correlation:0.052182
  |
Component:237; Jaccard:0.082364
 |
Component:312; Jaccard:0.088037
 |
Component:334; Jaccard:0.088418
 |
Component:287; Jaccard:0.085756
 |
Component:325; Jaccard:0.063098
 |
Component:044; Jaccard:0.040151
 |
Component:325; Jaccard:0.021408
 |
Correlation:0.13288
  |
Correlation:0.093349
  |
Correlation:0.17942
  |
Correlation:-0.0060951
  |
Correlation:-0.079974
  |
Correlation:0.052182
  |
Correlation:-0.079974
  |
Component:118; Jaccard:0.082033
 |
Component:237; Jaccard:0.084987
 |
Component:329; Jaccard:0.087176
 |
Component:329; Jaccard:0.078692
 |
Component:287; Jaccard:0.062505
 |
Component:325; Jaccard:0.038823
 |
Component:130; Jaccard:0.021141
 |
Correlation:0.12663
  |
Correlation:0.13288
  |
Correlation:0.026768
  |
Correlation:0.026768
  |
Correlation:-0.0060951
  |
Correlation:-0.079974
  |
Correlation:0.17156
  |
Component:012; Jaccard:0.080595
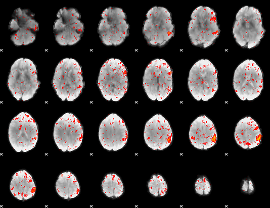 |
Component:012; Jaccard:0.08455
 |
Component:312; Jaccard:0.086213
 |
Component:044; Jaccard:0.078514
 |
Component:044; Jaccard:0.061118
 |
Component:242; Jaccard:0.037788
 |
Component:242; Jaccard:0.020936
 |
Correlation:0.17731
  |
Correlation:0.17731
  |
Correlation:0.093349
  |
Correlation:0.052182
  |
Correlation:0.052182
  |
Correlation:0.11084
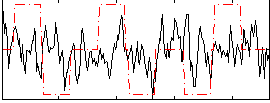  |
Correlation:0.11084
  |
Component:280; Jaccard:0.07927
 |
Component:044; Jaccard:0.082756
 |
Component:044; Jaccard:0.085893
 |
Component:325; Jaccard:0.078042
 |
Component:242; Jaccard:0.05954
 |
Component:287; Jaccard:0.036721
 |
Component:383; Jaccard:0.019461
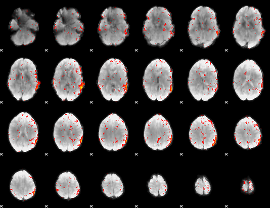 |
Correlation:0.06025
 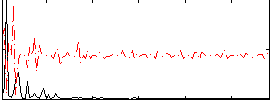 |
Correlation:0.052182
  |
Correlation:0.052182
  |
Correlation:-0.079974
  |
Correlation:0.11084
  |
Correlation:-0.0060951
  |
Correlation:0.016102
 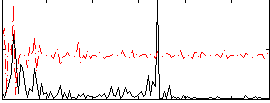 |
Component:154; Jaccard:0.078451
 |
Component:329; Jaccard:0.082721
 |
Component:242; Jaccard:0.084168
 |
Component:242; Jaccard:0.077556
 |
Component:329; Jaccard:0.059532
 |
Component:383; Jaccard:0.035228
 |
Component:287; Jaccard:0.018725
 |
Correlation:-0.0043034
  |
Correlation:0.026768
  |
Correlation:0.11084
  |
Correlation:0.11084
  |
Correlation:0.026768
  |
Correlation:0.016102
  |
Correlation:-0.0060951
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:175; Jaccard:0.14363
 |
Component:175; Jaccard:0.17637
 |
Component:175; Jaccard:0.21033
 |
Component:175; Jaccard:0.23835
 |
Component:175; Jaccard:0.23942
 |
Component:175; Jaccard:0.21217
 |
Component:175; Jaccard:0.1597
 |
Correlation:0.055742
  |
Correlation:0.055742
  |
Correlation:0.055742
  |
Correlation:0.055742
  |
Correlation:0.055742
  |
Correlation:0.055742
  |
Correlation:0.055742
  |
Component:139; Jaccard:0.12454
 |
Component:139; Jaccard:0.14189
 |
Component:139; Jaccard:0.15299
 |
Component:325; Jaccard:0.15597
 |
Component:325; Jaccard:0.15436
 |
Component:325; Jaccard:0.13856
 |
Component:325; Jaccard:0.11267
 |
Correlation:0.10223
  |
Correlation:0.10223
  |
Correlation:0.10223
  |
Correlation:0.09856
  |
Correlation:0.09856
  |
Correlation:0.09856
  |
Correlation:0.09856
  |
Component:044; Jaccard:0.11557
 |
Component:044; Jaccard:0.13245
 |
Component:044; Jaccard:0.14735
 |
Component:044; Jaccard:0.15389
 |
Component:044; Jaccard:0.14602
 |
Component:044; Jaccard:0.12206
 |
Component:044; Jaccard:0.095119
 |
Correlation:0.19036
  |
Correlation:0.19036
  |
Correlation:0.19036
  |
Correlation:0.19036
  |
Correlation:0.19036
  |
Correlation:0.19036
  |
Correlation:0.19036
  |
Component:218; Jaccard:0.10981
 |
Component:325; Jaccard:0.12698
 |
Component:325; Jaccard:0.14452
 |
Component:139; Jaccard:0.15166
 |
Component:139; Jaccard:0.13788
 |
Component:218; Jaccard:0.11267
 |
Component:218; Jaccard:0.084786
 |
Correlation:-0.065644
  |
Correlation:0.09856
  |
Correlation:0.09856
  |
Correlation:0.10223
  |
Correlation:0.10223
  |
Correlation:-0.065644
  |
Correlation:-0.065644
  |
Component:325; Jaccard:0.10758
 |
Component:218; Jaccard:0.126
 |
Component:218; Jaccard:0.13833
 |
Component:218; Jaccard:0.14312
 |
Component:218; Jaccard:0.13206
 |
Component:139; Jaccard:0.11001
 |
Component:139; Jaccard:0.080356
 |
Correlation:0.09856
  |
Correlation:-0.065644
  |
Correlation:-0.065644
  |
Correlation:-0.065644
  |
Correlation:-0.065644
  |
Correlation:0.10223
  |
Correlation:0.10223
  |
Component:171; Jaccard:0.10437
 |
Component:171; Jaccard:0.11187
 |
Component:171; Jaccard:0.11483
 |
Component:353; Jaccard:0.11723
 |
Component:353; Jaccard:0.10648
 |
Component:353; Jaccard:0.091
 |
Component:353; Jaccard:0.065913
 |
Correlation:0.035342
  |
Correlation:0.035342
  |
Correlation:0.035342
  |
Correlation:0.13111
  |
Correlation:0.13111
  |
Correlation:0.13111
  |
Correlation:0.13111
  |
Component:059; Jaccard:0.094683
 |
Component:353; Jaccard:0.10618
 |
Component:353; Jaccard:0.11462
 |
Component:118; Jaccard:0.11114
 |
Component:118; Jaccard:0.10456
 |
Component:118; Jaccard:0.087126
 |
Component:118; Jaccard:0.065859
 |
Correlation:0.069382
  |
Correlation:0.13111
  |
Correlation:0.13111
  |
Correlation:0.26696
  |
Correlation:0.26696
  |
Correlation:0.26696
  |
Correlation:0.26696
  |
Component:353; Jaccard:0.094634
 |
Component:118; Jaccard:0.10081
 |
Component:118; Jaccard:0.10802
 |
Component:171; Jaccard:0.10719
 |
Component:270; Jaccard:0.091772
 |
Component:270; Jaccard:0.077098
 |
Component:270; Jaccard:0.058009
 |
Correlation:0.13111
  |
Correlation:0.26696
  |
Correlation:0.26696
  |
Correlation:0.035342
  |
Correlation:0.066242
  |
Correlation:0.066242
  |
Correlation:0.066242
  |
Component:250; Jaccard:0.09275
 |
Component:059; Jaccard:0.099878
 |
Component:059; Jaccard:0.10196
 |
Component:270; Jaccard:0.097233
 |
Component:171; Jaccard:0.091221
 |
Component:106; Jaccard:0.071974
 |
Component:106; Jaccard:0.055219
 |
Correlation:-0.01891
  |
Correlation:0.069382
  |
Correlation:0.069382
  |
Correlation:0.066242
  |
Correlation:0.035342
  |
Correlation:0.28159
  |
Correlation:0.28159
  |
Component:118; Jaccard:0.091483
 |
Component:250; Jaccard:0.096589
 |
Component:270; Jaccard:0.098272
 |
Component:059; Jaccard:0.094645
 |
Component:106; Jaccard:0.085892
 |
Component:171; Jaccard:0.066558
 |
Component:081; Jaccard:0.046052
 |
Correlation:0.26696
  |
Correlation:-0.01891
  |
Correlation:0.066242
  |
Correlation:0.069382
  |
Correlation:0.28159
  |
Correlation:0.035342
  |
Correlation:-0.027665
  |
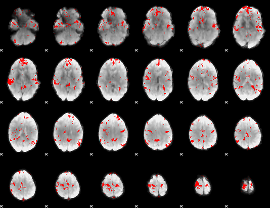
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
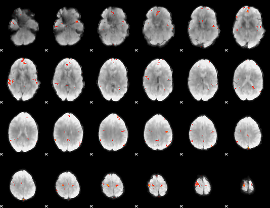
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
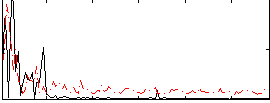
Component:175; Jaccard:0.15843
 |
Component:175; Jaccard:0.1834
 |
Component:175; Jaccard:0.19669
 |
Component:175; Jaccard:0.18963
 |
Component:175; Jaccard:0.16073
 |
Component:175; Jaccard:0.11823
 |
Component:175; Jaccard:0.073624
 |
Correlation:0.14662
  |
Correlation:0.14662
  |
Correlation:0.14662
  |
Correlation:0.14662
  |
Correlation:0.14662
  |
Correlation:0.14662
  |
Correlation:0.14662
  |
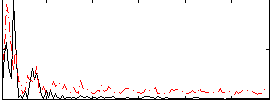
Component:171; Jaccard:0.13416
 |
Component:171; Jaccard:0.14701
 |
Component:171; Jaccard:0.15337
 |
Component:171; Jaccard:0.14106
 |
Component:171; Jaccard:0.11929
 |
Component:171; Jaccard:0.089064
 |
Component:171; Jaccard:0.059197
 |
Correlation:0.17344
  |
Correlation:0.17344
  |
Correlation:0.17344
  |
Correlation:0.17344
  |
Correlation:0.17344
  |
Correlation:0.17344
  |
Correlation:0.17344
  |
Component:139; Jaccard:0.10452
 |
Component:353; Jaccard:0.10756
 |
Component:068; Jaccard:0.10927
 |
Component:068; Jaccard:0.10706
 |
Component:068; Jaccard:0.09214
 |
Component:068; Jaccard:0.071234
 |
Component:068; Jaccard:0.051375
 |
Correlation:0.21677
  |
Correlation:0.051927
  |
Correlation:0.26468
  |
Correlation:0.26468
  |
Correlation:0.26468
  |
Correlation:0.26468
  |
Correlation:0.26468
  |
Component:353; Jaccard:0.10229
 |
Component:139; Jaccard:0.10373
 |
Component:353; Jaccard:0.10722
 |
Component:353; Jaccard:0.099612
 |
Component:353; Jaccard:0.083723
 |
Component:137; Jaccard:0.064609
 |
Component:137; Jaccard:0.044834
 |
Correlation:0.051927
  |
Correlation:0.21677
  |
Correlation:0.051927
  |
Correlation:0.051927
  |
Correlation:0.051927
  |
Correlation:0.2459
  |
Correlation:0.2459
  |
Component:218; Jaccard:0.094213
 |
Component:068; Jaccard:0.1008
 |
Component:218; Jaccard:0.096177
 |
Component:137; Jaccard:0.094313
 |
Component:137; Jaccard:0.08188
 |
Component:353; Jaccard:0.061241
 |
Component:353; Jaccard:0.040441
 |
Correlation:0.092333
  |
Correlation:0.26468
  |
Correlation:0.092333
  |
Correlation:0.2459
  |
Correlation:0.2459
  |
Correlation:0.051927
  |
Correlation:0.051927
  |
Component:044; Jaccard:0.09033
 |
Component:218; Jaccard:0.096438
 |
Component:137; Jaccard:0.095535
 |
Component:218; Jaccard:0.084945
 |
Component:024; Jaccard:0.071289
 |
Component:024; Jaccard:0.058178
 |
Component:024; Jaccard:0.038567
 |
Correlation:0.10232
  |
Correlation:0.092333
  |
Correlation:0.2459
  |
Correlation:0.092333
  |
Correlation:0.1837
  |
Correlation:0.1837
  |
Correlation:0.1837
  |
Component:068; Jaccard:0.08893
 |
Component:044; Jaccard:0.092466
 |
Component:139; Jaccard:0.095275
 |
Component:270; Jaccard:0.083659
 |
Component:270; Jaccard:0.070617
 |
Component:097; Jaccard:0.051783
 |
Component:001; Jaccard:0.036883
 |
Correlation:0.26468
  |
Correlation:0.10232
  |
Correlation:0.21677
  |
Correlation:0.15689
  |
Correlation:0.15689
  |
Correlation:0.2726
  |
Correlation:0.10157
  |
Component:160; Jaccard:0.08692
 |
Component:270; Jaccard:0.090395
 |
Component:270; Jaccard:0.089528
 |
Component:024; Jaccard:0.081733
 |
Component:218; Jaccard:0.069402
 |
Component:270; Jaccard:0.050714
 |
Component:097; Jaccard:0.036031
 |
Correlation:0.15201
  |
Correlation:0.15689
  |
Correlation:0.15689
  |
Correlation:0.1837
  |
Correlation:0.092333
  |
Correlation:0.15689
  |
Correlation:0.2726
  |
Component:270; Jaccard:0.08581
 |
Component:160; Jaccard:0.088318
 |
Component:325; Jaccard:0.087362
 |
Component:139; Jaccard:0.08134
 |
Component:325; Jaccard:0.065965
 |
Component:001; Jaccard:0.050184
 |
Component:270; Jaccard:0.034283
 |
Correlation:0.15689
  |
Correlation:0.15201
  |
Correlation:0.23349
  |
Correlation:0.21677
  |
Correlation:0.23349
  |
Correlation:0.10157
  |
Correlation:0.15689
  |
Component:325; Jaccard:0.085542
 |
Component:325; Jaccard:0.087721
 |
Component:044; Jaccard:0.08708
 |
Component:325; Jaccard:0.079777
 |
Component:097; Jaccard:0.065392
 |
Component:218; Jaccard:0.048174
 |
Component:325; Jaccard:0.031086
 |
Correlation:0.23349
  |
Correlation:0.23349
  |
Correlation:0.10232
  |
Correlation:0.23349
  |
Correlation:0.2726
  |
Correlation:0.092333
  |
Correlation:0.23349
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































