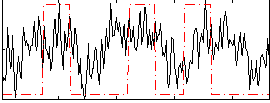
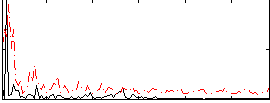
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
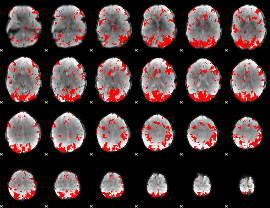 |
GLM binary result thresholded by 3
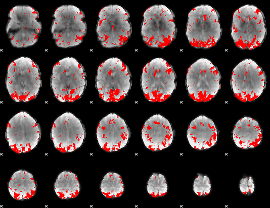 |
GLM binary result thresholded by 3.5
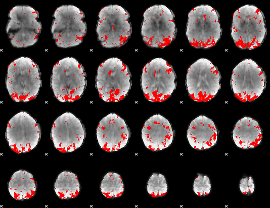 |
GLM binary result thresholded by 4
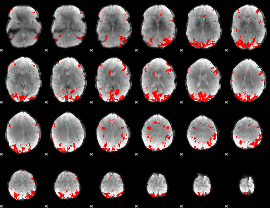 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
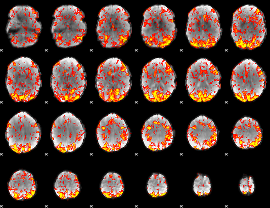 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
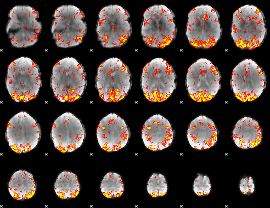 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
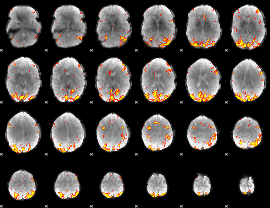 |
Component:019; Jaccard:0.12948
 |
Component:019; Jaccard:0.16634
 |
Component:019; Jaccard:0.21106
 |
Component:019; Jaccard:0.25962
 |
Component:019; Jaccard:0.30974
 |
Component:019; Jaccard:0.35437
 |
Component:019; Jaccard:0.38349
 |
Correlation:0.27844
 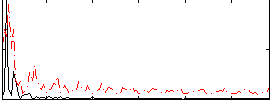 |
Correlation:0.27844
  |
Correlation:0.27844
  |
Correlation:0.27844
  |
Correlation:0.27844
  |
Correlation:0.27844
  |
Correlation:0.27844
  |
Component:162; Jaccard:0.12878
 |
Component:400; Jaccard:0.15309
 |
Component:400; Jaccard:0.18173
 |
Component:400; Jaccard:0.206
 |
Component:228; Jaccard:0.22845
 |
Component:228; Jaccard:0.25721
 |
Component:228; Jaccard:0.27734
 |
Correlation:0.21879
  |
Correlation:0.1609
  |
Correlation:0.1609
  |
Correlation:0.1609
  |
Correlation:0.1185
  |
Correlation:0.1185
  |
Correlation:0.1185
  |
Component:400; Jaccard:0.12651
 |
Component:162; Jaccard:0.14991
 |
Component:162; Jaccard:0.16966
 |
Component:228; Jaccard:0.19457
 |
Component:400; Jaccard:0.22705
 |
Component:400; Jaccard:0.23886
 |
Component:400; Jaccard:0.24024
 |
Correlation:0.1609
  |
Correlation:0.21879
  |
Correlation:0.21879
  |
Correlation:0.1185
  |
Correlation:0.1609
  |
Correlation:0.1609
  |
Correlation:0.1609
  |
Component:134; Jaccard:0.12246
 |
Component:134; Jaccard:0.14453
 |
Component:274; Jaccard:0.16605
 |
Component:274; Jaccard:0.19061
 |
Component:274; Jaccard:0.20787
 |
Component:274; Jaccard:0.21903
 |
Component:274; Jaccard:0.22134
 |
Correlation:0.22692
  |
Correlation:0.22692
  |
Correlation:0.1376
  |
Correlation:0.1376
  |
Correlation:0.1376
  |
Correlation:0.1376
  |
Correlation:0.1376
  |
Component:396; Jaccard:0.11714
 |
Component:274; Jaccard:0.13909
 |
Component:134; Jaccard:0.16562
 |
Component:162; Jaccard:0.18597
 |
Component:162; Jaccard:0.1974
 |
Component:338; Jaccard:0.20348
 |
Component:338; Jaccard:0.21036
 |
Correlation:0.068888
 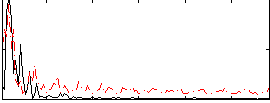 |
Correlation:0.1376
  |
Correlation:0.22692
  |
Correlation:0.21879
  |
Correlation:0.21879
  |
Correlation:0.19863
 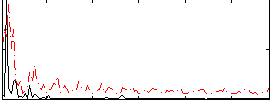 |
Correlation:0.19863
  |
Component:274; Jaccard:0.11342
 |
Component:396; Jaccard:0.13466
 |
Component:228; Jaccard:0.1616
 |
Component:134; Jaccard:0.18245
 |
Component:134; Jaccard:0.19478
 |
Component:134; Jaccard:0.20097
 |
Component:134; Jaccard:0.20016
 |
Correlation:0.1376
  |
Correlation:0.068888
  |
Correlation:0.1185
  |
Correlation:0.22692
  |
Correlation:0.22692
  |
Correlation:0.22692
  |
Correlation:0.22692
  |
Component:384; Jaccard:0.10999
 |
Component:228; Jaccard:0.13143
 |
Component:396; Jaccard:0.14978
 |
Component:338; Jaccard:0.17117
 |
Component:338; Jaccard:0.19049
 |
Component:162; Jaccard:0.19992
 |
Component:162; Jaccard:0.19563
 |
Correlation:0.052474
  |
Correlation:0.1185
  |
Correlation:0.068888
  |
Correlation:0.19863
  |
Correlation:0.19863
  |
Correlation:0.21879
  |
Correlation:0.21879
  |
Component:228; Jaccard:0.10565
 |
Component:071; Jaccard:0.12246
 |
Component:338; Jaccard:0.14651
 |
Component:071; Jaccard:0.16019
 |
Component:071; Jaccard:0.17437
 |
Component:071; Jaccard:0.18394
 |
Component:071; Jaccard:0.18384
 |
Correlation:0.1185
  |
Correlation:0.24811
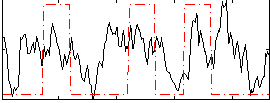 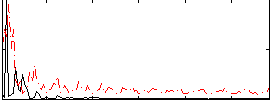 |
Correlation:0.19863
  |
Correlation:0.24811
  |
Correlation:0.24811
  |
Correlation:0.24811
  |
Correlation:0.24811
  |
Component:232; Jaccard:0.10489
 |
Component:338; Jaccard:0.12123
 |
Component:071; Jaccard:0.1423
 |
Component:396; Jaccard:0.15748
 |
Component:396; Jaccard:0.16085
 |
Component:396; Jaccard:0.15658
 |
Component:237; Jaccard:0.1588
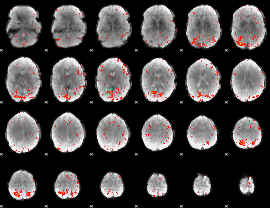 |
Correlation:0.16541
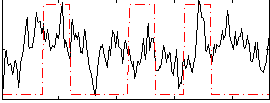 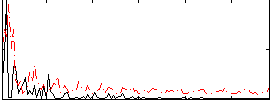 |
Correlation:0.19863
  |
Correlation:0.24811
  |
Correlation:0.068888
  |
Correlation:0.068888
  |
Correlation:0.068888
  |
Correlation:0.2683
  |
Component:071; Jaccard:0.10397
 |
Component:232; Jaccard:0.11807
 |
Component:232; Jaccard:0.12806
 |
Component:237; Jaccard:0.13708
 |
Component:237; Jaccard:0.1481
 |
Component:237; Jaccard:0.15653
 |
Component:396; Jaccard:0.14731
 |
Correlation:0.24811
  |
Correlation:0.16541
  |
Correlation:0.16541
  |
Correlation:0.2683
  |
Correlation:0.2683
  |
Correlation:0.2683
  |
Correlation:0.068888
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
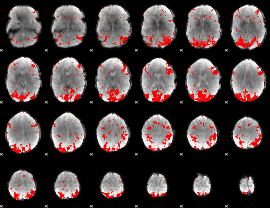 |
GLM binary result thresholded by 3
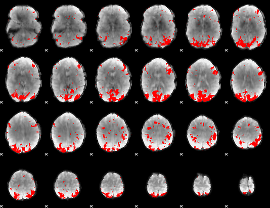 |
GLM binary result thresholded by 3.5
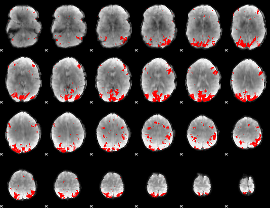 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
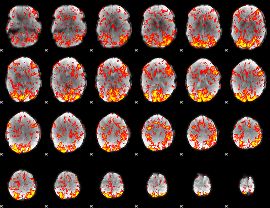 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
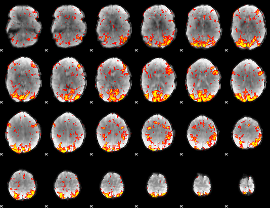 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
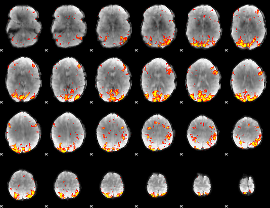 |
GLM Z-score result thresholded by 3.5
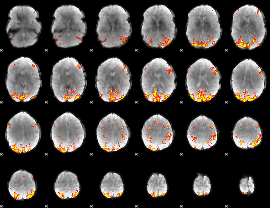 |
GLM Z-score result thresholded by 4
 |
Component:019; Jaccard:0.16015
 |
Component:019; Jaccard:0.20922
 |
Component:019; Jaccard:0.26293
 |
Component:019; Jaccard:0.31297
 |
Component:019; Jaccard:0.34697
 |
Component:019; Jaccard:0.36165
 |
Component:019; Jaccard:0.35976
 |
Correlation:0.17019
  |
Correlation:0.17019
  |
Correlation:0.17019
  |
Correlation:0.17019
  |
Correlation:0.17019
  |
Correlation:0.17019
  |
Correlation:0.17019
  |
Component:134; Jaccard:0.15016
 |
Component:228; Jaccard:0.18609
 |
Component:228; Jaccard:0.23635
 |
Component:228; Jaccard:0.28304
 |
Component:228; Jaccard:0.3178
 |
Component:228; Jaccard:0.33654
 |
Component:228; Jaccard:0.33721
 |
Correlation:0.26417
  |
Correlation:0.3442
  |
Correlation:0.3442
  |
Correlation:0.3442
  |
Correlation:0.3442
  |
Correlation:0.3442
  |
Correlation:0.3442
  |
Component:274; Jaccard:0.14527
 |
Component:274; Jaccard:0.18296
 |
Component:274; Jaccard:0.21767
 |
Component:274; Jaccard:0.23687
 |
Component:274; Jaccard:0.24512
 |
Component:274; Jaccard:0.23467
 |
Component:274; Jaccard:0.21561
 |
Correlation:0.20645
  |
Correlation:0.20645
  |
Correlation:0.20645
  |
Correlation:0.20645
  |
Correlation:0.20645
  |
Correlation:0.20645
  |
Correlation:0.20645
  |
Component:228; Jaccard:0.14111
 |
Component:134; Jaccard:0.17788
 |
Component:134; Jaccard:0.20265
 |
Component:134; Jaccard:0.21469
 |
Component:134; Jaccard:0.21976
 |
Component:338; Jaccard:0.21883
 |
Component:338; Jaccard:0.21068
 |
Correlation:0.3442
  |
Correlation:0.26417
  |
Correlation:0.26417
  |
Correlation:0.26417
  |
Correlation:0.26417
  |
Correlation:0.28563
  |
Correlation:0.28563
  |
Component:162; Jaccard:0.13997
 |
Component:400; Jaccard:0.1651
 |
Component:400; Jaccard:0.19157
 |
Component:400; Jaccard:0.20887
 |
Component:338; Jaccard:0.21941
 |
Component:134; Jaccard:0.21587
 |
Component:134; Jaccard:0.20319
 |
Correlation:0.062643
  |
Correlation:0.20958
  |
Correlation:0.20958
  |
Correlation:0.20958
  |
Correlation:0.28563
  |
Correlation:0.26417
  |
Correlation:0.26417
  |
Component:400; Jaccard:0.13882
 |
Component:162; Jaccard:0.16132
 |
Component:338; Jaccard:0.18445
 |
Component:338; Jaccard:0.20739
 |
Component:400; Jaccard:0.21339
 |
Component:400; Jaccard:0.2055
 |
Component:400; Jaccard:0.19151
 |
Correlation:0.20958
  |
Correlation:0.062643
  |
Correlation:0.28563
  |
Correlation:0.28563
  |
Correlation:0.20958
  |
Correlation:0.20958
  |
Correlation:0.20958
  |
Component:232; Jaccard:0.13527
 |
Component:232; Jaccard:0.15442
 |
Component:162; Jaccard:0.17883
 |
Component:071; Jaccard:0.18823
 |
Component:071; Jaccard:0.19465
 |
Component:071; Jaccard:0.19057
 |
Component:071; Jaccard:0.1794
 |
Correlation:0.22257
  |
Correlation:0.22257
  |
Correlation:0.062643
  |
Correlation:0.36099
  |
Correlation:0.36099
  |
Correlation:0.36099
  |
Correlation:0.36099
  |
Component:071; Jaccard:0.12509
 |
Component:338; Jaccard:0.15422
 |
Component:071; Jaccard:0.17333
 |
Component:162; Jaccard:0.18594
 |
Component:162; Jaccard:0.18348
 |
Component:162; Jaccard:0.17552
 |
Component:162; Jaccard:0.16229
 |
Correlation:0.36099
  |
Correlation:0.28563
  |
Correlation:0.36099
  |
Correlation:0.062643
  |
Correlation:0.062643
  |
Correlation:0.062643
  |
Correlation:0.062643
  |
Component:338; Jaccard:0.1227
 |
Component:071; Jaccard:0.14971
 |
Component:232; Jaccard:0.16661
 |
Component:232; Jaccard:0.16723
 |
Component:232; Jaccard:0.16157
 |
Component:237; Jaccard:0.15217
 |
Component:237; Jaccard:0.14579
 |
Correlation:0.28563
  |
Correlation:0.36099
  |
Correlation:0.22257
  |
Correlation:0.22257
  |
Correlation:0.22257
  |
Correlation:0.19392
  |
Correlation:0.19392
  |
Component:396; Jaccard:0.11046
 |
Component:396; Jaccard:0.12368
 |
Component:181; Jaccard:0.13498
 |
Component:237; Jaccard:0.14675
 |
Component:237; Jaccard:0.15261
 |
Component:232; Jaccard:0.14979
 |
Component:232; Jaccard:0.13832
 |
Correlation:0.15113
  |
Correlation:0.15113
  |
Correlation:0.20815
  |
Correlation:0.19392
  |
Correlation:0.19392
  |
Correlation:0.22257
  |
Correlation:0.22257
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
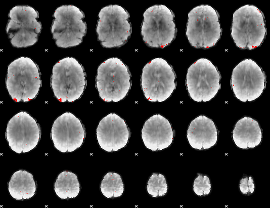 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:073; Jaccard:0.11151
 |
Component:073; Jaccard:0.14563
 |
Component:073; Jaccard:0.18115
 |
Component:073; Jaccard:0.18413
 |
Component:073; Jaccard:0.14113
 |
Component:073; Jaccard:0.087652
 |
Component:073; Jaccard:0.04636
 |
Correlation:0.3991
  |
Correlation:0.3991
  |
Correlation:0.3991
  |
Correlation:0.3991
  |
Correlation:0.3991
  |
Correlation:0.3991
  |
Correlation:0.3991
  |
Component:167; Jaccard:0.10136
 |
Component:167; Jaccard:0.11092
 |
Component:167; Jaccard:0.1075
 |
Component:375; Jaccard:0.0855
 |
Component:375; Jaccard:0.052059
 |
Component:218; Jaccard:0.027935
 |
Component:218; Jaccard:0.018247
 |
Correlation:0.10646
  |
Correlation:0.10646
  |
Correlation:0.10646
  |
Correlation:0.086224
  |
Correlation:0.086224
  |
Correlation:0.25958
  |
Correlation:0.25958
  |
Component:375; Jaccard:0.097562
 |
Component:375; Jaccard:0.10699
 |
Component:375; Jaccard:0.10487
 |
Component:167; Jaccard:0.083005
 |
Component:167; Jaccard:0.048381
 |
Component:375; Jaccard:0.026713
 |
Component:375; Jaccard:0.011596
 |
Correlation:0.086224
  |
Correlation:0.086224
  |
Correlation:0.086224
  |
Correlation:0.10646
  |
Correlation:0.10646
  |
Correlation:0.086224
  |
Correlation:0.086224
  |
Component:355; Jaccard:0.088346
 |
Component:355; Jaccard:0.092667
 |
Component:117; Jaccard:0.093454
 |
Component:117; Jaccard:0.075679
 |
Component:051; Jaccard:0.04459
 |
Component:167; Jaccard:0.025432
 |
Component:167; Jaccard:0.011197
 |
Correlation:0.11254
  |
Correlation:0.11254
  |
Correlation:0.26225
  |
Correlation:0.26225
  |
Correlation:-0.037834
  |
Correlation:0.10646
  |
Correlation:0.10646
  |
Component:396; Jaccard:0.083789
 |
Component:117; Jaccard:0.091826
 |
Component:355; Jaccard:0.084094
 |
Component:161; Jaccard:0.068897
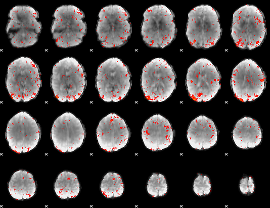 |
Component:117; Jaccard:0.044026
 |
Component:051; Jaccard:0.023612
 |
Component:384; Jaccard:0.010035
 |
Correlation:-0.048744
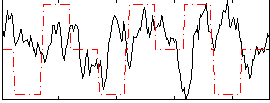  |
Correlation:0.26225
  |
Correlation:0.11254
  |
Correlation:2.6559e-05
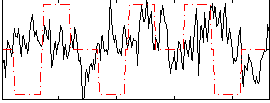 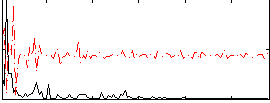 |
Correlation:0.26225
  |
Correlation:-0.037834
  |
Correlation:-0.076311
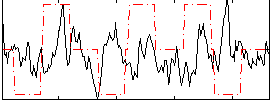  |
Component:117; Jaccard:0.080843
 |
Component:396; Jaccard:0.08878
 |
Component:400; Jaccard:0.083643
 |
Component:400; Jaccard:0.06772
 |
Component:218; Jaccard:0.043705
 |
Component:400; Jaccard:0.023135
 |
Component:051; Jaccard:0.009937
 |
Correlation:0.26225
  |
Correlation:-0.048744
  |
Correlation:-0.028852
  |
Correlation:-0.028852
  |
Correlation:0.25958
  |
Correlation:-0.028852
  |
Correlation:-0.037834
  |
Component:384; Jaccard:0.080267
 |
Component:131; Jaccard:0.087807
 |
Component:396; Jaccard:0.081513
 |
Component:051; Jaccard:0.067547
 |
Component:161; Jaccard:0.043522
 |
Component:161; Jaccard:0.022041
 |
Component:400; Jaccard:0.009894
 |
Correlation:-0.076311
  |
Correlation:0.28685
  |
Correlation:-0.048744
  |
Correlation:-0.037834
  |
Correlation:2.6559e-05
  |
Correlation:2.6559e-05
  |
Correlation:-0.028852
  |
Component:090; Jaccard:0.079163
 |
Component:400; Jaccard:0.083821
 |
Component:131; Jaccard:0.081257
 |
Component:019; Jaccard:0.064385
 |
Component:400; Jaccard:0.042519
 |
Component:019; Jaccard:0.020525
 |
Component:118; Jaccard:0.009076
 |
Correlation:0.33583
 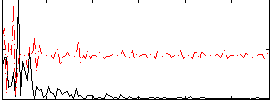 |
Correlation:-0.028852
  |
Correlation:0.28685
  |
Correlation:0.06416
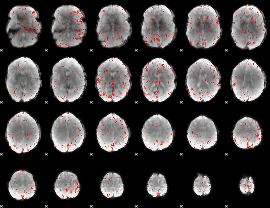
  |
Correlation:-0.028852
  |
Correlation:0.06416
  |
Correlation:0.13357
  |
Component:131; Jaccard:0.079118
 |
Component:019; Jaccard:0.080069
 |
Component:019; Jaccard:0.078508
 |
Component:355; Jaccard:0.063319
 |
Component:019; Jaccard:0.040323
 |
Component:117; Jaccard:0.019496
 |
Component:333; Jaccard:0.008794
 |
Correlation:0.28685
  |
Correlation:0.06416
  |
Correlation:0.06416
  |
Correlation:0.11254
  |
Correlation:0.06416
  |
Correlation:0.26225
  |
Correlation:-0.13049
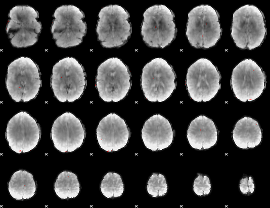
  |
Component:400; Jaccard:0.078215
 |
Component:090; Jaccard:0.079865
 |
Component:161; Jaccard:0.077916
 |
Component:396; Jaccard:0.059825
 |
Component:301; Jaccard:0.037656
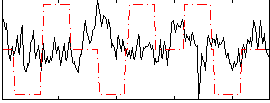
 |
Component:355; Jaccard:0.019156
 |
Component:161; Jaccard:0.008564
 |
Correlation:-0.028852
  |
Correlation:0.33583
  |
Correlation:2.6559e-05
  |
Correlation:-0.048744
  |
Correlation:0.18614
  |
Correlation:0.11254
  |
Correlation:2.6559e-05
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
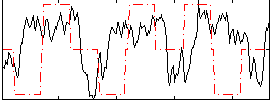
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
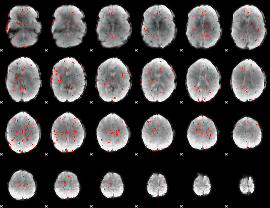 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:235; Jaccard:0.093685
 |
Component:235; Jaccard:0.10141
 |
Component:235; Jaccard:0.087205
 |
Component:235; Jaccard:0.056594
 |
Component:235; Jaccard:0.025278
 |
Component:235; Jaccard:0.007596
 |
Component:235; Jaccard:0.002753
 |
Correlation:0.30118
  |
Correlation:0.30118
  |
Correlation:0.30118
  |
Correlation:0.30118
  |
Correlation:0.30118
  |
Correlation:0.30118
  |
Correlation:0.30118
  |
Component:186; Jaccard:0.086124
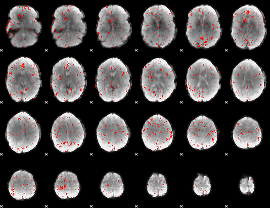 |
Component:186; Jaccard:0.082002
 |
Component:186; Jaccard:0.064793
 |
Component:186; Jaccard:0.039131
 |
Component:068; Jaccard:0.017918
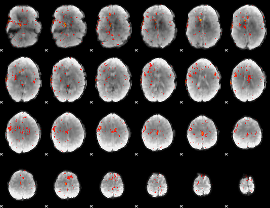 |
Component:068; Jaccard:0.005177
 |
Component:068; Jaccard:0.002316
 |
Correlation:0.44421
  |
Correlation:0.44421
  |
Correlation:0.44421
  |
Correlation:0.44421
  |
Correlation:0.24984
  |
Correlation:0.24984
  |
Correlation:0.24984
  |
Component:361; Jaccard:0.075956
 |
Component:361; Jaccard:0.068919
 |
Component:068; Jaccard:0.05297
 |
Component:068; Jaccard:0.035807
 |
Component:186; Jaccard:0.017813
 |
Component:186; Jaccard:0.004841
 |
Component:186; Jaccard:0.002009
 |
Correlation:0.20108
  |
Correlation:0.20108
  |
Correlation:0.24984
  |
Correlation:0.24984
  |
Correlation:0.44421
  |
Correlation:0.44421
  |
Correlation:0.44421
  |
Component:094; Jaccard:0.063078
 |
Component:068; Jaccard:0.058938
 |
Component:361; Jaccard:0.05025
 |
Component:361; Jaccard:0.026493
 |
Component:290; Jaccard:0.013095
 |
Component:228; Jaccard:0.00366
 |
Component:228; Jaccard:0.001689
 |
Correlation:0.25271
  |
Correlation:0.24984
  |
Correlation:0.20108
  |
Correlation:0.20108
  |
Correlation:0.24505
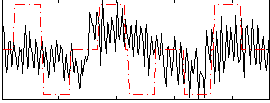  |
Correlation:0.13377
  |
Correlation:0.13377
  |
Component:300; Jaccard:0.061681
 |
Component:286; Jaccard:0.058595
 |
Component:194; Jaccard:0.04408
 |
Component:290; Jaccard:0.026451
 |
Component:194; Jaccard:0.010014
 |
Component:290; Jaccard:0.003618
 |
Component:035; Jaccard:0.001386
 |
Correlation:0.086303
 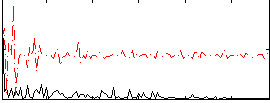 |
Correlation:0.082086
 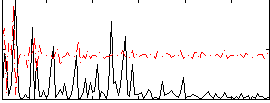 |
Correlation:0.12512
 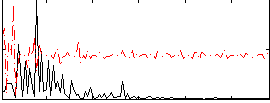 |
Correlation:0.24505
  |
Correlation:0.12512
  |
Correlation:0.24505
  |
Correlation:0.16299
 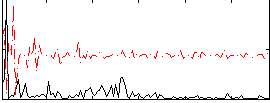 |
Component:286; Jaccard:0.061091
 |
Component:094; Jaccard:0.057004
 |
Component:286; Jaccard:0.043789
 |
Component:143; Jaccard:0.025548
 |
Component:361; Jaccard:0.009834
 |
Component:035; Jaccard:0.002987
 |
Component:173; Jaccard:0.001293
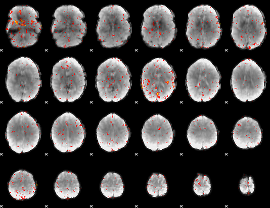 |
Correlation:0.082086
  |
Correlation:0.25271
  |
Correlation:0.082086
  |
Correlation:0.15436
  |
Correlation:0.20108
  |
Correlation:0.16299
  |
Correlation:-0.033985
  |
Component:241; Jaccard:0.060499
 |
Component:300; Jaccard:0.056502
 |
Component:143; Jaccard:0.041683
 |
Component:194; Jaccard:0.025239
 |
Component:081; Jaccard:0.009466
 |
Component:001; Jaccard:0.00292
 |
Component:290; Jaccard:0.001221
 |
Correlation:0.18789
  |
Correlation:0.086303
  |
Correlation:0.15436
  |
Correlation:0.12512
  |
Correlation:0.11299
 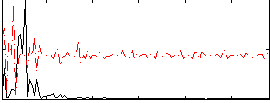 |
Correlation:0.2582
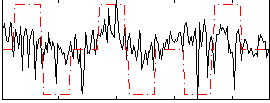 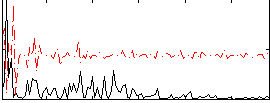 |
Correlation:0.24505
  |
Component:001; Jaccard:0.059116
 |
Component:143; Jaccard:0.053732
 |
Component:300; Jaccard:0.041374
 |
Component:370; Jaccard:0.02338
 |
Component:143; Jaccard:0.009445
 |
Component:112; Jaccard:0.002809
 |
Component:307; Jaccard:0.001161
 |
Correlation:0.2582
  |
Correlation:0.15436
  |
Correlation:0.086303
  |
Correlation:0.12638
  |
Correlation:0.15436
  |
Correlation:0.13481
  |
Correlation:0.13749
  |
Component:112; Jaccard:0.057911
 |
Component:370; Jaccard:0.053356
 |
Component:370; Jaccard:0.041099
 |
Component:081; Jaccard:0.023156
 |
Component:001; Jaccard:0.009354
 |
Component:081; Jaccard:0.002785
 |
Component:150; Jaccard:0.001136
 |
Correlation:0.13481
  |
Correlation:0.12638
  |
Correlation:0.12638
  |
Correlation:0.11299
  |
Correlation:0.2582
  |
Correlation:0.11299
  |
Correlation:0.11533
  |
Component:194; Jaccard:0.057764
 |
Component:194; Jaccard:0.053264
 |
Component:094; Jaccard:0.040333
 |
Component:286; Jaccard:0.023114
 |
Component:300; Jaccard:0.009327
 |
Component:173; Jaccard:0.002426
 |
Component:112; Jaccard:0.001022
 |
Correlation:0.12512
  |
Correlation:0.12512
  |
Correlation:0.25271
  |
Correlation:0.082086
  |
Correlation:0.086303
  |
Correlation:-0.033985
  |
Correlation:0.13481
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
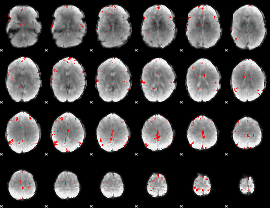 |
GLM binary result thresholded by 4
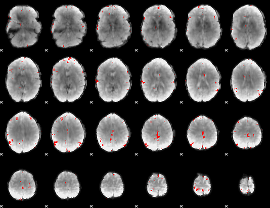 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
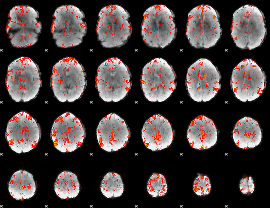 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
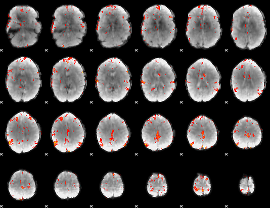 |
GLM Z-score result thresholded by 3.5
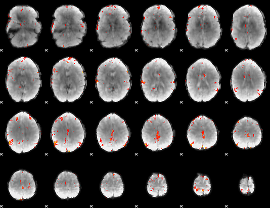 |
GLM Z-score result thresholded by 4
 |
Component:041; Jaccard:0.11543
 |
Component:041; Jaccard:0.13549
 |
Component:256; Jaccard:0.15882
 |
Component:256; Jaccard:0.17317
 |
Component:256; Jaccard:0.16595
 |
Component:256; Jaccard:0.13917
 |
Component:256; Jaccard:0.097778
 |
Correlation:0.0085581
  |
Correlation:0.0085581
  |
Correlation:0.21865
  |
Correlation:0.21865
  |
Correlation:0.21865
  |
Correlation:0.21865
  |
Correlation:0.21865
  |
Component:256; Jaccard:0.11145
 |
Component:256; Jaccard:0.13509
 |
Component:041; Jaccard:0.15455
 |
Component:041; Jaccard:0.1601
 |
Component:041; Jaccard:0.14758
 |
Component:041; Jaccard:0.11835
 |
Component:041; Jaccard:0.077367
 |
Correlation:0.21865
  |
Correlation:0.21865
  |
Correlation:0.0085581
  |
Correlation:0.0085581
  |
Correlation:0.0085581
  |
Correlation:0.0085581
  |
Correlation:0.0085581
  |
Component:368; Jaccard:0.10919
 |
Component:368; Jaccard:0.11911
 |
Component:368; Jaccard:0.12069
 |
Component:201; Jaccard:0.12037
 |
Component:201; Jaccard:0.11223
 |
Component:201; Jaccard:0.0839
 |
Component:331; Jaccard:0.058096
 |
Correlation:0.13441
  |
Correlation:0.13441
  |
Correlation:0.13441
  |
Correlation:0.056022
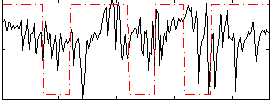  |
Correlation:0.056022
  |
Correlation:0.056022
  |
Correlation:0.22222
  |
Component:243; Jaccard:0.093302
 |
Component:243; Jaccard:0.10327
 |
Component:201; Jaccard:0.11205
 |
Component:368; Jaccard:0.1112
 |
Component:331; Jaccard:0.099855
 |
Component:331; Jaccard:0.081171
 |
Component:139; Jaccard:0.052449
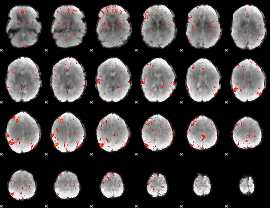 |
Correlation:0.14816
  |
Correlation:0.14816
  |
Correlation:0.056022
  |
Correlation:0.13441
  |
Correlation:0.22222
  |
Correlation:0.22222
  |
Correlation:0.21908
 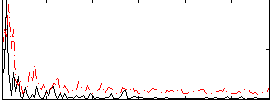 |
Component:148; Jaccard:0.092711
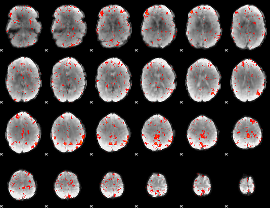 |
Component:283; Jaccard:0.098971
 |
Component:243; Jaccard:0.1082
 |
Component:183; Jaccard:0.10886
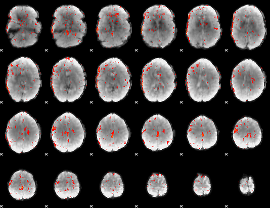 |
Component:183; Jaccard:0.098986
 |
Component:183; Jaccard:0.076028
 |
Component:201; Jaccard:0.049567
 |
Correlation:0.19897
 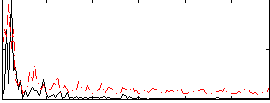 |
Correlation:0.29131
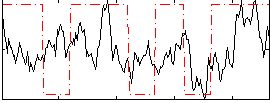  |
Correlation:0.14816
  |
Correlation:0.045439
  |
Correlation:0.045439
  |
Correlation:0.045439
  |
Correlation:0.056022
  |
Component:283; Jaccard:0.089504
 |
Component:148; Jaccard:0.096764
 |
Component:183; Jaccard:0.10662
 |
Component:331; Jaccard:0.10626
 |
Component:243; Jaccard:0.092728
 |
Component:139; Jaccard:0.074186
 |
Component:183; Jaccard:0.049228
 |
Correlation:0.29131
  |
Correlation:0.19897
  |
Correlation:0.045439
  |
Correlation:0.22222
  |
Correlation:0.14816
  |
Correlation:0.21908
  |
Correlation:0.045439
  |
Component:189; Jaccard:0.088752
 |
Component:201; Jaccard:0.096457
 |
Component:283; Jaccard:0.10514
 |
Component:243; Jaccard:0.10587
 |
Component:139; Jaccard:0.09077
 |
Component:235; Jaccard:0.07037
 |
Component:333; Jaccard:0.046764
 |
Correlation:0.084373
 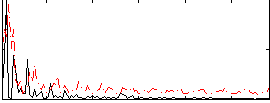 |
Correlation:0.056022
  |
Correlation:0.29131
  |
Correlation:0.14816
  |
Correlation:0.21908
  |
Correlation:0.25033
  |
Correlation:0.22179
  |
Component:340; Jaccard:0.084573
 |
Component:183; Jaccard:0.09634
 |
Component:331; Jaccard:0.10181
 |
Component:283; Jaccard:0.10109
 |
Component:235; Jaccard:0.090572
 |
Component:243; Jaccard:0.07013
 |
Component:235; Jaccard:0.046003
 |
Correlation:0.15978
  |
Correlation:0.045439
  |
Correlation:0.22222
  |
Correlation:0.29131
  |
Correlation:0.25033
  |
Correlation:0.14816
  |
Correlation:0.25033
  |
Component:183; Jaccard:0.084463
 |
Component:189; Jaccard:0.095911
 |
Component:235; Jaccard:0.09942
 |
Component:139; Jaccard:0.10017
 |
Component:368; Jaccard:0.090189
 |
Component:283; Jaccard:0.06876
 |
Component:283; Jaccard:0.04386
 |
Correlation:0.045439
  |
Correlation:0.084373
  |
Correlation:0.25033
  |
Correlation:0.21908
  |
Correlation:0.13441
  |
Correlation:0.29131
  |
Correlation:0.29131
  |
Component:222; Jaccard:0.081106
 |
Component:331; Jaccard:0.092347
 |
Component:189; Jaccard:0.097818
 |
Component:235; Jaccard:0.099616
 |
Component:283; Jaccard:0.088561
 |
Component:368; Jaccard:0.065202
 |
Component:368; Jaccard:0.042797
 |
Correlation:0.044752
  |
Correlation:0.22222
  |
Correlation:0.084373
  |
Correlation:0.25033
  |
Correlation:0.29131
  |
Correlation:0.13441
  |
Correlation:0.13441
  |
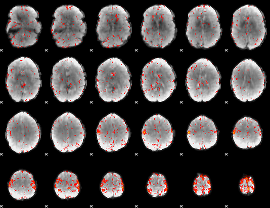
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
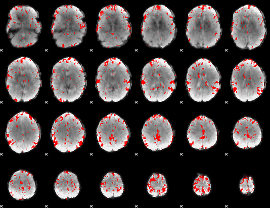 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
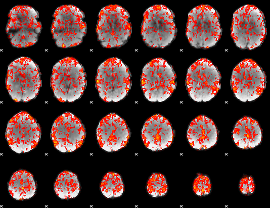 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:148; Jaccard:0.1206
 |
Component:148; Jaccard:0.13907
 |
Component:041; Jaccard:0.16242
 |
Component:041; Jaccard:0.18149
 |
Component:041; Jaccard:0.17884
 |
Component:041; Jaccard:0.14787
 |
Component:041; Jaccard:0.10008
 |
Correlation:0.10999
  |
Correlation:0.10999
  |
Correlation:0.24015
  |
Correlation:0.24015
  |
Correlation:0.24015
  |
Correlation:0.24015
  |
Correlation:0.24015
  |
Component:041; Jaccard:0.10678
 |
Component:041; Jaccard:0.13324
 |
Component:148; Jaccard:0.15198
 |
Component:148; Jaccard:0.15288
 |
Component:256; Jaccard:0.14434
 |
Component:256; Jaccard:0.11799
 |
Component:256; Jaccard:0.081889
 |
Correlation:0.24015
  |
Correlation:0.24015
  |
Correlation:0.10999
  |
Correlation:0.10999
  |
Correlation:0.075945
  |
Correlation:0.075945
  |
Correlation:0.075945
  |
Component:368; Jaccard:0.10364
 |
Component:368; Jaccard:0.11985
 |
Component:256; Jaccard:0.13439
 |
Component:256; Jaccard:0.14703
 |
Component:148; Jaccard:0.136
 |
Component:148; Jaccard:0.10747
 |
Component:148; Jaccard:0.072593
 |
Correlation:0.12386
  |
Correlation:0.12386
  |
Correlation:0.075945
  |
Correlation:0.075945
  |
Correlation:0.10999
  |
Correlation:0.10999
  |
Correlation:0.10999
  |
Component:147; Jaccard:0.10328
 |
Component:147; Jaccard:0.11834
 |
Component:147; Jaccard:0.12888
 |
Component:189; Jaccard:0.13133
 |
Component:189; Jaccard:0.12085
 |
Component:189; Jaccard:0.097035
 |
Component:189; Jaccard:0.068621
 |
Correlation:0.25742
  |
Correlation:0.25742
  |
Correlation:0.25742
  |
Correlation:0.14913
  |
Correlation:0.14913
  |
Correlation:0.14913
  |
Correlation:0.14913
  |
Component:340; Jaccard:0.10272
 |
Component:189; Jaccard:0.11375
 |
Component:368; Jaccard:0.12782
 |
Component:147; Jaccard:0.12781
 |
Component:147; Jaccard:0.11227
 |
Component:333; Jaccard:0.091559
 |
Component:333; Jaccard:0.064985
 |
Correlation:-0.078044
  |
Correlation:0.14913
  |
Correlation:0.12386
  |
Correlation:0.25742
  |
Correlation:0.25742
  |
Correlation:0.001627
  |
Correlation:0.001627
  |
Component:189; Jaccard:0.098593
 |
Component:256; Jaccard:0.11334
 |
Component:189; Jaccard:0.1267
 |
Component:368; Jaccard:0.12205
 |
Component:333; Jaccard:0.10825
 |
Component:331; Jaccard:0.086809
 |
Component:331; Jaccard:0.063568
 |
Correlation:0.14913
  |
Correlation:0.075945
  |
Correlation:0.14913
  |
Correlation:0.12386
  |
Correlation:0.001627
  |
Correlation:0.29678
  |
Correlation:0.29678
  |
Component:185; Jaccard:0.0942
 |
Component:340; Jaccard:0.11178
 |
Component:340; Jaccard:0.11594
 |
Component:243; Jaccard:0.11427
 |
Component:331; Jaccard:0.10487
 |
Component:147; Jaccard:0.085172
 |
Component:139; Jaccard:0.057333
 |
Correlation:0.087848
  |
Correlation:-0.078044
  |
Correlation:-0.078044
  |
Correlation:0.069279
  |
Correlation:0.29678
  |
Correlation:0.25742
  |
Correlation:0.16283
  |
Component:256; Jaccard:0.094185
 |
Component:243; Jaccard:0.10433
 |
Component:243; Jaccard:0.1145
 |
Component:333; Jaccard:0.11058
 |
Component:368; Jaccard:0.10257
 |
Component:139; Jaccard:0.077996
 |
Component:147; Jaccard:0.055786
 |
Correlation:0.075945
  |
Correlation:0.069279
  |
Correlation:0.069279
  |
Correlation:0.001627
  |
Correlation:0.12386
  |
Correlation:0.16283
  |
Correlation:0.25742
  |
Component:243; Jaccard:0.091664
 |
Component:185; Jaccard:0.09737
 |
Component:333; Jaccard:0.10535
 |
Component:340; Jaccard:0.10965
 |
Component:243; Jaccard:0.099655
 |
Component:243; Jaccard:0.073657
 |
Component:243; Jaccard:0.048798
 |
Correlation:0.069279
  |
Correlation:0.087848
  |
Correlation:0.001627
  |
Correlation:-0.078044
  |
Correlation:0.069279
  |
Correlation:0.069279
  |
Correlation:0.069279
  |
Component:090; Jaccard:0.089754
 |
Component:222; Jaccard:0.09709
 |
Component:222; Jaccard:0.10135
 |
Component:331; Jaccard:0.10795
 |
Component:139; Jaccard:0.094358
 |
Component:368; Jaccard:0.072233
 |
Component:368; Jaccard:0.048404
 |
Correlation:0.35068
  |
Correlation:-0.10444
  |
Correlation:-0.10444
  |
Correlation:0.29678
  |
Correlation:0.16283
  |
Correlation:0.12386
  |
Correlation:0.12386
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































