Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
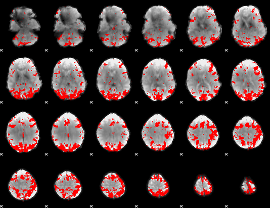 |
GLM binary result thresholded by 4
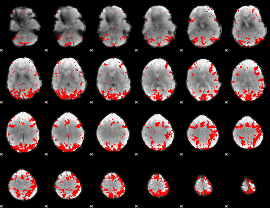 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
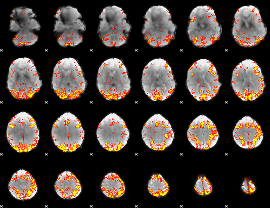 |
GLM Z-score result thresholded by 4
 |
Component:343; Jaccard:0.12785
 |
Component:343; Jaccard:0.15214
 |
Component:343; Jaccard:0.18054
 |
Component:343; Jaccard:0.21226
 |
Component:343; Jaccard:0.24481
 |
Component:343; Jaccard:0.27503
 |
Component:343; Jaccard:0.30184
 |
Correlation:0.2897
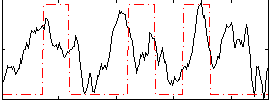  |
Correlation:0.2897
  |
Correlation:0.2897
  |
Correlation:0.2897
  |
Correlation:0.2897
  |
Correlation:0.2897
  |
Correlation:0.2897
  |
Component:339; Jaccard:0.11981
 |
Component:339; Jaccard:0.14233
 |
Component:339; Jaccard:0.16832
 |
Component:339; Jaccard:0.19788
 |
Component:104; Jaccard:0.22919
 |
Component:104; Jaccard:0.25952
 |
Component:104; Jaccard:0.28393
 |
Correlation:0.21636
  |
Correlation:0.21636
  |
Correlation:0.21636
  |
Correlation:0.21636
  |
Correlation:0.23482
  |
Correlation:0.23482
  |
Correlation:0.23482
  |
Component:329; Jaccard:0.11699
 |
Component:329; Jaccard:0.13862
 |
Component:104; Jaccard:0.16527
 |
Component:104; Jaccard:0.19681
 |
Component:339; Jaccard:0.22726
 |
Component:339; Jaccard:0.25283
 |
Component:339; Jaccard:0.2738
 |
Correlation:0.2787
  |
Correlation:0.2787
  |
Correlation:0.23482
  |
Correlation:0.23482
  |
Correlation:0.21636
  |
Correlation:0.21636
  |
Correlation:0.21636
  |
Component:301; Jaccard:0.11579
 |
Component:104; Jaccard:0.13755
 |
Component:329; Jaccard:0.16389
 |
Component:329; Jaccard:0.19202
 |
Component:329; Jaccard:0.22037
 |
Component:329; Jaccard:0.24452
 |
Component:329; Jaccard:0.2624
 |
Correlation:0.23445
  |
Correlation:0.23482
  |
Correlation:0.2787
  |
Correlation:0.2787
  |
Correlation:0.2787
  |
Correlation:0.2787
  |
Correlation:0.2787
  |
Component:104; Jaccard:0.11429
 |
Component:301; Jaccard:0.13626
 |
Component:301; Jaccard:0.15983
 |
Component:301; Jaccard:0.18324
 |
Component:301; Jaccard:0.20471
 |
Component:301; Jaccard:0.21936
 |
Component:301; Jaccard:0.22849
 |
Correlation:0.23482
  |
Correlation:0.23445
  |
Correlation:0.23445
  |
Correlation:0.23445
  |
Correlation:0.23445
  |
Correlation:0.23445
  |
Correlation:0.23445
  |
Component:241; Jaccard:0.1056
 |
Component:241; Jaccard:0.12279
 |
Component:241; Jaccard:0.14239
 |
Component:241; Jaccard:0.16168
 |
Component:241; Jaccard:0.18036
 |
Component:241; Jaccard:0.19437
 |
Component:328; Jaccard:0.20673
 |
Correlation:0.3088
  |
Correlation:0.3088
  |
Correlation:0.3088
  |
Correlation:0.3088
  |
Correlation:0.3088
  |
Correlation:0.3088
  |
Correlation:0.19849
  |
Component:384; Jaccard:0.10479
 |
Component:328; Jaccard:0.11843
 |
Component:328; Jaccard:0.13849
 |
Component:328; Jaccard:0.15889
 |
Component:328; Jaccard:0.17772
 |
Component:328; Jaccard:0.19419
 |
Component:241; Jaccard:0.20297
 |
Correlation:0.13531
  |
Correlation:0.19849
  |
Correlation:0.19849
  |
Correlation:0.19849
  |
Correlation:0.19849
  |
Correlation:0.19849
  |
Correlation:0.3088
  |
Component:326; Jaccard:0.10316
 |
Component:384; Jaccard:0.1183
 |
Component:384; Jaccard:0.13262
 |
Component:384; Jaccard:0.14522
 |
Component:384; Jaccard:0.15509
 |
Component:384; Jaccard:0.16133
 |
Component:300; Jaccard:0.15882
 |
Correlation:0.11405
  |
Correlation:0.13531
  |
Correlation:0.13531
  |
Correlation:0.13531
  |
Correlation:0.13531
  |
Correlation:0.13531
  |
Correlation:0.24664
  |
Component:259; Jaccard:0.10124
 |
Component:326; Jaccard:0.11635
 |
Component:259; Jaccard:0.12861
 |
Component:259; Jaccard:0.14019
 |
Component:326; Jaccard:0.14875
 |
Component:326; Jaccard:0.15552
 |
Component:384; Jaccard:0.15785
 |
Correlation:0.27881
  |
Correlation:0.11405
  |
Correlation:0.27881
  |
Correlation:0.27881
  |
Correlation:0.11405
  |
Correlation:0.11405
  |
Correlation:0.13531
  |
Component:328; Jaccard:0.10086
 |
Component:259; Jaccard:0.11562
 |
Component:326; Jaccard:0.12847
 |
Component:326; Jaccard:0.1386
 |
Component:300; Jaccard:0.14835
 |
Component:300; Jaccard:0.15487
 |
Component:326; Jaccard:0.15655
 |
Correlation:0.19849
  |
Correlation:0.27881
  |
Correlation:0.11405
  |
Correlation:0.11405
  |
Correlation:0.24664
  |
Correlation:0.24664
  |
Correlation:0.11405
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
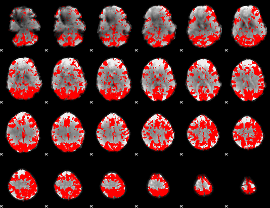 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
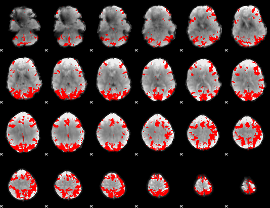 |
GLM Z-score result thresholded by 1
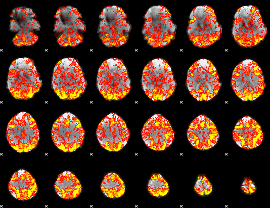 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
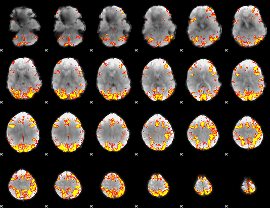 |
Component:343; Jaccard:0.12234
 |
Component:343; Jaccard:0.14502
 |
Component:343; Jaccard:0.17093
 |
Component:343; Jaccard:0.19938
 |
Component:343; Jaccard:0.22941
 |
Component:343; Jaccard:0.25871
 |
Component:343; Jaccard:0.28706
 |
Correlation:0.25936
  |
Correlation:0.25936
  |
Correlation:0.25936
  |
Correlation:0.25936
  |
Correlation:0.25936
  |
Correlation:0.25936
  |
Correlation:0.25936
  |
Component:339; Jaccard:0.11393
 |
Component:339; Jaccard:0.13412
 |
Component:339; Jaccard:0.15708
 |
Component:329; Jaccard:0.18307
 |
Component:329; Jaccard:0.21128
 |
Component:329; Jaccard:0.23701
 |
Component:329; Jaccard:0.25942
 |
Correlation:0.16841
  |
Correlation:0.16841
  |
Correlation:0.16841
  |
Correlation:0.10052
  |
Correlation:0.10052
  |
Correlation:0.10052
  |
Correlation:0.10052
  |
Component:329; Jaccard:0.11158
 |
Component:329; Jaccard:0.13216
 |
Component:329; Jaccard:0.15638
 |
Component:339; Jaccard:0.1817
 |
Component:339; Jaccard:0.20608
 |
Component:339; Jaccard:0.22898
 |
Component:104; Jaccard:0.25168
 |
Correlation:0.10052
  |
Correlation:0.10052
  |
Correlation:0.10052
  |
Correlation:0.16841
  |
Correlation:0.16841
  |
Correlation:0.16841
  |
Correlation:0.27159
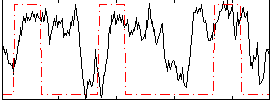 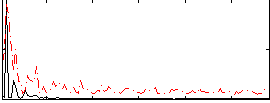 |
Component:301; Jaccard:0.111
 |
Component:301; Jaccard:0.13068
 |
Component:301; Jaccard:0.15282
 |
Component:301; Jaccard:0.1766
 |
Component:104; Jaccard:0.20181
 |
Component:104; Jaccard:0.22727
 |
Component:339; Jaccard:0.25073
 |
Correlation:0.14816
  |
Correlation:0.14816
  |
Correlation:0.14816
  |
Correlation:0.14816
  |
Correlation:0.27159
  |
Correlation:0.27159
  |
Correlation:0.16841
  |
Component:104; Jaccard:0.10739
 |
Component:104; Jaccard:0.12722
 |
Component:104; Jaccard:0.14976
 |
Component:104; Jaccard:0.17539
 |
Component:301; Jaccard:0.19865
 |
Component:301; Jaccard:0.21913
 |
Component:301; Jaccard:0.23456
 |
Correlation:0.27159
  |
Correlation:0.27159
  |
Correlation:0.27159
  |
Correlation:0.27159
  |
Correlation:0.14816
  |
Correlation:0.14816
  |
Correlation:0.14816
  |
Component:326; Jaccard:0.10536
 |
Component:326; Jaccard:0.11978
 |
Component:241; Jaccard:0.13526
 |
Component:328; Jaccard:0.1537
 |
Component:328; Jaccard:0.17373
 |
Component:328; Jaccard:0.19143
 |
Component:328; Jaccard:0.20744
 |
Correlation:0.31042
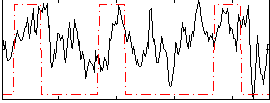  |
Correlation:0.31042
  |
Correlation:0.048995
 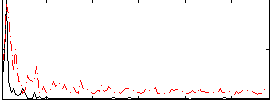 |
Correlation:0.33311
 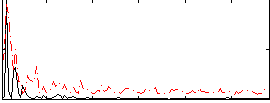 |
Correlation:0.33311
  |
Correlation:0.33311
  |
Correlation:0.33311
  |
Component:384; Jaccard:0.10466
 |
Component:384; Jaccard:0.1186
 |
Component:326; Jaccard:0.13505
 |
Component:241; Jaccard:0.15364
 |
Component:241; Jaccard:0.16961
 |
Component:241; Jaccard:0.18402
 |
Component:241; Jaccard:0.19342
 |
Correlation:0.14136
  |
Correlation:0.14136
  |
Correlation:0.31042
  |
Correlation:0.048995
  |
Correlation:0.048995
  |
Correlation:0.048995
  |
Correlation:0.048995
  |
Component:241; Jaccard:0.10127
 |
Component:241; Jaccard:0.11769
 |
Component:384; Jaccard:0.13371
 |
Component:326; Jaccard:0.1501
 |
Component:326; Jaccard:0.16228
 |
Component:326; Jaccard:0.17122
 |
Component:326; Jaccard:0.17652
 |
Correlation:0.048995
  |
Correlation:0.048995
  |
Correlation:0.14136
  |
Correlation:0.31042
  |
Correlation:0.31042
  |
Correlation:0.31042
  |
Correlation:0.31042
  |
Component:328; Jaccard:0.097732
 |
Component:328; Jaccard:0.11436
 |
Component:328; Jaccard:0.13334
 |
Component:384; Jaccard:0.14843
 |
Component:384; Jaccard:0.16091
 |
Component:384; Jaccard:0.16966
 |
Component:384; Jaccard:0.17499
 |
Correlation:0.33311
  |
Correlation:0.33311
  |
Correlation:0.33311
  |
Correlation:0.14136
  |
Correlation:0.14136
  |
Correlation:0.14136
  |
Correlation:0.14136
  |
Component:182; Jaccard:0.097404
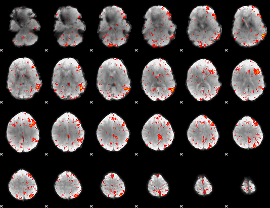 |
Component:300; Jaccard:0.11115
 |
Component:300; Jaccard:0.12663
 |
Component:300; Jaccard:0.14143
 |
Component:300; Jaccard:0.15444
 |
Component:300; Jaccard:0.1637
 |
Component:300; Jaccard:0.17011
 |
Correlation:0.18495
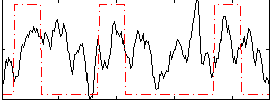  |
Correlation:0.18291
  |
Correlation:0.18291
  |
Correlation:0.18291
  |
Correlation:0.18291
  |
Correlation:0.18291
  |
Correlation:0.18291
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
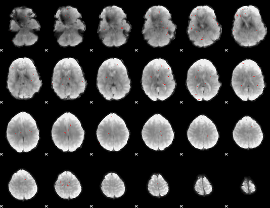 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
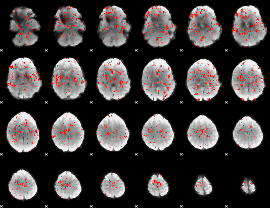 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:316; Jaccard:0.090397
 |
Component:316; Jaccard:0.087329
 |
Component:316; Jaccard:0.063526
 |
Component:028; Jaccard:0.031381
 |
Component:028; Jaccard:0.01252
 |
Component:148; Jaccard:0.004891
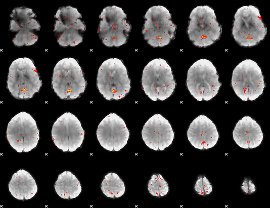 |
Component:148; Jaccard:0.001859
 |
Correlation:0.084812
  |
Correlation:0.084812
  |
Correlation:0.084812
  |
Correlation:0.14963
 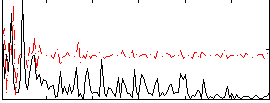 |
Correlation:0.14963
  |
Correlation:0.00062891
  |
Correlation:0.00062891
  |
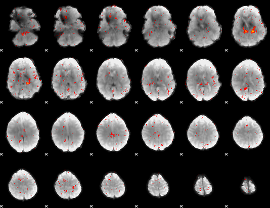
Component:138; Jaccard:0.075837
 |
Component:028; Jaccard:0.072109
 |
Component:028; Jaccard:0.057692
 |
Component:316; Jaccard:0.03092
 |
Component:148; Jaccard:0.01073
 |
Component:293; Jaccard:0.003077
 |
Component:216; Jaccard:0.001563
 |
Correlation:0.21745
  |
Correlation:0.14963
  |
Correlation:0.14963
  |
Correlation:0.084812
  |
Correlation:0.00062891
  |
Correlation:0.19911
  |
Correlation:-0.12182
  |
Component:028; Jaccard:0.075277
 |
Component:242; Jaccard:0.068066
 |
Component:293; Jaccard:0.051503
 |
Component:293; Jaccard:0.026902
 |
Component:316; Jaccard:0.009337
 |
Component:028; Jaccard:0.00259
 |
Component:237; Jaccard:0.000948
 |
Correlation:0.14963
  |
Correlation:0.19473
 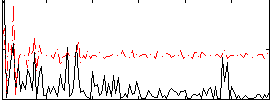 |
Correlation:0.19911
  |
Correlation:0.19911
  |
Correlation:0.084812
  |
Correlation:0.14963
  |
Correlation:0.15432
 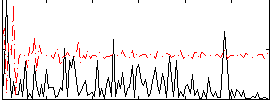 |
Component:242; Jaccard:0.074086
 |
Component:138; Jaccard:0.065489
 |
Component:242; Jaccard:0.047552
 |
Component:148; Jaccard:0.024505
 |
Component:293; Jaccard:0.009266
 |
Component:316; Jaccard:0.00238
 |
Component:293; Jaccard:0.000888
 |
Correlation:0.19473
  |
Correlation:0.21745
  |
Correlation:0.19473
  |
Correlation:0.00062891
  |
Correlation:0.19911
  |
Correlation:0.084812
  |
Correlation:0.19911
  |
Component:285; Jaccard:0.068958
 |
Component:293; Jaccard:0.064153
 |
Component:138; Jaccard:0.043654
 |
Component:242; Jaccard:0.024318
 |
Component:242; Jaccard:0.007927
 |
Component:138; Jaccard:0.002228
 |
Component:242; Jaccard:0.000879
 |
Correlation:0.20353
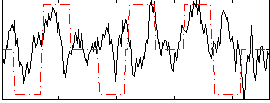  |
Correlation:0.19911
  |
Correlation:0.21745
  |
Correlation:0.19473
  |
Correlation:0.19473
  |
Correlation:0.21745
  |
Correlation:0.19473
  |
Component:218; Jaccard:0.068763
 |
Component:285; Jaccard:0.062442
 |
Component:148; Jaccard:0.043424
 |
Component:288; Jaccard:0.020695
 |
Component:173; Jaccard:0.007639
 |
Component:237; Jaccard:0.001986
 |
Component:341; Jaccard:0.000657
 |
Correlation:0.037066
 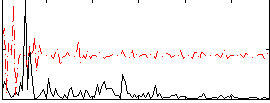 |
Correlation:0.20353
  |
Correlation:0.00062891
  |
Correlation:0.085358
  |
Correlation:0.18357
  |
Correlation:0.15432
  |
Correlation:0.066317
  |
Component:293; Jaccard:0.066897
 |
Component:218; Jaccard:0.060668
 |
Component:218; Jaccard:0.041677
 |
Component:007; Jaccard:0.020599
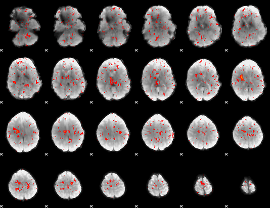 |
Component:138; Jaccard:0.007512
 |
Component:015; Jaccard:0.001977
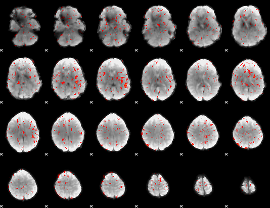 |
Component:138; Jaccard:0.000641
 |
Correlation:0.19911
  |
Correlation:0.037066
  |
Correlation:0.037066
  |
Correlation:0.18413
  |
Correlation:0.21745
  |
Correlation:-0.01959
  |
Correlation:0.21745
  |
Component:157; Jaccard:0.066598
 |
Component:005; Jaccard:0.059161
 |
Component:285; Jaccard:0.041099
 |
Component:005; Jaccard:0.020469
 |
Component:288; Jaccard:0.007005
 |
Component:346; Jaccard:0.001944
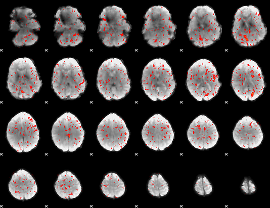 |
Component:306; Jaccard:0.000635
 |
Correlation:0.18485
 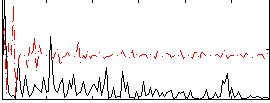 |
Correlation:0.1115
  |
Correlation:0.20353
  |
Correlation:0.1115
  |
Correlation:0.085358
  |
Correlation:0.20423
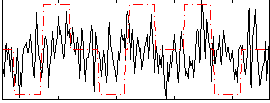 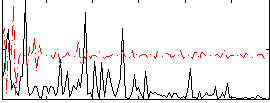 |
Correlation:0.016433
  |
Component:209; Jaccard:0.065947
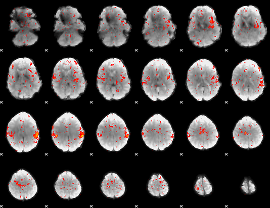 |
Component:007; Jaccard:0.057655
 |
Component:005; Jaccard:0.040128
 |
Component:138; Jaccard:0.019582
 |
Component:099; Jaccard:0.006853
 |
Component:173; Jaccard:0.001918
 |
Component:127; Jaccard:0.000632
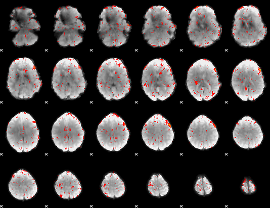 |
Correlation:-0.025722
  |
Correlation:0.18413
  |
Correlation:0.1115
  |
Correlation:0.21745
  |
Correlation:0.099479
  |
Correlation:0.18357
  |
Correlation:0.038029
 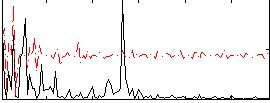 |
Component:005; Jaccard:0.065374
 |
Component:157; Jaccard:0.056773
 |
Component:007; Jaccard:0.040045
 |
Component:173; Jaccard:0.01947
 |
Component:218; Jaccard:0.005939
 |
Component:400; Jaccard:0.001901
 |
Component:239; Jaccard:0.000615
 |
Correlation:0.1115
  |
Correlation:0.18485
  |
Correlation:0.18413
  |
Correlation:0.18357
  |
Correlation:0.037066
  |
Correlation:0.19212
  |
Correlation:0.11297
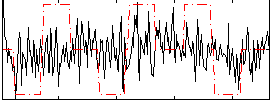 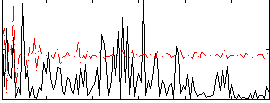 |
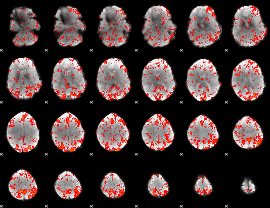
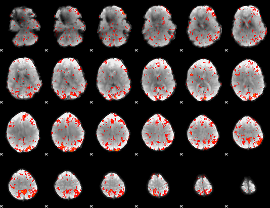
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
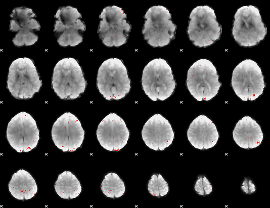 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
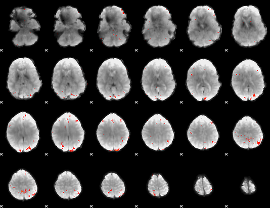 |
GLM Z-score result thresholded by 4
 |
Component:343; Jaccard:0.13455
 |
Component:343; Jaccard:0.15674
 |
Component:343; Jaccard:0.1589
 |
Component:343; Jaccard:0.12687
 |
Component:118; Jaccard:0.084335
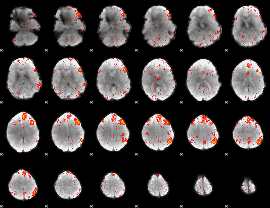 |
Component:343; Jaccard:0.04065
 |
Component:343; Jaccard:0.020387
 |
Correlation:-0.017983
  |
Correlation:-0.017983
  |
Correlation:-0.017983
  |
Correlation:-0.017983
  |
Correlation:0.055955
  |
Correlation:-0.017983
  |
Correlation:-0.017983
  |
Component:118; Jaccard:0.1138
 |
Component:118; Jaccard:0.13439
 |
Component:118; Jaccard:0.14351
 |
Component:118; Jaccard:0.12437
 |
Component:343; Jaccard:0.078327
 |
Component:118; Jaccard:0.039917
 |
Component:328; Jaccard:0.019404
 |
Correlation:0.055955
  |
Correlation:0.055955
  |
Correlation:0.055955
  |
Correlation:0.055955
  |
Correlation:-0.017983
  |
Correlation:0.055955
  |
Correlation:0.079792
  |
Component:329; Jaccard:0.11337
 |
Component:326; Jaccard:0.12569
 |
Component:326; Jaccard:0.12481
 |
Component:326; Jaccard:0.10204
 |
Component:328; Jaccard:0.067395
 |
Component:328; Jaccard:0.038904
 |
Component:118; Jaccard:0.015643
 |
Correlation:-0.10561
  |
Correlation:0.11639
  |
Correlation:0.11639
  |
Correlation:0.11639
  |
Correlation:0.079792
  |
Correlation:0.079792
  |
Correlation:0.055955
  |
Component:326; Jaccard:0.11203
 |
Component:329; Jaccard:0.12305
 |
Component:328; Jaccard:0.11796
 |
Component:328; Jaccard:0.099418
 |
Component:326; Jaccard:0.065877
 |
Component:326; Jaccard:0.032332
 |
Component:326; Jaccard:0.013229
 |
Correlation:0.11639
  |
Correlation:-0.10561
  |
Correlation:0.079792
  |
Correlation:0.079792
  |
Correlation:0.11639
  |
Correlation:0.11639
  |
Correlation:0.11639
  |
Component:301; Jaccard:0.1098
 |
Component:301; Jaccard:0.11801
 |
Component:329; Jaccard:0.11741
 |
Component:329; Jaccard:0.091872
 |
Component:355; Jaccard:0.056739
 |
Component:393; Jaccard:0.026302
 |
Component:393; Jaccard:0.010941
 |
Correlation:-0.051146
  |
Correlation:-0.051146
  |
Correlation:-0.10561
  |
Correlation:-0.10561
  |
Correlation:0.044908
  |
Correlation:-0.05025
  |
Correlation:-0.05025
  |
Component:372; Jaccard:0.10542
 |
Component:328; Jaccard:0.11743
 |
Component:372; Jaccard:0.10997
 |
Component:355; Jaccard:0.090698
 |
Component:393; Jaccard:0.051269
 |
Component:355; Jaccard:0.026156
 |
Component:355; Jaccard:0.009924
 |
Correlation:0.0045182
  |
Correlation:0.079792
  |
Correlation:0.0045182
  |
Correlation:0.044908
  |
Correlation:-0.05025
  |
Correlation:0.044908
  |
Correlation:0.044908
  |
Component:328; Jaccard:0.10388
 |
Component:372; Jaccard:0.11443
 |
Component:301; Jaccard:0.10971
 |
Component:372; Jaccard:0.084429
 |
Component:372; Jaccard:0.050852
 |
Component:372; Jaccard:0.02249
 |
Component:301; Jaccard:0.009798
 |
Correlation:0.079792
  |
Correlation:0.0045182
  |
Correlation:-0.051146
  |
Correlation:0.0045182
  |
Correlation:0.0045182
  |
Correlation:0.0045182
  |
Correlation:-0.051146
  |
Component:339; Jaccard:0.10169
 |
Component:355; Jaccard:0.10798
 |
Component:355; Jaccard:0.10963
 |
Component:301; Jaccard:0.083415
 |
Component:329; Jaccard:0.05028
 |
Component:301; Jaccard:0.022267
 |
Component:372; Jaccard:0.009474
 |
Correlation:-0.028419
  |
Correlation:0.044908
  |
Correlation:0.044908
  |
Correlation:-0.051146
  |
Correlation:-0.10561
  |
Correlation:-0.051146
  |
Correlation:0.0045182
  |
Component:384; Jaccard:0.095326
 |
Component:339; Jaccard:0.10441
 |
Component:339; Jaccard:0.094445
 |
Component:393; Jaccard:0.073685
 |
Component:301; Jaccard:0.04552
 |
Component:329; Jaccard:0.021613
 |
Component:329; Jaccard:0.009451
 |
Correlation:0.003586
  |
Correlation:-0.028419
  |
Correlation:-0.028419
  |
Correlation:-0.05025
  |
Correlation:-0.051146
  |
Correlation:-0.10561
  |
Correlation:-0.10561
  |
Component:182; Jaccard:0.094883
 |
Component:300; Jaccard:0.10054
 |
Component:300; Jaccard:0.092382
 |
Component:300; Jaccard:0.070062
 |
Component:217; Jaccard:0.045429
 |
Component:217; Jaccard:0.020184
 |
Component:339; Jaccard:0.009018
 |
Correlation:-0.075449
  |
Correlation:-0.037774
  |
Correlation:-0.037774
  |
Correlation:-0.037774
  |
Correlation:-0.16403
  |
Correlation:-0.16403
  |
Correlation:-0.028419
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
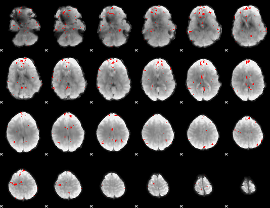 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
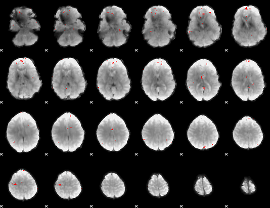 |
GLM Z-score result thresholded by 1
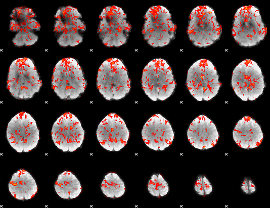 |
GLM Z-score result thresholded by 1.5
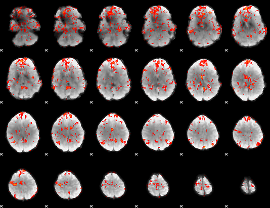 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
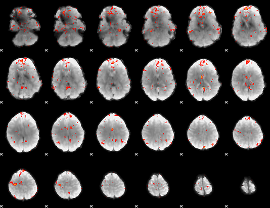 |
GLM Z-score result thresholded by 3
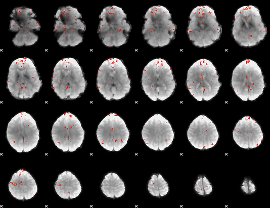 |
GLM Z-score result thresholded by 3.5
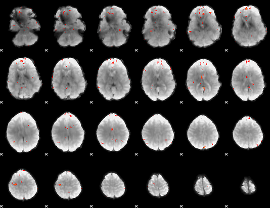 |
GLM Z-score result thresholded by 4
 |
Component:319; Jaccard:0.1286
 |
Component:319; Jaccard:0.13895
 |
Component:319; Jaccard:0.13589
 |
Component:319; Jaccard:0.11881
 |
Component:319; Jaccard:0.090228
 |
Component:319; Jaccard:0.059697
 |
Component:319; Jaccard:0.031895
 |
Correlation:0.073792
  |
Correlation:0.073792
  |
Correlation:0.073792
  |
Correlation:0.073792
  |
Correlation:0.073792
  |
Correlation:0.073792
  |
Correlation:0.073792
  |
Component:229; Jaccard:0.11142
 |
Component:229; Jaccard:0.11988
 |
Component:229; Jaccard:0.12314
 |
Component:229; Jaccard:0.10802
 |
Component:229; Jaccard:0.080776
 |
Component:229; Jaccard:0.051858
 |
Component:229; Jaccard:0.027598
 |
Correlation:0.024228
  |
Correlation:0.024228
  |
Correlation:0.024228
  |
Correlation:0.024228
  |
Correlation:0.024228
  |
Correlation:0.024228
  |
Correlation:0.024228
  |
Component:153; Jaccard:0.11093
 |
Component:153; Jaccard:0.11729
 |
Component:153; Jaccard:0.11733
 |
Component:153; Jaccard:0.10308
 |
Component:153; Jaccard:0.075228
 |
Component:153; Jaccard:0.048198
 |
Component:207; Jaccard:0.02528
 |
Correlation:0.078348
  |
Correlation:0.078348
  |
Correlation:0.078348
  |
Correlation:0.078348
  |
Correlation:0.078348
  |
Correlation:0.078348
  |
Correlation:0.11202
  |
Component:294; Jaccard:0.10462
 |
Component:294; Jaccard:0.11012
 |
Component:294; Jaccard:0.10799
 |
Component:294; Jaccard:0.09104
 |
Component:294; Jaccard:0.065198
 |
Component:207; Jaccard:0.044044
 |
Component:153; Jaccard:0.025036
 |
Correlation:0.024778
  |
Correlation:0.024778
  |
Correlation:0.024778
  |
Correlation:0.024778
  |
Correlation:0.024778
  |
Correlation:0.11202
  |
Correlation:0.078348
  |
Component:247; Jaccard:0.095112
 |
Component:247; Jaccard:0.095416
 |
Component:393; Jaccard:0.08571
 |
Component:207; Jaccard:0.074143
 |
Component:207; Jaccard:0.061897
 |
Component:294; Jaccard:0.040433
 |
Component:075; Jaccard:0.023575
 |
Correlation:-0.12476
  |
Correlation:-0.12476
  |
Correlation:-0.1411
  |
Correlation:0.11202
  |
Correlation:0.11202
  |
Correlation:0.024778
  |
Correlation:0.235
  |
Component:393; Jaccard:0.093099
 |
Component:393; Jaccard:0.09423
 |
Component:247; Jaccard:0.085511
 |
Component:247; Jaccard:0.071067
 |
Component:120; Jaccard:0.051046
 |
Component:120; Jaccard:0.036341
 |
Component:120; Jaccard:0.020969
 |
Correlation:-0.1411
  |
Correlation:-0.1411
  |
Correlation:-0.12476
  |
Correlation:-0.12476
  |
Correlation:0.18574
 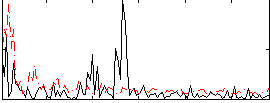 |
Correlation:0.18574
  |
Correlation:0.18574
  |
Component:010; Jaccard:0.085205
 |
Component:207; Jaccard:0.084835
 |
Component:207; Jaccard:0.084625
 |
Component:393; Jaccard:0.07012
 |
Component:314; Jaccard:0.050399
 |
Component:075; Jaccard:0.034945
 |
Component:314; Jaccard:0.01886
 |
Correlation:-0.049803
 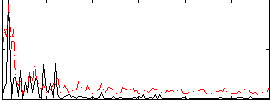 |
Correlation:0.11202
  |
Correlation:0.11202
  |
Correlation:-0.1411
  |
Correlation:0.25434
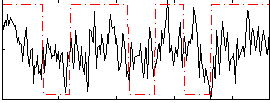 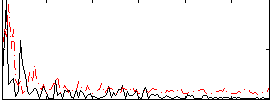 |
Correlation:0.235
  |
Correlation:0.25434
  |
Component:207; Jaccard:0.082177
 |
Component:010; Jaccard:0.083486
 |
Component:314; Jaccard:0.076996
 |
Component:314; Jaccard:0.064333
 |
Component:247; Jaccard:0.050329
 |
Component:314; Jaccard:0.034221
 |
Component:294; Jaccard:0.018773
 |
Correlation:0.11202
  |
Correlation:-0.049803
  |
Correlation:0.25434
  |
Correlation:0.25434
  |
Correlation:-0.12476
  |
Correlation:0.25434
  |
Correlation:0.024778
  |
Component:314; Jaccard:0.081411
 |
Component:314; Jaccard:0.083108
 |
Component:010; Jaccard:0.076509
 |
Component:120; Jaccard:0.063749
 |
Component:393; Jaccard:0.047644
 |
Component:247; Jaccard:0.03089
 |
Component:053; Jaccard:0.017466
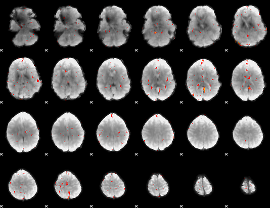 |
Correlation:0.25434
  |
Correlation:0.25434
  |
Correlation:-0.049803
  |
Correlation:0.18574
  |
Correlation:-0.1411
  |
Correlation:-0.12476
  |
Correlation:0.24082
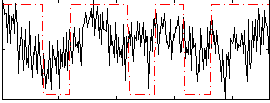  |
Component:120; Jaccard:0.071012
 |
Component:120; Jaccard:0.07317
 |
Component:120; Jaccard:0.0713
 |
Component:076; Jaccard:0.062254
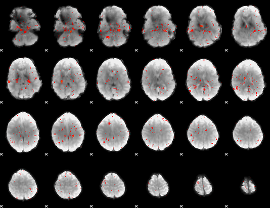 |
Component:076; Jaccard:0.046193
 |
Component:053; Jaccard:0.030592
 |
Component:247; Jaccard:0.01564
 |
Correlation:0.18574
  |
Correlation:0.18574
  |
Correlation:0.18574
  |
Correlation:0.067246
  |
Correlation:0.067246
  |
Correlation:0.24082
  |
Correlation:-0.12476
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
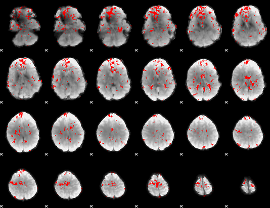 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
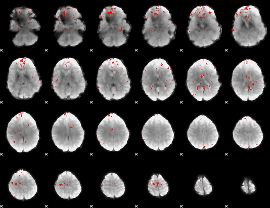 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
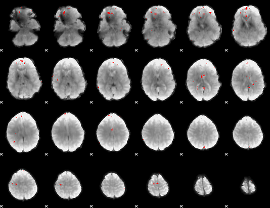 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
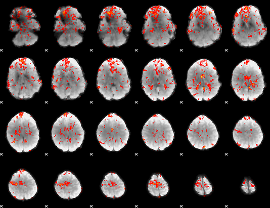 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
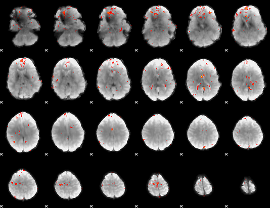 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
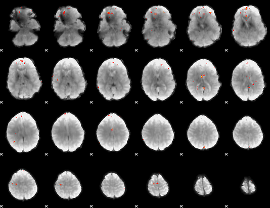 |
Component:319; Jaccard:0.12334
 |
Component:319; Jaccard:0.13222
 |
Component:319; Jaccard:0.12947
 |
Component:319; Jaccard:0.11107
 |
Component:319; Jaccard:0.081983
 |
Component:075; Jaccard:0.060035
 |
Component:075; Jaccard:0.040479
 |
Correlation:0.17822
  |
Correlation:0.17822
  |
Correlation:0.17822
  |
Correlation:0.17822
  |
Correlation:0.17822
  |
Correlation:0.0013903
  |
Correlation:0.0013903
  |
Component:229; Jaccard:0.11218
 |
Component:229; Jaccard:0.12075
 |
Component:229; Jaccard:0.11923
 |
Component:229; Jaccard:0.10275
 |
Component:229; Jaccard:0.077549
 |
Component:319; Jaccard:0.05514
 |
Component:319; Jaccard:0.034313
 |
Correlation:0.15472
  |
Correlation:0.15472
  |
Correlation:0.15472
  |
Correlation:0.15472
  |
Correlation:0.15472
  |
Correlation:0.17822
  |
Correlation:0.17822
  |
Component:153; Jaccard:0.10963
 |
Component:153; Jaccard:0.11788
 |
Component:153; Jaccard:0.11575
 |
Component:153; Jaccard:0.098508
 |
Component:075; Jaccard:0.075703
 |
Component:207; Jaccard:0.054191
 |
Component:207; Jaccard:0.032188
 |
Correlation:0.060839
  |
Correlation:0.060839
  |
Correlation:0.060839
  |
Correlation:0.060839
  |
Correlation:0.0013903
  |
Correlation:0.075767
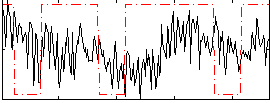  |
Correlation:0.075767
  |
Component:207; Jaccard:0.090397
 |
Component:207; Jaccard:0.095496
 |
Component:207; Jaccard:0.097691
 |
Component:207; Jaccard:0.090647
 |
Component:207; Jaccard:0.07544
 |
Component:229; Jaccard:0.050951
 |
Component:229; Jaccard:0.03167
 |
Correlation:0.075767
  |
Correlation:0.075767
  |
Correlation:0.075767
  |
Correlation:0.075767
  |
Correlation:0.075767
  |
Correlation:0.15472
  |
Correlation:0.15472
  |
Component:247; Jaccard:0.089535
 |
Component:247; Jaccard:0.090418
 |
Component:247; Jaccard:0.083726
 |
Component:075; Jaccard:0.086804
 |
Component:153; Jaccard:0.071277
 |
Component:153; Jaccard:0.049542
 |
Component:153; Jaccard:0.030666
 |
Correlation:0.029474
 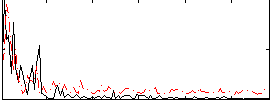 |
Correlation:0.029474
  |
Correlation:0.029474
  |
Correlation:0.0013903
  |
Correlation:0.060839
  |
Correlation:0.060839
  |
Correlation:0.060839
  |
Component:316; Jaccard:0.088098
 |
Component:316; Jaccard:0.088
 |
Component:075; Jaccard:0.083657
 |
Component:120; Jaccard:0.075299
 |
Component:120; Jaccard:0.063579
 |
Component:120; Jaccard:0.045403
 |
Component:120; Jaccard:0.027743
 |
Correlation:0.18014
  |
Correlation:0.18014
  |
Correlation:0.0013903
  |
Correlation:0.1649
  |
Correlation:0.1649
  |
Correlation:0.1649
  |
Correlation:0.1649
  |
Component:136; Jaccard:0.086611
 |
Component:136; Jaccard:0.086348
 |
Component:120; Jaccard:0.081645
 |
Component:247; Jaccard:0.071365
 |
Component:053; Jaccard:0.054614
 |
Component:053; Jaccard:0.038697
 |
Component:053; Jaccard:0.022706
 |
Correlation:0.17993
  |
Correlation:0.17993
  |
Correlation:0.1649
  |
Correlation:0.029474
  |
Correlation:0.13714
  |
Correlation:0.13714
  |
Correlation:0.13714
  |
Component:130; Jaccard:0.085626
 |
Component:130; Jaccard:0.085761
 |
Component:136; Jaccard:0.080175
 |
Component:136; Jaccard:0.067003
 |
Component:247; Jaccard:0.052574
 |
Component:247; Jaccard:0.033553
 |
Component:247; Jaccard:0.021069
 |
Correlation:0.070348
  |
Correlation:0.070348
  |
Correlation:0.17993
  |
Correlation:0.17993
  |
Correlation:0.029474
  |
Correlation:0.029474
  |
Correlation:0.029474
  |
Component:294; Jaccard:0.07848
 |
Component:120; Jaccard:0.083143
 |
Component:316; Jaccard:0.078746
 |
Component:130; Jaccard:0.062419
 |
Component:136; Jaccard:0.045987
 |
Component:379; Jaccard:0.032442
 |
Component:379; Jaccard:0.020593
 |
Correlation:0.077955
  |
Correlation:0.1649
  |
Correlation:0.18014
  |
Correlation:0.070348
  |
Correlation:0.17993
  |
Correlation:-0.2024
  |
Correlation:-0.2024
  |
Component:120; Jaccard:0.077833
 |
Component:294; Jaccard:0.077628
 |
Component:130; Jaccard:0.078042
 |
Component:316; Jaccard:0.061002
 |
Component:379; Jaccard:0.045803
 |
Component:004; Jaccard:0.031565
 |
Component:004; Jaccard:0.020177
 |
Correlation:0.1649
  |
Correlation:0.077955
  |
Correlation:0.070348
  |
Correlation:0.18014
  |
Correlation:-0.2024
  |
Correlation:0.064068
  |
Correlation:0.064068
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































