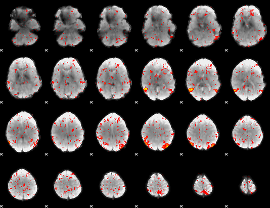
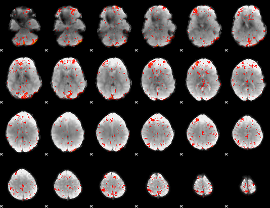
Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
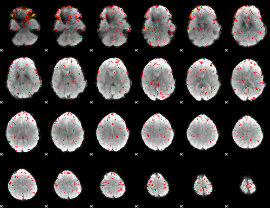
GLM result group-wise
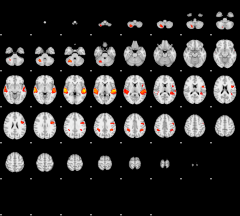 |
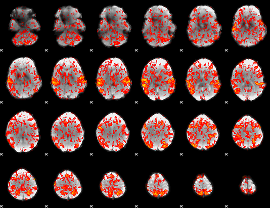
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
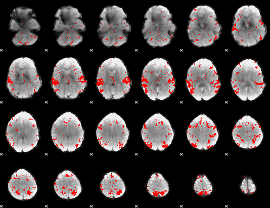 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
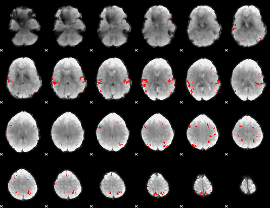 |
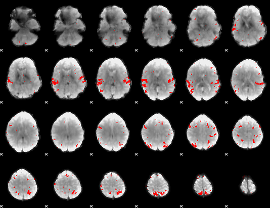
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
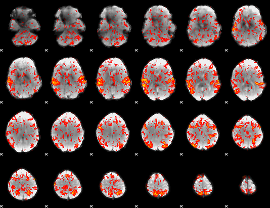 |
GLM Z-score result thresholded by 2
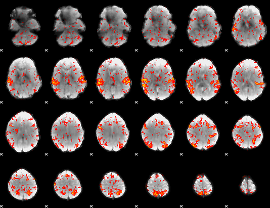 |
GLM Z-score result thresholded by 2.5
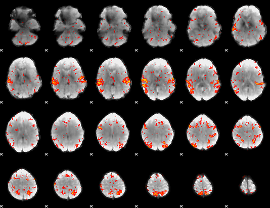 |
GLM Z-score result thresholded by 3
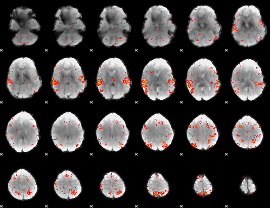 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
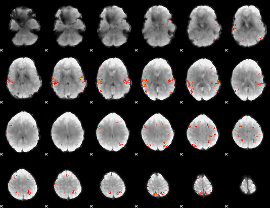 |
Component:140; Jaccard:0.13052
 |
Component:376; Jaccard:0.15867
 |
Component:376; Jaccard:0.18211
 |
Component:376; Jaccard:0.18164
 |
Component:156; Jaccard:0.16258
 |
Component:156; Jaccard:0.14074
 |
Component:156; Jaccard:0.10818
 |
Correlation:0.58538
 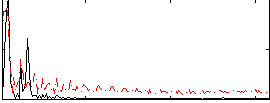 |
Correlation:0.5667
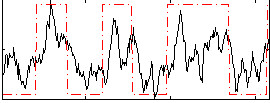 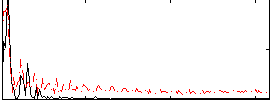 |
Correlation:0.5667
  |
Correlation:0.5667
  |
Correlation:-0.14912
 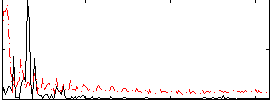 |
Correlation:-0.14912
  |
Correlation:-0.14912
  |
Component:376; Jaccard:0.12755
 |
Component:140; Jaccard:0.15407
 |
Component:140; Jaccard:0.16732
 |
Component:156; Jaccard:0.16705
 |
Component:376; Jaccard:0.15427
 |
Component:376; Jaccard:0.11937
 |
Component:394; Jaccard:0.088812
 |
Correlation:0.5667
  |
Correlation:0.58538
  |
Correlation:0.58538
  |
Correlation:-0.14912
  |
Correlation:0.5667
  |
Correlation:0.5667
  |
Correlation:-0.14765
 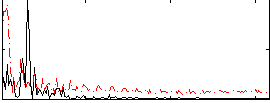 |
Component:319; Jaccard:0.12264
 |
Component:319; Jaccard:0.14012
 |
Component:156; Jaccard:0.15618
 |
Component:394; Jaccard:0.15846
 |
Component:394; Jaccard:0.14635
 |
Component:394; Jaccard:0.11832
 |
Component:376; Jaccard:0.081684
 |
Correlation:0.55904
  |
Correlation:0.55904
  |
Correlation:-0.14912
  |
Correlation:-0.14765
  |
Correlation:-0.14765
  |
Correlation:-0.14765
  |
Correlation:0.5667
  |
Component:311; Jaccard:0.11484
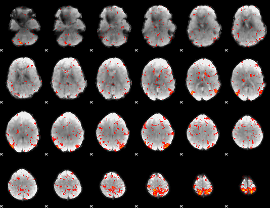 |
Component:394; Jaccard:0.13654
 |
Component:394; Jaccard:0.15255
 |
Component:140; Jaccard:0.15808
 |
Component:140; Jaccard:0.12862
 |
Component:140; Jaccard:0.092652
 |
Component:140; Jaccard:0.064032
 |
Correlation:0.22137
  |
Correlation:-0.14765
  |
Correlation:-0.14765
  |
Correlation:0.58538
  |
Correlation:0.58538
  |
Correlation:0.58538
  |
Correlation:0.58538
  |
Component:394; Jaccard:0.11463
 |
Component:156; Jaccard:0.13448
 |
Component:319; Jaccard:0.14748
 |
Component:311; Jaccard:0.13908
 |
Component:311; Jaccard:0.11764
 |
Component:311; Jaccard:0.089146
 |
Component:311; Jaccard:0.057278
 |
Correlation:-0.14765
  |
Correlation:-0.14912
  |
Correlation:0.55904
  |
Correlation:0.22137
  |
Correlation:0.22137
  |
Correlation:0.22137
  |
Correlation:0.22137
  |
Component:156; Jaccard:0.1098
 |
Component:311; Jaccard:0.1319
 |
Component:311; Jaccard:0.14236
 |
Component:319; Jaccard:0.13807
 |
Component:319; Jaccard:0.11416
 |
Component:319; Jaccard:0.083872
 |
Component:319; Jaccard:0.056193
 |
Correlation:-0.14912
  |
Correlation:0.22137
  |
Correlation:0.22137
  |
Correlation:0.55904
  |
Correlation:0.55904
  |
Correlation:0.55904
  |
Correlation:0.55904
  |
Component:329; Jaccard:0.10966
 |
Component:026; Jaccard:0.12705
 |
Component:026; Jaccard:0.13847
 |
Component:026; Jaccard:0.13296
 |
Component:026; Jaccard:0.11215
 |
Component:026; Jaccard:0.083752
 |
Component:026; Jaccard:0.05513
 |
Correlation:0.45744
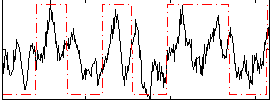 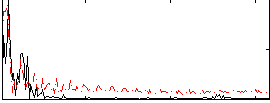 |
Correlation:0.54239
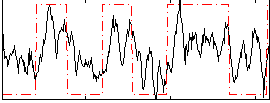 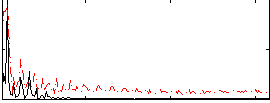 |
Correlation:0.54239
  |
Correlation:0.54239
  |
Correlation:0.54239
  |
Correlation:0.54239
  |
Correlation:0.54239
  |
Component:026; Jaccard:0.10918
 |
Component:329; Jaccard:0.11801
 |
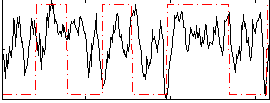
Component:002; Jaccard:0.11848
 |
Component:002; Jaccard:0.11078
 |
Component:002; Jaccard:0.092342
 |
Component:002; Jaccard:0.070254
 |
Component:002; Jaccard:0.047474
 |
Correlation:0.54239
  |
Correlation:0.45744
  |
Correlation:0.16772
 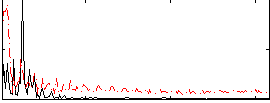 |
Correlation:0.16772
  |
Correlation:0.16772
  |
Correlation:0.16772
  |
Correlation:0.16772
  |
Component:002; Jaccard:0.10406
 |
Component:002; Jaccard:0.1148
 |
Component:329; Jaccard:0.11357
 |
Component:270; Jaccard:0.098832
 |
Component:270; Jaccard:0.080966
 |
Component:270; Jaccard:0.061244
 |
Component:270; Jaccard:0.043668
 |
Correlation:0.16772
  |
Correlation:0.16772
  |
Correlation:0.45744
  |
Correlation:0.024937
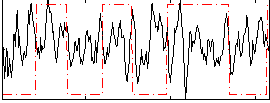 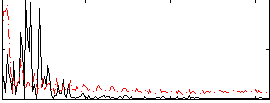 |
Correlation:0.024937
  |
Correlation:0.024937
  |
Correlation:0.024937
  |
Component:270; Jaccard:0.098923
 |
Component:270; Jaccard:0.10638
 |
Component:270; Jaccard:0.10772
 |
Component:329; Jaccard:0.098728
 |
Component:329; Jaccard:0.076397
 |
Component:361; Jaccard:0.055091
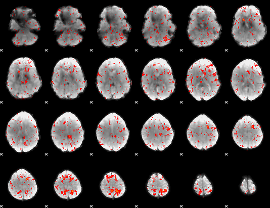 |
Component:361; Jaccard:0.036914
 |
Correlation:0.024937
  |
Correlation:0.024937
  |
Correlation:0.024937
  |
Correlation:0.45744
  |
Correlation:0.45744
  |
Correlation:0.40703
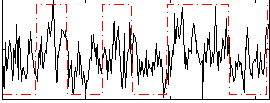 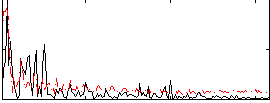 |
Correlation:0.40703
  |
Cope: 02:STORY BACK
|
GLM result group-wise
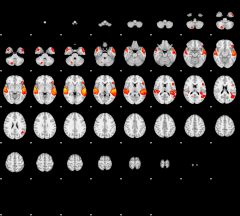 |
GLM binary result thresholded by 1
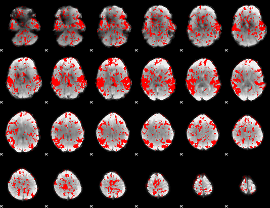 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
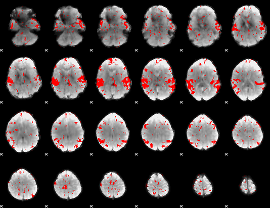 |
GLM binary result thresholded by 2.5
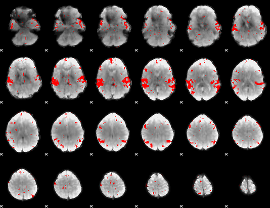 |
GLM binary result thresholded by 3
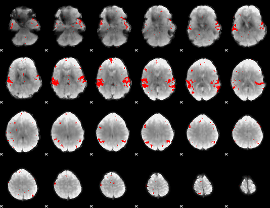 |
GLM binary result thresholded by 3.5
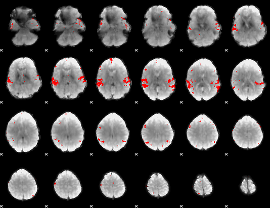 |
GLM binary result thresholded by 4
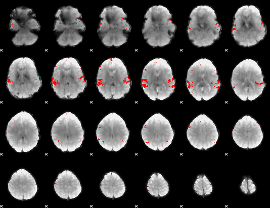 |
GLM Z-score result thresholded by 1
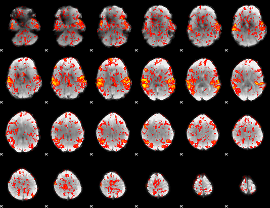 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
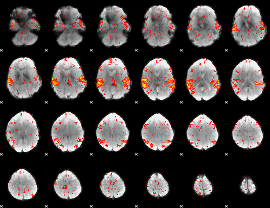 |
GLM Z-score result thresholded by 2.5
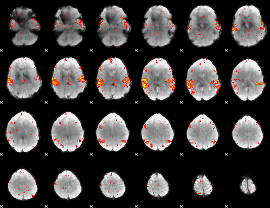 |
GLM Z-score result thresholded by 3
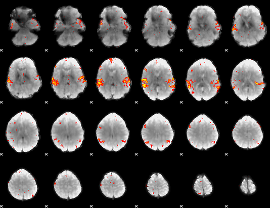 |
GLM Z-score result thresholded by 3.5
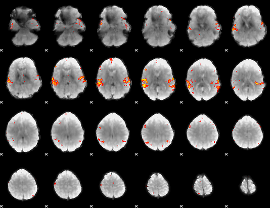 |
GLM Z-score result thresholded by 4
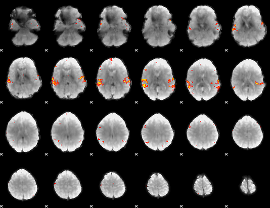 |
Component:394; Jaccard:0.17505
 |
Component:394; Jaccard:0.21475
 |
Component:394; Jaccard:0.24212
 |
Component:394; Jaccard:0.24337
 |
Component:394; Jaccard:0.22563
 |
Component:394; Jaccard:0.18845
 |
Component:394; Jaccard:0.1458
 |
Correlation:0.12191
  |
Correlation:0.12191
  |
Correlation:0.12191
  |
Correlation:0.12191
  |
Correlation:0.12191
  |
Correlation:0.12191
  |
Correlation:0.12191
  |
Component:156; Jaccard:0.15338
 |
Component:156; Jaccard:0.19178
 |
Component:156; Jaccard:0.22044
 |
Component:156; Jaccard:0.22696
 |
Component:156; Jaccard:0.20973
 |
Component:156; Jaccard:0.17684
 |
Component:156; Jaccard:0.14059
 |
Correlation:0.15637
  |
Correlation:0.15637
  |
Correlation:0.15637
  |
Correlation:0.15637
  |
Correlation:0.15637
  |
Correlation:0.15637
  |
Correlation:0.15637
  |
Component:275; Jaccard:0.13626
 |
Component:275; Jaccard:0.17075
 |
Component:275; Jaccard:0.19442
 |
Component:275; Jaccard:0.19181
 |
Component:275; Jaccard:0.16203
 |
Component:275; Jaccard:0.11998
 |
Component:117; Jaccard:0.088507
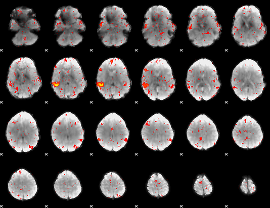 |
Correlation:0.45477
  |
Correlation:0.45477
  |
Correlation:0.45477
  |
Correlation:0.45477
  |
Correlation:0.45477
  |
Correlation:0.45477
  |
Correlation:0.27828
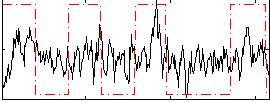  |
Component:117; Jaccard:0.11705
 |
Component:117; Jaccard:0.14031
 |
Component:117; Jaccard:0.15109
 |
Component:117; Jaccard:0.1493
 |
Component:117; Jaccard:0.13483
 |
Component:117; Jaccard:0.10834
 |
Component:275; Jaccard:0.082354
 |
Correlation:0.27828
  |
Correlation:0.27828
  |
Correlation:0.27828
  |
Correlation:0.27828
  |
Correlation:0.27828
  |
Correlation:0.27828
  |
Correlation:0.45477
  |
Component:270; Jaccard:0.10456
 |
Component:270; Jaccard:0.10781
 |
Component:270; Jaccard:0.10538
 |
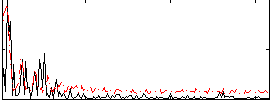
Component:120; Jaccard:0.095509
 |
Component:270; Jaccard:0.080935
 |
Component:270; Jaccard:0.063695
 |
Component:270; Jaccard:0.047157
 |
Correlation:-0.06638
 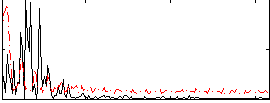 |
Correlation:-0.06638
  |
Correlation:-0.06638
  |
Correlation:0.27287
  |
Correlation:-0.06638
  |
Correlation:-0.06638
  |
Correlation:-0.06638
  |
Component:346; Jaccard:0.096002
 |
Component:120; Jaccard:0.10235
 |
Component:120; Jaccard:0.10381
 |
Component:270; Jaccard:0.094683
 |
Component:120; Jaccard:0.0791
 |
Component:120; Jaccard:0.062393
 |
Component:120; Jaccard:0.045918
 |
Correlation:0.20446
  |
Correlation:0.27287
  |
Correlation:0.27287
  |
Correlation:-0.06638
  |
Correlation:0.27287
  |
Correlation:0.27287
  |
Correlation:0.27287
  |
Component:120; Jaccard:0.09202
 |
Component:346; Jaccard:0.09902
 |
Component:262; Jaccard:0.095675
 |
Component:262; Jaccard:0.093262
 |
Component:262; Jaccard:0.078922
 |
Component:262; Jaccard:0.061397
 |
Component:262; Jaccard:0.044865
 |
Correlation:0.27287
  |
Correlation:0.20446
  |
Correlation:0.45339
  |
Correlation:0.45339
  |
Correlation:0.45339
  |
Correlation:0.45339
  |
Correlation:0.45339
  |
Component:037; Jaccard:0.089895
 |
Component:365; Jaccard:0.092985
 |
Component:346; Jaccard:0.092717
 |
Component:365; Jaccard:0.082666
 |
Component:365; Jaccard:0.066578
 |
Component:365; Jaccard:0.05304
 |
Component:365; Jaccard:0.039707
 |
Correlation:-0.028156
  |
Correlation:0.31463
  |
Correlation:0.20446
  |
Correlation:0.31463
  |
Correlation:0.31463
  |
Correlation:0.31463
  |
Correlation:0.31463
  |
Component:365; Jaccard:0.088186
 |
Component:262; Jaccard:0.090418
 |
Component:365; Jaccard:0.091312
 |
Component:346; Jaccard:0.076577
 |
Component:190; Jaccard:0.063697
 |
Component:190; Jaccard:0.047544
 |
Component:190; Jaccard:0.035653
 |
Correlation:0.31463
  |
Correlation:0.45339
  |
Correlation:0.31463
  |
Correlation:0.20446
  |
Correlation:0.43608
  |
Correlation:0.43608
  |
Correlation:0.43608
  |
Component:062; Jaccard:0.081793
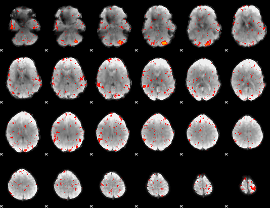 |
Component:062; Jaccard:0.084994
 |
Component:190; Jaccard:0.083846
 |
Component:190; Jaccard:0.076385
 |
Component:062; Jaccard:0.059237
 |
Component:134; Jaccard:0.046025
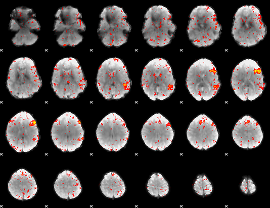 |
Component:134; Jaccard:0.035436
 |
Correlation:0.23525
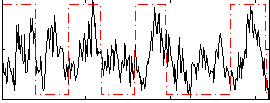 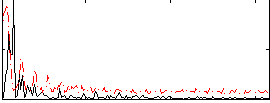 |
Correlation:0.23525
  |
Correlation:0.43608
  |
Correlation:0.43608
  |
Correlation:0.23525
  |
Correlation:-0.12162
 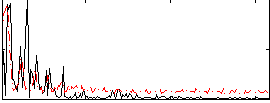 |
Correlation:-0.12162
  |
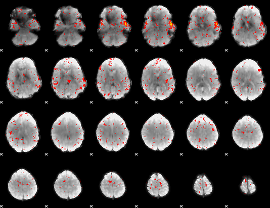
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
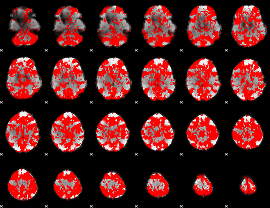 |
GLM binary result thresholded by 1.5
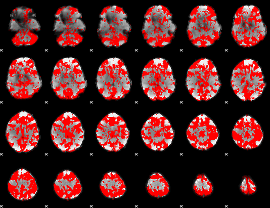 |
GLM binary result thresholded by 2
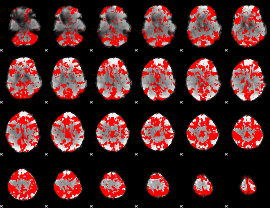 |
GLM binary result thresholded by 2.5
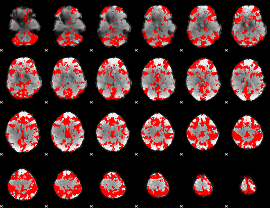 |
GLM binary result thresholded by 3
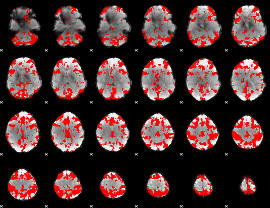 |
GLM binary result thresholded by 3.5
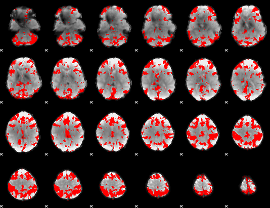 |
GLM binary result thresholded by 4
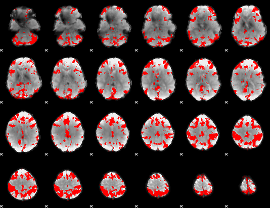 |
GLM Z-score result thresholded by 1
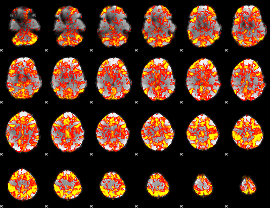 |
GLM Z-score result thresholded by 1.5
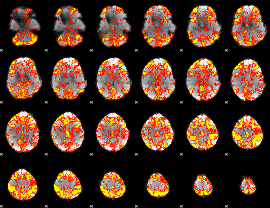 |
GLM Z-score result thresholded by 2
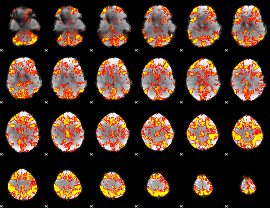 |
GLM Z-score result thresholded by 2.5
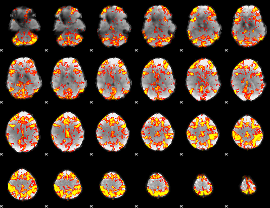 |
GLM Z-score result thresholded by 3
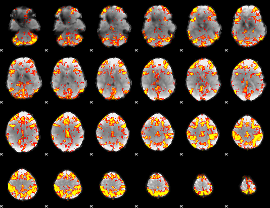 |
GLM Z-score result thresholded by 3.5
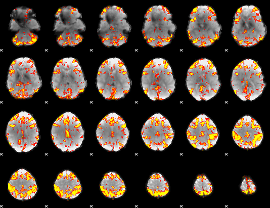 |
GLM Z-score result thresholded by 4
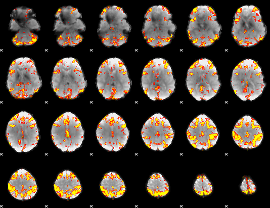 |
Component:319; Jaccard:0.11965
 |
Component:319; Jaccard:0.13843
 |
Component:319; Jaccard:0.16129
 |
Component:319; Jaccard:0.18709
 |
Component:319; Jaccard:0.21424
 |
Component:319; Jaccard:0.24042
 |
Component:319; Jaccard:0.26461
 |
Correlation:0.5877
  |
Correlation:0.5877
  |
Correlation:0.5877
  |
Correlation:0.5877
  |
Correlation:0.5877
  |
Correlation:0.5877
  |
Correlation:0.5877
  |
Component:321; Jaccard:0.11685
 |
Component:321; Jaccard:0.13445
 |
Component:321; Jaccard:0.15563
 |
Component:321; Jaccard:0.17814
 |
Component:321; Jaccard:0.20156
 |
Component:321; Jaccard:0.22366
 |
Component:321; Jaccard:0.24362
 |
Correlation:0.58971
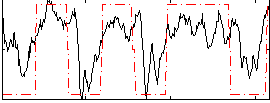  |
Correlation:0.58971
  |
Correlation:0.58971
  |
Correlation:0.58971
  |
Correlation:0.58971
  |
Correlation:0.58971
  |
Correlation:0.58971
  |
Component:329; Jaccard:0.11596
 |
Component:329; Jaccard:0.1309
 |
Component:329; Jaccard:0.14741
 |
Component:329; Jaccard:0.16442
 |
Component:329; Jaccard:0.1806
 |
Component:255; Jaccard:0.19714
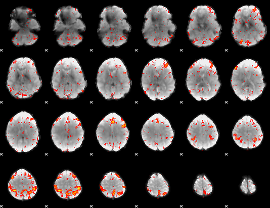 |
Component:255; Jaccard:0.21267
 |
Correlation:0.47778
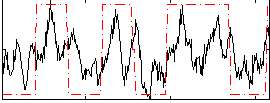  |
Correlation:0.47778
  |
Correlation:0.47778
  |
Correlation:0.47778
  |
Correlation:0.47778
  |
Correlation:0.52337
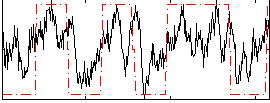 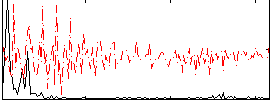 |
Correlation:0.52337
  |
Component:255; Jaccard:0.1086
 |
Component:255; Jaccard:0.1237
 |
Component:255; Jaccard:0.14065
 |
Component:255; Jaccard:0.15941
 |
Component:255; Jaccard:0.17876
 |
Component:329; Jaccard:0.19515
 |
Component:329; Jaccard:0.20748
 |
Correlation:0.52337
  |
Correlation:0.52337
  |
Correlation:0.52337
  |
Correlation:0.52337
  |
Correlation:0.52337
  |
Correlation:0.47778
  |
Correlation:0.47778
  |
Component:140; Jaccard:0.10848
 |
Component:140; Jaccard:0.12338
 |
Component:140; Jaccard:0.14037
 |
Component:140; Jaccard:0.15784
 |
Component:140; Jaccard:0.17452
 |
Component:140; Jaccard:0.18934
 |
Component:140; Jaccard:0.2027
 |
Correlation:0.62151
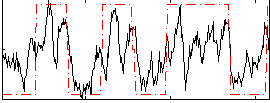 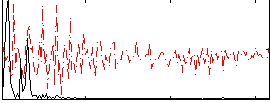 |
Correlation:0.62151
  |
Correlation:0.62151
  |
Correlation:0.62151
  |
Correlation:0.62151
  |
Correlation:0.62151
  |
Correlation:0.62151
  |
Component:369; Jaccard:0.10782
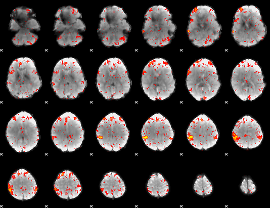 |
Component:369; Jaccard:0.11988
 |
Component:369; Jaccard:0.13308
 |
Component:369; Jaccard:0.14571
 |
Component:369; Jaccard:0.15752
 |
Component:376; Jaccard:0.17475
 |
Component:376; Jaccard:0.19042
 |
Correlation:0.47056
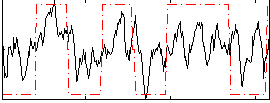 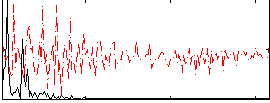 |
Correlation:0.47056
  |
Correlation:0.47056
  |
Correlation:0.47056
  |
Correlation:0.47056
  |
Correlation:0.59409
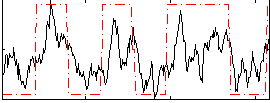 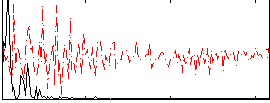 |
Correlation:0.59409
  |
Component:043; Jaccard:0.10418
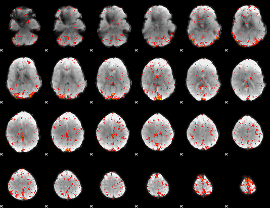 |
Component:043; Jaccard:0.11362
 |
Component:239; Jaccard:0.12638
 |
Component:239; Jaccard:0.1406
 |
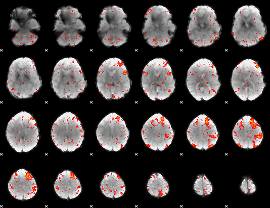
Component:376; Jaccard:0.15699
 |
Component:369; Jaccard:0.16588
 |
Component:026; Jaccard:0.17297
 |
Correlation:0.38403
 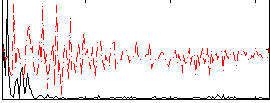 |
Correlation:0.38403
  |
Correlation:0.45589
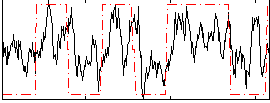  |
Correlation:0.45589
  |
Correlation:0.59409
  |
Correlation:0.47056
  |
Correlation:0.5489
  |
Component:286; Jaccard:0.10311
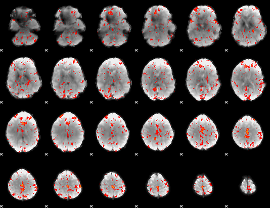 |
Component:239; Jaccard:0.11227
 |
Component:174; Jaccard:0.12362
 |
Component:376; Jaccard:0.13905
 |
Component:239; Jaccard:0.15359
 |
Component:239; Jaccard:0.16382
 |
Component:239; Jaccard:0.17231
 |
Correlation:0.38782
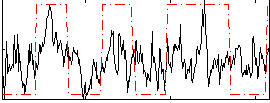 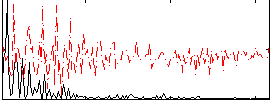 |
Correlation:0.45589
  |
Correlation:0.52898
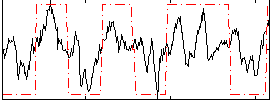 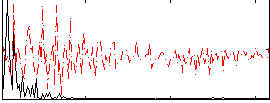 |
Correlation:0.59409
  |
Correlation:0.45589
  |
Correlation:0.45589
  |
Correlation:0.45589
  |
Component:306; Jaccard:0.10165
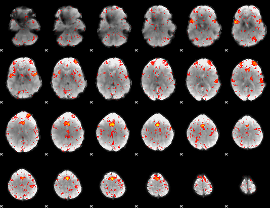 |
Component:286; Jaccard:0.11221
 |
Component:043; Jaccard:0.12273
 |
Component:026; Jaccard:0.13615
 |
Component:026; Jaccard:0.15059
 |
Component:026; Jaccard:0.16211
 |
Component:369; Jaccard:0.17162
 |
Correlation:0.41523
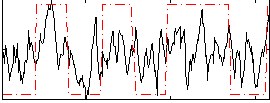 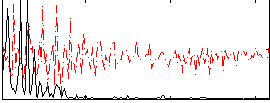 |
Correlation:0.38782
  |
Correlation:0.38403
  |
Correlation:0.5489
  |
Correlation:0.5489
  |
Correlation:0.5489
  |
Correlation:0.47056
  |
Component:150; Jaccard:0.10021
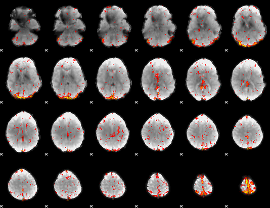 |
Component:174; Jaccard:0.11122
 |
Component:376; Jaccard:0.12182
 |
Component:174; Jaccard:0.1355
 |
Component:174; Jaccard:0.14662
 |
Component:174; Jaccard:0.15487
 |
Component:174; Jaccard:0.16012
 |
Correlation:0.41367
  |
Correlation:0.52898
  |
Correlation:0.59409
  |
Correlation:0.52898
  |
Correlation:0.52898
  |
Correlation:0.52898
  |
Correlation:0.52898
  |
Cope: 04:STORY-MATH BACK
|
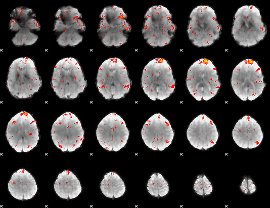
GLM result group-wise
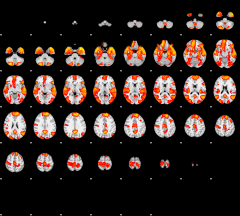 |
GLM binary result thresholded by 1
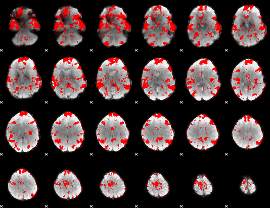 |
GLM binary result thresholded by 1.5
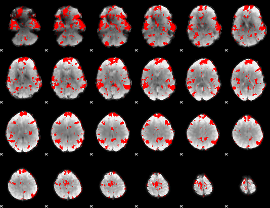 |
GLM binary result thresholded by 2
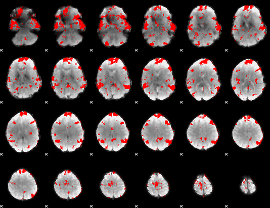 |
GLM binary result thresholded by 2.5
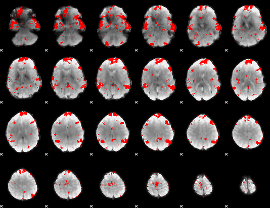 |
GLM binary result thresholded by 3
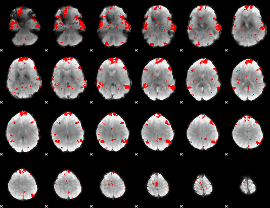 |
GLM binary result thresholded by 3.5
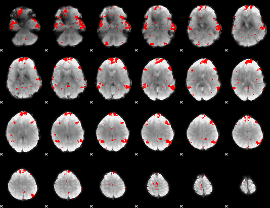 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
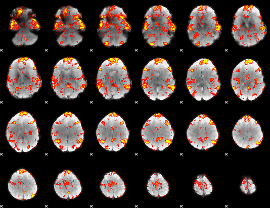 |
GLM Z-score result thresholded by 1.5
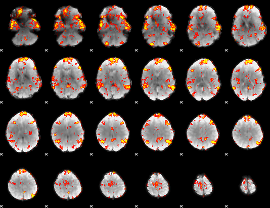 |
GLM Z-score result thresholded by 2
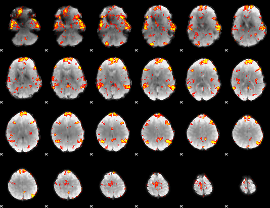 |
GLM Z-score result thresholded by 2.5
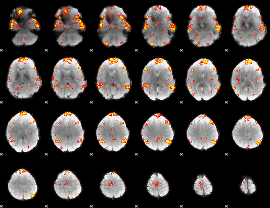 |
GLM Z-score result thresholded by 3
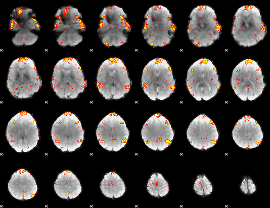 |
GLM Z-score result thresholded by 3.5
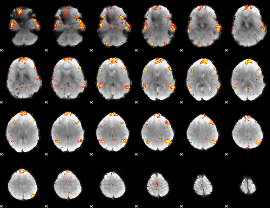 |
GLM Z-score result thresholded by 4
 |
Component:275; Jaccard:0.16895
 |
Component:275; Jaccard:0.2045
 |
Component:275; Jaccard:0.24213
 |
Component:275; Jaccard:0.27805
 |
Component:275; Jaccard:0.30886
 |
Component:275; Jaccard:0.33092
 |
Component:275; Jaccard:0.34234
 |
Correlation:0.45418
  |
Correlation:0.45418
  |
Correlation:0.45418
  |
Correlation:0.45418
  |
Correlation:0.45418
  |
Correlation:0.45418
  |
Correlation:0.45418
  |
Component:383; Jaccard:0.16024
 |
Component:383; Jaccard:0.18269
 |
Component:383; Jaccard:0.20422
 |
Component:383; Jaccard:0.22195
 |
Component:262; Jaccard:0.23727
 |
Component:262; Jaccard:0.25223
 |
Component:262; Jaccard:0.25992
 |
Correlation:0.24999
  |
Correlation:0.24999
  |
Correlation:0.24999
  |
Correlation:0.24999
  |
Correlation:0.45725
  |
Correlation:0.45725
  |
Correlation:0.45725
  |
Component:190; Jaccard:0.13913
 |
Component:262; Jaccard:0.15976
 |
Component:262; Jaccard:0.18784
 |
Component:262; Jaccard:0.2135
 |
Component:383; Jaccard:0.23311
 |
Component:383; Jaccard:0.23542
 |
Component:383; Jaccard:0.23231
 |
Correlation:0.43623
  |
Correlation:0.45725
  |
Correlation:0.45725
  |
Correlation:0.45725
  |
Correlation:0.24999
  |
Correlation:0.24999
  |
Correlation:0.24999
  |
Component:166; Jaccard:0.13835
 |
Component:106; Jaccard:0.15915
 |
Component:106; Jaccard:0.17863
 |
Component:106; Jaccard:0.19534
 |
Component:106; Jaccard:0.20619
 |
Component:106; Jaccard:0.21081
 |
Component:106; Jaccard:0.21176
 |
Correlation:0.32107
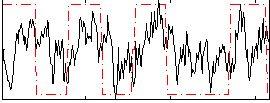 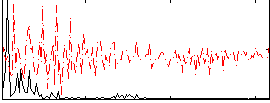 |
Correlation:0.42982
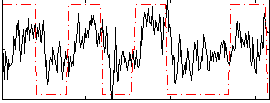 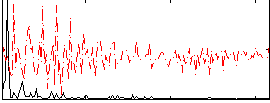 |
Correlation:0.42982
  |
Correlation:0.42982
  |
Correlation:0.42982
  |
Correlation:0.42982
  |
Correlation:0.42982
  |
Component:106; Jaccard:0.13722
 |
Component:166; Jaccard:0.15558
 |
Component:166; Jaccard:0.17024
 |
Component:166; Jaccard:0.18227
 |
Component:166; Jaccard:0.1914
 |
Component:166; Jaccard:0.19251
 |
Component:166; Jaccard:0.18699
 |
Correlation:0.42982
  |
Correlation:0.32107
  |
Correlation:0.32107
  |
Correlation:0.32107
  |
Correlation:0.32107
  |
Correlation:0.32107
  |
Correlation:0.32107
  |
Component:262; Jaccard:0.13387
 |
Component:190; Jaccard:0.15455
 |
Component:190; Jaccard:0.16942
 |
Component:190; Jaccard:0.1799
 |
Component:190; Jaccard:0.18436
 |
Component:190; Jaccard:0.18159
 |
Component:190; Jaccard:0.17431
 |
Correlation:0.45725
  |
Correlation:0.43623
  |
Correlation:0.43623
  |
Correlation:0.43623
  |
Correlation:0.43623
  |
Correlation:0.43623
  |
Correlation:0.43623
  |
Component:191; Jaccard:0.13087
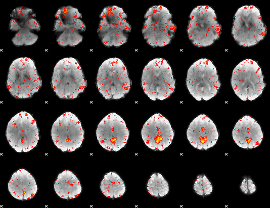 |
Component:271; Jaccard:0.14271
 |
Component:271; Jaccard:0.15433
 |
Component:271; Jaccard:0.16069
 |
Component:271; Jaccard:0.16348
 |
Component:271; Jaccard:0.15812
 |
Component:271; Jaccard:0.15161
 |
Correlation:0.11295
  |
Correlation:0.17704
  |
Correlation:0.17704
  |
Correlation:0.17704
  |
Correlation:0.17704
  |
Correlation:0.17704
  |
Correlation:0.17704
  |
Component:271; Jaccard:0.13015
 |
Component:191; Jaccard:0.13807
 |
Component:191; Jaccard:0.14189
 |
Component:097; Jaccard:0.14033
 |
Component:191; Jaccard:0.1355
 |
Component:332; Jaccard:0.13368
 |
Component:332; Jaccard:0.13038
 |
Correlation:0.17704
  |
Correlation:0.11295
  |
Correlation:0.11295
  |
Correlation:0.12836
  |
Correlation:0.11295
  |
Correlation:0.15945
  |
Correlation:0.15945
  |
Component:097; Jaccard:0.12846
 |
Component:097; Jaccard:0.1352
 |
Component:097; Jaccard:0.13952
 |
Component:191; Jaccard:0.13977
 |
Component:097; Jaccard:0.13453
 |
Component:191; Jaccard:0.12681
 |
Component:191; Jaccard:0.11618
 |
Correlation:0.12836
  |
Correlation:0.12836
  |
Correlation:0.12836
  |
Correlation:0.11295
  |
Correlation:0.12836
  |
Correlation:0.11295
  |
Correlation:0.11295
  |
Component:365; Jaccard:0.11069
 |
Component:332; Jaccard:0.11823
 |
Component:332; Jaccard:0.1269
 |
Component:332; Jaccard:0.13158
 |
Component:332; Jaccard:0.13429
 |
Component:097; Jaccard:0.12466
 |
Component:097; Jaccard:0.11386
 |
Correlation:0.30842
  |
Correlation:0.15945
  |
Correlation:0.15945
  |
Correlation:0.15945
  |
Correlation:0.15945
  |
Correlation:0.12836
  |
Correlation:0.12836
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
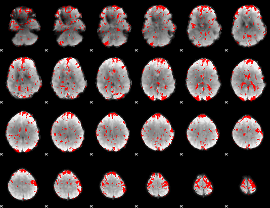 |
GLM binary result thresholded by 2
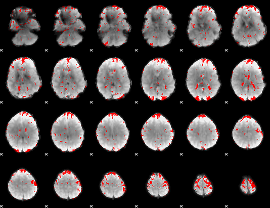 |
GLM binary result thresholded by 2.5
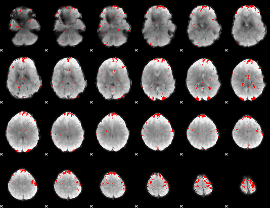 |
GLM binary result thresholded by 3
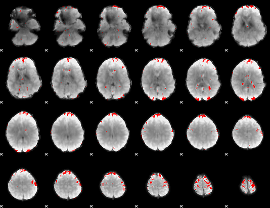 |
GLM binary result thresholded by 3.5
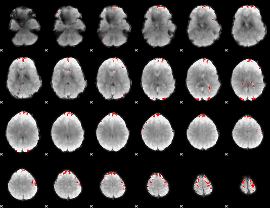 |
GLM binary result thresholded by 4
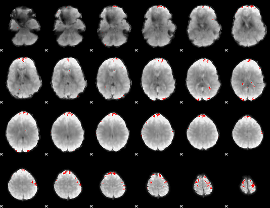 |
GLM Z-score result thresholded by 1
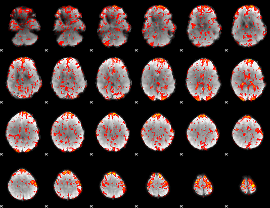 |
GLM Z-score result thresholded by 1.5
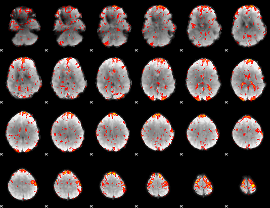 |
GLM Z-score result thresholded by 2
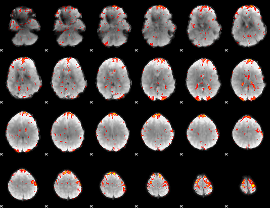 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
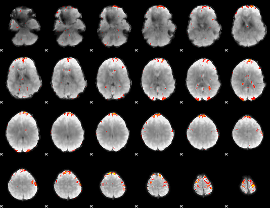 |
GLM Z-score result thresholded by 3.5
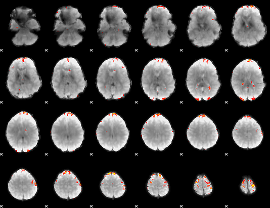 |
GLM Z-score result thresholded by 4
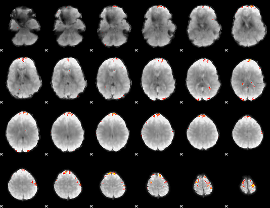 |
Component:091; Jaccard:0.17416
 |
Component:091; Jaccard:0.20853
 |
Component:091; Jaccard:0.22779
 |
Component:091; Jaccard:0.21423
 |
Component:091; Jaccard:0.18308
 |
Component:091; Jaccard:0.14239
 |
Component:091; Jaccard:0.10684
 |
Correlation:0.055594
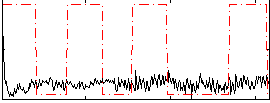 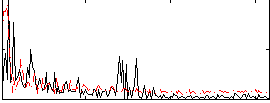 |
Correlation:0.055594
  |
Correlation:0.055594
  |
Correlation:0.055594
  |
Correlation:0.055594
  |
Correlation:0.055594
  |
Correlation:0.055594
  |
Component:380; Jaccard:0.15531
 |
Component:380; Jaccard:0.16201
 |
Component:078; Jaccard:0.17061
 |
Component:078; Jaccard:0.17037
 |
Component:078; Jaccard:0.15088
 |
Component:078; Jaccard:0.1254
 |
Component:078; Jaccard:0.10067
 |
Correlation:-0.046038
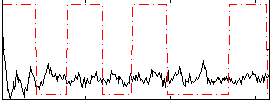 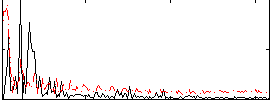 |
Correlation:-0.046038
  |
Correlation:0.12901
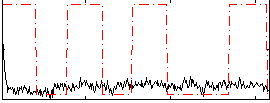 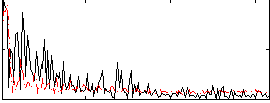 |
Correlation:0.12901
  |
Correlation:0.12901
  |
Correlation:0.12901
  |
Correlation:0.12901
  |
Component:078; Jaccard:0.12235
 |
Component:078; Jaccard:0.15051
 |
Component:380; Jaccard:0.15188
 |
Component:380; Jaccard:0.12658
 |
Component:380; Jaccard:0.09766
 |
Component:380; Jaccard:0.071391
 |
Component:380; Jaccard:0.049581
 |
Correlation:0.12901
  |
Correlation:0.12901
  |
Correlation:-0.046038
  |
Correlation:-0.046038
  |
Correlation:-0.046038
  |
Correlation:-0.046038
  |
Correlation:-0.046038
  |
Component:340; Jaccard:0.11776
 |
Component:340; Jaccard:0.12439
 |
Component:340; Jaccard:0.11902
 |
Component:340; Jaccard:0.097351
 |
Component:340; Jaccard:0.074889
 |
Component:166; Jaccard:0.051871
 |
Component:166; Jaccard:0.040891
 |
Correlation:-0.088768
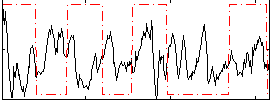 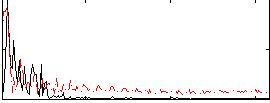 |
Correlation:-0.088768
  |
Correlation:-0.088768
  |
Correlation:-0.088768
  |
Correlation:-0.088768
  |
Correlation:0.28621
 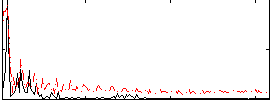 |
Correlation:0.28621
  |
Component:383; Jaccard:0.092053
 |
Component:383; Jaccard:0.094666
 |
Component:383; Jaccard:0.084406
 |
Component:166; Jaccard:0.073034
 |
Component:166; Jaccard:0.062205
 |
Component:179; Jaccard:0.051314
 |
Component:179; Jaccard:0.040035
 |
Correlation:0.24106
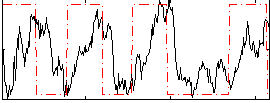  |
Correlation:0.24106
  |
Correlation:0.24106
  |
Correlation:0.28621
  |
Correlation:0.28621
  |
Correlation:0.22806
 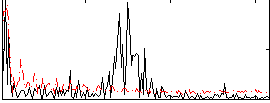 |
Correlation:0.22806
  |
Component:217; Jaccard:0.08973
 |
Component:264; Jaccard:0.089786
 |
Component:217; Jaccard:0.083814
 |
Component:383; Jaccard:0.070726
 |
Component:179; Jaccard:0.059872
 |
Component:340; Jaccard:0.050725
 |
Component:383; Jaccard:0.032568
 |
Correlation:-0.19301
  |
Correlation:-0.078865
  |
Correlation:-0.19301
  |
Correlation:0.24106
  |
Correlation:0.22806
  |
Correlation:-0.088768
  |
Correlation:0.24106
  |
Component:264; Jaccard:0.088481
 |
Component:217; Jaccard:0.089617
 |
Component:264; Jaccard:0.083053
 |
Component:264; Jaccard:0.070177
 |
Component:271; Jaccard:0.056601
 |
Component:271; Jaccard:0.04362
 |
Component:271; Jaccard:0.03253
 |
Correlation:-0.078865
  |
Correlation:-0.19301
  |
Correlation:-0.078865
  |
Correlation:-0.078865
  |
Correlation:0.15624
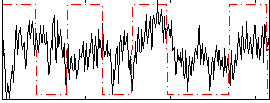 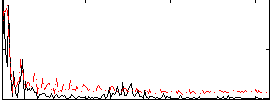 |
Correlation:0.15624
  |
Correlation:0.15624
  |
Component:285; Jaccard:0.088088
 |
Component:285; Jaccard:0.082378
 |
Component:166; Jaccard:0.079629
 |
Component:271; Jaccard:0.069903
 |
Component:264; Jaccard:0.054854
 |
Component:383; Jaccard:0.042625
 |
Component:340; Jaccard:0.032311
 |
Correlation:-0.25014
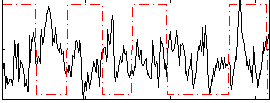 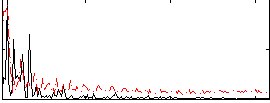 |
Correlation:-0.25014
  |
Correlation:0.28621
  |
Correlation:0.15624
  |
Correlation:-0.078865
  |
Correlation:0.24106
  |
Correlation:-0.088768
  |
Component:148; Jaccard:0.082644
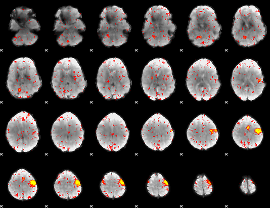 |
Component:166; Jaccard:0.080581
 |
Component:271; Jaccard:0.077543
 |
Component:217; Jaccard:0.068859
 |
Component:383; Jaccard:0.054388
 |
Component:217; Jaccard:0.04048
 |
Component:190; Jaccard:0.029485
 |
Correlation:-0.1985
  |
Correlation:0.28621
  |
Correlation:0.15624
  |
Correlation:-0.19301
  |
Correlation:0.24106
  |
Correlation:-0.19301
  |
Correlation:0.42026
 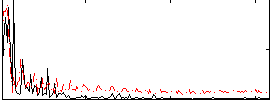 |
Component:258; Jaccard:0.078356
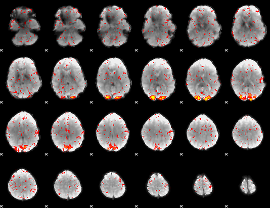 |
Component:148; Jaccard:0.079741
 |
Component:285; Jaccard:0.071722
 |
Component:179; Jaccard:0.067833
 |
Component:217; Jaccard:0.053832
 |
Component:264; Jaccard:0.040201
 |
Component:264; Jaccard:0.02914
 |
Correlation:-0.060375
 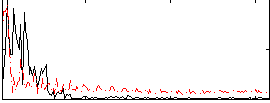 |
Correlation:-0.1985
  |
Correlation:-0.25014
  |
Correlation:0.22806
  |
Correlation:-0.19301
  |
Correlation:-0.078865
  |
Correlation:-0.078865
  |
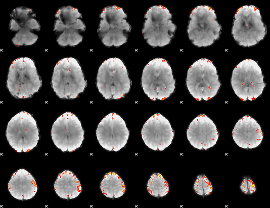
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
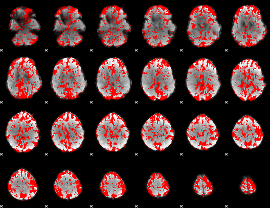 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
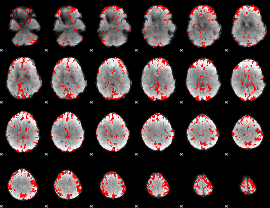 |
GLM binary result thresholded by 2.5
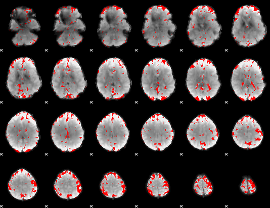 |
GLM binary result thresholded by 3
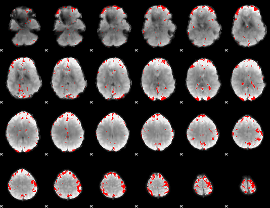 |
GLM binary result thresholded by 3.5
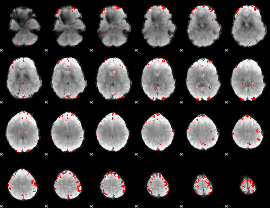 |
GLM binary result thresholded by 4
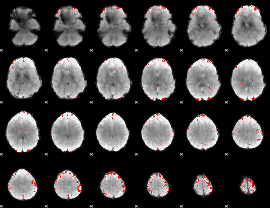 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
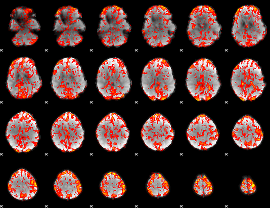
GLM Z-score result thresholded by 2
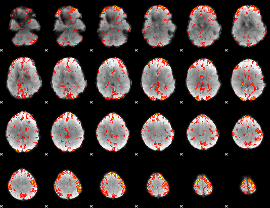 |
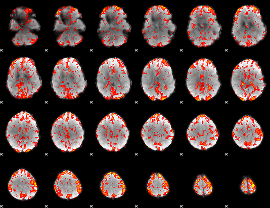
GLM Z-score result thresholded by 2.5
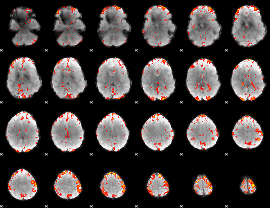 |
GLM Z-score result thresholded by 3
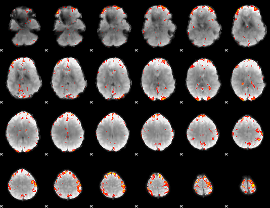 |
GLM Z-score result thresholded by 3.5
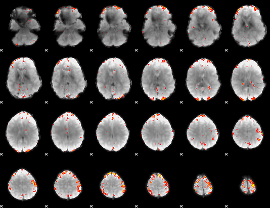 |
GLM Z-score result thresholded by 4
 |
Component:321; Jaccard:0.15837
 |
Component:321; Jaccard:0.19182
 |
Component:321; Jaccard:0.21689
 |
Component:321; Jaccard:0.21449
 |
Component:321; Jaccard:0.18496
 |
Component:091; Jaccard:0.15693
 |
Component:091; Jaccard:0.12884
 |
Correlation:0.59314
  |
Correlation:0.59314
  |
Correlation:0.59314
  |
Correlation:0.59314
  |
Correlation:0.59314
  |
Correlation:-0.042746
  |
Correlation:-0.042746
  |
Component:380; Jaccard:0.1458
 |
Component:380; Jaccard:0.16051
 |
Component:091; Jaccard:0.17895
 |
Component:091; Jaccard:0.18827
 |
Component:091; Jaccard:0.18207
 |
Component:321; Jaccard:0.1416
 |
Component:078; Jaccard:0.10015
 |
Correlation:0.024177
  |
Correlation:0.024177
  |
Correlation:-0.042746
  |
Correlation:-0.042746
  |
Correlation:-0.042746
  |
Correlation:0.59314
  |
Correlation:-0.13976
  |
Component:091; Jaccard:0.13238
 |
Component:091; Jaccard:0.15698
 |
Component:380; Jaccard:0.16729
 |
Component:380; Jaccard:0.15771
 |
Component:380; Jaccard:0.13857
 |
Component:380; Jaccard:0.11218
 |
Component:321; Jaccard:0.098775
 |
Correlation:-0.042746
  |
Correlation:-0.042746
  |
Correlation:0.024177
  |
Correlation:0.024177
  |
Correlation:0.024177
  |
Correlation:0.024177
  |
Correlation:0.59314
  |
Component:251; Jaccard:0.12339
 |
Component:251; Jaccard:0.13937
 |
Component:251; Jaccard:0.14765
 |
Component:251; Jaccard:0.1409
 |
Component:078; Jaccard:0.12309
 |
Component:078; Jaccard:0.11193
 |
Component:380; Jaccard:0.088841
 |
Correlation:0.40639
  |
Correlation:0.40639
  |
Correlation:0.40639
  |
Correlation:0.40639
  |
Correlation:-0.13976
  |
Correlation:-0.13976
  |
Correlation:0.024177
  |
Component:217; Jaccard:0.11657
 |
Component:217; Jaccard:0.13257
 |
Component:217; Jaccard:0.14015
 |
Component:217; Jaccard:0.13538
 |
Component:251; Jaccard:0.12158
 |
Component:251; Jaccard:0.097238
 |
Component:340; Jaccard:0.073435
 |
Correlation:0.17378
  |
Correlation:0.17378
  |
Correlation:0.17378
  |
Correlation:0.17378
  |
Correlation:0.40639
  |
Correlation:0.40639
  |
Correlation:0.068469
  |
Component:340; Jaccard:0.11512
 |
Component:340; Jaccard:0.13015
 |
Component:340; Jaccard:0.13728
 |
Component:340; Jaccard:0.13156
 |
Component:217; Jaccard:0.11784
 |
Component:340; Jaccard:0.094362
 |
Component:217; Jaccard:0.073096
 |
Correlation:0.068469
  |
Correlation:0.068469
  |
Correlation:0.068469
  |
Correlation:0.068469
  |
Correlation:0.17378
  |
Correlation:0.068469
  |
Correlation:0.17378
  |
Component:019; Jaccard:0.10662
 |
Component:174; Jaccard:0.11267
 |
Component:174; Jaccard:0.11501
 |
Component:078; Jaccard:0.12424
 |
Component:340; Jaccard:0.11504
 |
Component:217; Jaccard:0.092954
 |
Component:251; Jaccard:0.072888
 |
Correlation:0.29838
  |
Correlation:0.52436
  |
Correlation:0.52436
  |
Correlation:-0.13976
  |
Correlation:0.068469
  |
Correlation:0.17378
  |
Correlation:0.40639
  |
Component:124; Jaccard:0.10561
 |
Component:124; Jaccard:0.11
 |
Component:078; Jaccard:0.11434
 |
Component:174; Jaccard:0.10277
 |
Component:285; Jaccard:0.082562
 |
Component:148; Jaccard:0.063294
 |
Component:148; Jaccard:0.046668
 |
Correlation:0.34594
  |
Correlation:0.34594
  |
Correlation:-0.13976
  |
Correlation:0.52436
  |
Correlation:0.2268
  |
Correlation:0.24915
  |
Correlation:0.24915
  |
Component:174; Jaccard:0.105
 |
Component:019; Jaccard:0.10747
 |
Component:124; Jaccard:0.10716
 |
Component:124; Jaccard:0.097787
 |
Component:174; Jaccard:0.080212
 |
Component:285; Jaccard:0.062948
 |
Component:285; Jaccard:0.045624
 |
Correlation:0.52436
  |
Correlation:0.29838
  |
Correlation:0.34594
  |
Correlation:0.34594
  |
Correlation:0.52436
  |
Correlation:0.2268
  |
Correlation:0.2268
  |
Component:356; Jaccard:0.09965
 |
Component:285; Jaccard:0.1054
 |
Component:285; Jaccard:0.10392
 |
Component:285; Jaccard:0.097096
 |
Component:255; Jaccard:0.080037
 |
Component:255; Jaccard:0.06166
 |
Component:255; Jaccard:0.044267
 |
Correlation:0.40876
  |
Correlation:0.2268
  |
Correlation:0.2268
  |
Correlation:0.2268
  |
Correlation:0.53846
  |
Correlation:0.53846
  |
Correlation:0.53846
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































