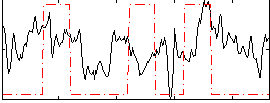
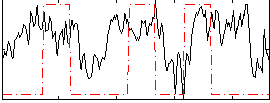
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
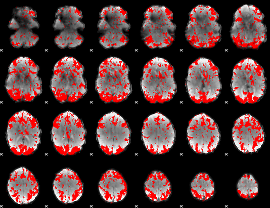 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
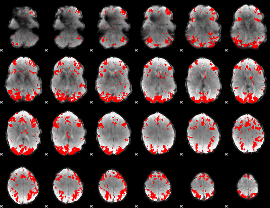 |
GLM binary result thresholded by 3.5
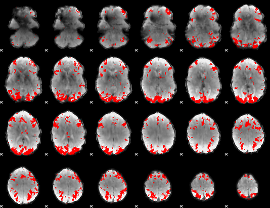 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
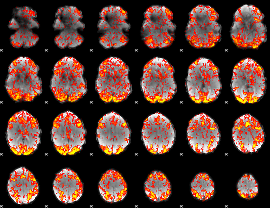 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
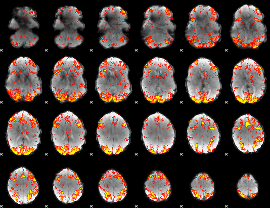 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
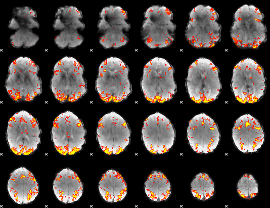 |
GLM Z-score result thresholded by 4
 |
Component:340; Jaccard:0.11247
 |
Component:340; Jaccard:0.13859
 |
Component:340; Jaccard:0.16676
 |
Component:343; Jaccard:0.19766
 |
Component:303; Jaccard:0.23145
 |
Component:303; Jaccard:0.26478
 |
Component:303; Jaccard:0.29098
 |
Correlation:0.17458
  |
Correlation:0.17458
  |
Correlation:0.17458
  |
Correlation:0.12005
  |
Correlation:0.17645
  |
Correlation:0.17645
  |
Correlation:0.17645
  |
Component:252; Jaccard:0.10811
 |
Component:343; Jaccard:0.12948
 |
Component:343; Jaccard:0.16184
 |
Component:303; Jaccard:0.19392
 |
Component:343; Jaccard:0.22816
 |
Component:343; Jaccard:0.25126
 |
Component:343; Jaccard:0.26388
 |
Correlation:0.24213
  |
Correlation:0.12005
  |
Correlation:0.12005
  |
Correlation:0.17645
  |
Correlation:0.12005
  |
Correlation:0.12005
  |
Correlation:0.12005
  |
Component:097; Jaccard:0.10687
 |
Component:097; Jaccard:0.12735
 |
Component:303; Jaccard:0.15634
 |
Component:340; Jaccard:0.19334
 |
Component:294; Jaccard:0.22441
 |
Component:294; Jaccard:0.24827
 |
Component:294; Jaccard:0.26202
 |
Correlation:0.13866
  |
Correlation:0.13866
  |
Correlation:0.17645
  |
Correlation:0.17458
  |
Correlation:0.22724
  |
Correlation:0.22724
  |
Correlation:0.22724
  |
Component:378; Jaccard:0.10685
 |
Component:252; Jaccard:0.12627
 |
Component:294; Jaccard:0.15462
 |
Component:294; Jaccard:0.19137
 |
Component:340; Jaccard:0.21387
 |
Component:340; Jaccard:0.22444
 |
Component:340; Jaccard:0.22537
 |
Correlation:0.24172
  |
Correlation:0.24213
  |
Correlation:0.22724
  |
Correlation:0.22724
  |
Correlation:0.17458
  |
Correlation:0.17458
  |
Correlation:0.17458
  |
Component:343; Jaccard:0.1021
 |
Component:378; Jaccard:0.12461
 |
Component:097; Jaccard:0.14613
 |
Component:221; Jaccard:0.16404
 |
Component:221; Jaccard:0.18165
 |
Component:221; Jaccard:0.19271
 |
Component:221; Jaccard:0.19723
 |
Correlation:0.12005
  |
Correlation:0.24172
  |
Correlation:0.13866
  |
Correlation:0.26981
  |
Correlation:0.26981
  |
Correlation:0.26981
  |
Correlation:0.26981
  |
Component:099; Jaccard:0.099635
 |
Component:303; Jaccard:0.12301
 |
Component:221; Jaccard:0.14285
 |
Component:097; Jaccard:0.16029
 |
Component:097; Jaccard:0.16817
 |
Component:097; Jaccard:0.16959
 |
Component:097; Jaccard:0.16557
 |
Correlation:0.1852
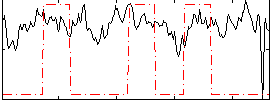  |
Correlation:0.17645
  |
Correlation:0.26981
  |
Correlation:0.13866
  |
Correlation:0.13866
  |
Correlation:0.13866
  |
Correlation:0.13866
  |
Component:181; Jaccard:0.099164
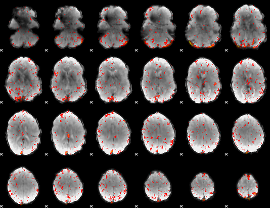 |
Component:294; Jaccard:0.1217
 |
Component:252; Jaccard:0.14179
 |
Component:252; Jaccard:0.15395
 |
Component:218; Jaccard:0.16497
 |
Component:218; Jaccard:0.1665
 |
Component:218; Jaccard:0.16138
 |
Correlation:0.37531
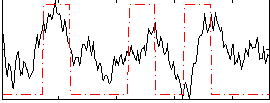  |
Correlation:0.22724
  |
Correlation:0.24213
  |
Correlation:0.24213
  |
Correlation:0.42088
  |
Correlation:0.42088
  |
Correlation:0.42088
  |
Component:221; Jaccard:0.098344
 |
Component:221; Jaccard:0.11947
 |
Component:378; Jaccard:0.14067
 |
Component:218; Jaccard:0.15358
 |
Component:252; Jaccard:0.16014
 |
Component:378; Jaccard:0.16089
 |
Component:378; Jaccard:0.15458
 |
Correlation:0.26981
  |
Correlation:0.26981
  |
Correlation:0.24172
  |
Correlation:0.42088
  |
Correlation:0.24213
  |
Correlation:0.24172
  |
Correlation:0.24172
  |
Component:303; Jaccard:0.095677
 |
Component:218; Jaccard:0.11537
 |
Component:218; Jaccard:0.13579
 |
Component:378; Jaccard:0.15329
 |
Component:378; Jaccard:0.15994
 |
Component:252; Jaccard:0.15722
 |
Component:252; Jaccard:0.1455
 |
Correlation:0.17645
  |
Correlation:0.42088
  |
Correlation:0.42088
  |
Correlation:0.24172
  |
Correlation:0.24172
  |
Correlation:0.24213
  |
Correlation:0.24213
  |
Component:218; Jaccard:0.09552
 |
Component:099; Jaccard:0.11516
 |
Component:099; Jaccard:0.12983
 |
Component:099; Jaccard:0.1405
 |
Component:199; Jaccard:0.14632
 |
Component:199; Jaccard:0.14749
 |
Component:390; Jaccard:0.14466
 |
Correlation:0.42088
  |
Correlation:0.1852
  |
Correlation:0.1852
  |
Correlation:0.1852
  |
Correlation:0.31893
 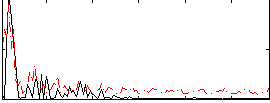 |
Correlation:0.31893
  |
Correlation:0.11549
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
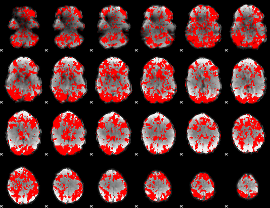 |
GLM binary result thresholded by 1.5
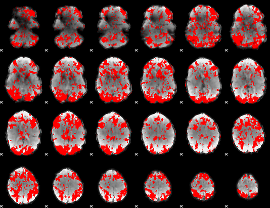 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
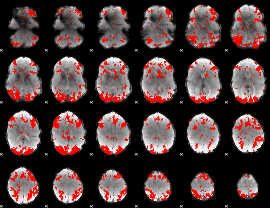 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
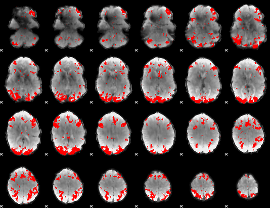 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
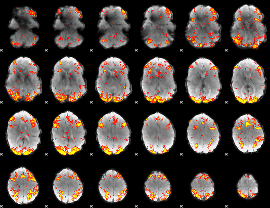
GLM Z-score result thresholded by 3
 |
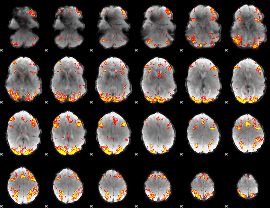
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:340; Jaccard:0.12548
 |
Component:303; Jaccard:0.15055
 |
Component:303; Jaccard:0.18976
 |
Component:303; Jaccard:0.23366
 |
Component:303; Jaccard:0.27804
 |
Component:303; Jaccard:0.31943
 |
Component:303; Jaccard:0.35265
 |
Correlation:0.042899
  |
Correlation:0.25751
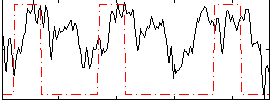 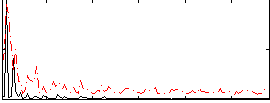 |
Correlation:0.25751
  |
Correlation:0.25751
  |
Correlation:0.25751
  |
Correlation:0.25751
  |
Correlation:0.25751
  |
Component:343; Jaccard:0.1221
 |
Component:343; Jaccard:0.14985
 |
Component:343; Jaccard:0.1818
 |
Component:343; Jaccard:0.21462
 |
Component:343; Jaccard:0.24762
 |
Component:343; Jaccard:0.27459
 |
Component:343; Jaccard:0.29212
 |
Correlation:0.17067
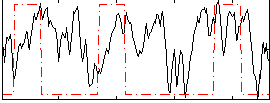  |
Correlation:0.17067
  |
Correlation:0.17067
  |
Correlation:0.17067
  |
Correlation:0.17067
  |
Correlation:0.17067
  |
Correlation:0.17067
  |
Component:303; Jaccard:0.11898
 |
Component:340; Jaccard:0.14709
 |
Component:294; Jaccard:0.17367
 |
Component:294; Jaccard:0.2083
 |
Component:294; Jaccard:0.24231
 |
Component:294; Jaccard:0.26645
 |
Component:294; Jaccard:0.28063
 |
Correlation:0.25751
  |
Correlation:0.042899
  |
Correlation:0.26675
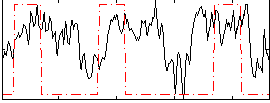  |
Correlation:0.26675
  |
Correlation:0.26675
  |
Correlation:0.26675
  |
Correlation:0.26675
  |
Component:099; Jaccard:0.11432
 |
Component:294; Jaccard:0.14106
 |
Component:340; Jaccard:0.16961
 |
Component:340; Jaccard:0.19087
 |
Component:340; Jaccard:0.20688
 |
Component:340; Jaccard:0.21578
 |
Component:340; Jaccard:0.21893
 |
Correlation:0.00040984
  |
Correlation:0.26675
  |
Correlation:0.042899
  |
Correlation:0.042899
  |
Correlation:0.042899
  |
Correlation:0.042899
  |
Correlation:0.042899
  |
Component:294; Jaccard:0.1134
 |
Component:099; Jaccard:0.12981
 |
Component:221; Jaccard:0.14758
 |
Component:221; Jaccard:0.16683
 |
Component:221; Jaccard:0.1826
 |
Component:221; Jaccard:0.19179
 |
Component:221; Jaccard:0.19411
 |
Correlation:0.26675
  |
Correlation:0.00040984
  |
Correlation:0.17682
 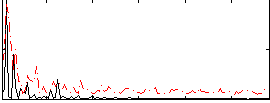 |
Correlation:0.17682
  |
Correlation:0.17682
  |
Correlation:0.17682
  |
Correlation:0.17682
  |
Component:252; Jaccard:0.11154
 |
Component:221; Jaccard:0.12838
 |
Component:099; Jaccard:0.14456
 |
Component:099; Jaccard:0.15715
 |
Component:099; Jaccard:0.16534
 |
Component:099; Jaccard:0.16591
 |
Component:099; Jaccard:0.16009
 |
Correlation:-0.1046
 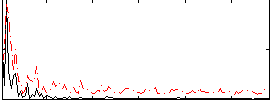 |
Correlation:0.17682
  |
Correlation:0.00040984
  |
Correlation:0.00040984
  |
Correlation:0.00040984
  |
Correlation:0.00040984
  |
Correlation:0.00040984
  |
Component:221; Jaccard:0.11077
 |
Component:252; Jaccard:0.12311
 |
Component:252; Jaccard:0.134
 |
Component:382; Jaccard:0.14157
 |
Component:382; Jaccard:0.14888
 |
Component:382; Jaccard:0.15088
 |
Component:382; Jaccard:0.14426
 |
Correlation:0.17682
  |
Correlation:-0.1046
  |
Correlation:-0.1046
  |
Correlation:0.14007
  |
Correlation:0.14007
  |
Correlation:0.14007
  |
Correlation:0.14007
  |
Component:097; Jaccard:0.10594
 |
Component:097; Jaccard:0.11915
 |
Component:097; Jaccard:0.13176
 |
Component:097; Jaccard:0.14059
 |
Component:097; Jaccard:0.14501
 |
Component:097; Jaccard:0.14442
 |
Component:390; Jaccard:0.1427
 |
Correlation:0.12421
  |
Correlation:0.12421
  |
Correlation:0.12421
  |
Correlation:0.12421
  |
Correlation:0.12421
  |
Correlation:0.12421
  |
Correlation:0.07916
  |
Component:382; Jaccard:0.10231
 |
Component:382; Jaccard:0.11657
 |
Component:382; Jaccard:0.13032
 |
Component:252; Jaccard:0.14047
 |
Component:252; Jaccard:0.14448
 |
Component:252; Jaccard:0.14297
 |
Component:097; Jaccard:0.14192
 |
Correlation:0.14007
  |
Correlation:0.14007
  |
Correlation:0.14007
  |
Correlation:-0.1046
  |
Correlation:-0.1046
  |
Correlation:-0.1046
  |
Correlation:0.12421
  |
Component:219; Jaccard:0.098648
 |
Component:219; Jaccard:0.10766
 |
Component:390; Jaccard:0.11717
 |
Component:390; Jaccard:0.12855
 |
Component:390; Jaccard:0.13715
 |
Component:390; Jaccard:0.14183
 |
Component:252; Jaccard:0.13689
 |
Correlation:-0.012902
  |
Correlation:-0.012902
  |
Correlation:0.07916
  |
Correlation:0.07916
  |
Correlation:0.07916
  |
Correlation:0.07916
  |
Correlation:-0.1046
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
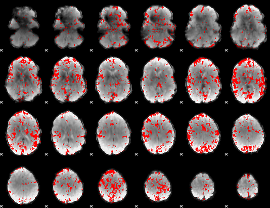
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
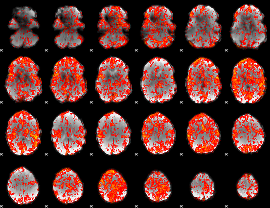 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:150; Jaccard:0.099293
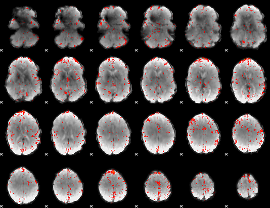 |
Component:150; Jaccard:0.1194
 |
Component:150; Jaccard:0.13961
 |
Component:150; Jaccard:0.15301
 |
Component:150; Jaccard:0.14967
 |
Component:150; Jaccard:0.12556
 |
Component:150; Jaccard:0.090135
 |
Correlation:0.34345
  |
Correlation:0.34345
  |
Correlation:0.34345
  |
Correlation:0.34345
  |
Correlation:0.34345
  |
Correlation:0.34345
  |
Correlation:0.34345
  |
Component:166; Jaccard:0.096021
 |
Component:166; Jaccard:0.10993
 |
Component:166; Jaccard:0.12261
 |
Component:166; Jaccard:0.12844
 |
Component:166; Jaccard:0.12261
 |
Component:107; Jaccard:0.10886
 |
Component:107; Jaccard:0.084686
 |
Correlation:0.19371
  |
Correlation:0.19371
  |
Correlation:0.19371
  |
Correlation:0.19371
  |
Correlation:0.19371
  |
Correlation:0.29953
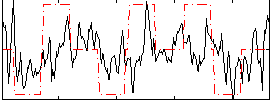  |
Correlation:0.29953
  |
Component:181; Jaccard:0.091014
 |
Component:181; Jaccard:0.10237
 |
Component:181; Jaccard:0.10981
 |
Component:385; Jaccard:0.11887
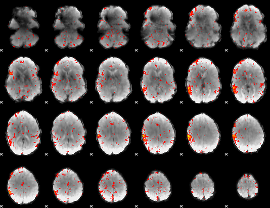 |
Component:385; Jaccard:0.11905
 |
Component:385; Jaccard:0.10544
 |
Component:385; Jaccard:0.081165
 |
Correlation:0.34789
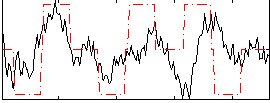  |
Correlation:0.34789
  |
Correlation:0.34789
  |
Correlation:0.36553
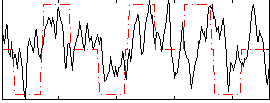  |
Correlation:0.36553
  |
Correlation:0.36553
  |
Correlation:0.36553
  |
Component:056; Jaccard:0.08906
 |
Component:056; Jaccard:0.099728
 |
Component:385; Jaccard:0.10928
 |
Component:107; Jaccard:0.11474
 |
Component:107; Jaccard:0.11797
 |
Component:166; Jaccard:0.10048
 |
Component:166; Jaccard:0.071911
 |
Correlation:0.4386
  |
Correlation:0.4386
  |
Correlation:0.36553
  |
Correlation:0.29953
  |
Correlation:0.29953
  |
Correlation:0.19371
  |
Correlation:0.19371
  |
Component:385; Jaccard:0.080578
 |
Component:385; Jaccard:0.094719
 |
Component:056; Jaccard:0.10738
 |
Component:181; Jaccard:0.11144
 |
Component:181; Jaccard:0.1043
 |
Component:181; Jaccard:0.083906
 |
Component:144; Jaccard:0.059747
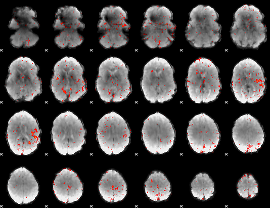 |
Correlation:0.36553
  |
Correlation:0.36553
  |
Correlation:0.4386
  |
Correlation:0.34789
  |
Correlation:0.34789
  |
Correlation:0.34789
  |
Correlation:0.31402
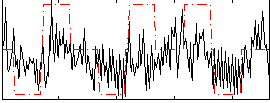  |
Component:012; Jaccard:0.079486
 |
Component:107; Jaccard:0.089303
 |
Component:107; Jaccard:0.1033
 |
Component:056; Jaccard:0.10785
 |
Component:056; Jaccard:0.097184
 |
Component:372; Jaccard:0.083111
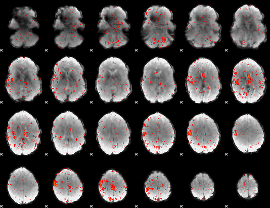 |
Component:372; Jaccard:0.059408
 |
Correlation:0.27009
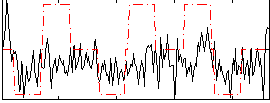 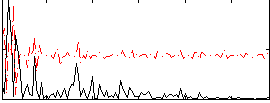 |
Correlation:0.29953
  |
Correlation:0.29953
  |
Correlation:0.4386
  |
Correlation:0.4386
  |
Correlation:0.4426
  |
Correlation:0.4426
  |
Component:107; Jaccard:0.07589
 |
Component:012; Jaccard:0.087212
 |
Component:012; Jaccard:0.092126
 |
Component:372; Jaccard:0.09386
 |
Component:372; Jaccard:0.094741
 |
Component:144; Jaccard:0.074834
 |
Component:181; Jaccard:0.058976
 |
Correlation:0.29953
  |
Correlation:0.27009
  |
Correlation:0.27009
  |
Correlation:0.4426
  |
Correlation:0.4426
  |
Correlation:0.31402
  |
Correlation:0.34789
  |
Component:239; Jaccard:0.07413
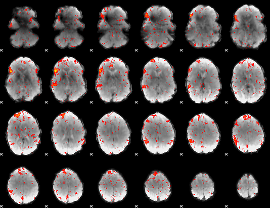 |
Component:008; Jaccard:0.080729
 |
Component:372; Jaccard:0.087276
 |
Component:012; Jaccard:0.08961
 |
Component:228; Jaccard:0.08678
 |
Component:228; Jaccard:0.07308
 |
Component:228; Jaccard:0.056144
 |
Correlation:0.23479
  |
Correlation:0.13244
  |
Correlation:0.4426
  |
Correlation:0.27009
  |
Correlation:0.42382
  |
Correlation:0.42382
  |
Correlation:0.42382
  |
Component:008; Jaccard:0.073361
 |
Component:239; Jaccard:0.080298
 |
Component:008; Jaccard:0.086258
 |
Component:008; Jaccard:0.087865
 |
Component:008; Jaccard:0.082559
 |
Component:056; Jaccard:0.072138
 |
Component:034; Jaccard:0.052692
 |
Correlation:0.13244
  |
Correlation:0.23479
  |
Correlation:0.13244
  |
Correlation:0.13244
  |
Correlation:0.13244
  |
Correlation:0.4386
  |
Correlation:0.30264
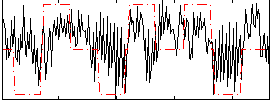  |
Component:387; Jaccard:0.07242
 |
Component:372; Jaccard:0.076941
 |
Component:239; Jaccard:0.082443
 |
Component:228; Jaccard:0.084676
 |
Component:144; Jaccard:0.081426
 |
Component:034; Jaccard:0.067308
 |
Component:046; Jaccard:0.052295
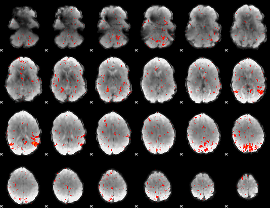 |
Correlation:0.26181
  |
Correlation:0.4426
  |
Correlation:0.23479
  |
Correlation:0.42382
  |
Correlation:0.31402
  |
Correlation:0.30264
  |
Correlation:0.29794
 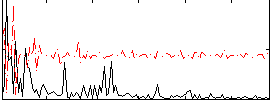 |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
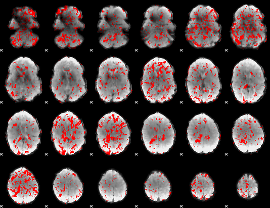 |
GLM binary result thresholded by 1.5
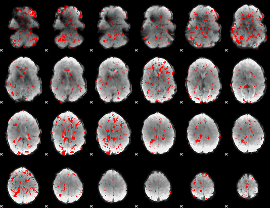 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
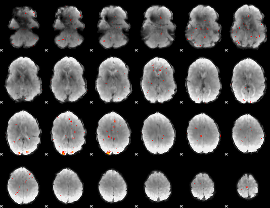 |
GLM Z-score result thresholded by 3.5
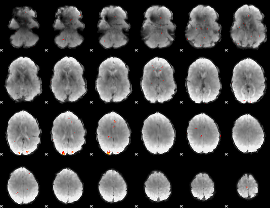 |
GLM Z-score result thresholded by 4
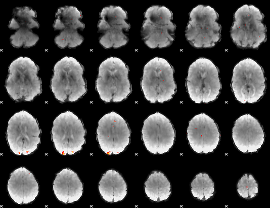 |
Component:071; Jaccard:0.088204
 |
Component:071; Jaccard:0.090302
 |
Component:071; Jaccard:0.078792
 |
Component:071; Jaccard:0.052617
 |
Component:303; Jaccard:0.033836
 |
Component:303; Jaccard:0.022311
 |
Component:303; Jaccard:0.016004
 |
Correlation:0.32827
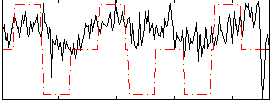  |
Correlation:0.32827
  |
Correlation:0.32827
  |
Correlation:0.32827
  |
Correlation:0.048044
  |
Correlation:0.048044
  |
Correlation:0.048044
  |
Component:341; Jaccard:0.07902
 |
Component:195; Jaccard:0.076736
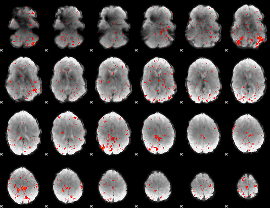 |
Component:195; Jaccard:0.065072
 |
Component:303; Jaccard:0.049176
 |
Component:071; Jaccard:0.028743
 |
Component:341; Jaccard:0.012295
 |
Component:341; Jaccard:0.008276
 |
Correlation:0.18723
 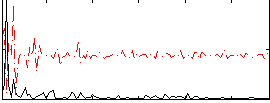 |
Correlation:0.18443
  |
Correlation:0.18443
  |
Correlation:0.048044
  |
Correlation:0.32827
  |
Correlation:0.18723
  |
Correlation:0.18723
  |
Component:172; Jaccard:0.075585
 |
Component:341; Jaccard:0.073747
 |
Component:303; Jaccard:0.063462
 |
Component:195; Jaccard:0.046947
 |
Component:195; Jaccard:0.023171
 |
Component:195; Jaccard:0.010036
 |
Component:099; Jaccard:0.004862
 |
Correlation:0.11616
 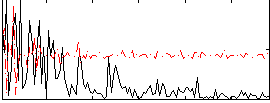 |
Correlation:0.18723
  |
Correlation:0.048044
  |
Correlation:0.18443
  |
Correlation:0.18443
  |
Correlation:0.18443
  |
Correlation:-0.10952
  |
Component:303; Jaccard:0.073314
 |
Component:303; Jaccard:0.070831
 |
Component:341; Jaccard:0.057963
 |
Component:341; Jaccard:0.039305
 |
Component:341; Jaccard:0.022811
 |
Component:071; Jaccard:0.009553
 |
Component:312; Jaccard:0.004785
 |
Correlation:0.048044
  |
Correlation:0.048044
  |
Correlation:0.18723
  |
Correlation:0.18723
  |
Correlation:0.18723
  |
Correlation:0.32827
  |
Correlation:0.27648
  |
Component:195; Jaccard:0.073005
 |
Component:307; Jaccard:0.068326
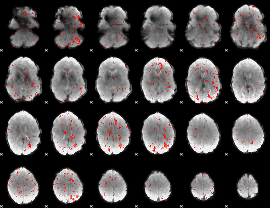 |
Component:307; Jaccard:0.057623
 |
Component:307; Jaccard:0.037475
 |
Component:373; Jaccard:0.018903
 |
Component:373; Jaccard:0.008076
 |
Component:195; Jaccard:0.004718
 |
Correlation:0.18443
  |
Correlation:0.21923
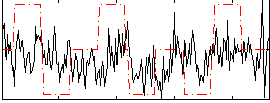  |
Correlation:0.21923
  |
Correlation:0.21923
  |
Correlation:0.12096
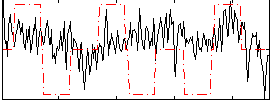  |
Correlation:0.12096
  |
Correlation:0.18443
  |
Component:348; Jaccard:0.069494
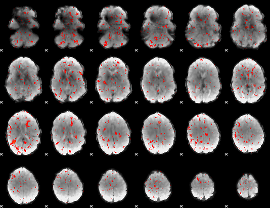 |
Component:348; Jaccard:0.065317
 |
Component:373; Jaccard:0.053968
 |
Component:373; Jaccard:0.03681
 |
Component:063; Jaccard:0.018628
 |
Component:099; Jaccard:0.007468
 |
Component:193; Jaccard:0.004191
 |
Correlation:0.15398
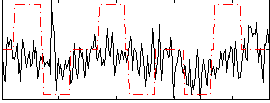  |
Correlation:0.15398
  |
Correlation:0.12096
  |
Correlation:0.12096
  |
Correlation:0.24889
  |
Correlation:-0.10952
  |
Correlation:0.15458
  |
Component:307; Jaccard:0.069366
 |
Component:373; Jaccard:0.064202
 |
Component:348; Jaccard:0.053193
 |
Component:318; Jaccard:0.034582
 |
Component:307; Jaccard:0.018365
 |
Component:312; Jaccard:0.007425
 |
Component:288; Jaccard:0.004169
 |
Correlation:0.21923
  |
Correlation:0.12096
  |
Correlation:0.15398
  |
Correlation:0.061438
  |
Correlation:0.21923
  |
Correlation:0.27648
  |
Correlation:0.26799
 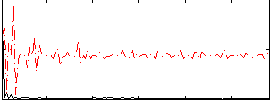 |
Component:139; Jaccard:0.06842
 |
Component:172; Jaccard:0.062446
 |
Component:318; Jaccard:0.052481
 |
Component:007; Jaccard:0.033489
 |
Component:312; Jaccard:0.016422
 |
Component:063; Jaccard:0.007389
 |
Component:343; Jaccard:0.00397
 |
Correlation:0.23222
  |
Correlation:0.11616
  |
Correlation:0.061438
  |
Correlation:0.020613
 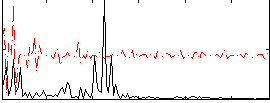 |
Correlation:0.27648
  |
Correlation:0.24889
  |
Correlation:0.030004
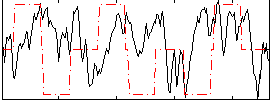 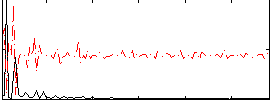 |
Component:313; Jaccard:0.06766
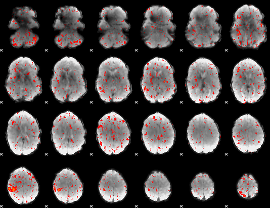 |
Component:318; Jaccard:0.06214
 |
Component:007; Jaccard:0.049915
 |
Component:348; Jaccard:0.033207
 |
Component:318; Jaccard:0.016247
 |
Component:307; Jaccard:0.007374
 |
Component:370; Jaccard:0.003939
 |
Correlation:0.079276
  |
Correlation:0.061438
  |
Correlation:0.020613
  |
Correlation:0.15398
  |
Correlation:0.061438
  |
Correlation:0.21923
  |
Correlation:0.05555
  |
Component:004; Jaccard:0.066089
 |
Component:004; Jaccard:0.061099
 |
Component:132; Jaccard:0.04947
 |
Component:132; Jaccard:0.032606
 |
Component:132; Jaccard:0.016208
 |
Component:193; Jaccard:0.007316
 |
Component:118; Jaccard:0.003894
 |
Correlation:0.23739
  |
Correlation:0.23739
  |
Correlation:0.018804
  |
Correlation:0.018804
  |
Correlation:0.018804
  |
Correlation:0.15458
  |
Correlation:0.19154
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:209; Jaccard:0.095825
 |
Component:209; Jaccard:0.10609
 |
Component:209; Jaccard:0.10234
 |
Component:209; Jaccard:0.082955
 |
Component:209; Jaccard:0.048587
 |
Component:209; Jaccard:0.021785
 |
Component:209; Jaccard:0.007489
 |
Correlation:-0.11792
  |
Correlation:-0.11792
  |
Correlation:-0.11792
  |
Correlation:-0.11792
  |
Correlation:-0.11792
  |
Correlation:-0.11792
  |
Correlation:-0.11792
  |
Component:042; Jaccard:0.095339
 |
Component:042; Jaccard:0.091402
 |
Component:042; Jaccard:0.0756
 |
Component:042; Jaccard:0.049835
 |
Component:042; Jaccard:0.025508
 |
Component:042; Jaccard:0.010227
 |
Component:062; Jaccard:0.003714
 |
Correlation:0.0007872
  |
Correlation:0.0007872
  |
Correlation:0.0007872
  |
Correlation:0.0007872
  |
Correlation:0.0007872
  |
Correlation:0.0007872
  |
Correlation:0.18664
 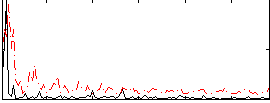 |
Component:035; Jaccard:0.077235
 |
Component:035; Jaccard:0.074455
 |
Component:331; Jaccard:0.060338
 |
Component:066; Jaccard:0.04
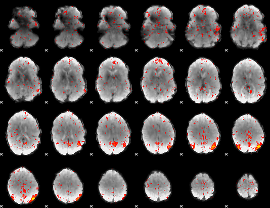 |
Component:066; Jaccard:0.018863
 |
Component:062; Jaccard:0.008964
 |
Component:290; Jaccard:0.003488
 |
Correlation:0.097471
  |
Correlation:0.097471
  |
Correlation:0.096988
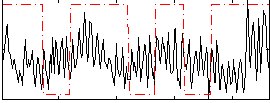  |
Correlation:-0.036435
  |
Correlation:-0.036435
  |
Correlation:0.18664
  |
Correlation:0.15529
  |
Component:066; Jaccard:0.072133
 |
Component:066; Jaccard:0.069541
 |
Component:066; Jaccard:0.059006
 |
Component:035; Jaccard:0.037995
 |
Component:367; Jaccard:0.018835
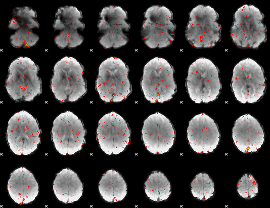 |
Component:290; Jaccard:0.008541
 |
Component:113; Jaccard:0.003314
 |
Correlation:-0.036435
  |
Correlation:-0.036435
  |
Correlation:-0.036435
  |
Correlation:0.097471
  |
Correlation:0.17631
  |
Correlation:0.15529
  |
Correlation:0.079278
  |
Component:032; Jaccard:0.070803
 |
Component:228; Jaccard:0.068912
 |
Component:228; Jaccard:0.058556
 |
Component:331; Jaccard:0.036941
 |
Component:062; Jaccard:0.018788
 |
Component:032; Jaccard:0.008524
 |
Component:042; Jaccard:0.003018
 |
Correlation:0.10784
  |
Correlation:-0.18317
  |
Correlation:-0.18317
  |
Correlation:0.096988
  |
Correlation:0.18664
  |
Correlation:0.10784
  |
Correlation:0.0007872
  |
Component:228; Jaccard:0.070248
 |
Component:331; Jaccard:0.068812
 |
Component:035; Jaccard:0.05823
 |
Component:212; Jaccard:0.036258
 |
Component:032; Jaccard:0.018556
 |
Component:113; Jaccard:0.007627
 |
Component:367; Jaccard:0.002736
 |
Correlation:-0.18317
  |
Correlation:0.096988
  |
Correlation:0.097471
  |
Correlation:-0.022497
  |
Correlation:0.10784
  |
Correlation:0.079278
  |
Correlation:0.17631
  |
Component:388; Jaccard:0.068995
 |
Component:186; Jaccard:0.068019
 |
Component:212; Jaccard:0.05586
 |
Component:186; Jaccard:0.036257
 |
Component:290; Jaccard:0.018075
 |
Component:066; Jaccard:0.007575
 |
Component:212; Jaccard:0.00262
 |
Correlation:0.041503
  |
Correlation:-0.06122
  |
Correlation:-0.022497
  |
Correlation:-0.06122
  |
Correlation:0.15529
  |
Correlation:-0.036435
  |
Correlation:-0.022497
  |
Component:186; Jaccard:0.068298
 |
Component:032; Jaccard:0.066318
 |
Component:186; Jaccard:0.055542
 |
Component:032; Jaccard:0.035023
 |
Component:133; Jaccard:0.018065
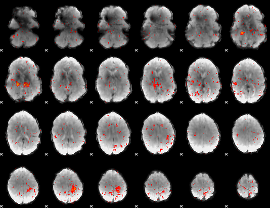 |
Component:240; Jaccard:0.00743
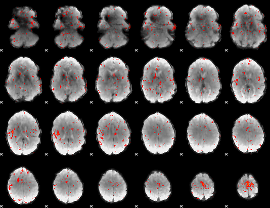 |
Component:073; Jaccard:0.002604
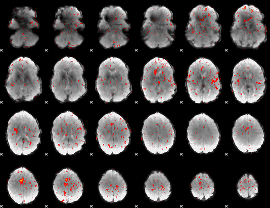 |
Correlation:-0.06122
  |
Correlation:0.10784
  |
Correlation:-0.06122
  |
Correlation:0.10784
  |
Correlation:-0.060035
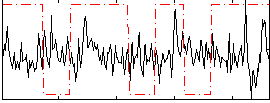 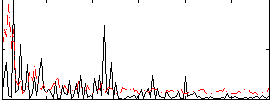 |
Correlation:0.17851
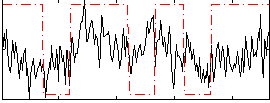  |
Correlation:0.1778
 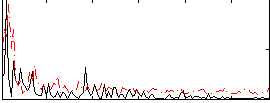 |
Component:212; Jaccard:0.06582
 |
Component:212; Jaccard:0.065282
 |
Component:032; Jaccard:0.053837
 |
Component:133; Jaccard:0.034403
 |
Component:035; Jaccard:0.018005
 |
Component:331; Jaccard:0.007107
 |
Component:032; Jaccard:0.002451
 |
Correlation:-0.022497
  |
Correlation:-0.022497
  |
Correlation:0.10784
  |
Correlation:-0.060035
  |
Correlation:0.097471
  |
Correlation:0.096988
  |
Correlation:0.10784
  |
Component:290; Jaccard:0.065277
 |
Component:217; Jaccard:0.062299
 |
Component:133; Jaccard:0.050663
 |
Component:228; Jaccard:0.033971
 |
Component:186; Jaccard:0.017993
 |
Component:355; Jaccard:0.00708
 |
Component:240; Jaccard:0.002404
 |
Correlation:0.15529
  |
Correlation:-0.019072
  |
Correlation:-0.060035
  |
Correlation:-0.18317
  |
Correlation:-0.06122
  |
Correlation:0.15969
  |
Correlation:0.17851
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
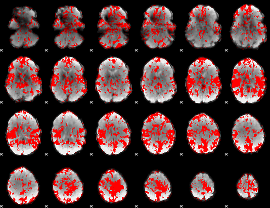 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
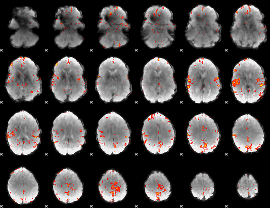 |
GLM Z-score result thresholded by 4
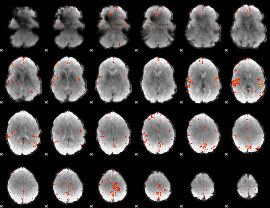 |
Component:166; Jaccard:0.094019
 |
Component:228; Jaccard:0.1014
 |
Component:228; Jaccard:0.11713
 |
Component:228; Jaccard:0.12958
 |
Component:209; Jaccard:0.13419
 |
Component:209; Jaccard:0.13173
 |
Component:209; Jaccard:0.11766
 |
Correlation:0.21704
  |
Correlation:0.5319
 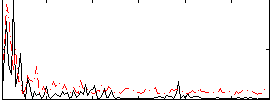 |
Correlation:0.5319
  |
Correlation:0.5319
  |
Correlation:0.21199
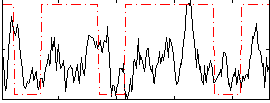  |
Correlation:0.21199
  |
Correlation:0.21199
  |
Component:388; Jaccard:0.088613
 |
Component:166; Jaccard:0.099902
 |
Component:209; Jaccard:0.10903
 |
Component:209; Jaccard:0.12352
 |
Component:228; Jaccard:0.13235
 |
Component:228; Jaccard:0.12301
 |
Component:228; Jaccard:0.10691
 |
Correlation:0.1657
  |
Correlation:0.21704
  |
Correlation:0.21199
  |
Correlation:0.21199
  |
Correlation:0.5319
  |
Correlation:0.5319
  |
Correlation:0.5319
  |
Component:042; Jaccard:0.087092
 |
Component:042; Jaccard:0.093364
 |
Component:212; Jaccard:0.10408
 |
Component:212; Jaccard:0.11418
 |
Component:212; Jaccard:0.11643
 |
Component:212; Jaccard:0.11076
 |
Component:212; Jaccard:0.094338
 |
Correlation:0.047607
  |
Correlation:0.047607
  |
Correlation:0.35505
  |
Correlation:0.35505
  |
Correlation:0.35505
  |
Correlation:0.35505
  |
Correlation:0.35505
  |
Component:228; Jaccard:0.086813
 |
Component:209; Jaccard:0.092721
 |
Component:166; Jaccard:0.10043
 |
Component:331; Jaccard:0.10749
 |
Component:331; Jaccard:0.11148
 |
Component:331; Jaccard:0.10246
 |
Component:331; Jaccard:0.084681
 |
Correlation:0.5319
  |
Correlation:0.21199
  |
Correlation:0.21704
  |
Correlation:0.30732
  |
Correlation:0.30732
  |
Correlation:0.30732
  |
Correlation:0.30732
  |
Component:008; Jaccard:0.082894
 |
Component:388; Jaccard:0.092578
 |
Component:290; Jaccard:0.10001
 |
Component:290; Jaccard:0.10423
 |
Component:290; Jaccard:0.10247
 |
Component:144; Jaccard:0.094065
 |
Component:144; Jaccard:0.080884
 |
Correlation:0.12458
  |
Correlation:0.1657
  |
Correlation:0.12846
  |
Correlation:0.12846
  |
Correlation:0.12846
  |
Correlation:0.37385
  |
Correlation:0.37385
  |
Component:290; Jaccard:0.082499
 |
Component:212; Jaccard:0.092298
 |
Component:331; Jaccard:0.099545
 |
Component:186; Jaccard:0.10296
 |
Component:186; Jaccard:0.099638
 |
Component:290; Jaccard:0.092288
 |
Component:290; Jaccard:0.075489
 |
Correlation:0.12846
  |
Correlation:0.35505
  |
Correlation:0.30732
  |
Correlation:0.43193
  |
Correlation:0.43193
  |
Correlation:0.12846
  |
Correlation:0.12846
  |
Component:212; Jaccard:0.081319
 |
Component:290; Jaccard:0.092082
 |
Component:186; Jaccard:0.097951
 |
Component:075; Jaccard:0.10042
 |
Component:144; Jaccard:0.098714
 |
Component:186; Jaccard:0.090353
 |
Component:186; Jaccard:0.073621
 |
Correlation:0.35505
  |
Correlation:0.12846
  |
Correlation:0.43193
  |
Correlation:0.48484
  |
Correlation:0.37385
  |
Correlation:0.43193
  |
Correlation:0.43193
  |
Component:107; Jaccard:0.080338
 |
Component:186; Jaccard:0.089891
 |
Component:042; Jaccard:0.097302
 |
Component:107; Jaccard:0.099401
 |
Component:075; Jaccard:0.09836
 |
Component:075; Jaccard:0.087486
 |
Component:107; Jaccard:0.071268
 |
Correlation:0.39858
  |
Correlation:0.43193
  |
Correlation:0.047607
  |
Correlation:0.39858
  |
Correlation:0.48484
  |
Correlation:0.48484
  |
Correlation:0.39858
  |
Component:150; Jaccard:0.079457
 |
Component:008; Jaccard:0.089756
 |
Component:075; Jaccard:0.09587
 |
Component:166; Jaccard:0.099253
 |
Component:107; Jaccard:0.096932
 |
Component:107; Jaccard:0.086916
 |
Component:075; Jaccard:0.069641
 |
Correlation:0.30599
  |
Correlation:0.12458
  |
Correlation:0.48484
  |
Correlation:0.21704
  |
Correlation:0.39858
  |
Correlation:0.39858
  |
Correlation:0.48484
  |
Component:186; Jaccard:0.079172
 |
Component:075; Jaccard:0.088467
 |
Component:008; Jaccard:0.095441
 |
Component:042; Jaccard:0.098162
 |
Component:042; Jaccard:0.094272
 |
Component:042; Jaccard:0.083652
 |
Component:042; Jaccard:0.0673
 |
Correlation:0.43193
  |
Correlation:0.48484
  |
Correlation:0.12458
  |
Correlation:0.047607
  |
Correlation:0.047607
  |
Correlation:0.047607
  |
Correlation:0.047607
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































