Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
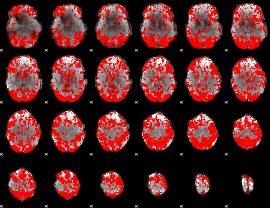 |
GLM binary result thresholded by 1.5
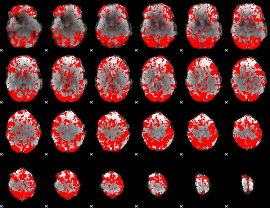 |
GLM binary result thresholded by 2
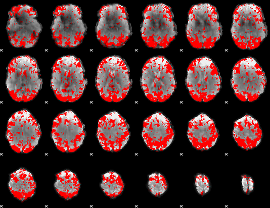 |
GLM binary result thresholded by 2.5
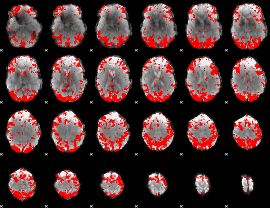 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
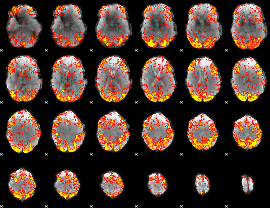 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
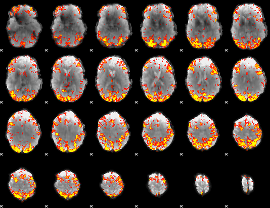 |
GLM Z-score result thresholded by 3.5
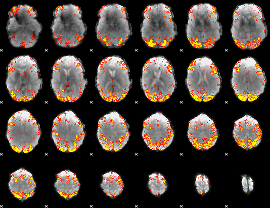 |
GLM Z-score result thresholded by 4
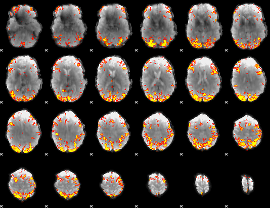 |
Component:206; Jaccard:0.12011
 |
Component:206; Jaccard:0.14688
 |
Component:206; Jaccard:0.17929
 |
Component:206; Jaccard:0.21615
 |
Component:206; Jaccard:0.25377
 |
Component:206; Jaccard:0.28975
 |
Component:206; Jaccard:0.32086
 |
Correlation:0.20153
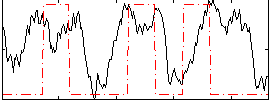 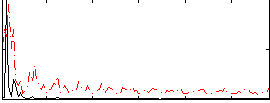 |
Correlation:0.20153
  |
Correlation:0.20153
  |
Correlation:0.20153
  |
Correlation:0.20153
  |
Correlation:0.20153
  |
Correlation:0.20153
  |
Component:331; Jaccard:0.11419
 |
Component:331; Jaccard:0.13819
 |
Component:331; Jaccard:0.16519
 |
Component:331; Jaccard:0.1939
 |
Component:331; Jaccard:0.21961
 |
Component:331; Jaccard:0.24161
 |
Component:331; Jaccard:0.25685
 |
Correlation:0.23867
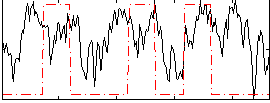 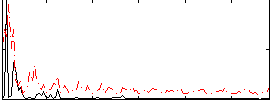 |
Correlation:0.23867
  |
Correlation:0.23867
  |
Correlation:0.23867
  |
Correlation:0.23867
  |
Correlation:0.23867
  |
Correlation:0.23867
  |
Component:163; Jaccard:0.11352
 |
Component:163; Jaccard:0.13594
 |
Component:163; Jaccard:0.15999
 |
Component:163; Jaccard:0.18325
 |
Component:163; Jaccard:0.20137
 |
Component:163; Jaccard:0.21306
 |
Component:237; Jaccard:0.22804
 |
Correlation:0.14671
 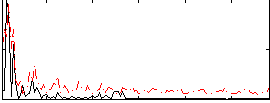 |
Correlation:0.14671
  |
Correlation:0.14671
  |
Correlation:0.14671
  |
Correlation:0.14671
  |
Correlation:0.14671
  |
Correlation:0.15241
 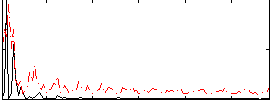 |
Component:255; Jaccard:0.10402
 |
Component:255; Jaccard:0.12249
 |
Component:255; Jaccard:0.14095
 |
Component:242; Jaccard:0.16458
 |
Component:237; Jaccard:0.18688
 |
Component:237; Jaccard:0.21051
 |
Component:242; Jaccard:0.21749
 |
Correlation:0.027077
 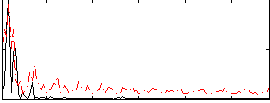 |
Correlation:0.027077
  |
Correlation:0.027077
  |
Correlation:0.19234
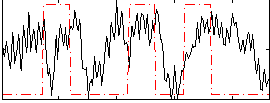  |
Correlation:0.15241
  |
Correlation:0.15241
  |
Correlation:0.19234
  |
Component:354; Jaccard:0.10125
 |
Component:354; Jaccard:0.11898
 |
Component:242; Jaccard:0.14072
 |
Component:237; Jaccard:0.1633
 |
Component:242; Jaccard:0.18664
 |
Component:242; Jaccard:0.2056
 |
Component:163; Jaccard:0.21682
 |
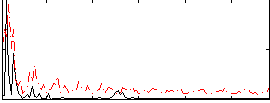
Correlation:0.094991
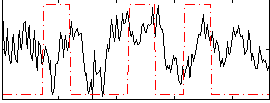 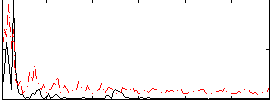 |
Correlation:0.094991
  |
Correlation:0.19234
  |
Correlation:0.15241
  |
Correlation:0.19234
  |
Correlation:0.19234
  |
Correlation:0.14671
  |
Component:149; Jaccard:0.098358
 |
Component:242; Jaccard:0.11839
 |
Component:149; Jaccard:0.13922
 |
Component:149; Jaccard:0.16216
 |
Component:149; Jaccard:0.18371
 |
Component:149; Jaccard:0.20169
 |
Component:149; Jaccard:0.21532
 |
Correlation:0.22685
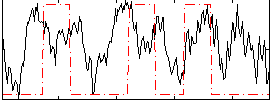 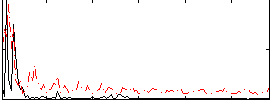 |
Correlation:0.19234
  |
Correlation:0.22685
  |
Correlation:0.22685
  |
Correlation:0.22685
  |
Correlation:0.22685
  |
Correlation:0.22685
  |
Component:242; Jaccard:0.098228
 |
Component:149; Jaccard:0.11741
 |
Component:237; Jaccard:0.13893
 |
Component:255; Jaccard:0.15796
 |
Component:255; Jaccard:0.17109
 |
Component:354; Jaccard:0.18238
 |
Component:354; Jaccard:0.18675
 |
Correlation:0.19234
  |
Correlation:0.22685
  |
Correlation:0.15241
  |
Correlation:0.027077
  |
Correlation:0.027077
  |
Correlation:0.094991
  |
Correlation:0.094991
  |
Component:327; Jaccard:0.095954
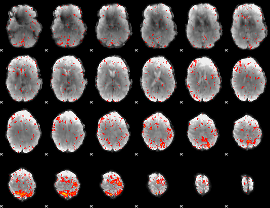 |
Component:237; Jaccard:0.1159
 |
Component:354; Jaccard:0.13804
 |
Component:354; Jaccard:0.156
 |
Component:354; Jaccard:0.1702
 |
Component:255; Jaccard:0.17625
 |
Component:255; Jaccard:0.17473
 |
Correlation:-0.017401
 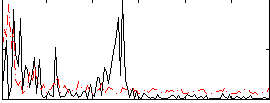 |
Correlation:0.15241
  |
Correlation:0.094991
  |
Correlation:0.094991
  |
Correlation:0.094991
  |
Correlation:0.027077
  |
Correlation:0.027077
  |
Component:237; Jaccard:0.095841
 |
Component:273; Jaccard:0.10714
 |
Component:273; Jaccard:0.11974
 |
Component:273; Jaccard:0.13209
 |
Component:273; Jaccard:0.14343
 |
Component:273; Jaccard:0.15187
 |
Component:273; Jaccard:0.15672
 |
Correlation:0.15241
  |
Correlation:-0.075987
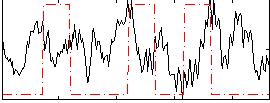 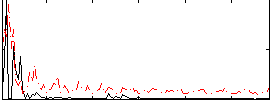 |
Correlation:-0.075987
  |
Correlation:-0.075987
  |
Correlation:-0.075987
  |
Correlation:-0.075987
  |
Correlation:-0.075987
  |
Component:273; Jaccard:0.094521
 |
Component:327; Jaccard:0.10712
 |
Component:116; Jaccard:0.11941
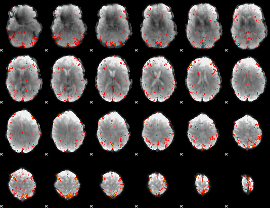 |
Component:116; Jaccard:0.13092
 |
Component:116; Jaccard:0.14039
 |
Component:116; Jaccard:0.14645
 |
Component:116; Jaccard:0.14956
 |
Correlation:-0.075987
  |
Correlation:-0.017401
  |
Correlation:-0.12608
  |
Correlation:-0.12608
  |
Correlation:-0.12608
  |
Correlation:-0.12608
  |
Correlation:-0.12608
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
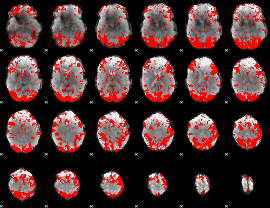 |
GLM binary result thresholded by 2.5
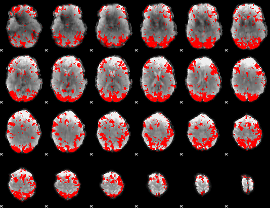 |
GLM binary result thresholded by 3
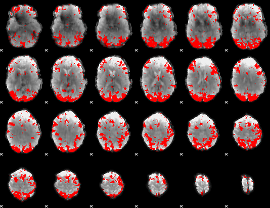 |
GLM binary result thresholded by 3.5
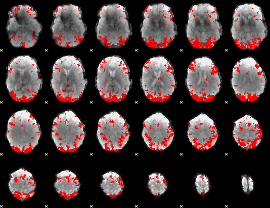 |
GLM binary result thresholded by 4
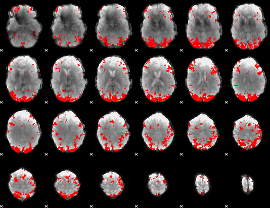 |
GLM Z-score result thresholded by 1
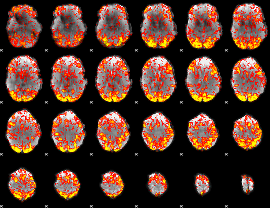 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
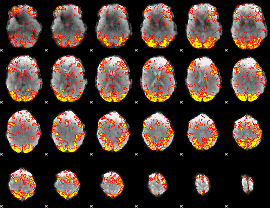 |
GLM Z-score result thresholded by 2.5
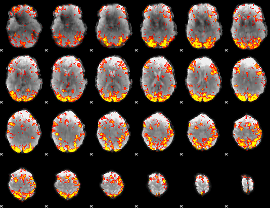 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:206; Jaccard:0.1325
 |
Component:206; Jaccard:0.16349
 |
Component:206; Jaccard:0.19956
 |
Component:206; Jaccard:0.23936
 |
Component:206; Jaccard:0.27817
 |
Component:206; Jaccard:0.31037
 |
Component:206; Jaccard:0.3349
 |
Correlation:0.11582
  |
Correlation:0.11582
  |
Correlation:0.11582
  |
Correlation:0.11582
  |
Correlation:0.11582
  |
Correlation:0.11582
  |
Correlation:0.11582
  |
Component:331; Jaccard:0.12661
 |
Component:331; Jaccard:0.15512
 |
Component:331; Jaccard:0.18728
 |
Component:331; Jaccard:0.22066
 |
Component:331; Jaccard:0.25088
 |
Component:331; Jaccard:0.2755
 |
Component:331; Jaccard:0.29049
 |
Correlation:0.29253
  |
Correlation:0.29253
  |
Correlation:0.29253
  |
Correlation:0.29253
  |
Correlation:0.29253
  |
Correlation:0.29253
  |
Correlation:0.29253
  |
Component:163; Jaccard:0.11342
 |
Component:149; Jaccard:0.13574
 |
Component:149; Jaccard:0.16391
 |
Component:149; Jaccard:0.19285
 |
Component:237; Jaccard:0.22329
 |
Component:237; Jaccard:0.25094
 |
Component:237; Jaccard:0.27381
 |
Correlation:0.11973
  |
Correlation:0.069162
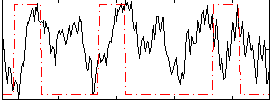 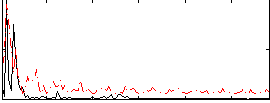 |
Correlation:0.069162
  |
Correlation:0.069162
  |
Correlation:0.31109
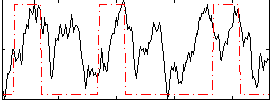 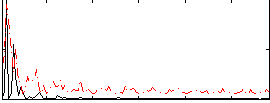 |
Correlation:0.31109
  |
Correlation:0.31109
  |
Component:149; Jaccard:0.11156
 |
Component:237; Jaccard:0.13235
 |
Component:237; Jaccard:0.16133
 |
Component:237; Jaccard:0.19253
 |
Component:149; Jaccard:0.21808
 |
Component:149; Jaccard:0.24006
 |
Component:149; Jaccard:0.2549
 |
Correlation:0.069162
  |
Correlation:0.31109
  |
Correlation:0.31109
  |
Correlation:0.31109
  |
Correlation:0.069162
  |
Correlation:0.069162
  |
Correlation:0.069162
  |
Component:273; Jaccard:0.11049
 |
Component:163; Jaccard:0.13081
 |
Component:163; Jaccard:0.14849
 |
Component:242; Jaccard:0.16906
 |
Component:242; Jaccard:0.18553
 |
Component:242; Jaccard:0.19821
 |
Component:242; Jaccard:0.20374
 |
Correlation:0.20572
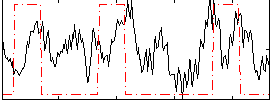 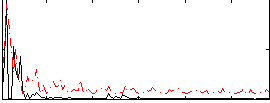 |
Correlation:0.11973
  |
Correlation:0.11973
  |
Correlation:0.23654
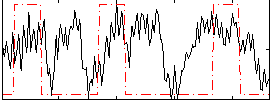 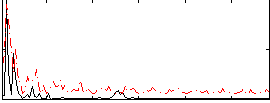 |
Correlation:0.23654
  |
Correlation:0.23654
  |
Correlation:0.23654
  |
Component:354; Jaccard:0.10908
 |
Component:273; Jaccard:0.1286
 |
Component:242; Jaccard:0.14727
 |
Component:163; Jaccard:0.16323
 |
Component:273; Jaccard:0.17629
 |
Component:273; Jaccard:0.18385
 |
Component:273; Jaccard:0.18724
 |
Correlation:0.084184
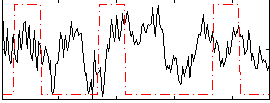 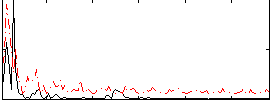 |
Correlation:0.20572
  |
Correlation:0.23654
  |
Correlation:0.11973
  |
Correlation:0.20572
  |
Correlation:0.20572
  |
Correlation:0.20572
  |
Component:237; Jaccard:0.10804
 |
Component:354; Jaccard:0.12689
 |
Component:273; Jaccard:0.14699
 |
Component:273; Jaccard:0.1626
 |
Component:163; Jaccard:0.17238
 |
Component:354; Jaccard:0.17572
 |
Component:052; Jaccard:0.17963
 |
Correlation:0.31109
  |
Correlation:0.084184
  |
Correlation:0.20572
  |
Correlation:0.20572
  |
Correlation:0.11973
  |
Correlation:0.084184
  |
Correlation:0.27935
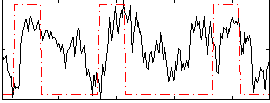 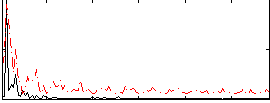 |
Component:255; Jaccard:0.10802
 |
Component:242; Jaccard:0.1253
 |
Component:354; Jaccard:0.14526
 |
Component:354; Jaccard:0.16095
 |
Component:354; Jaccard:0.17194
 |
Component:163; Jaccard:0.17293
 |
Component:354; Jaccard:0.1733
 |
Correlation:0.15862
  |
Correlation:0.23654
  |
Correlation:0.084184
  |
Correlation:0.084184
  |
Correlation:0.084184
  |
Correlation:0.11973
  |
Correlation:0.084184
  |
Component:242; Jaccard:0.10486
 |
Component:255; Jaccard:0.12311
 |
Component:255; Jaccard:0.13772
 |
Component:255; Jaccard:0.14783
 |
Component:052; Jaccard:0.16017
 |
Component:052; Jaccard:0.17201
 |
Component:163; Jaccard:0.17153
 |
Correlation:0.23654
  |
Correlation:0.15862
  |
Correlation:0.15862
  |
Correlation:0.15862
  |
Correlation:0.27935
  |
Correlation:0.27935
  |
Correlation:0.11973
  |
Component:327; Jaccard:0.102
 |
Component:327; Jaccard:0.11306
 |
Component:052; Jaccard:0.12674
 |
Component:052; Jaccard:0.14396
 |
Component:255; Jaccard:0.15008
 |
Component:255; Jaccard:0.14694
 |
Component:055; Jaccard:0.13925
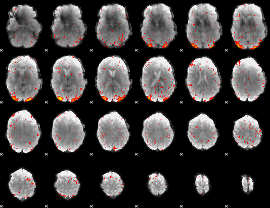 |
Correlation:0.037097
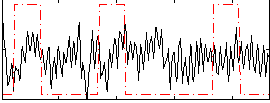 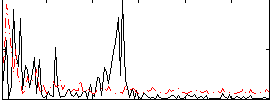 |
Correlation:0.037097
  |
Correlation:0.27935
  |
Correlation:0.27935
  |
Correlation:0.15862
  |
Correlation:0.15862
  |
Correlation:0.026426
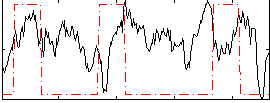  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
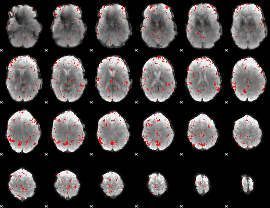 |
GLM binary result thresholded by 3
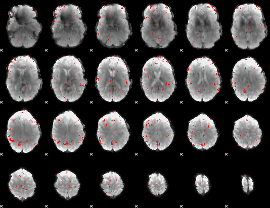 |
GLM binary result thresholded by 3.5
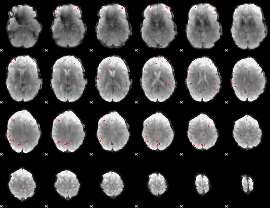 |
GLM binary result thresholded by 4
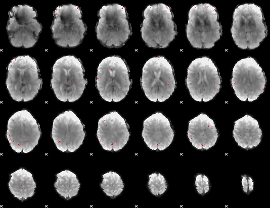 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
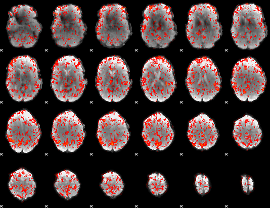 |
GLM Z-score result thresholded by 2
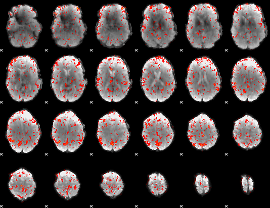 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
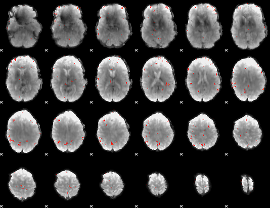 |
GLM Z-score result thresholded by 4
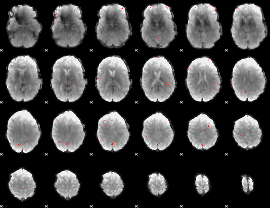 |
Component:148; Jaccard:0.12254
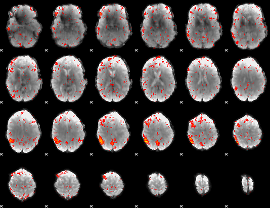 |
Component:148; Jaccard:0.14863
 |
Component:148; Jaccard:0.15986
 |
Component:148; Jaccard:0.14213
 |
Component:148; Jaccard:0.09568
 |
Component:148; Jaccard:0.048615
 |
Component:148; Jaccard:0.02122
 |
Correlation:0.12907
 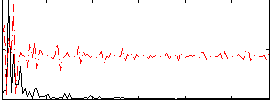 |
Correlation:0.12907
  |
Correlation:0.12907
  |
Correlation:0.12907
  |
Correlation:0.12907
  |
Correlation:0.12907
  |
Correlation:0.12907
  |
Component:163; Jaccard:0.091014
 |
Component:163; Jaccard:0.092382
 |
Component:163; Jaccard:0.085723
 |
Component:129; Jaccard:0.074618
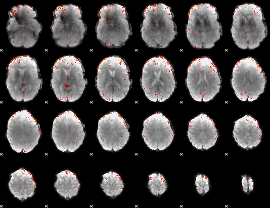 |
Component:129; Jaccard:0.055666
 |
Component:129; Jaccard:0.033877
 |
Component:129; Jaccard:0.015722
 |
Correlation:0.015997
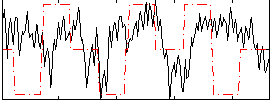  |
Correlation:0.015997
  |
Correlation:0.015997
  |
Correlation:0.3378
 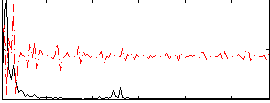 |
Correlation:0.3378
  |
Correlation:0.3378
  |
Correlation:0.3378
  |
Component:391; Jaccard:0.077679
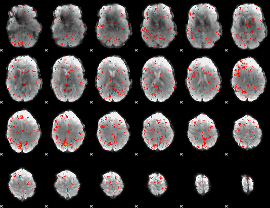 |
Component:391; Jaccard:0.084412
 |
Component:391; Jaccard:0.079941
 |
Component:163; Jaccard:0.068433
 |
Component:391; Jaccard:0.044178
 |
Component:391; Jaccard:0.022948
 |
Component:347; Jaccard:0.010996
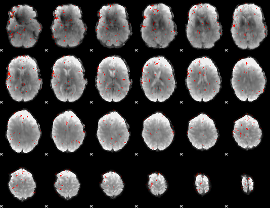 |
Correlation:0.092567
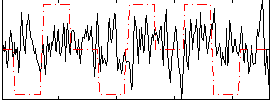  |
Correlation:0.092567
  |
Correlation:0.092567
  |
Correlation:0.015997
  |
Correlation:0.092567
  |
Correlation:0.092567
  |
Correlation:0.1383
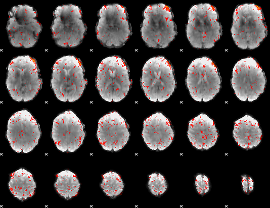
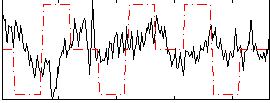
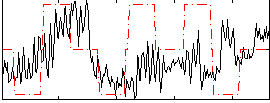  |
Component:194; Jaccard:0.07349
 |
Component:194; Jaccard:0.0776
 |
Component:129; Jaccard:0.07684
 |
Component:391; Jaccard:0.065891
 |
Component:163; Jaccard:0.043636
 |
Component:347; Jaccard:0.022506
 |
Component:391; Jaccard:0.010454
 |
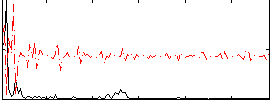
Correlation:-0.072879
  |
Correlation:-0.072879
  |
Correlation:0.3378
  |
Correlation:0.092567
  |
Correlation:0.015997
  |
Correlation:0.1383
  |
Correlation:0.092567
  |
Component:019; Jaccard:0.072684
 |
Component:246; Jaccard:0.074261
 |
Component:225; Jaccard:0.07491
 |
Component:225; Jaccard:0.065799
 |
Component:225; Jaccard:0.042231
 |
Component:163; Jaccard:0.022259
 |
Component:163; Jaccard:0.010132
 |
Correlation:0.018422
  |
Correlation:0.1461
  |
Correlation:0.27496
  |
Correlation:0.27496
  |
Correlation:0.27496
  |
Correlation:0.015997
  |
Correlation:0.015997
  |
Component:255; Jaccard:0.071672
 |
Component:225; Jaccard:0.073649
 |
Component:194; Jaccard:0.073667
 |
Component:335; Jaccard:0.06104
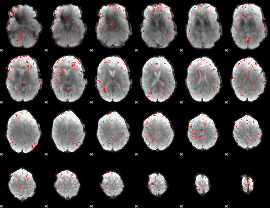 |
Component:335; Jaccard:0.040801
 |
Component:225; Jaccard:0.021697
 |
Component:335; Jaccard:0.009718
 |
Correlation:-0.077965
  |
Correlation:0.27496
  |
Correlation:-0.072879
  |
Correlation:0.26647
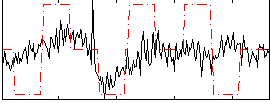 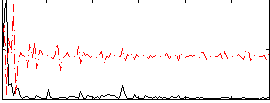 |
Correlation:0.26647
  |
Correlation:0.27496
  |
Correlation:0.26647
  |
Component:392; Jaccard:0.07043
 |
Component:392; Jaccard:0.072207
 |
Component:246; Jaccard:0.071621
 |
Component:246; Jaccard:0.058567
 |
Component:173; Jaccard:0.038816
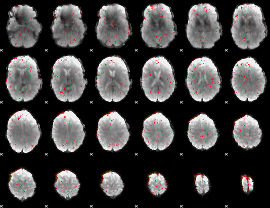 |
Component:173; Jaccard:0.021001
 |
Component:225; Jaccard:0.009456
 |
Correlation:0.1889
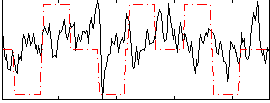 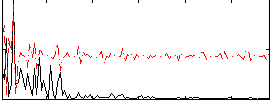 |
Correlation:0.1889
  |
Correlation:0.1461
  |
Correlation:0.1461
  |
Correlation:0.13832
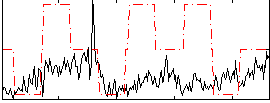 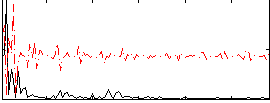 |
Correlation:0.13832
  |
Correlation:0.27496
  |
Component:355; Jaccard:0.070233
 |
Component:019; Jaccard:0.071862
 |
Component:335; Jaccard:0.067674
 |
Component:194; Jaccard:0.057279
 |
Component:246; Jaccard:0.037402
 |
Component:335; Jaccard:0.020769
 |
Component:246; Jaccard:0.00826
 |
Correlation:-0.12971
  |
Correlation:0.018422
  |
Correlation:0.26647
  |
Correlation:-0.072879
  |
Correlation:0.1461
  |
Correlation:0.26647
  |
Correlation:0.1461
  |
Component:246; Jaccard:0.070195
 |
Component:355; Jaccard:0.07034
 |
Component:392; Jaccard:0.06562
 |
Component:392; Jaccard:0.052621
 |
Component:194; Jaccard:0.035223
 |
Component:246; Jaccard:0.020363
 |
Component:136; Jaccard:0.00792
 |
Correlation:0.1461
  |
Correlation:-0.12971
  |
Correlation:0.1889
  |
Correlation:0.1889
  |
Correlation:-0.072879
  |
Correlation:0.1461
  |
Correlation:0.1121
  |
Component:306; Jaccard:0.069218
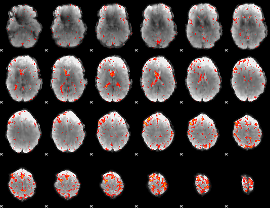 |
Component:255; Jaccard:0.06906
 |
Component:387; Jaccard:0.06346
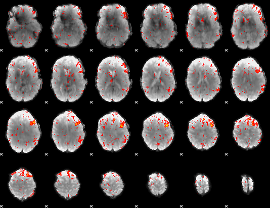 |
Component:173; Jaccard:0.052177
 |
Component:347; Jaccard:0.03433
 |
Component:136; Jaccard:0.018617
 |
Component:387; Jaccard:0.007382
 |
Correlation:0.025584
 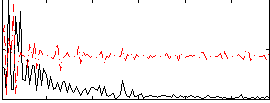 |
Correlation:-0.077965
  |
Correlation:-0.042545
 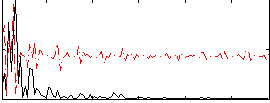 |
Correlation:0.13832
  |
Correlation:0.1383
  |
Correlation:0.1121
  |
Correlation:-0.042545
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
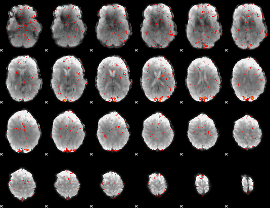
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
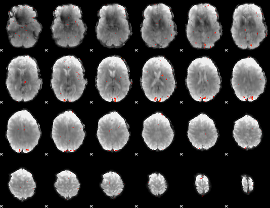
GLM Z-score result thresholded by 3
 |
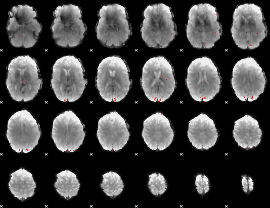
GLM Z-score result thresholded by 3.5
 |
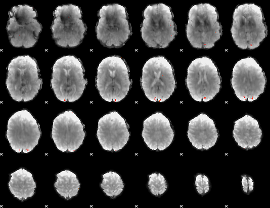
GLM Z-score result thresholded by 4
 |
Component:222; Jaccard:0.10817
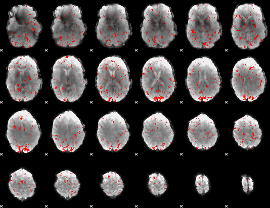 |
Component:222; Jaccard:0.11391
 |
Component:222; Jaccard:0.10466
 |
Component:222; Jaccard:0.074452
 |
Component:222; Jaccard:0.044718
 |
Component:222; Jaccard:0.027414
 |
Component:052; Jaccard:0.015693
 |
Correlation:0.24139
  |
Correlation:0.24139
  |
Correlation:0.24139
  |
Correlation:0.24139
  |
Correlation:0.24139
  |
Correlation:0.24139
  |
Correlation:0.050348
 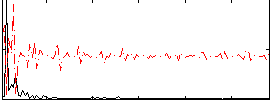 |
Component:052; Jaccard:0.083844
 |
Component:052; Jaccard:0.087767
 |
Component:052; Jaccard:0.080342
 |
Component:052; Jaccard:0.063698
 |
Component:052; Jaccard:0.042734
 |
Component:052; Jaccard:0.025912
 |
Component:222; Jaccard:0.015333
 |
Correlation:0.050348
  |
Correlation:0.050348
  |
Correlation:0.050348
  |
Correlation:0.050348
  |
Correlation:0.050348
  |
Correlation:0.050348
  |
Correlation:0.24139
  |
Component:042; Jaccard:0.080439
 |
Component:357; Jaccard:0.080868
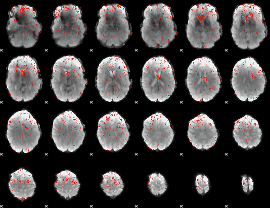 |
Component:357; Jaccard:0.065271
 |
Component:042; Jaccard:0.041855
 |
Component:149; Jaccard:0.029062
 |
Component:237; Jaccard:0.017556
 |
Component:237; Jaccard:0.010543
 |
Correlation:0.34663
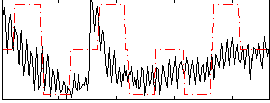 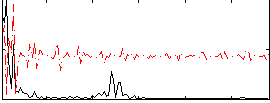 |
Correlation:0.35134
  |
Correlation:0.35134
  |
Correlation:0.34663
  |
Correlation:-0.093464
  |
Correlation:0.094051
  |
Correlation:0.094051
  |
Component:357; Jaccard:0.079187
 |
Component:042; Jaccard:0.079934
 |
Component:042; Jaccard:0.065059
 |
Component:357; Jaccard:0.041651
 |
Component:237; Jaccard:0.028807
 |
Component:149; Jaccard:0.017349
 |
Component:149; Jaccard:0.009967
 |
Correlation:0.35134
  |
Correlation:0.34663
  |
Correlation:0.34663
  |
Correlation:0.35134
  |
Correlation:0.094051
  |
Correlation:-0.093464
  |
Correlation:-0.093464
  |
Component:273; Jaccard:0.075038
 |
Component:273; Jaccard:0.070569
 |
Component:273; Jaccard:0.05788
 |
Component:149; Jaccard:0.041605
 |
Component:273; Jaccard:0.023105
 |
Component:273; Jaccard:0.01223
 |
Component:273; Jaccard:0.006202
 |
Correlation:0.16697
  |
Correlation:0.16697
  |
Correlation:0.16697
  |
Correlation:-0.093464
  |
Correlation:0.16697
  |
Correlation:0.16697
  |
Correlation:0.16697
  |
Component:104; Jaccard:0.062107
 |
Component:149; Jaccard:0.060854
 |
Component:229; Jaccard:0.055508
 |
Component:237; Jaccard:0.040934
 |
Component:357; Jaccard:0.020176
 |
Component:331; Jaccard:0.009795
 |
Component:331; Jaccard:0.005473
 |
Correlation:0.3257
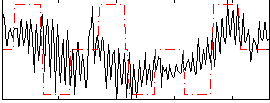 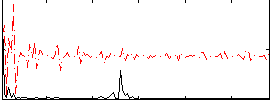 |
Correlation:-0.093464
  |
Correlation:0.094883
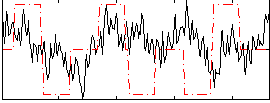 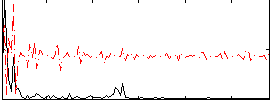 |
Correlation:0.094051
  |
Correlation:0.35134
  |
Correlation:0.031925
  |
Correlation:0.031925
  |
Component:149; Jaccard:0.061888
 |
Component:229; Jaccard:0.059293
 |
Component:149; Jaccard:0.054692
 |
Component:273; Jaccard:0.040065
 |
Component:042; Jaccard:0.018531
 |
Component:204; Jaccard:0.008768
 |
Component:204; Jaccard:0.005392
 |
Correlation:-0.093464
  |
Correlation:0.094883
  |
Correlation:-0.093464
  |
Correlation:0.16697
  |
Correlation:0.34663
  |
Correlation:0.086906
 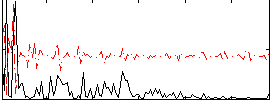 |
Correlation:0.086906
  |
Component:237; Jaccard:0.057751
 |
Component:104; Jaccard:0.058412
 |
Component:237; Jaccard:0.051574
 |
Component:229; Jaccard:0.038737
 |
Component:229; Jaccard:0.017741
 |
Component:106; Jaccard:0.008678
 |
Component:351; Jaccard:0.003548
 |
Correlation:0.094051
  |
Correlation:0.3257
  |
Correlation:0.094051
  |
Correlation:0.094883
  |
Correlation:0.094883
  |
Correlation:0.010149
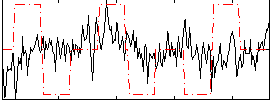 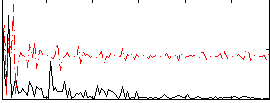 |
Correlation:0.0059607
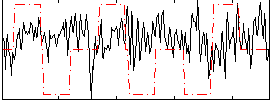 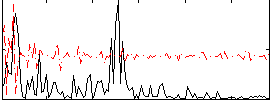 |
Component:106; Jaccard:0.057571
 |
Component:237; Jaccard:0.056453
 |
Component:106; Jaccard:0.048202
 |
Component:106; Jaccard:0.030353
 |
Component:331; Jaccard:0.017452
 |
Component:357; Jaccard:0.008068
 |
Component:106; Jaccard:0.003533
 |
Correlation:0.010149
  |
Correlation:0.094051
  |
Correlation:0.010149
  |
Correlation:0.010149
  |
Correlation:0.031925
  |
Correlation:0.35134
  |
Correlation:0.010149
  |
Component:229; Jaccard:0.056635
 |
Component:106; Jaccard:0.055776
 |
Component:104; Jaccard:0.045302
 |
Component:014; Jaccard:0.029788
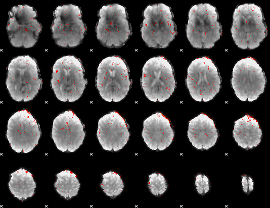 |
Component:106; Jaccard:0.015677
 |
Component:229; Jaccard:0.007452
 |
Component:357; Jaccard:0.0033
 |
Correlation:0.094883
  |
Correlation:0.010149
  |
Correlation:0.3257
  |
Correlation:0.012862
 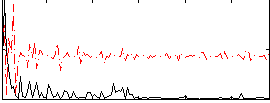 |
Correlation:0.010149
  |
Correlation:0.094883
  |
Correlation:0.35134
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
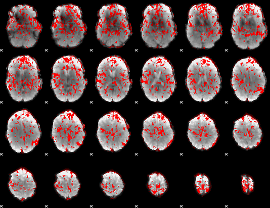 |
GLM binary result thresholded by 1.5
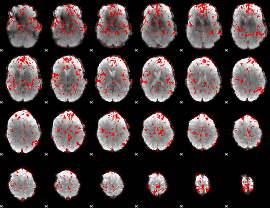 |
GLM binary result thresholded by 2
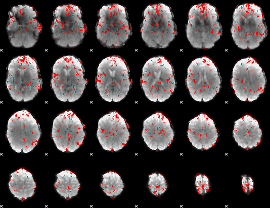 |
GLM binary result thresholded by 2.5
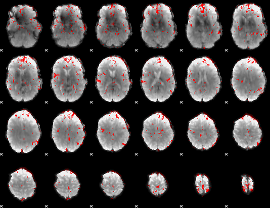 |
GLM binary result thresholded by 3
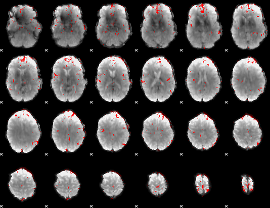 |
GLM binary result thresholded by 3.5
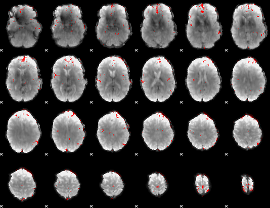 |
GLM binary result thresholded by 4
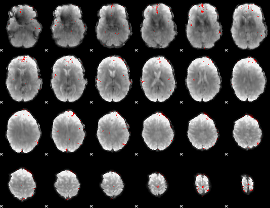 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
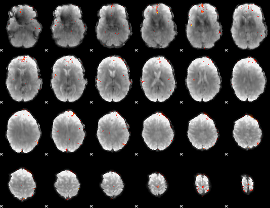 |
Component:042; Jaccard:0.11718
 |
Component:090; Jaccard:0.13939
 |
Component:090; Jaccard:0.17344
 |
Component:090; Jaccard:0.20261
 |
Component:090; Jaccard:0.20682
 |
Component:090; Jaccard:0.18533
 |
Component:090; Jaccard:0.14474
 |
Correlation:0.39959
  |
Correlation:0.32056
  |
Correlation:0.32056
  |
Correlation:0.32056
  |
Correlation:0.32056
  |
Correlation:0.32056
  |
Correlation:0.32056
  |
Component:090; Jaccard:0.10902
 |
Component:042; Jaccard:0.13468
 |
Component:042; Jaccard:0.14882
 |
Component:042; Jaccard:0.14973
 |
Component:042; Jaccard:0.13554
 |
Component:042; Jaccard:0.11258
 |
Component:042; Jaccard:0.081515
 |
Correlation:0.32056
  |
Correlation:0.39959
  |
Correlation:0.39959
  |
Correlation:0.39959
  |
Correlation:0.39959
  |
Correlation:0.39959
  |
Correlation:0.39959
  |
Component:345; Jaccard:0.095487
 |
Component:241; Jaccard:0.10744
 |
Component:241; Jaccard:0.11612
 |
Component:241; Jaccard:0.11684
 |
Component:241; Jaccard:0.11008
 |
Component:241; Jaccard:0.091903
 |
Component:241; Jaccard:0.068708
 |
Correlation:0.34583
  |
Correlation:0.23326
  |
Correlation:0.23326
  |
Correlation:0.23326
  |
Correlation:0.23326
  |
Correlation:0.23326
  |
Correlation:0.23326
  |
Component:241; Jaccard:0.094621
 |
Component:345; Jaccard:0.10158
 |
Component:345; Jaccard:0.10047
 |
Component:267; Jaccard:0.10643
 |
Component:267; Jaccard:0.10004
 |
Component:267; Jaccard:0.085753
 |
Component:267; Jaccard:0.065961
 |
Correlation:0.23326
  |
Correlation:0.34583
  |
Correlation:0.34583
  |
Correlation:0.3179
 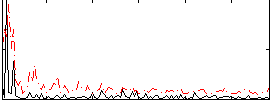 |
Correlation:0.3179
  |
Correlation:0.3179
  |
Correlation:0.3179
  |
Component:341; Jaccard:0.090358
 |
Component:341; Jaccard:0.09518
 |
Component:267; Jaccard:0.10038
 |
Component:091; Jaccard:0.09837
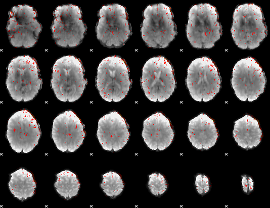 |
Component:091; Jaccard:0.086435
 |
Component:091; Jaccard:0.067797
 |
Component:135; Jaccard:0.051138
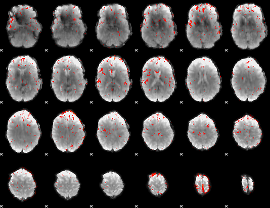 |
Correlation:0.23222
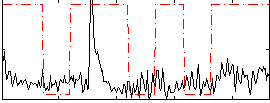  |
Correlation:0.23222
  |
Correlation:0.3179
  |
Correlation:0.1805
  |
Correlation:0.1805
  |
Correlation:0.1805
  |
Correlation:0.080395
  |
Component:085; Jaccard:0.087189
 |
Component:091; Jaccard:0.094409
 |
Component:091; Jaccard:0.099429
 |
Component:345; Jaccard:0.089718
 |
Component:135; Jaccard:0.08061
 |
Component:135; Jaccard:0.067457
 |
Component:091; Jaccard:0.04975
 |
Correlation:0.060833
  |
Correlation:0.1805
  |
Correlation:0.1805
  |
Correlation:0.34583
  |
Correlation:0.080395
  |
Correlation:0.080395
  |
Correlation:0.1805
  |
Component:091; Jaccard:0.084993
 |
Component:085; Jaccard:0.09095
 |
Component:341; Jaccard:0.094404
 |
Component:341; Jaccard:0.085936
 |
Component:085; Jaccard:0.072132
 |
Component:085; Jaccard:0.055858
 |
Component:014; Jaccard:0.046178
 |
Correlation:0.1805
  |
Correlation:0.060833
  |
Correlation:0.23222
  |
Correlation:0.23222
  |
Correlation:0.060833
  |
Correlation:0.060833
  |
Correlation:0.11583
  |
Component:369; Jaccard:0.080486
 |
Component:267; Jaccard:0.08959
 |
Component:085; Jaccard:0.090097
 |
Component:085; Jaccard:0.084526
 |
Component:345; Jaccard:0.071358
 |
Component:014; Jaccard:0.055792
 |
Component:211; Jaccard:0.040773
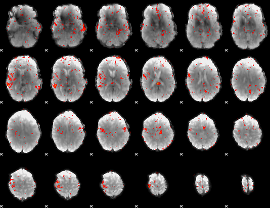 |
Correlation:0.12043
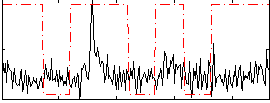 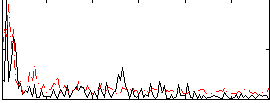 |
Correlation:0.3179
  |
Correlation:0.060833
  |
Correlation:0.060833
  |
Correlation:0.34583
  |
Correlation:0.11583
  |
Correlation:0.2143
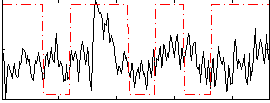 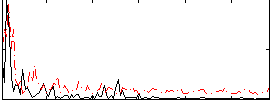 |
Component:189; Jaccard:0.079693
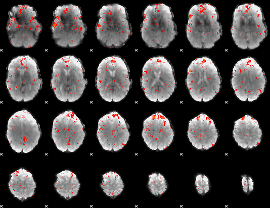 |
Component:369; Jaccard:0.085953
 |
Component:369; Jaccard:0.086867
 |
Component:135; Jaccard:0.084464
 |
Component:341; Jaccard:0.071013
 |
Component:369; Jaccard:0.054785
 |
Component:085; Jaccard:0.03819
 |
Correlation:0.09322
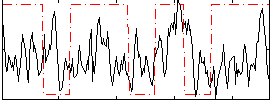 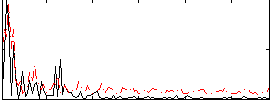 |
Correlation:0.12043
  |
Correlation:0.12043
  |
Correlation:0.080395
  |
Correlation:0.23222
  |
Correlation:0.12043
  |
Correlation:0.060833
  |
Component:381; Jaccard:0.079103
 |
Component:189; Jaccard:0.083659
 |
Component:189; Jaccard:0.084029
 |
Component:369; Jaccard:0.081591
 |
Component:369; Jaccard:0.070275
 |
Component:341; Jaccard:0.053433
 |
Component:141; Jaccard:0.037431
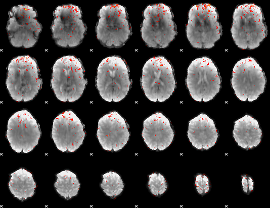 |
Correlation:0.22415
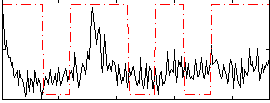 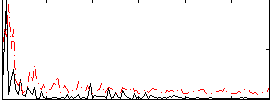 |
Correlation:0.09322
  |
Correlation:0.09322
  |
Correlation:0.12043
  |
Correlation:0.12043
  |
Correlation:0.23222
  |
Correlation:0.2823
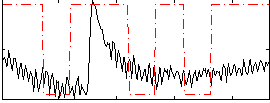  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
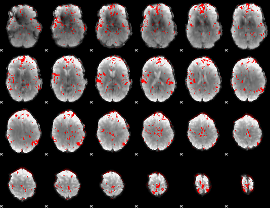 |
GLM binary result thresholded by 3
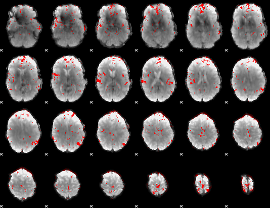 |
GLM binary result thresholded by 3.5
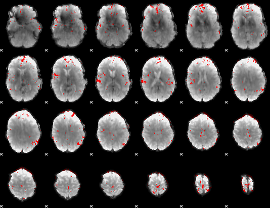 |
GLM binary result thresholded by 4
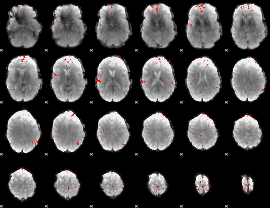 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
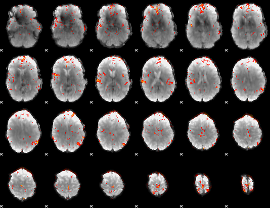 |
GLM Z-score result thresholded by 3.5
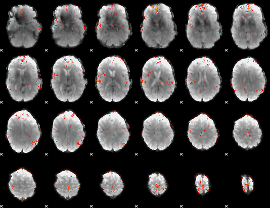 |
GLM Z-score result thresholded by 4
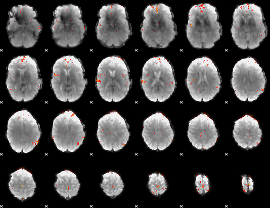 |
Component:345; Jaccard:0.104
 |
Component:090; Jaccard:0.12743
 |
Component:090; Jaccard:0.1626
 |
Component:090; Jaccard:0.19767
 |
Component:090; Jaccard:0.22012
 |
Component:090; Jaccard:0.21313
 |
Component:090; Jaccard:0.17279
 |
Correlation:-0.050316
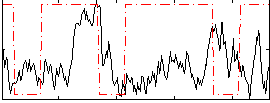  |
Correlation:0.12127
  |
Correlation:0.12127
  |
Correlation:0.12127
  |
Correlation:0.12127
  |
Correlation:0.12127
  |
Correlation:0.12127
  |
Component:085; Jaccard:0.10239
 |
Component:345; Jaccard:0.11551
 |
Component:345; Jaccard:0.12353
 |
Component:345; Jaccard:0.12182
 |
Component:173; Jaccard:0.11842
 |
Component:173; Jaccard:0.10483
 |
Component:173; Jaccard:0.077831
 |
Correlation:0.17972
  |
Correlation:-0.050316
  |
Correlation:-0.050316
  |
Correlation:-0.050316
  |
Correlation:0.25912
  |
Correlation:0.25912
  |
Correlation:0.25912
  |
Component:090; Jaccard:0.098899
 |
Component:085; Jaccard:0.11495
 |
Component:085; Jaccard:0.12183
 |
Component:085; Jaccard:0.12143
 |
Component:345; Jaccard:0.11145
 |
Component:345; Jaccard:0.092504
 |
Component:345; Jaccard:0.068135
 |
Correlation:0.12127
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Correlation:-0.050316
  |
Correlation:-0.050316
  |
Correlation:-0.050316
  |
Component:381; Jaccard:0.09624
 |
Component:341; Jaccard:0.10457
 |
Component:173; Jaccard:0.1093
 |
Component:173; Jaccard:0.11819
 |
Component:085; Jaccard:0.11098
 |
Component:085; Jaccard:0.091893
 |
Component:085; Jaccard:0.066687
 |
Correlation:0.11312
  |
Correlation:0.027576
  |
Correlation:0.25912
  |
Correlation:0.25912
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Component:341; Jaccard:0.095057
 |
Component:381; Jaccard:0.10408
 |
Component:381; Jaccard:0.10902
 |
Component:341; Jaccard:0.10631
 |
Component:369; Jaccard:0.096767
 |
Component:267; Jaccard:0.081612
 |
Component:267; Jaccard:0.065453
 |
Correlation:0.027576
  |
Correlation:0.11312
  |
Correlation:0.11312
  |
Correlation:0.027576
  |
Correlation:0.14149
  |
Correlation:0.29163
  |
Correlation:0.29163
  |
Component:369; Jaccard:0.083445
 |
Component:173; Jaccard:0.094538
 |
Component:341; Jaccard:0.1077
 |
Component:369; Jaccard:0.10533
 |
Component:267; Jaccard:0.094606
 |
Component:369; Jaccard:0.0784
 |
Component:241; Jaccard:0.060942
 |
Correlation:0.14149
  |
Correlation:0.25912
  |
Correlation:0.027576
  |
Correlation:0.14149
  |
Correlation:0.29163
  |
Correlation:0.14149
  |
Correlation:0.045314
  |
Component:019; Jaccard:0.08313
 |
Component:369; Jaccard:0.093131
 |
Component:369; Jaccard:0.10167
 |
Component:381; Jaccard:0.1052
 |
Component:381; Jaccard:0.092189
 |
Component:241; Jaccard:0.078345
 |
Component:135; Jaccard:0.057477
 |
Correlation:0.089817
  |
Correlation:0.14149
  |
Correlation:0.14149
  |
Correlation:0.11312
  |
Correlation:0.11312
  |
Correlation:0.045314
  |
Correlation:0.14121
  |
Component:241; Jaccard:0.081774
 |
Component:241; Jaccard:0.091169
 |
Component:241; Jaccard:0.098285
 |
Component:241; Jaccard:0.096825
 |
Component:341; Jaccard:0.091535
 |
Component:381; Jaccard:0.075248
 |
Component:369; Jaccard:0.056256
 |
Correlation:0.045314
  |
Correlation:0.045314
  |
Correlation:0.045314
  |
Correlation:0.045314
  |
Correlation:0.027576
  |
Correlation:0.11312
  |
Correlation:0.14149
  |
Component:042; Jaccard:0.080924
 |
Component:042; Jaccard:0.086778
 |
Component:267; Jaccard:0.088611
 |
Component:267; Jaccard:0.093854
 |
Component:241; Jaccard:0.091049
 |
Component:341; Jaccard:0.07215
 |
Component:347; Jaccard:0.053875
 |
Correlation:-0.18525
  |
Correlation:-0.18525
  |
Correlation:0.29163
  |
Correlation:0.29163
  |
Correlation:0.045314
  |
Correlation:0.027576
  |
Correlation:0.22389
  |
Component:173; Jaccard:0.078945
 |
Component:091; Jaccard:0.084456
 |
Component:042; Jaccard:0.088461
 |
Component:091; Jaccard:0.087358
 |
Component:135; Jaccard:0.08356
 |
Component:135; Jaccard:0.072065
 |
Component:234; Jaccard:0.052169
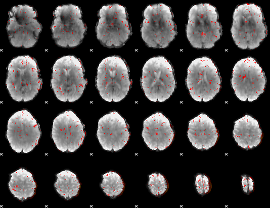 |
Correlation:0.25912
  |
Correlation:0.019979
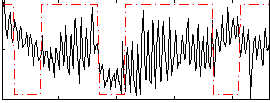 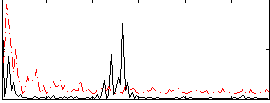 |
Correlation:-0.18525
  |
Correlation:0.019979
  |
Correlation:0.14121
  |
Correlation:0.14121
  |
Correlation:0.15988
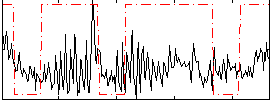  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































