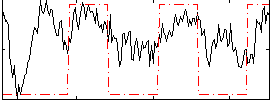
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
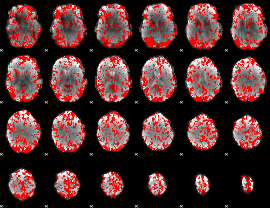 |
GLM binary result thresholded by 1.5
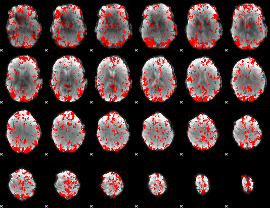 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
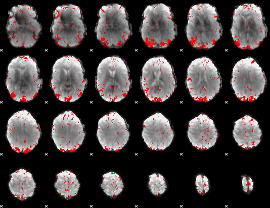 |
GLM binary result thresholded by 3
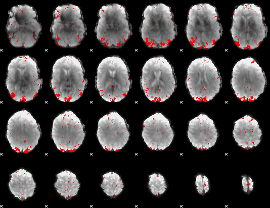 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
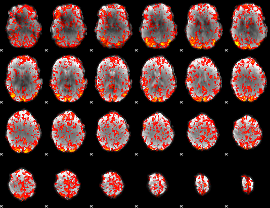 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
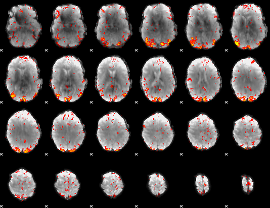 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
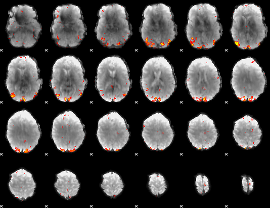 |
GLM Z-score result thresholded by 4
 |
Component:173; Jaccard:0.11933
 |
Component:173; Jaccard:0.16592
 |
Component:173; Jaccard:0.21997
 |
Component:173; Jaccard:0.25368
 |
Component:173; Jaccard:0.24504
 |
Component:173; Jaccard:0.2051
 |
Component:173; Jaccard:0.15912
 |
Correlation:0.53053
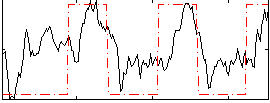  |
Correlation:0.53053
  |
Correlation:0.53053
  |
Correlation:0.53053
  |
Correlation:0.53053
  |
Correlation:0.53053
  |
Correlation:0.53053
  |
Component:271; Jaccard:0.1145
 |
Component:271; Jaccard:0.14674
 |
Component:271; Jaccard:0.17839
 |
Component:271; Jaccard:0.19428
 |
Component:271; Jaccard:0.18271
 |
Component:271; Jaccard:0.15232
 |
Component:271; Jaccard:0.1204
 |
Correlation:0.042775
  |
Correlation:0.042775
  |
Correlation:0.042775
  |
Correlation:0.042775
  |
Correlation:0.042775
  |
Correlation:0.042775
  |
Correlation:0.042775
  |
Component:055; Jaccard:0.09862
 |
Component:055; Jaccard:0.12403
 |
Component:055; Jaccard:0.1466
 |
Component:055; Jaccard:0.1522
 |
Component:055; Jaccard:0.13547
 |
Component:044; Jaccard:0.11322
 |
Component:044; Jaccard:0.08941
 |
Correlation:0.44672
  |
Correlation:0.44672
  |
Correlation:0.44672
  |
Correlation:0.44672
  |
Correlation:0.44672
  |
Correlation:-0.18155
  |
Correlation:-0.18155
  |
Component:307; Jaccard:0.095431
 |
Component:044; Jaccard:0.11361
 |
Component:044; Jaccard:0.13239
 |
Component:044; Jaccard:0.14292
 |
Component:044; Jaccard:0.1336
 |
Component:055; Jaccard:0.10848
 |
Component:055; Jaccard:0.082961
 |
Correlation:-0.03659
  |
Correlation:-0.18155
  |
Correlation:-0.18155
  |
Correlation:-0.18155
  |
Correlation:-0.18155
  |
Correlation:0.44672
  |
Correlation:0.44672
  |
Component:044; Jaccard:0.093406
 |
Component:362; Jaccard:0.10623
 |
Component:362; Jaccard:0.11155
 |
Component:101; Jaccard:0.1033
 |
Component:101; Jaccard:0.088885
 |
Component:101; Jaccard:0.072752
 |
Component:101; Jaccard:0.054129
 |
Correlation:-0.18155
  |
Correlation:0.16354
  |
Correlation:0.16354
  |
Correlation:0.16668
  |
Correlation:0.16668
  |
Correlation:0.16668
  |
Correlation:0.16668
  |
Component:101; Jaccard:0.092404
 |
Component:307; Jaccard:0.10353
 |
Component:101; Jaccard:0.10839
 |
Component:362; Jaccard:0.10283
 |
Component:362; Jaccard:0.077375
 |
Component:389; Jaccard:0.055261
 |
Component:254; Jaccard:0.044015
 |
Correlation:0.16668
  |
Correlation:-0.03659
  |
Correlation:0.16668
  |
Correlation:0.16354
  |
Correlation:0.16354
  |
Correlation:0.2847
  |
Correlation:0.090271
  |
Component:362; Jaccard:0.090025
 |
Component:101; Jaccard:0.1019
 |
Component:307; Jaccard:0.10447
 |
Component:389; Jaccard:0.09425
 |
Component:389; Jaccard:0.076997
 |
Component:254; Jaccard:0.054999
 |
Component:034; Jaccard:0.042263
 |
Correlation:0.16354
  |
Correlation:0.16668
  |
Correlation:-0.03659
  |
Correlation:0.2847
  |
Correlation:0.2847
  |
Correlation:0.090271
  |
Correlation:0.28106
  |
Component:378; Jaccard:0.083259
 |
Component:280; Jaccard:0.089749
 |
Component:389; Jaccard:0.096491
 |
Component:307; Jaccard:0.091426
 |
Component:254; Jaccard:0.07284
 |
Component:034; Jaccard:0.054495
 |
Component:307; Jaccard:0.040227
 |
Correlation:0.072629
  |
Correlation:0.14692
  |
Correlation:0.2847
  |
Correlation:-0.03659
  |
Correlation:0.090271
  |
Correlation:0.28106
  |
Correlation:-0.03659
  |
Component:280; Jaccard:0.08191
 |
Component:389; Jaccard:0.08679
 |
Component:254; Jaccard:0.088152
 |
Component:254; Jaccard:0.085242
 |
Component:307; Jaccard:0.071635
 |
Component:307; Jaccard:0.054423
 |
Component:389; Jaccard:0.039638
 |
Correlation:0.14692
  |
Correlation:0.2847
  |
Correlation:0.090271
  |
Correlation:0.090271
  |
Correlation:-0.03659
  |
Correlation:-0.03659
  |
Correlation:0.2847
  |
Component:033; Jaccard:0.08181
 |
Component:105; Jaccard:0.086731
 |
Component:280; Jaccard:0.087246
 |
Component:105; Jaccard:0.077127
 |
Component:034; Jaccard:0.065947
 |
Component:362; Jaccard:0.052318
 |
Component:362; Jaccard:0.038834
 |
Correlation:0.13237
  |
Correlation:0.48171
  |
Correlation:0.14692
  |
Correlation:0.48171
  |
Correlation:0.28106
  |
Correlation:0.16354
  |
Correlation:0.16354
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
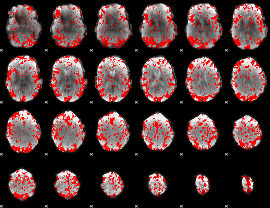 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
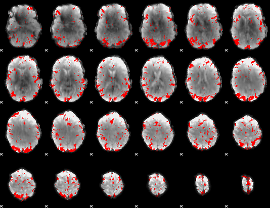 |
GLM binary result thresholded by 3
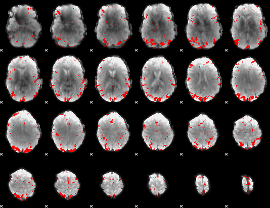 |
GLM binary result thresholded by 3.5
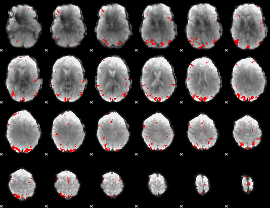 |
GLM binary result thresholded by 4
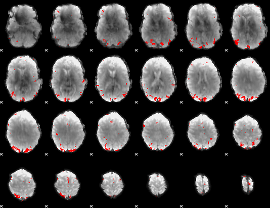 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
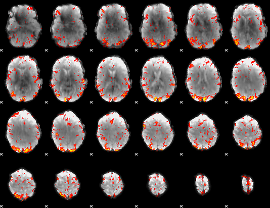 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
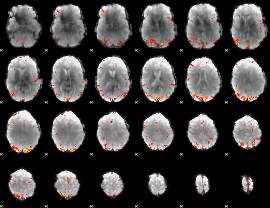 |
GLM Z-score result thresholded by 4
 |
Component:324; Jaccard:0.11514
 |
Component:044; Jaccard:0.15084
 |
Component:044; Jaccard:0.20157
 |
Component:044; Jaccard:0.24246
 |
Component:044; Jaccard:0.24864
 |
Component:044; Jaccard:0.21869
 |
Component:044; Jaccard:0.17706
 |
Correlation:0.27937
  |
Correlation:0.40496
  |
Correlation:0.40496
  |
Correlation:0.40496
  |
Correlation:0.40496
  |
Correlation:0.40496
  |
Correlation:0.40496
  |
Component:044; Jaccard:0.10781
 |
Component:324; Jaccard:0.14824
 |
Component:324; Jaccard:0.17882
 |
Component:324; Jaccard:0.19735
 |
Component:324; Jaccard:0.18571
 |
Component:271; Jaccard:0.16311
 |
Component:271; Jaccard:0.13501
 |
Correlation:0.40496
  |
Correlation:0.27937
  |
Correlation:0.27937
  |
Correlation:0.27937
  |
Correlation:0.27937
  |
Correlation:0.26338
  |
Correlation:0.26338
  |
Component:271; Jaccard:0.10656
 |
Component:271; Jaccard:0.13139
 |
Component:271; Jaccard:0.15886
 |
Component:271; Jaccard:0.17956
 |
Component:271; Jaccard:0.18111
 |
Component:324; Jaccard:0.15204
 |
Component:324; Jaccard:0.10921
 |
Correlation:0.26338
  |
Correlation:0.26338
  |
Correlation:0.26338
  |
Correlation:0.26338
  |
Correlation:0.26338
  |
Correlation:0.27937
  |
Correlation:0.27937
  |
Component:101; Jaccard:0.10491
 |
Component:101; Jaccard:0.12725
 |
Component:101; Jaccard:0.14731
 |
Component:101; Jaccard:0.15772
 |
Component:110; Jaccard:0.14896
 |
Component:110; Jaccard:0.1262
 |
Component:110; Jaccard:0.097004
 |
Correlation:0.14341
  |
Correlation:0.14341
  |
Correlation:0.14341
  |
Correlation:0.14341
  |
Correlation:0.466
  |
Correlation:0.466
  |
Correlation:0.466
  |
Component:110; Jaccard:0.099306
 |
Component:110; Jaccard:0.12095
 |
Component:110; Jaccard:0.14094
 |
Component:110; Jaccard:0.15373
 |
Component:101; Jaccard:0.14744
 |
Component:101; Jaccard:0.11822
 |
Component:101; Jaccard:0.089275
 |
Correlation:0.466
  |
Correlation:0.466
  |
Correlation:0.466
  |
Correlation:0.466
  |
Correlation:0.14341
  |
Correlation:0.14341
  |
Correlation:0.14341
  |
Component:307; Jaccard:0.097274
 |
Component:307; Jaccard:0.10783
 |
Component:362; Jaccard:0.11723
 |
Component:362; Jaccard:0.11647
 |
Component:173; Jaccard:0.10723
 |
Component:173; Jaccard:0.09491
 |
Component:173; Jaccard:0.076451
 |
Correlation:0.061236
  |
Correlation:0.061236
  |
Correlation:0.079325
  |
Correlation:0.079325
  |
Correlation:-0.32193
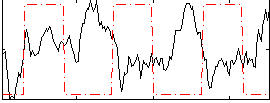  |
Correlation:-0.32193
  |
Correlation:-0.32193
  |
Component:385; Jaccard:0.092071
 |
Component:362; Jaccard:0.10508
 |
Component:307; Jaccard:0.11238
 |
Component:173; Jaccard:0.1094
 |
Component:362; Jaccard:0.096254
 |
Component:254; Jaccard:0.076689
 |
Component:254; Jaccard:0.060964
 |
Correlation:0.16484
  |
Correlation:0.079325
  |
Correlation:0.061236
  |
Correlation:-0.32193
  |
Correlation:0.079325
  |
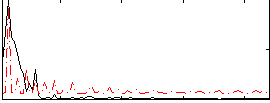
Correlation:0.070377
  |
Correlation:0.070377
  |
Component:232; Jaccard:0.090624
 |
Component:325; Jaccard:0.099243
 |
Component:325; Jaccard:0.10633
 |
Component:307; Jaccard:0.10496
 |
Component:254; Jaccard:0.092616
 |
Component:307; Jaccard:0.073872
 |
Component:055; Jaccard:0.057166
 |
Correlation:0.08324
  |
Correlation:0.23548
  |
Correlation:0.23548
  |
Correlation:0.061236
  |
Correlation:0.070377
  |
Correlation:0.061236
  |
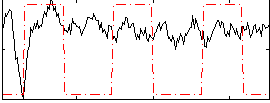
Correlation:-0.1811
 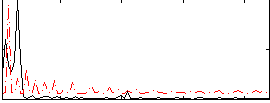 |
Component:362; Jaccard:0.088523
 |
Component:232; Jaccard:0.096927
 |
Component:173; Jaccard:0.10237
 |
Component:325; Jaccard:0.10131
 |
Component:307; Jaccard:0.091583
 |
Component:362; Jaccard:0.071617
 |
Component:307; Jaccard:0.055749
 |
Correlation:0.079325
  |
Correlation:0.08324
  |
Correlation:-0.32193
  |
Correlation:0.23548
  |
Correlation:0.061236
  |
Correlation:0.079325
  |
Correlation:0.061236
  |
Component:033; Jaccard:0.087122
 |
Component:033; Jaccard:0.096026
 |
Component:033; Jaccard:0.098958
 |
Component:254; Jaccard:0.10115
 |
Component:325; Jaccard:0.083333
 |
Component:055; Jaccard:0.069753
 |
Component:362; Jaccard:0.050209
 |
Correlation:0.19781
  |
Correlation:0.19781
  |
Correlation:0.19781
  |
Correlation:0.070377
  |
Correlation:0.23548
  |
Correlation:-0.1811
  |
Correlation:0.079325
  |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
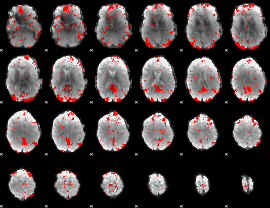 |
GLM binary result thresholded by 2.5
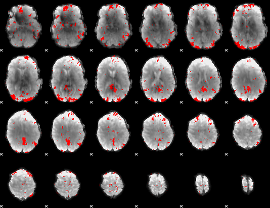 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
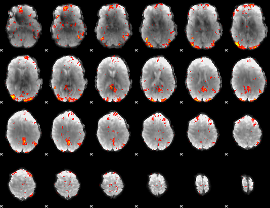 |
GLM Z-score result thresholded by 3
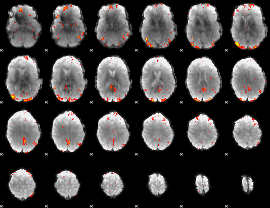 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:231; Jaccard:0.14372
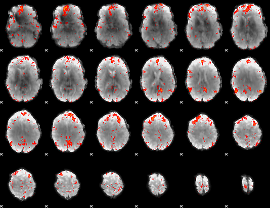 |
Component:231; Jaccard:0.16719
 |
Component:173; Jaccard:0.18911
 |
Component:173; Jaccard:0.19796
 |
Component:173; Jaccard:0.1821
 |
Component:173; Jaccard:0.15174
 |
Component:173; Jaccard:0.11592
 |
Correlation:0.22848
  |
Correlation:0.22848
  |
Correlation:0.46239
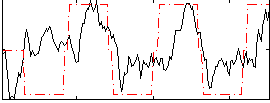 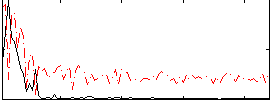 |
Correlation:0.46239
  |
Correlation:0.46239
  |
Correlation:0.46239
  |
Correlation:0.46239
  |
Component:173; Jaccard:0.12933
 |
Component:173; Jaccard:0.16152
 |
Component:231; Jaccard:0.17317
 |
Component:247; Jaccard:0.17319
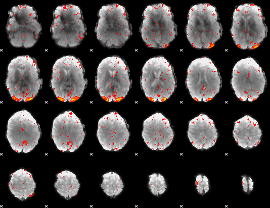 |
Component:247; Jaccard:0.16075
 |
Component:247; Jaccard:0.13478
 |
Component:247; Jaccard:0.098379
 |
Correlation:0.46239
  |
Correlation:0.46239
  |
Correlation:0.22848
  |
Correlation:0.25997
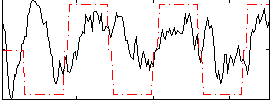  |
Correlation:0.25997
  |
Correlation:0.25997
  |
Correlation:0.25997
  |
Component:332; Jaccard:0.11699
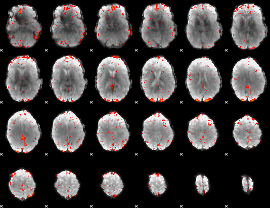 |
Component:247; Jaccard:0.13859
 |
Component:247; Jaccard:0.16311
 |
Component:231; Jaccard:0.15616
 |
Component:231; Jaccard:0.12044
 |
Component:332; Jaccard:0.091635
 |
Component:332; Jaccard:0.064408
 |
Correlation:0.072955
  |
Correlation:0.25997
  |
Correlation:0.25997
  |
Correlation:0.22848
  |
Correlation:0.22848
  |
Correlation:0.072955
  |
Correlation:0.072955
  |
Component:247; Jaccard:0.11281
 |
Component:332; Jaccard:0.13584
 |
Component:332; Jaccard:0.14667
 |
Component:332; Jaccard:0.14125
 |
Component:332; Jaccard:0.12014
 |
Component:208; Jaccard:0.088253
 |
Component:208; Jaccard:0.063538
 |
Correlation:0.25997
  |
Correlation:0.072955
  |
Correlation:0.072955
  |
Correlation:0.072955
  |
Correlation:0.072955
  |
Correlation:0.48344
  |
Correlation:0.48344
  |
Component:091; Jaccard:0.11228
 |
Component:091; Jaccard:0.12563
 |
Component:208; Jaccard:0.13722
 |
Component:208; Jaccard:0.13284
 |
Component:208; Jaccard:0.11308
 |
Component:231; Jaccard:0.083883
 |
Component:389; Jaccard:0.062141
 |
Correlation:0.097112
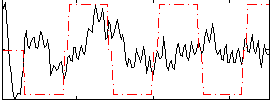 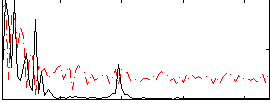 |
Correlation:0.097112
  |
Correlation:0.48344
  |
Correlation:0.48344
  |
Correlation:0.48344
  |
Correlation:0.22848
  |
Correlation:0.22043
  |
Component:208; Jaccard:0.10682
 |
Component:208; Jaccard:0.12441
 |
Component:091; Jaccard:0.13116
 |
Component:013; Jaccard:0.12233
 |
Component:013; Jaccard:0.10548
 |
Component:013; Jaccard:0.083023
 |
Component:013; Jaccard:0.059841
 |
Correlation:0.48344
  |
Correlation:0.48344
  |
Correlation:0.097112
  |
Correlation:0.13259
  |
Correlation:0.13259
  |
Correlation:0.13259
  |
Correlation:0.13259
  |
Component:105; Jaccard:0.10279
 |
Component:105; Jaccard:0.11546
 |
Component:013; Jaccard:0.12467
 |
Component:389; Jaccard:0.11825
 |
Component:389; Jaccard:0.10187
 |
Component:389; Jaccard:0.082324
 |
Component:105; Jaccard:0.058369
 |
Correlation:0.37405
  |
Correlation:0.37405
  |
Correlation:0.13259
  |
Correlation:0.22043
  |
Correlation:0.22043
  |
Correlation:0.22043
  |
Correlation:0.37405
  |
Component:389; Jaccard:0.093607
 |
Component:013; Jaccard:0.11164
 |
Component:105; Jaccard:0.12096
 |
Component:091; Jaccard:0.11819
 |
Component:105; Jaccard:0.099166
 |
Component:105; Jaccard:0.07935
 |
Component:231; Jaccard:0.052934
 |
Correlation:0.22043
  |
Correlation:0.13259
  |
Correlation:0.37405
  |
Correlation:0.097112
  |
Correlation:0.37405
  |
Correlation:0.37405
  |
Correlation:0.22848
  |
Component:013; Jaccard:0.092126
 |
Component:389; Jaccard:0.1102
 |
Component:389; Jaccard:0.11908
 |
Component:105; Jaccard:0.11624
 |
Component:091; Jaccard:0.093846
 |
Component:055; Jaccard:0.065759
 |
Component:055; Jaccard:0.048275
 |
Correlation:0.13259
  |
Correlation:0.22043
  |
Correlation:0.22043
  |
Correlation:0.37405
  |
Correlation:0.097112
  |
Correlation:0.34054
  |
Correlation:0.34054
  |
Component:055; Jaccard:0.087017
 |
Component:055; Jaccard:0.096465
 |
Component:055; Jaccard:0.10236
 |
Component:055; Jaccard:0.098114
 |
Component:055; Jaccard:0.083416
 |
Component:091; Jaccard:0.064924
 |
Component:091; Jaccard:0.03956
 |
Correlation:0.34054
  |
Correlation:0.34054
  |
Correlation:0.34054
  |
Correlation:0.34054
  |
Correlation:0.34054
  |
Correlation:0.097112
  |
Correlation:0.097112
  |
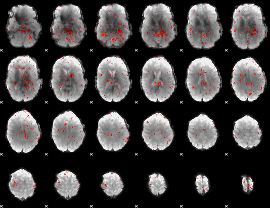
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
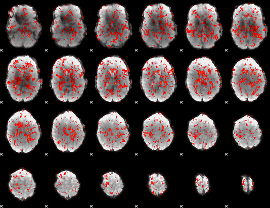 |
GLM Z-score result thresholded by 1.5
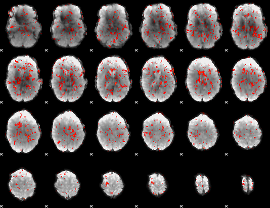 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:238; Jaccard:0.079852
 |
Component:335; Jaccard:0.085652
 |
Component:335; Jaccard:0.075592
 |
Component:335; Jaccard:0.049971
 |
Component:335; Jaccard:0.022782
 |
Component:214; Jaccard:0.008924
 |
Component:214; Jaccard:0.002894
 |
Correlation:0.080806
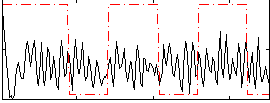  |
Correlation:0.029058
  |
Correlation:0.029058
  |
Correlation:0.029058
  |
Correlation:0.029058
  |
Correlation:0.049951
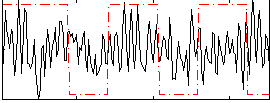  |
Correlation:0.049951
  |
Component:335; Jaccard:0.078523
 |
Component:007; Jaccard:0.077334
 |
Component:007; Jaccard:0.063059
 |
Component:214; Jaccard:0.041818
 |
Component:214; Jaccard:0.020878
 |
Component:067; Jaccard:0.007176
 |
Component:067; Jaccard:0.002224
 |
Correlation:0.029058
  |
Correlation:0.22883
  |
Correlation:0.22883
  |
Correlation:0.049951
  |
Correlation:0.049951
  |
Correlation:0.10871
  |
Correlation:0.10871
  |
Component:007; Jaccard:0.077269
 |
Component:238; Jaccard:0.072145
 |
Component:187; Jaccard:0.06258
 |
Component:187; Jaccard:0.040968
 |
Component:187; Jaccard:0.018621
 |
Component:335; Jaccard:0.00699
 |
Component:089; Jaccard:0.001955
 |
Correlation:0.22883
  |
Correlation:0.080806
  |
Correlation:-0.14997
  |
Correlation:-0.14997
  |
Correlation:-0.14997
  |
Correlation:0.029058
  |
Correlation:0.1237
  |
Component:177; Jaccard:0.073749
 |
Component:187; Jaccard:0.071743
 |
Component:214; Jaccard:0.061677
 |
Component:007; Jaccard:0.038555
 |
Component:067; Jaccard:0.01739
 |
Component:089; Jaccard:0.006054
 |
Component:335; Jaccard:0.00156
 |
Correlation:0.12627
  |
Correlation:-0.14997
  |
Correlation:0.049951
  |
Correlation:0.22883
  |
Correlation:0.10871
  |
Correlation:0.1237
  |
Correlation:0.029058
  |
Component:097; Jaccard:0.072416
 |
Component:214; Jaccard:0.070661
 |
Component:177; Jaccard:0.055504
 |
Component:067; Jaccard:0.036504
 |
Component:089; Jaccard:0.01561
 |
Component:312; Jaccard:0.005452
 |
Component:310; Jaccard:0.001406
 |
Correlation:0.11349
  |
Correlation:0.049951
  |
Correlation:0.12627
  |
Correlation:0.10871
  |
Correlation:0.1237
  |
Correlation:0.045089
  |
Correlation:0.16699
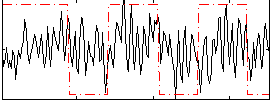  |
Component:093; Jaccard:0.072226
 |
Component:177; Jaccard:0.070102
 |
Component:067; Jaccard:0.052294
 |
Component:136; Jaccard:0.03183
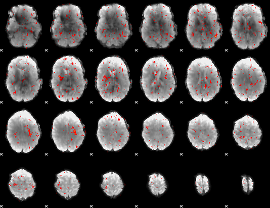 |
Component:007; Jaccard:0.014754
 |
Component:187; Jaccard:0.005283
 |
Component:154; Jaccard:0.001374
 |
Correlation:0.075458
 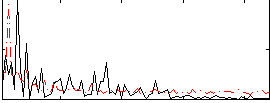 |
Correlation:0.12627
  |
Correlation:0.10871
  |
Correlation:0.14556
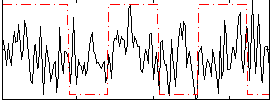  |
Correlation:0.22883
  |
Correlation:-0.14997
  |
Correlation:-0.021892
  |
Component:136; Jaccard:0.068151
 |
Component:097; Jaccard:0.067006
 |
Component:136; Jaccard:0.051819
 |
Component:312; Jaccard:0.031824
 |
Component:312; Jaccard:0.014672
 |
Component:007; Jaccard:0.005108
 |
Component:007; Jaccard:0.001358
 |
Correlation:0.14556
  |
Correlation:0.11349
  |
Correlation:0.14556
  |
Correlation:0.045089
  |
Correlation:0.045089
  |
Correlation:0.22883
  |
Correlation:0.22883
  |
Component:187; Jaccard:0.06732
 |
Component:093; Jaccard:0.065849
 |
Component:097; Jaccard:0.050569
 |
Component:089; Jaccard:0.030771
 |
Component:311; Jaccard:0.012663
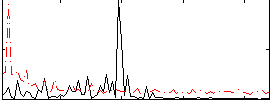
 |
Component:154; Jaccard:0.004535
 |
Component:085; Jaccard:0.001201
 |
Correlation:-0.14997
  |
Correlation:0.075458
  |
Correlation:0.11349
  |
Correlation:0.1237
  |
Correlation:0.060256
  |
Correlation:-0.021892
  |
Correlation:0.080048
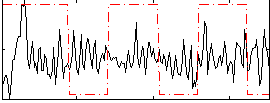  |
Component:214; Jaccard:0.067051
 |
Component:136; Jaccard:0.065423
 |
Component:312; Jaccard:0.050269
 |
Component:177; Jaccard:0.029514
 |
Component:136; Jaccard:0.012477
 |
Component:299; Jaccard:0.004245
 |
Component:234; Jaccard:0.001159
 |
Correlation:0.049951
  |
Correlation:0.14556
  |
Correlation:0.045089
  |
Correlation:0.12627
  |
Correlation:0.14556
  |
Correlation:0.19145
  |
Correlation:0.077173
  |
Component:310; Jaccard:0.065638
 |
Component:310; Jaccard:0.063196
 |
Component:310; Jaccard:0.049607
 |
Component:311; Jaccard:0.028163
 |
Component:234; Jaccard:0.012388
 |
Component:234; Jaccard:0.004233
 |
Component:177; Jaccard:0.001122
 |
Correlation:0.16699
  |
Correlation:0.16699
  |
Correlation:0.16699
  |
Correlation:0.060256
  |
Correlation:0.077173
  |
Correlation:0.077173
  |
Correlation:0.12627
  |
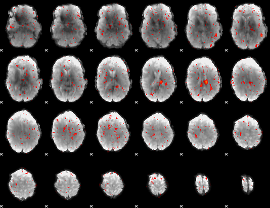
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
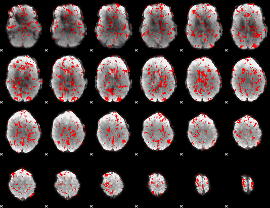 |
GLM binary result thresholded by 1.5
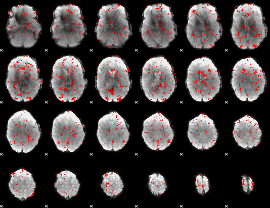 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
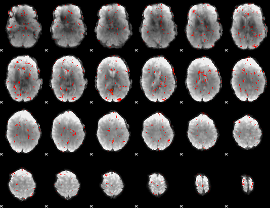 |
GLM Z-score result thresholded by 2.5
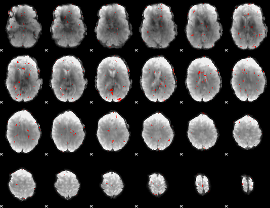 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
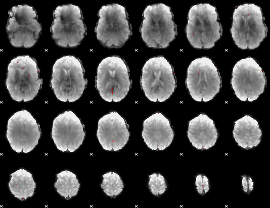 |
GLM Z-score result thresholded by 4
 |
Component:177; Jaccard:0.086604
 |
Component:177; Jaccard:0.086131
 |
Component:177; Jaccard:0.06931
 |
Component:177; Jaccard:0.041488
 |
Component:139; Jaccard:0.024438
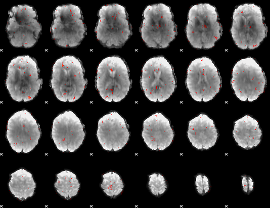 |
Component:139; Jaccard:0.012117
 |
Component:139; Jaccard:0.004708
 |
Correlation:0.034898
  |
Correlation:0.034898
  |
Correlation:0.034898
  |
Correlation:0.034898
  |
Correlation:0.16295
  |
Correlation:0.16295
  |
Correlation:0.16295
  |
Component:013; Jaccard:0.081073
 |
Component:013; Jaccard:0.07862
 |
Component:013; Jaccard:0.063407
 |
Component:067; Jaccard:0.041101
 |
Component:067; Jaccard:0.023573
 |
Component:013; Jaccard:0.01005
 |
Component:177; Jaccard:0.003592
 |
Correlation:0.2019
  |
Correlation:0.2019
  |
Correlation:0.2019
  |
Correlation:0.23109
  |
Correlation:0.23109
  |
Correlation:0.2019
  |
Correlation:0.034898
  |
Component:274; Jaccard:0.071397
 |
Component:063; Jaccard:0.064548
 |
Component:067; Jaccard:0.055024
 |
Component:013; Jaccard:0.041089
 |
Component:013; Jaccard:0.0225
 |
Component:067; Jaccard:0.00982
 |
Component:013; Jaccard:0.003341
 |
Correlation:0.14439
  |
Correlation:0.16573
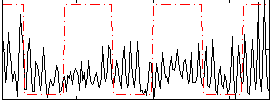  |
Correlation:0.23109
  |
Correlation:0.2019
  |
Correlation:0.2019
  |
Correlation:0.23109
  |
Correlation:0.2019
  |
Component:247; Jaccard:0.066413
 |
Component:149; Jaccard:0.06141
 |
Component:063; Jaccard:0.05405
 |
Component:139; Jaccard:0.037864
 |
Component:177; Jaccard:0.019744
 |
Component:177; Jaccard:0.009353
 |
Component:067; Jaccard:0.002588
 |
Correlation:0.10854
  |
Correlation:-0.065653
  |
Correlation:0.16573
  |
Correlation:0.16295
  |
Correlation:0.034898
  |
Correlation:0.034898
  |
Correlation:0.23109
  |
Component:149; Jaccard:0.066309
 |
Component:067; Jaccard:0.059952
 |
Component:187; Jaccard:0.047127
 |
Component:063; Jaccard:0.034683
 |
Component:089; Jaccard:0.015882
 |
Component:089; Jaccard:0.007044
 |
Component:089; Jaccard:0.002165
 |
Correlation:-0.065653
  |
Correlation:0.23109
  |
Correlation:0.1195
 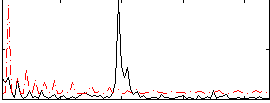 |
Correlation:0.16573
  |
Correlation:0.036652
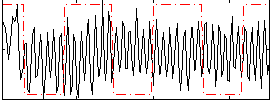 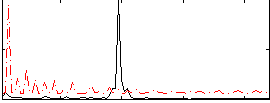 |
Correlation:0.036652
  |
Correlation:0.036652
  |
Component:208; Jaccard:0.066139
 |
Component:247; Jaccard:0.059901
 |
Component:149; Jaccard:0.046278
 |
Component:089; Jaccard:0.030916
 |
Component:063; Jaccard:0.015871
 |
Component:063; Jaccard:0.006586
 |
Component:142; Jaccard:0.002101
 |
Correlation:0.31109
  |
Correlation:0.10854
  |
Correlation:-0.065653
  |
Correlation:0.036652
  |
Correlation:0.16573
  |
Correlation:0.16573
  |
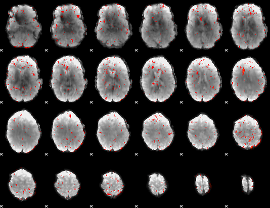
Correlation:0.045815
  |
Component:063; Jaccard:0.061488
 |
Component:187; Jaccard:0.057396
 |
Component:139; Jaccard:0.04569
 |
Component:187; Jaccard:0.030226
 |
Component:187; Jaccard:0.014227
 |
Component:142; Jaccard:0.00574
 |
Component:155; Jaccard:0.001674
 |
Correlation:0.16573
  |
Correlation:0.1195
  |
Correlation:0.16295
  |
Correlation:0.1195
  |
Correlation:0.1195
  |
Correlation:0.045815
  |
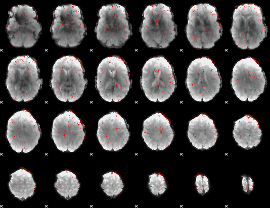
Correlation:0.12956
  |
Component:259; Jaccard:0.060272
 |
Component:208; Jaccard:0.057095
 |
Component:234; Jaccard:0.044101
 |
Component:142; Jaccard:0.026915
 |
Component:142; Jaccard:0.012897
 |
Component:155; Jaccard:0.004892
 |
Component:063; Jaccard:0.00138
 |
Correlation:-0.0082305
  |
Correlation:0.31109
  |
Correlation:0.034774
  |
Correlation:0.045815
  |
Correlation:0.045815
  |
Correlation:0.12956
  |
Correlation:0.16573
  |
Component:091; Jaccard:0.059884
 |
Component:161; Jaccard:0.053646
 |
Component:027; Jaccard:0.043994
 |
Component:234; Jaccard:0.026529
 |
Component:241; Jaccard:0.011714
 |
Component:241; Jaccard:0.004675
 |
Component:299; Jaccard:0.001357
 |
Correlation:0.030331
  |
Correlation:0.030827
  |
Correlation:0.073934
  |
Correlation:0.034774
  |
Correlation:-0.0022418
  |
Correlation:-0.0022418
  |
Correlation:-0.032033
  |
Component:047; Jaccard:0.059211
 |
Component:234; Jaccard:0.05358
 |
Component:247; Jaccard:0.043307
 |
Component:027; Jaccard:0.026476
 |
Component:027; Jaccard:0.011218
 |
Component:299; Jaccard:0.004089
 |
Component:241; Jaccard:0.001132
 |
Correlation:-0.079443
  |
Correlation:0.034774
  |
Correlation:0.10854
  |
Correlation:0.073934
  |
Correlation:0.073934
  |
Correlation:-0.032033
  |
Correlation:-0.0022418
  |
Cope: 06:SHAPES-FACES BACK
|
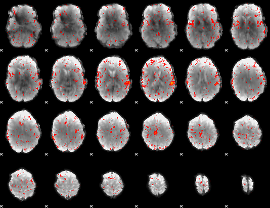
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
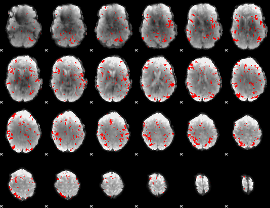 |
GLM binary result thresholded by 3
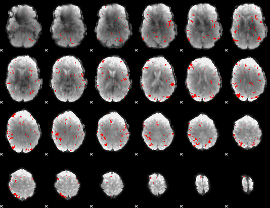 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
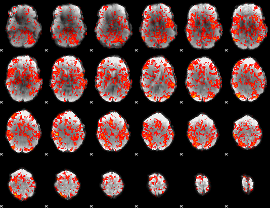 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
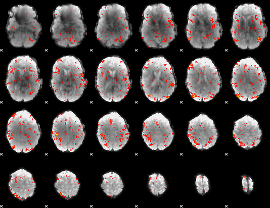 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
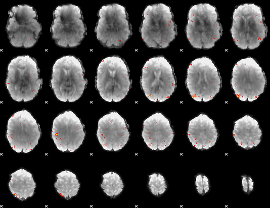 |
Component:110; Jaccard:0.1287
 |
Component:110; Jaccard:0.16299
 |
Component:110; Jaccard:0.19365
 |
Component:110; Jaccard:0.20159
 |
Component:110; Jaccard:0.16823
 |
Component:110; Jaccard:0.11384
 |
Component:110; Jaccard:0.067438
 |
Correlation:0.41823
 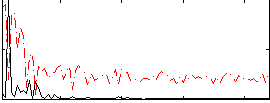 |
Correlation:0.41823
  |
Correlation:0.41823
  |
Correlation:0.41823
  |
Correlation:0.41823
  |
Correlation:0.41823
  |
Correlation:0.41823
  |
Component:324; Jaccard:0.12119
 |
Component:324; Jaccard:0.14637
 |
Component:324; Jaccard:0.16478
 |
Component:324; Jaccard:0.16121
 |
Component:324; Jaccard:0.1277
 |
Component:324; Jaccard:0.086322
 |
Component:324; Jaccard:0.050517
 |
Correlation:0.19907
  |
Correlation:0.19907
  |
Correlation:0.19907
  |
Correlation:0.19907
  |
Correlation:0.19907
  |
Correlation:0.19907
  |
Correlation:0.19907
  |
Component:250; Jaccard:0.11251
 |
Component:250; Jaccard:0.12088
 |
Component:284; Jaccard:0.12439
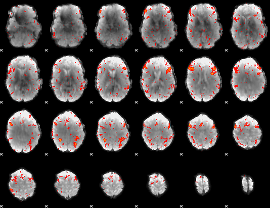 |
Component:284; Jaccard:0.11144
 |
Component:284; Jaccard:0.082595
 |
Component:360; Jaccard:0.051618
 |
Component:360; Jaccard:0.031716
 |
Correlation:0.23368
 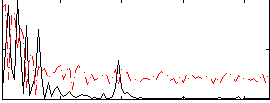 |
Correlation:0.23368
  |
Correlation:0.052335
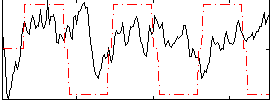 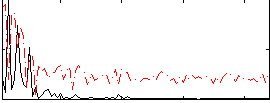 |
Correlation:0.052335
  |
Correlation:0.052335
  |
Correlation:0.28927
  |
Correlation:0.28927
  |
Component:284; Jaccard:0.10373
 |
Component:284; Jaccard:0.11926
 |
Component:250; Jaccard:0.1176
 |
Component:250; Jaccard:0.099064
 |
Component:360; Jaccard:0.071358
 |
Component:284; Jaccard:0.045104
 |
Component:044; Jaccard:0.024778
 |
Correlation:0.052335
  |
Correlation:0.052335
  |
Correlation:0.23368
  |
Correlation:0.23368
  |
Correlation:0.28927
  |
Correlation:0.052335
  |
Correlation:0.31813
  |
Component:232; Jaccard:0.10208
 |
Component:232; Jaccard:0.11029
 |
Component:080; Jaccard:0.11046
 |
Component:080; Jaccard:0.09452
 |
Component:250; Jaccard:0.068151
 |
Component:044; Jaccard:0.042202
 |
Component:215; Jaccard:0.022028
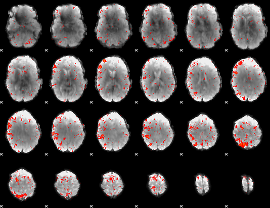 |
Correlation:-0.0081035
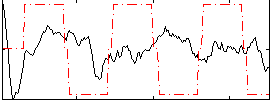  |
Correlation:-0.0081035
  |
Correlation:0.30556
  |
Correlation:0.30556
  |
Correlation:0.23368
  |
Correlation:0.31813
  |
Correlation:0.14125
  |
Component:080; Jaccard:0.10083
 |
Component:080; Jaccard:0.10978
 |
Component:232; Jaccard:0.1067
 |
Component:232; Jaccard:0.088366
 |
Component:080; Jaccard:0.066216
 |
Component:080; Jaccard:0.03775
 |
Component:284; Jaccard:0.020682
 |
Correlation:0.30556
  |
Correlation:0.30556
  |
Correlation:-0.0081035
  |
Correlation:-0.0081035
  |
Correlation:0.30556
  |
Correlation:0.30556
  |
Correlation:0.052335
  |
Component:385; Jaccard:0.088471
 |
Component:127; Jaccard:0.095979
 |
Component:127; Jaccard:0.096767
 |
Component:127; Jaccard:0.086227
 |
Component:044; Jaccard:0.065387
 |
Component:250; Jaccard:0.037364
 |
Component:250; Jaccard:0.019232
 |
Correlation:0.10698
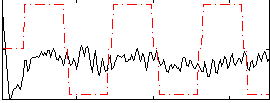  |
Correlation:0.13371
  |
Correlation:0.13371
  |
Correlation:0.13371
  |
Correlation:0.31813
  |
Correlation:0.23368
  |
Correlation:0.23368
  |
Component:127; Jaccard:0.086955
 |
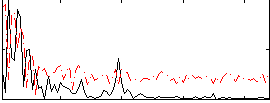
Component:184; Jaccard:0.093044
 |
Component:184; Jaccard:0.090218
 |
Component:044; Jaccard:0.084164
 |
Component:232; Jaccard:0.059546
 |
Component:215; Jaccard:0.037228
 |
Component:080; Jaccard:0.018338
 |
Correlation:0.13371
  |
Correlation:0.36883
 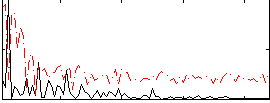 |
Correlation:0.36883
  |
Correlation:0.31813
  |
Correlation:-0.0081035
  |
Correlation:0.14125
  |
Correlation:0.30556
  |
Component:184; Jaccard:0.086423
 |
Component:190; Jaccard:0.089423
 |
Component:190; Jaccard:0.089451
 |
Component:360; Jaccard:0.080815
 |
Component:127; Jaccard:0.058389
 |
Component:108; Jaccard:0.035025
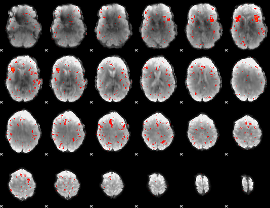 |
Component:108; Jaccard:0.018071
 |
Correlation:0.36883
  |
Correlation:0.17096
  |
Correlation:0.17096
  |
Correlation:0.28927
  |
Correlation:0.13371
  |
Correlation:0.23979
  |
Correlation:0.23979
  |
Component:190; Jaccard:0.081796
 |
Component:385; Jaccard:0.08919
 |
Component:310; Jaccard:0.087893
 |
Component:190; Jaccard:0.079628
 |
Component:215; Jaccard:0.058321
 |
Component:232; Jaccard:0.033149
 |
Component:232; Jaccard:0.0178
 |
Correlation:0.17096
  |
Correlation:0.10698
  |
Correlation:0.2109
  |
Correlation:0.17096
  |
Correlation:0.14125
  |
Correlation:-0.0081035
  |
Correlation:-0.0081035
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































