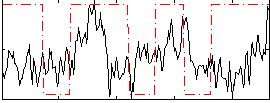
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
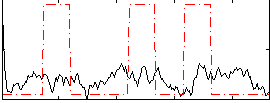
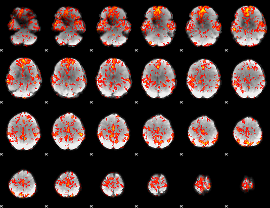
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
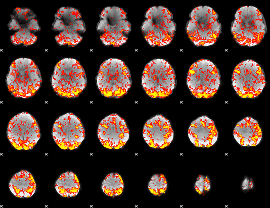 |
GLM Z-score result thresholded by 2
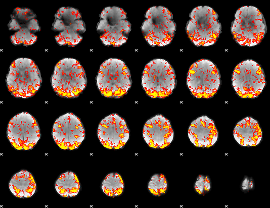 |
GLM Z-score result thresholded by 2.5
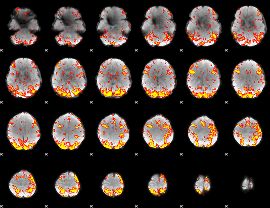 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
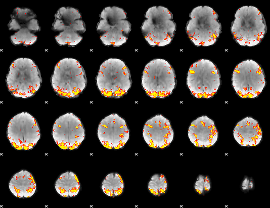 |
Component:246; Jaccard:0.14122
 |
Component:246; Jaccard:0.17732
 |
Component:246; Jaccard:0.22079
 |
Component:246; Jaccard:0.27035
 |
Component:246; Jaccard:0.31774
 |
Component:246; Jaccard:0.35964
 |
Component:246; Jaccard:0.38943
 |
Correlation:0.18151
  |
Correlation:0.18151
  |
Correlation:0.18151
  |
Correlation:0.18151
  |
Correlation:0.18151
  |
Correlation:0.18151
  |
Correlation:0.18151
  |
Component:053; Jaccard:0.14039
 |
Component:053; Jaccard:0.17367
 |
Component:053; Jaccard:0.21211
 |
Component:053; Jaccard:0.25101
 |
Component:053; Jaccard:0.2871
 |
Component:053; Jaccard:0.31447
 |
Component:053; Jaccard:0.33062
 |
Correlation:0.20374
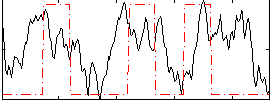  |
Correlation:0.20374
  |
Correlation:0.20374
  |
Correlation:0.20374
  |
Correlation:0.20374
  |
Correlation:0.20374
  |
Correlation:0.20374
  |
Component:242; Jaccard:0.1275
 |
Component:242; Jaccard:0.15507
 |
Component:242; Jaccard:0.18537
 |
Component:242; Jaccard:0.21459
 |
Component:242; Jaccard:0.23878
 |
Component:242; Jaccard:0.25551
 |
Component:242; Jaccard:0.26613
 |
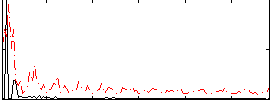
Correlation:0.1867
  |
Correlation:0.1867
  |
Correlation:0.1867
  |
Correlation:0.1867
  |
Correlation:0.1867
  |
Correlation:0.1867
  |
Correlation:0.1867
  |
Component:375; Jaccard:0.12727
 |
Component:363; Jaccard:0.14729
 |
Component:363; Jaccard:0.17376
 |
Component:363; Jaccard:0.19928
 |
Component:363; Jaccard:0.21724
 |
Component:363; Jaccard:0.22794
 |
Component:363; Jaccard:0.22784
 |
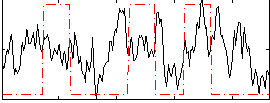
Correlation:0.056342
 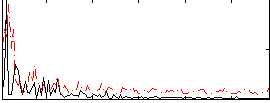 |
Correlation:0.037126
  |
Correlation:0.037126
  |
Correlation:0.037126
  |
Correlation:0.037126
  |
Correlation:0.037126
  |
Correlation:0.037126
  |
Component:298; Jaccard:0.12656
 |
Component:298; Jaccard:0.14562
 |
Component:298; Jaccard:0.16377
 |
Component:298; Jaccard:0.17874
 |
Component:153; Jaccard:0.18884
 |
Component:197; Jaccard:0.20055
 |
Component:197; Jaccard:0.20878
 |
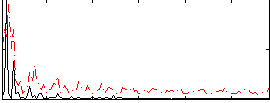
Correlation:0.17359
  |
Correlation:0.17359
  |
Correlation:0.17359
  |
Correlation:0.17359
  |
Correlation:0.064822
  |
Correlation:0.15745
  |
Correlation:0.15745
  |
Component:147; Jaccard:0.12432
 |
Component:375; Jaccard:0.14184
 |
Component:153; Jaccard:0.15527
 |
Component:153; Jaccard:0.17458
 |
Component:298; Jaccard:0.18629
 |
Component:153; Jaccard:0.19854
 |
Component:153; Jaccard:0.20041
 |
Correlation:0.20814
  |
Correlation:0.056342
  |
Correlation:0.064822
  |
Correlation:0.064822
  |
Correlation:0.17359
  |
Correlation:0.064822
  |
Correlation:0.064822
  |
Component:363; Jaccard:0.12217
 |
Component:147; Jaccard:0.14078
 |
Component:375; Jaccard:0.15459
 |
Component:336; Jaccard:0.16953
 |
Component:197; Jaccard:0.18491
 |
Component:336; Jaccard:0.1895
 |
Component:336; Jaccard:0.18783
 |
Correlation:0.037126
  |
Correlation:0.20814
  |
Correlation:0.056342
  |
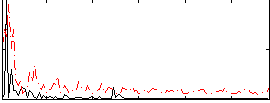
Correlation:0.2273
 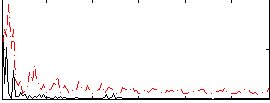 |
Correlation:0.15745
  |
Correlation:0.2273
  |
Correlation:0.2273
  |
Component:153; Jaccard:0.11151
 |
Component:153; Jaccard:0.13309
 |
Component:147; Jaccard:0.15205
 |
Component:197; Jaccard:0.166
 |
Component:336; Jaccard:0.18375
 |
Component:298; Jaccard:0.18617
 |
Component:298; Jaccard:0.18217
 |
Correlation:0.064822
  |
Correlation:0.064822
  |
Correlation:0.20814
  |
Correlation:0.15745
  |
Correlation:0.2273
  |
Correlation:0.17359
  |
Correlation:0.17359
  |
Component:336; Jaccard:0.10456
 |
Component:336; Jaccard:0.12653
 |
Component:336; Jaccard:0.14884
 |
Component:375; Jaccard:0.16231
 |
Component:375; Jaccard:0.16619
 |
Component:315; Jaccard:0.16787
 |
Component:219; Jaccard:0.17151
 |
Correlation:0.2273
  |
Correlation:0.2273
  |
Correlation:0.2273
  |
Correlation:0.056342
  |
Correlation:0.056342
  |
Correlation:0.12254
  |
Correlation:0.26008
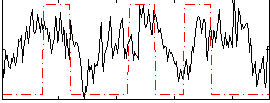  |
Component:062; Jaccard:0.1021
 |
Component:197; Jaccard:0.12112
 |
Component:197; Jaccard:0.14322
 |
Component:147; Jaccard:0.15821
 |
Component:315; Jaccard:0.16045
 |
Component:219; Jaccard:0.16621
 |
Component:315; Jaccard:0.16898
 |
Correlation:-0.051963
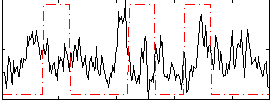  |
Correlation:0.15745
  |
Correlation:0.15745
  |
Correlation:0.20814
  |
Correlation:0.12254
  |
Correlation:0.26008
  |
Correlation:0.12254
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
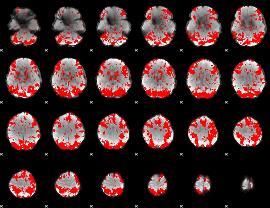 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
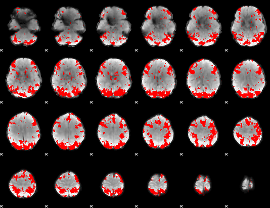 |
GLM binary result thresholded by 3.5
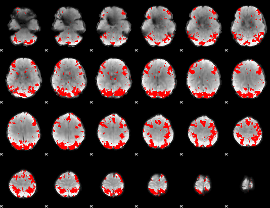 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
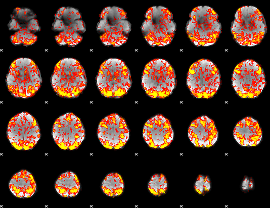 |
GLM Z-score result thresholded by 1.5
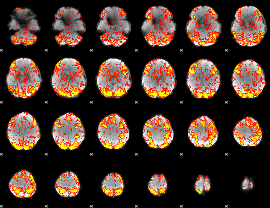 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
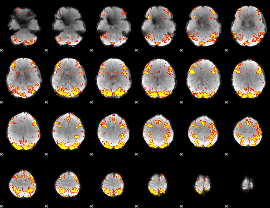 |
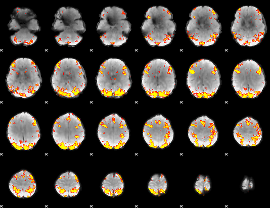
GLM Z-score result thresholded by 4
 |
Component:053; Jaccard:0.13849
 |
Component:246; Jaccard:0.1675
 |
Component:246; Jaccard:0.20418
 |
Component:246; Jaccard:0.24386
 |
Component:246; Jaccard:0.28537
 |
Component:246; Jaccard:0.3229
 |
Component:246; Jaccard:0.35858
 |
Correlation:0.18407
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Component:246; Jaccard:0.1372
 |
Component:053; Jaccard:0.16701
 |
Component:053; Jaccard:0.20032
 |
Component:053; Jaccard:0.23639
 |
Component:053; Jaccard:0.27151
 |
Component:053; Jaccard:0.30142
 |
Component:053; Jaccard:0.32601
 |
Correlation:0.368
  |
Correlation:0.18407
  |
Correlation:0.18407
  |
Correlation:0.18407
  |
Correlation:0.18407
  |
Correlation:0.18407
  |
Correlation:0.18407
  |
Component:375; Jaccard:0.1261
 |
Component:363; Jaccard:0.146
 |
Component:363; Jaccard:0.17239
 |
Component:363; Jaccard:0.19916
 |
Component:363; Jaccard:0.22235
 |
Component:363; Jaccard:0.24053
 |
Component:363; Jaccard:0.25413
 |
Correlation:0.20089
  |
Correlation:0.47895
  |
Correlation:0.47895
  |
Correlation:0.47895
  |
Correlation:0.47895
  |
Correlation:0.47895
  |
Correlation:0.47895
  |
Component:298; Jaccard:0.12317
 |
Component:242; Jaccard:0.14273
 |
Component:242; Jaccard:0.16548
 |
Component:242; Jaccard:0.18773
 |
Component:242; Jaccard:0.20686
 |
Component:242; Jaccard:0.22196
 |
Component:242; Jaccard:0.23139
 |
Correlation:0.16918
  |
Correlation:0.23588
  |
Correlation:0.23588
  |
Correlation:0.23588
  |
Correlation:0.23588
  |
Correlation:0.23588
  |
Correlation:0.23588
  |
Component:363; Jaccard:0.12282
 |
Component:375; Jaccard:0.14004
 |
Component:298; Jaccard:0.15612
 |
Component:232; Jaccard:0.17587
 |
Component:232; Jaccard:0.19455
 |
Component:232; Jaccard:0.20864
 |
Component:232; Jaccard:0.21994
 |
Correlation:0.47895
  |
Correlation:0.20089
  |
Correlation:0.16918
  |
Correlation:0.39516
  |
Correlation:0.39516
  |
Correlation:0.39516
  |
Correlation:0.39516
  |
Component:242; Jaccard:0.12111
 |
Component:298; Jaccard:0.1392
 |
Component:232; Jaccard:0.15529
 |
Component:153; Jaccard:0.1757
 |
Component:153; Jaccard:0.19333
 |
Component:153; Jaccard:0.20717
 |
Component:153; Jaccard:0.21865
 |
Correlation:0.23588
  |
Correlation:0.16918
  |
Correlation:0.39516
  |
Correlation:0.42514
  |
Correlation:0.42514
  |
Correlation:0.42514
  |
Correlation:0.42514
  |
Component:153; Jaccard:0.11275
 |
Component:232; Jaccard:0.13292
 |
Component:153; Jaccard:0.15455
 |
Component:298; Jaccard:0.16953
 |
Component:197; Jaccard:0.18188
 |
Component:197; Jaccard:0.1971
 |
Component:197; Jaccard:0.20931
 |
Correlation:0.42514
  |
Correlation:0.39516
  |
Correlation:0.42514
  |
Correlation:0.16918
  |
Correlation:0.43089
  |
Correlation:0.43089
  |
Correlation:0.43089
  |
Component:232; Jaccard:0.11254
 |
Component:153; Jaccard:0.13268
 |
Component:375; Jaccard:0.15332
 |
Component:375; Jaccard:0.16414
 |
Component:298; Jaccard:0.17935
 |
Component:298; Jaccard:0.18282
 |
Component:298; Jaccard:0.18368
 |
Correlation:0.39516
  |
Correlation:0.42514
  |
Correlation:0.20089
  |
Correlation:0.20089
  |
Correlation:0.16918
  |
Correlation:0.16918
  |
Correlation:0.16918
  |
Component:251; Jaccard:0.11038
 |
Component:197; Jaccard:0.12136
 |
Component:197; Jaccard:0.1425
 |
Component:197; Jaccard:0.16334
 |
Component:375; Jaccard:0.17041
 |
Component:375; Jaccard:0.173
 |
Component:375; Jaccard:0.17174
 |
Correlation:0.14545
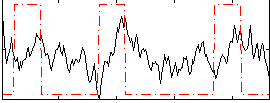  |
Correlation:0.43089
  |
Correlation:0.43089
  |
Correlation:0.43089
  |
Correlation:0.20089
  |
Correlation:0.20089
  |
Correlation:0.20089
  |
Component:062; Jaccard:0.10488
 |
Component:251; Jaccard:0.12086
 |
Component:315; Jaccard:0.13075
 |
Component:315; Jaccard:0.14529
 |
Component:315; Jaccard:0.15593
 |
Component:315; Jaccard:0.16495
 |
Component:315; Jaccard:0.17122
 |
Correlation:0.18369
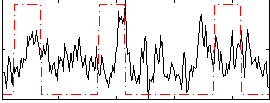  |
Correlation:0.14545
  |
Correlation:0.19585
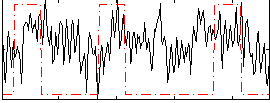  |
Correlation:0.19585
  |
Correlation:0.19585
  |
Correlation:0.19585
  |
Correlation:0.19585
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
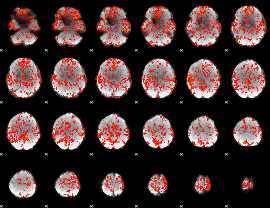 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
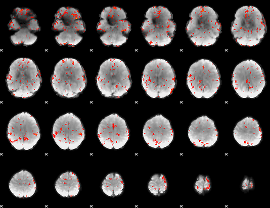 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:196; Jaccard:0.10501
 |
Component:196; Jaccard:0.10664
 |
Component:398; Jaccard:0.10656
 |
Component:019; Jaccard:0.092194
 |
Component:270; Jaccard:0.074563
 |
Component:270; Jaccard:0.049827
 |
Component:270; Jaccard:0.024809
 |
Correlation:0.21169
  |
Correlation:0.21169
  |
Correlation:0.080706
  |
Correlation:0.26065
  |
Correlation:0.16365
 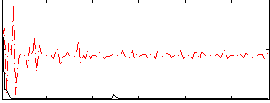 |
Correlation:0.16365
  |
Correlation:0.16365
  |
Component:398; Jaccard:0.095449
 |
Component:398; Jaccard:0.10484
 |
Component:019; Jaccard:0.10013
 |
Component:398; Jaccard:0.090731
 |
Component:019; Jaccard:0.070115
 |
Component:019; Jaccard:0.045687
 |
Component:019; Jaccard:0.022083
 |
Correlation:0.080706
  |
Correlation:0.080706
  |
Correlation:0.26065
  |
Correlation:0.080706
  |
Correlation:0.26065
  |
Correlation:0.26065
  |
Correlation:0.26065
  |
Component:019; Jaccard:0.087015
 |
Component:019; Jaccard:0.096772
 |
Component:196; Jaccard:0.099588
 |
Component:270; Jaccard:0.085892
 |
Component:398; Jaccard:0.061987
 |
Component:129; Jaccard:0.039511
 |
Component:042; Jaccard:0.020161
 |
Correlation:0.26065
  |
Correlation:0.26065
  |
Correlation:0.21169
  |
Correlation:0.16365
  |
Correlation:0.080706
  |
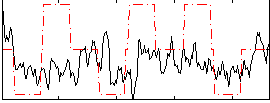
Correlation:0.21469
 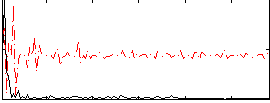 |
Correlation:0.13799
  |
Component:358; Jaccard:0.085305
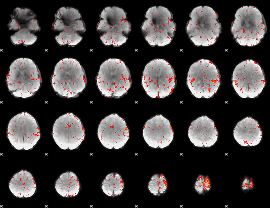 |
Component:358; Jaccard:0.090949
 |
Component:042; Jaccard:0.093446
 |
Component:042; Jaccard:0.082993
 |
Component:042; Jaccard:0.061443
 |
Component:042; Jaccard:0.039402
 |
Component:129; Jaccard:0.019152
 |
Correlation:0.26768
  |
Correlation:0.26768
  |
Correlation:0.13799
  |
Correlation:0.13799
  |
Correlation:0.13799
  |
Correlation:0.13799
  |
Correlation:0.21469
  |
Component:042; Jaccard:0.081482
 |
Component:042; Jaccard:0.090085
 |
Component:358; Jaccard:0.08903
 |
Component:386; Jaccard:0.080032
 |
Component:386; Jaccard:0.059722
 |
Component:098; Jaccard:0.03877
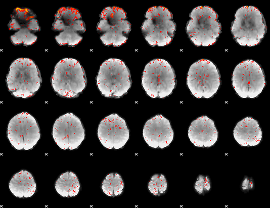 |
Component:098; Jaccard:0.018227
 |
Correlation:0.13799
  |
Correlation:0.13799
  |
Correlation:0.26768
  |
Correlation:0.23004
 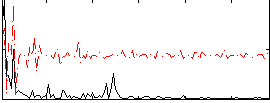 |
Correlation:0.23004
  |
Correlation:0.293
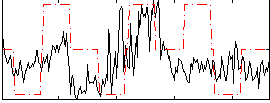  |
Correlation:0.293
  |
Component:332; Jaccard:0.081281
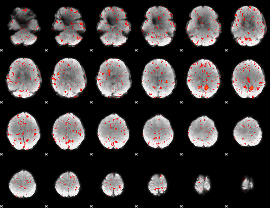 |
Component:098; Jaccard:0.085853
 |
Component:386; Jaccard:0.088388
 |
Component:358; Jaccard:0.077425
 |
Component:358; Jaccard:0.058936
 |
Component:386; Jaccard:0.035865
 |
Component:322; Jaccard:0.017396
 |
Correlation:0.23233
  |
Correlation:0.293
  |
Correlation:0.23004
  |
Correlation:0.26768
  |
Correlation:0.26768
  |
Correlation:0.23004
  |
Correlation:0.35053
  |
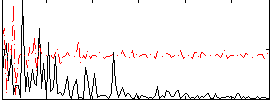
Component:104; Jaccard:0.079836
 |
Component:386; Jaccard:0.08559
 |
Component:098; Jaccard:0.086121
 |
Component:196; Jaccard:0.077403
 |
Component:098; Jaccard:0.058509
 |
Component:322; Jaccard:0.035583
 |
Component:386; Jaccard:0.015899
 |
Correlation:0.10991
  |
Correlation:0.23004
  |
Correlation:0.293
  |
Correlation:0.21169
  |
Correlation:0.293
  |
Correlation:0.35053
  |
Correlation:0.23004
  |
Component:098; Jaccard:0.079093
 |
Component:104; Jaccard:0.081356
 |
Component:270; Jaccard:0.083266
 |
Component:098; Jaccard:0.075894
 |
Component:129; Jaccard:0.0585
 |
Component:358; Jaccard:0.034538
 |
Component:358; Jaccard:0.015826
 |
Correlation:0.293
  |
Correlation:0.10991
  |
Correlation:0.16365
  |
Correlation:0.293
  |
Correlation:0.21469
  |
Correlation:0.26768
  |
Correlation:0.26768
  |
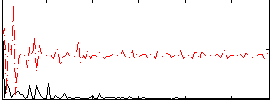
Component:386; Jaccard:0.078301
 |
Component:322; Jaccard:0.081093
 |
Component:312; Jaccard:0.081514
 |
Component:322; Jaccard:0.07312
 |
Component:322; Jaccard:0.05695
 |
Component:312; Jaccard:0.033104
 |
Component:312; Jaccard:0.015668
 |
Correlation:0.23004
  |
Correlation:0.35053
  |
Correlation:0.30874
  |
Correlation:0.35053
  |
Correlation:0.35053
  |
Correlation:0.30874
  |
Correlation:0.30874
  |
Component:312; Jaccard:0.077466
 |
Component:312; Jaccard:0.080862
 |
Component:322; Jaccard:0.0808
 |
Component:129; Jaccard:0.072132
 |
Component:312; Jaccard:0.054933
 |
Component:398; Jaccard:0.032453
 |
Component:079; Jaccard:0.013944
 |
Correlation:0.30874
  |
Correlation:0.30874
  |
Correlation:0.35053
  |
Correlation:0.21469
  |
Correlation:0.30874
  |
Correlation:0.080706
  |
Correlation:0.13474
  |
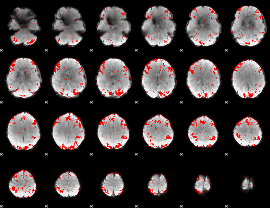
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
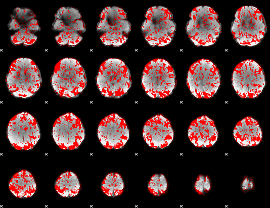 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
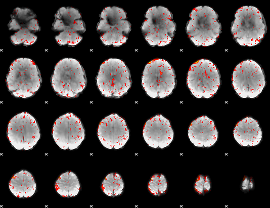
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:251; Jaccard:0.13203
 |
Component:251; Jaccard:0.14872
 |
Component:232; Jaccard:0.16898
 |
Component:232; Jaccard:0.18275
 |
Component:232; Jaccard:0.18505
 |
Component:232; Jaccard:0.17076
 |
Component:232; Jaccard:0.14373
 |
Correlation:0.073625
  |
Correlation:0.073625
  |
Correlation:0.23416
  |
Correlation:0.23416
  |
Correlation:0.23416
  |
Correlation:0.23416
  |
Correlation:0.23416
  |
Component:232; Jaccard:0.12198
 |
Component:232; Jaccard:0.14588
 |
Component:251; Jaccard:0.15997
 |
Component:251; Jaccard:0.16199
 |
Component:251; Jaccard:0.15001
 |
Component:251; Jaccard:0.12953
 |
Component:251; Jaccard:0.10223
 |
Correlation:0.23416
  |
Correlation:0.23416
  |
Correlation:0.073625
  |
Correlation:0.073625
  |
Correlation:0.073625
  |
Correlation:0.073625
  |
Correlation:0.073625
  |
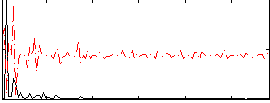
Component:246; Jaccard:0.12143
 |
Component:246; Jaccard:0.13787
 |
Component:246; Jaccard:0.14763
 |
Component:246; Jaccard:0.14827
 |
Component:246; Jaccard:0.14047
 |
Component:246; Jaccard:0.11808
 |
Component:246; Jaccard:0.092844
 |
Correlation:0.11054
  |
Correlation:0.11054
  |
Correlation:0.11054
  |
Correlation:0.11054
  |
Correlation:0.11054
  |
Correlation:0.11054
  |
Correlation:0.11054
  |
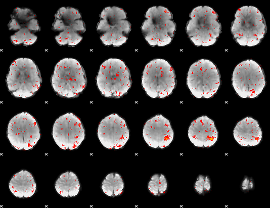
Component:053; Jaccard:0.11427
 |
Component:053; Jaccard:0.12599
 |
Component:220; Jaccard:0.13244
 |
Component:220; Jaccard:0.13329
 |
Component:220; Jaccard:0.12524
 |
Component:220; Jaccard:0.10982
 |
Component:282; Jaccard:0.090044
 |
Correlation:-0.011658
  |
Correlation:-0.011658
  |
Correlation:-0.022531
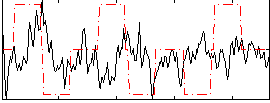  |
Correlation:-0.022531
  |
Correlation:-0.022531
  |
Correlation:-0.022531
  |
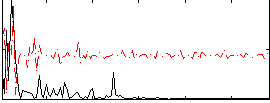
Correlation:0.23612
  |
Component:220; Jaccard:0.11227
 |
Component:220; Jaccard:0.12355
 |
Component:053; Jaccard:0.13206
 |
Component:053; Jaccard:0.13118
 |
Component:053; Jaccard:0.12211
 |
Component:282; Jaccard:0.10484
 |
Component:220; Jaccard:0.089805
 |
Correlation:-0.022531
  |
Correlation:-0.022531
  |
Correlation:-0.011658
  |
Correlation:-0.011658
  |
Correlation:-0.011658
  |
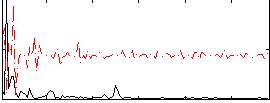
Correlation:0.23612
  |
Correlation:-0.022531
  |
Component:181; Jaccard:0.1084
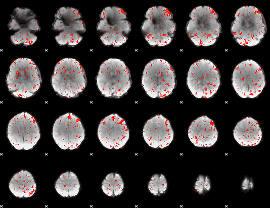 |
Component:181; Jaccard:0.11811
 |
Component:181; Jaccard:0.12105
 |
Component:363; Jaccard:0.12141
 |
Component:313; Jaccard:0.11517
 |
Component:053; Jaccard:0.10324
 |
Component:363; Jaccard:0.08544
 |
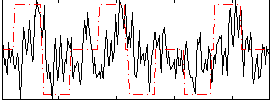
Correlation:0.29259
  |
Correlation:0.29259
  |
Correlation:0.29259
  |
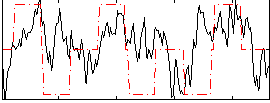
Correlation:0.26187
  |
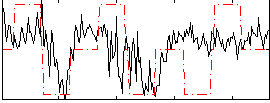
Correlation:0.3385
  |
Correlation:-0.011658
  |
Correlation:0.26187
  |
Component:363; Jaccard:0.10124
 |
Component:363; Jaccard:0.11189
 |
Component:363; Jaccard:0.11952
 |
Component:313; Jaccard:0.11705
 |
Component:363; Jaccard:0.11484
 |
Component:363; Jaccard:0.10118
 |
Component:053; Jaccard:0.082402
 |
Correlation:0.26187
  |
Correlation:0.26187
  |
Correlation:0.26187
  |
Correlation:0.3385
  |
Correlation:0.26187
  |
Correlation:0.26187
  |
Correlation:-0.011658
  |
Component:187; Jaccard:0.099232
 |
Component:187; Jaccard:0.1063
 |
Component:313; Jaccard:0.1102
 |
Component:181; Jaccard:0.11697
 |
Component:291; Jaccard:0.11137
 |
Component:291; Jaccard:0.097561
 |
Component:064; Jaccard:0.078391
 |
Correlation:0.032739
 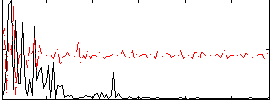 |
Correlation:0.032739
  |
Correlation:0.3385
  |
Correlation:0.29259
  |
Correlation:0.30398
  |
Correlation:0.30398
  |
Correlation:0.16816
  |
Component:328; Jaccard:0.097221
 |
Component:153; Jaccard:0.10236
 |
Component:291; Jaccard:0.10915
 |
Component:291; Jaccard:0.1153
 |
Component:282; Jaccard:0.1108
 |
Component:313; Jaccard:0.096619
 |
Component:291; Jaccard:0.076
 |
Correlation:0.26909
  |
Correlation:0.21356
  |
Correlation:0.30398
  |
Correlation:0.30398
  |
Correlation:0.23612
  |
Correlation:0.3385
  |
Correlation:0.30398
  |
Component:153; Jaccard:0.091933
 |
Component:328; Jaccard:0.10135
 |
Component:153; Jaccard:0.10822
 |
Component:282; Jaccard:0.11003
 |
Component:181; Jaccard:0.10431
 |
Component:064; Jaccard:0.089516
 |
Component:313; Jaccard:0.071132
 |
Correlation:0.21356
  |
Correlation:0.26909
  |
Correlation:0.21356
  |
Correlation:0.23612
  |
Correlation:0.29259
  |
Correlation:0.16816
  |
Correlation:0.3385
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:369; Jaccard:0.12323
 |
Component:369; Jaccard:0.14448
 |
Component:369; Jaccard:0.1599
 |
Component:369; Jaccard:0.16043
 |
Component:369; Jaccard:0.1467
 |
Component:369; Jaccard:0.11656
 |
Component:369; Jaccard:0.081852
 |
Correlation:0.12167
  |
Correlation:0.12167
  |
Correlation:0.12167
  |
Correlation:0.12167
  |
Correlation:0.12167
  |
Correlation:0.12167
  |
Correlation:0.12167
  |
Component:042; Jaccard:0.10609
 |
Component:313; Jaccard:0.12633
 |
Component:313; Jaccard:0.14757
 |
Component:313; Jaccard:0.15642
 |
Component:313; Jaccard:0.14504
 |
Component:313; Jaccard:0.11642
 |
Component:313; Jaccard:0.076162
 |
Correlation:0.066432
  |
Correlation:0.43865
  |
Correlation:0.43865
  |
Correlation:0.43865
  |
Correlation:0.43865
  |
Correlation:0.43865
  |
Correlation:0.43865
  |
Component:177; Jaccard:0.10294
 |
Component:042; Jaccard:0.11878
 |
Component:042; Jaccard:0.12392
 |
Component:042; Jaccard:0.12396
 |
Component:042; Jaccard:0.11107
 |
Component:257; Jaccard:0.092092
 |
Component:042; Jaccard:0.062889
 |
Correlation:0.13821
  |
Correlation:0.066432
  |
Correlation:0.066432
  |
Correlation:0.066432
  |
Correlation:0.066432
  |
Correlation:0.22395
  |
Correlation:0.066432
  |
Component:313; Jaccard:0.10282
 |
Component:177; Jaccard:0.11368
 |
Component:177; Jaccard:0.11988
 |
Component:342; Jaccard:0.12194
 |
Component:257; Jaccard:0.10816
 |
Component:042; Jaccard:0.087994
 |
Component:257; Jaccard:0.061868
 |
Correlation:0.43865
  |
Correlation:0.13821
  |
Correlation:0.13821
  |
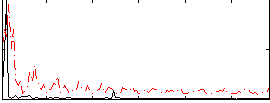
Correlation:0.2187
  |
Correlation:0.22395
  |
Correlation:0.066432
  |
Correlation:0.22395
  |
Component:342; Jaccard:0.094754
 |
Component:342; Jaccard:0.11061
 |
Component:342; Jaccard:0.11957
 |
Component:177; Jaccard:0.11612
 |
Component:342; Jaccard:0.10616
 |
Component:342; Jaccard:0.083827
 |
Component:342; Jaccard:0.051463
 |
Correlation:0.2187
  |
Correlation:0.2187
  |
Correlation:0.2187
  |
Correlation:0.13821
  |
Correlation:0.2187
  |
Correlation:0.2187
  |
Correlation:0.2187
  |
Component:237; Jaccard:0.094605
 |
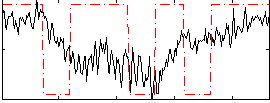
Component:146; Jaccard:0.10704
 |
Component:146; Jaccard:0.11168
 |
Component:257; Jaccard:0.1122
 |
Component:177; Jaccard:0.097217
 |
Component:177; Jaccard:0.073702
 |
Component:326; Jaccard:0.046283
 |
Correlation:0.018177
  |
Correlation:0.29112
  |
Correlation:0.29112
  |
Correlation:0.22395
  |
Correlation:0.13821
  |
Correlation:0.13821
  |
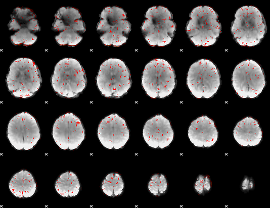
Correlation:0.27242
  |
Component:146; Jaccard:0.09351
 |
Component:078; Jaccard:0.10129
 |
Component:078; Jaccard:0.10753
 |
Component:078; Jaccard:0.10442
 |
Component:078; Jaccard:0.088206
 |
Component:146; Jaccard:0.063416
 |
Component:177; Jaccard:0.045664
 |
Correlation:0.29112
  |
Correlation:0.12085
  |
Correlation:0.12085
  |
Correlation:0.12085
  |
Correlation:0.12085
  |
Correlation:0.29112
  |
Correlation:0.13821
  |
Component:078; Jaccard:0.091772
 |
Component:237; Jaccard:0.09544
 |
Component:257; Jaccard:0.10448
 |
Component:146; Jaccard:0.10415
 |
Component:146; Jaccard:0.086234
 |
Component:078; Jaccard:0.062683
 |
Component:146; Jaccard:0.037294
 |
Correlation:0.12085
  |
Correlation:0.018177
  |
Correlation:0.22395
  |
Correlation:0.29112
  |
Correlation:0.29112
  |
Correlation:0.12085
  |
Correlation:0.29112
  |
Component:140; Jaccard:0.083814
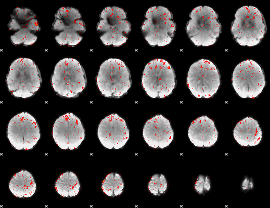 |
Component:257; Jaccard:0.090998
 |
Component:237; Jaccard:0.090026
 |
Component:131; Jaccard:0.084909
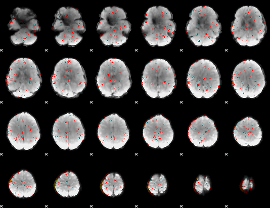 |
Component:131; Jaccard:0.075527
 |
Component:326; Jaccard:0.058312
 |
Component:078; Jaccard:0.036398
 |
Correlation:0.088943
  |
Correlation:0.22395
  |
Correlation:0.018177
  |
Correlation:0.16242
  |
Correlation:0.16242
  |
Correlation:0.27242
  |
Correlation:0.12085
  |
Component:299; Jaccard:0.082967
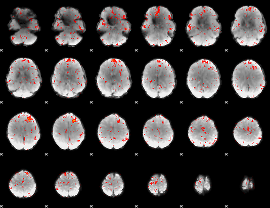 |
Component:140; Jaccard:0.088743
 |
Component:291; Jaccard:0.088552
 |
Component:291; Jaccard:0.084858
 |
Component:291; Jaccard:0.071072
 |
Component:131; Jaccard:0.056431
 |
Component:079; Jaccard:0.035359
 |
Correlation:-0.031564
  |
Correlation:0.088943
  |
Correlation:0.35545
  |
Correlation:0.35545
  |
Correlation:0.35545
  |
Correlation:0.16242
  |
Correlation:0.11252
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:042; Jaccard:0.13006
 |
Component:042; Jaccard:0.16172
 |
Component:042; Jaccard:0.19746
 |
Component:042; Jaccard:0.2256
 |
Component:042; Jaccard:0.23024
 |
Component:042; Jaccard:0.21126
 |
Component:042; Jaccard:0.17398
 |
Correlation:0.29924
  |
Correlation:0.29924
  |
Correlation:0.29924
  |
Correlation:0.29924
  |
Correlation:0.29924
  |
Correlation:0.29924
  |
Correlation:0.29924
  |
Component:237; Jaccard:0.10903
 |
Component:369; Jaccard:0.12147
 |
Component:369; Jaccard:0.13978
 |
Component:369; Jaccard:0.14939
 |
Component:369; Jaccard:0.14771
 |
Component:369; Jaccard:0.13486
 |
Component:369; Jaccard:0.11129
 |
Correlation:0.24102
  |
Correlation:0.28475
  |
Correlation:0.28475
  |
Correlation:0.28475
  |
Correlation:0.28475
  |
Correlation:0.28475
  |
Correlation:0.28475
  |
Component:369; Jaccard:0.10422
 |
Component:237; Jaccard:0.11559
 |
Component:386; Jaccard:0.1228
 |
Component:079; Jaccard:0.12419
 |
Component:079; Jaccard:0.12301
 |
Component:079; Jaccard:0.11465
 |
Component:079; Jaccard:0.091127
 |
Correlation:0.28475
  |
Correlation:0.24102
  |
Correlation:0.22458
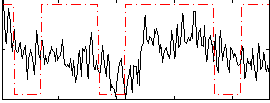 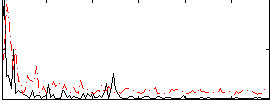 |
Correlation:0.33986
  |
Correlation:0.33986
  |
Correlation:0.33986
  |
Correlation:0.33986
  |
Component:386; Jaccard:0.09983
 |
Component:386; Jaccard:0.11283
 |
Component:237; Jaccard:0.11674
 |
Component:386; Jaccard:0.11968
 |
Component:386; Jaccard:0.10922
 |
Component:326; Jaccard:0.089775
 |
Component:326; Jaccard:0.083346
 |
Correlation:0.22458
  |
Correlation:0.22458
  |
Correlation:0.24102
  |
Correlation:0.22458
  |
Correlation:0.22458
  |
Correlation:0.16139
  |
Correlation:0.16139
  |
Component:258; Jaccard:0.092298
 |
Component:079; Jaccard:0.10154
 |
Component:079; Jaccard:0.11527
 |
Component:237; Jaccard:0.11122
 |
Component:237; Jaccard:0.10017
 |
Component:386; Jaccard:0.089113
 |
Component:237; Jaccard:0.067998
 |
Correlation:0.41877
  |
Correlation:0.33986
  |
Correlation:0.33986
  |
Correlation:0.24102
  |
Correlation:0.24102
  |
Correlation:0.22458
  |
Correlation:0.24102
  |
Component:061; Jaccard:0.088763
 |
Component:322; Jaccard:0.097133
 |
Component:322; Jaccard:0.10365
 |
Component:322; Jaccard:0.1053
 |
Component:322; Jaccard:0.099416
 |
Component:237; Jaccard:0.086533
 |
Component:386; Jaccard:0.064637
 |
Correlation:0.056188
  |
Correlation:0.37336
  |
Correlation:0.37336
  |
Correlation:0.37336
  |
Correlation:0.37336
  |
Correlation:0.24102
  |
Correlation:0.22458
  |
Component:322; Jaccard:0.087926
 |
Component:258; Jaccard:0.096896
 |
Component:292; Jaccard:0.099993
 |
Component:292; Jaccard:0.10081
 |
Component:292; Jaccard:0.093876
 |
Component:322; Jaccard:0.084783
 |
Component:322; Jaccard:0.062746
 |
Correlation:0.37336
  |
Correlation:0.41877
  |
Correlation:0.13464
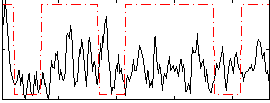 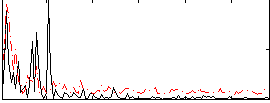 |
Correlation:0.13464
  |
Correlation:0.13464
  |
Correlation:0.37336
  |
Correlation:0.37336
  |
Component:335; Jaccard:0.087545
 |
Component:292; Jaccard:0.093173
 |
Component:258; Jaccard:0.098347
 |
Component:258; Jaccard:0.092671
 |
Component:326; Jaccard:0.087967
 |
Component:292; Jaccard:0.077653
 |
Component:292; Jaccard:0.05732
 |
Correlation:0.13414
  |
Correlation:0.13464
  |
Correlation:0.41877
  |
Correlation:0.41877
  |
Correlation:0.16139
  |
Correlation:0.13464
  |
Correlation:0.13464
  |
Component:079; Jaccard:0.087128
 |
Component:061; Jaccard:0.093057
 |
Component:398; Jaccard:0.096015
 |
Component:398; Jaccard:0.092481
 |
Component:258; Jaccard:0.081567
 |
Component:254; Jaccard:0.069899
 |
Component:254; Jaccard:0.057016
 |
Correlation:0.33986
  |
Correlation:0.056188
  |
Correlation:0.17909
  |
Correlation:0.17909
  |
Correlation:0.41877
  |
Correlation:0.19713
  |
Correlation:0.19713
  |
Component:292; Jaccard:0.086089
 |
Component:398; Jaccard:0.092201
 |
Component:061; Jaccard:0.093284
 |
Component:061; Jaccard:0.088974
 |
Component:061; Jaccard:0.080942
 |
Component:258; Jaccard:0.065801
 |
Component:100; Jaccard:0.051977
 |
Correlation:0.13464
  |
Correlation:0.17909
  |
Correlation:0.056188
  |
Correlation:0.056188
  |
Correlation:0.056188
  |
Correlation:0.41877
  |
Correlation:0.075538
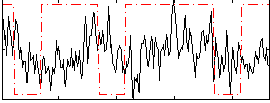  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































