Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
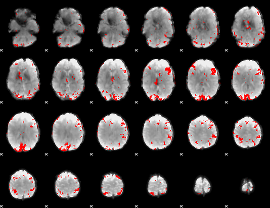 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
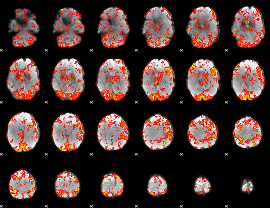 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
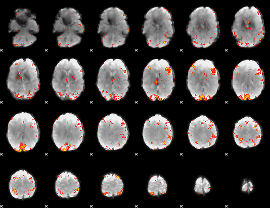 |
GLM Z-score result thresholded by 4
 |
Component:153; Jaccard:0.14223
 |
Component:153; Jaccard:0.176
 |
Component:153; Jaccard:0.21096
 |
Component:153; Jaccard:0.24238
 |
Component:153; Jaccard:0.26104
 |
Component:153; Jaccard:0.26498
 |
Component:153; Jaccard:0.24736
 |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Component:023; Jaccard:0.13952
 |
Component:023; Jaccard:0.15929
 |
Component:023; Jaccard:0.17167
 |
Component:280; Jaccard:0.1837
 |
Component:280; Jaccard:0.18918
 |
Component:280; Jaccard:0.17989
 |
Component:280; Jaccard:0.15673
 |
Correlation:0.16204
  |
Correlation:0.16204
  |
Correlation:0.16204
  |
Correlation:0.31272
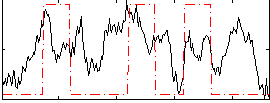 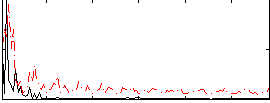 |
Correlation:0.31272
  |
Correlation:0.31272
  |
Correlation:0.31272
  |
Component:355; Jaccard:0.12639
 |
Component:280; Jaccard:0.14729
 |
Component:280; Jaccard:0.16843
 |
Component:023; Jaccard:0.17544
 |
Component:023; Jaccard:0.1676
 |
Component:236; Jaccard:0.15421
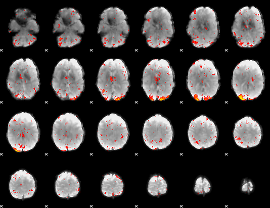 |
Component:236; Jaccard:0.13876
 |
Correlation:0.14014
 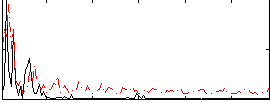 |
Correlation:0.31272
  |
Correlation:0.31272
  |
Correlation:0.16204
  |
Correlation:0.16204
  |
Correlation:0.32428
 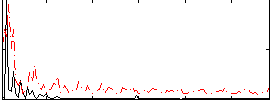 |
Correlation:0.32428
  |
Component:280; Jaccard:0.1244
 |
Component:355; Jaccard:0.14216
 |
Component:355; Jaccard:0.15656
 |
Component:355; Jaccard:0.16286
 |
Component:236; Jaccard:0.16421
 |
Component:023; Jaccard:0.14948
 |
Component:355; Jaccard:0.12718
 |
Correlation:0.31272
  |
Correlation:0.14014
  |
Correlation:0.14014
  |
Correlation:0.14014
  |
Correlation:0.32428
  |
Correlation:0.16204
  |
Correlation:0.14014
  |
Component:064; Jaccard:0.11252
 |
Component:064; Jaccard:0.12545
 |
Component:236; Jaccard:0.14233
 |
Component:236; Jaccard:0.15801
 |
Component:355; Jaccard:0.15848
 |
Component:355; Jaccard:0.14732
 |
Component:023; Jaccard:0.12001
 |
Correlation:0.21354
  |
Correlation:0.21354
  |
Correlation:0.32428
  |
Correlation:0.32428
  |
Correlation:0.14014
  |
Correlation:0.14014
  |
Correlation:0.16204
  |
Component:088; Jaccard:0.10887
 |
Component:236; Jaccard:0.12389
 |
Component:064; Jaccard:0.13237
 |
Component:064; Jaccard:0.13713
 |
Component:017; Jaccard:0.13601
 |
Component:017; Jaccard:0.13011
 |
Component:017; Jaccard:0.11838
 |
Correlation:0.049596
  |
Correlation:0.32428
  |
Correlation:0.21354
  |
Correlation:0.21354
  |
Correlation:0.22754
  |
Correlation:0.22754
  |
Correlation:0.22754
  |
Component:236; Jaccard:0.1052
 |
Component:088; Jaccard:0.12114
 |
Component:017; Jaccard:0.12888
 |
Component:017; Jaccard:0.13511
 |
Component:396; Jaccard:0.13426
 |
Component:396; Jaccard:0.13008
 |
Component:396; Jaccard:0.11547
 |
Correlation:0.32428
  |
Correlation:0.049596
  |
Correlation:0.22754
  |
Correlation:0.22754
  |
Correlation:0.3476
  |
Correlation:0.3476
  |
Correlation:0.3476
  |
Component:234; Jaccard:0.10419
 |
Component:017; Jaccard:0.11734
 |
Component:088; Jaccard:0.12834
 |
Component:088; Jaccard:0.13008
 |
Component:064; Jaccard:0.13304
 |
Component:064; Jaccard:0.12298
 |
Component:064; Jaccard:0.10442
 |
Correlation:0.22481
  |
Correlation:0.22754
  |
Correlation:0.049596
  |
Correlation:0.049596
  |
Correlation:0.21354
  |
Correlation:0.21354
  |
Correlation:0.21354
  |
Component:017; Jaccard:0.10395
 |
Component:279; Jaccard:0.11475
 |
Component:059; Jaccard:0.1247
 |
Component:396; Jaccard:0.13001
 |
Component:059; Jaccard:0.13045
 |
Component:059; Jaccard:0.11918
 |
Component:059; Jaccard:0.10235
 |
Correlation:0.22754
  |
Correlation:0.12684
  |
Correlation:-0.0099935
  |
Correlation:0.3476
  |
Correlation:-0.0099935
  |
Correlation:-0.0099935
  |
Correlation:-0.0099935
  |
Component:279; Jaccard:0.10356
 |
Component:059; Jaccard:0.1127
 |
Component:279; Jaccard:0.12152
 |
Component:059; Jaccard:0.12976
 |
Component:088; Jaccard:0.12398
 |
Component:088; Jaccard:0.1098
 |
Component:279; Jaccard:0.090027
 |
Correlation:0.12684
  |
Correlation:-0.0099935
  |
Correlation:0.12684
  |
Correlation:-0.0099935
  |
Correlation:0.049596
  |
Correlation:0.049596
  |
Correlation:0.12684
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
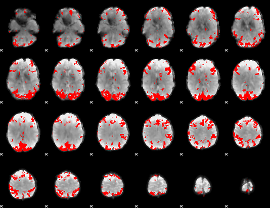 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
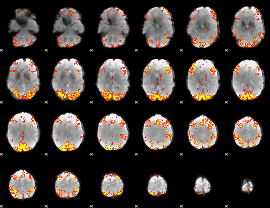 |
GLM Z-score result thresholded by 4
 |
Component:023; Jaccard:0.1319
 |
Component:023; Jaccard:0.1522
 |
Component:023; Jaccard:0.17198
 |
Component:153; Jaccard:0.19778
 |
Component:153; Jaccard:0.22651
 |
Component:153; Jaccard:0.25295
 |
Component:153; Jaccard:0.27581
 |
Correlation:0.15066
  |
Correlation:0.15066
  |
Correlation:0.15066
  |
Correlation:0.27404
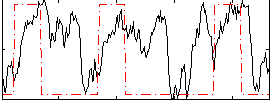 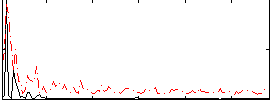 |
Correlation:0.27404
  |
Correlation:0.27404
  |
Correlation:0.27404
  |
Component:355; Jaccard:0.12178
 |
Component:153; Jaccard:0.14416
 |
Component:153; Jaccard:0.17047
 |
Component:023; Jaccard:0.18821
 |
Component:280; Jaccard:0.20127
 |
Component:280; Jaccard:0.21882
 |
Component:280; Jaccard:0.23036
 |
Correlation:0.21913
  |
Correlation:0.27404
  |
Correlation:0.27404
  |
Correlation:0.15066
  |
Correlation:0.034443
  |
Correlation:0.034443
  |
Correlation:0.034443
  |
Component:153; Jaccard:0.12069
 |
Component:355; Jaccard:0.13831
 |
Component:280; Jaccard:0.15893
 |
Component:280; Jaccard:0.18082
 |
Component:023; Jaccard:0.19994
 |
Component:023; Jaccard:0.20411
 |
Component:059; Jaccard:0.20945
 |
Correlation:0.27404
  |
Correlation:0.21913
  |
Correlation:0.034443
  |
Correlation:0.034443
  |
Correlation:0.15066
  |
Correlation:0.15066
  |
Correlation:0.3534
  |
Component:280; Jaccard:0.11694
 |
Component:280; Jaccard:0.13731
 |
Component:355; Jaccard:0.15462
 |
Component:355; Jaccard:0.17005
 |
Component:355; Jaccard:0.18223
 |
Component:059; Jaccard:0.19452
 |
Component:023; Jaccard:0.20243
 |
Correlation:0.034443
  |
Correlation:0.034443
  |
Correlation:0.21913
  |
Correlation:0.21913
  |
Correlation:0.21913
  |
Correlation:0.3534
  |
Correlation:0.15066
  |
Component:251; Jaccard:0.11667
 |
Component:251; Jaccard:0.13116
 |
Component:088; Jaccard:0.14523
 |
Component:088; Jaccard:0.16142
 |
Component:059; Jaccard:0.17779
 |
Component:355; Jaccard:0.18917
 |
Component:355; Jaccard:0.19334
 |
Correlation:0.25039
 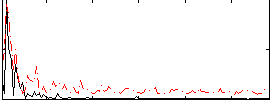 |
Correlation:0.25039
  |
Correlation:0.29194
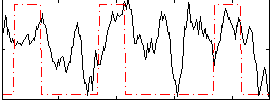  |
Correlation:0.29194
  |
Correlation:0.3534
  |
Correlation:0.21913
  |
Correlation:0.21913
  |
Component:234; Jaccard:0.11428
 |
Component:088; Jaccard:0.12932
 |
Component:251; Jaccard:0.14457
 |
Component:059; Jaccard:0.15962
 |
Component:088; Jaccard:0.17335
 |
Component:088; Jaccard:0.18491
 |
Component:088; Jaccard:0.1909
 |
Correlation:-0.086953
  |
Correlation:0.29194
  |
Correlation:0.25039
  |
Correlation:0.3534
  |
Correlation:0.29194
  |
Correlation:0.29194
  |
Correlation:0.29194
  |
Component:088; Jaccard:0.11197
 |
Component:076; Jaccard:0.12695
 |
Component:076; Jaccard:0.14171
 |
Component:251; Jaccard:0.15771
 |
Component:251; Jaccard:0.16945
 |
Component:251; Jaccard:0.17876
 |
Component:251; Jaccard:0.18537
 |
Correlation:0.29194
  |
Correlation:0.30101
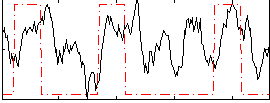 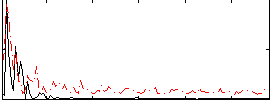 |
Correlation:0.30101
  |
Correlation:0.25039
  |
Correlation:0.25039
  |
Correlation:0.25039
  |
Correlation:0.25039
  |
Component:076; Jaccard:0.11165
 |
Component:234; Jaccard:0.12434
 |
Component:059; Jaccard:0.1408
 |
Component:076; Jaccard:0.15447
 |
Component:076; Jaccard:0.16472
 |
Component:076; Jaccard:0.1709
 |
Component:076; Jaccard:0.17325
 |
Correlation:0.30101
  |
Correlation:-0.086953
  |
Correlation:0.3534
  |
Correlation:0.30101
  |
Correlation:0.30101
  |
Correlation:0.30101
  |
Correlation:0.30101
  |
Component:064; Jaccard:0.10886
 |
Component:064; Jaccard:0.12289
 |
Component:064; Jaccard:0.13607
 |
Component:064; Jaccard:0.14584
 |
Component:279; Jaccard:0.15206
 |
Component:279; Jaccard:0.16051
 |
Component:279; Jaccard:0.16373
 |
Correlation:0.17158
  |
Correlation:0.17158
  |
Correlation:0.17158
  |
Correlation:0.17158
  |
Correlation:0.35736
  |
Correlation:0.35736
  |
Correlation:0.35736
  |
Component:059; Jaccard:0.10614
 |
Component:059; Jaccard:0.12262
 |
Component:279; Jaccard:0.13152
 |
Component:279; Jaccard:0.14247
 |
Component:064; Jaccard:0.15148
 |
Component:017; Jaccard:0.15356
 |
Component:017; Jaccard:0.15871
 |
Correlation:0.3534
  |
Correlation:0.3534
  |
Correlation:0.35736
  |
Correlation:0.35736
  |
Correlation:0.17158
  |
Correlation:0.26045
  |
Correlation:0.26045
  |
Cope: 03:MATCH-REL BACK
|
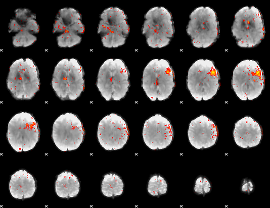
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
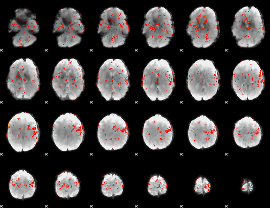 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
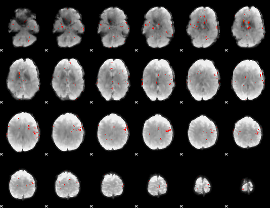 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:244; Jaccard:0.10016
 |
Component:244; Jaccard:0.12249
 |
Component:244; Jaccard:0.13317
 |
Component:089; Jaccard:0.1197
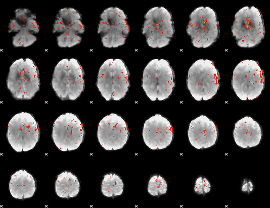 |
Component:089; Jaccard:0.082215
 |
Component:089; Jaccard:0.04158
 |
Component:089; Jaccard:0.010149
 |
Correlation:0.20997
  |
Correlation:0.20997
  |
Correlation:0.20997
  |
Correlation:0.20608
  |
Correlation:0.20608
  |
Correlation:0.20608
  |
Correlation:0.20608
  |
Component:089; Jaccard:0.093034
 |
Component:089; Jaccard:0.11719
 |
Component:089; Jaccard:0.13139
 |
Component:244; Jaccard:0.11775
 |
Component:244; Jaccard:0.071591
 |
Component:244; Jaccard:0.029893
 |
Component:353; Jaccard:0.008117
 |
Correlation:0.20608
  |
Correlation:0.20608
  |
Correlation:0.20608
  |
Correlation:0.20997
  |
Correlation:0.20997
  |
Correlation:0.20997
  |
Correlation:0.21185
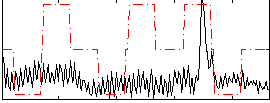  |
Component:208; Jaccard:0.08996
 |
Component:168; Jaccard:0.089791
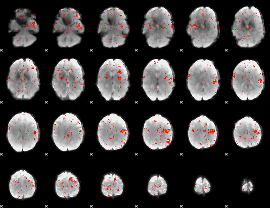 |
Component:353; Jaccard:0.096178
 |
Component:353; Jaccard:0.085797
 |
Component:353; Jaccard:0.051339
 |
Component:353; Jaccard:0.021893
 |
Component:346; Jaccard:0.00649
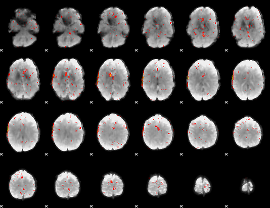 |
Correlation:0.27668
 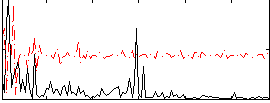 |
Correlation:0.19848
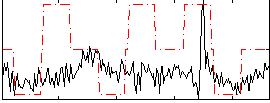  |
Correlation:0.21185
  |
Correlation:0.21185
  |
Correlation:0.21185
  |
Correlation:0.21185
  |
Correlation:0.13279
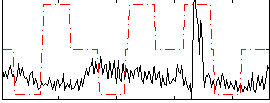  |
Component:014; Jaccard:0.083063
 |
Component:353; Jaccard:0.089399
 |
Component:058; Jaccard:0.091112
 |
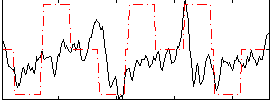
Component:347; Jaccard:0.073408
 |
Component:347; Jaccard:0.047214
 |
Component:347; Jaccard:0.02112
 |
Component:244; Jaccard:0.006455
 |
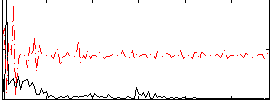
Correlation:0.23462
 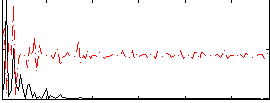 |
Correlation:0.21185
  |
Correlation:0.12501
  |
Correlation:0.096408
 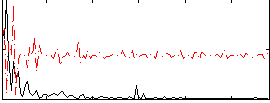 |
Correlation:0.096408
  |
Correlation:0.096408
  |
Correlation:0.20997
  |
Component:168; Jaccard:0.081859
 |
Component:058; Jaccard:0.088097
 |
Component:168; Jaccard:0.089631
 |
Component:168; Jaccard:0.073363
 |
Component:168; Jaccard:0.044161
 |
Component:168; Jaccard:0.020638
 |
Component:347; Jaccard:0.006119
 |
Correlation:0.19848
  |
Correlation:0.12501
  |
Correlation:0.19848
  |
Correlation:0.19848
  |
Correlation:0.19848
  |
Correlation:0.19848
  |
Correlation:0.096408
  |
Component:069; Jaccard:0.080844
 |
Component:208; Jaccard:0.088048
 |
Component:347; Jaccard:0.086259
 |
Component:058; Jaccard:0.070931
 |
Component:058; Jaccard:0.043877
 |
Component:082; Jaccard:0.015838
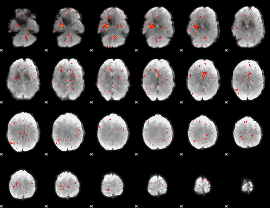 |
Component:168; Jaccard:0.00494
 |
Correlation:0.13613
  |
Correlation:0.27668
  |
Correlation:0.096408
  |
Correlation:0.12501
  |
Correlation:0.12501
  |
Correlation:0.19639
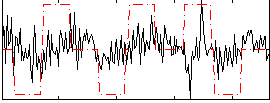  |
Correlation:0.19848
  |
Component:058; Jaccard:0.079166
 |
Component:069; Jaccard:0.085486
 |
Component:069; Jaccard:0.077139
 |
Component:208; Jaccard:0.053295
 |
Component:082; Jaccard:0.034222
 |
Component:058; Jaccard:0.015757
 |
Component:082; Jaccard:0.004565
 |
Correlation:0.12501
  |
Correlation:0.13613
  |
Correlation:0.13613
  |
Correlation:0.27668
  |
Correlation:0.19639
  |
Correlation:0.12501
  |
Correlation:0.19639
  |
Component:123; Jaccard:0.077541
 |
Component:347; Jaccard:0.081128
 |
Component:208; Jaccard:0.074384
 |
Component:082; Jaccard:0.053139
 |
Component:208; Jaccard:0.031271
 |
Component:346; Jaccard:0.014398
 |
Component:108; Jaccard:0.004103
 |
Correlation:0.23105
  |
Correlation:0.096408
  |
Correlation:0.27668
  |
Correlation:0.19639
  |
Correlation:0.27668
  |
Correlation:0.13279
  |
Correlation:0.15301
  |
Component:057; Jaccard:0.075647
 |
Component:123; Jaccard:0.077663
 |
Component:123; Jaccard:0.069202
 |
Component:069; Jaccard:0.052767
 |
Component:069; Jaccard:0.026934
 |
Component:208; Jaccard:0.013528
 |
Component:058; Jaccard:0.00394
 |
Correlation:0.20183
  |
Correlation:0.23105
  |
Correlation:0.23105
  |
Correlation:0.13613
  |
Correlation:0.13613
  |
Correlation:0.27668
  |
Correlation:0.12501
  |
Component:353; Jaccard:0.07554
 |
Component:014; Jaccard:0.073534
 |
Component:082; Jaccard:0.061429
 |
Component:123; Jaccard:0.047205
 |
Component:396; Jaccard:0.025786
 |
Component:069; Jaccard:0.012309
 |
Component:094; Jaccard:0.003807
 |
Correlation:0.21185
  |
Correlation:0.23462
  |
Correlation:0.19639
  |
Correlation:0.23105
  |
Correlation:0.11839
  |
Correlation:0.13613
  |
Correlation:-0.053038
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:251; Jaccard:0.11303
 |
Component:251; Jaccard:0.14035
 |
Component:251; Jaccard:0.17335
 |
Component:251; Jaccard:0.20457
 |
Component:251; Jaccard:0.22437
 |
Component:251; Jaccard:0.22082
 |
Component:251; Jaccard:0.19464
 |
Correlation:0.11675
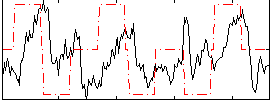  |
Correlation:0.11675
  |
Correlation:0.11675
  |
Correlation:0.11675
  |
Correlation:0.11675
  |
Correlation:0.11675
  |
Correlation:0.11675
  |
Component:076; Jaccard:0.10362
 |
Component:076; Jaccard:0.12866
 |
Component:076; Jaccard:0.15426
 |
Component:076; Jaccard:0.17399
 |
Component:059; Jaccard:0.19078
 |
Component:059; Jaccard:0.20255
 |
Component:059; Jaccard:0.18831
 |
Correlation:0.091526
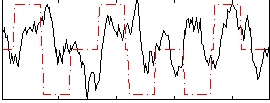  |
Correlation:0.091526
  |
Correlation:0.091526
  |
Correlation:0.091526
  |
Correlation:0.21538
 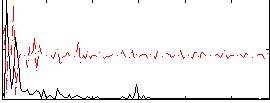 |
Correlation:0.21538
  |
Correlation:0.21538
  |
Component:307; Jaccard:0.099332
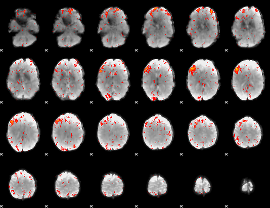 |
Component:307; Jaccard:0.11726
 |
Component:059; Jaccard:0.13725
 |
Component:059; Jaccard:0.16586
 |
Component:076; Jaccard:0.17548
 |
Component:076; Jaccard:0.15731
 |
Component:076; Jaccard:0.12525
 |
Correlation:0.33558
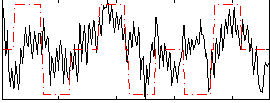  |
Correlation:0.33558
  |
Correlation:0.21538
  |
Correlation:0.21538
  |
Correlation:0.091526
  |
Correlation:0.091526
  |
Correlation:0.091526
  |
Component:138; Jaccard:0.089352
 |
Component:059; Jaccard:0.11053
 |
Component:307; Jaccard:0.13085
 |
Component:138; Jaccard:0.14434
 |
Component:138; Jaccard:0.15278
 |
Component:138; Jaccard:0.14123
 |
Component:138; Jaccard:0.11546
 |
Correlation:0.19596
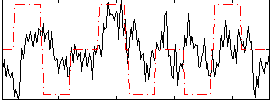  |
Correlation:0.21538
  |
Correlation:0.33558
  |
Correlation:0.19596
  |
Correlation:0.19596
  |
Correlation:0.19596
  |
Correlation:0.19596
  |
Component:088; Jaccard:0.088617
 |
Component:138; Jaccard:0.10784
 |
Component:138; Jaccard:0.12753
 |
Component:088; Jaccard:0.13751
 |
Component:088; Jaccard:0.14144
 |
Component:088; Jaccard:0.13267
 |
Component:088; Jaccard:0.11099
 |
Correlation:0.14364
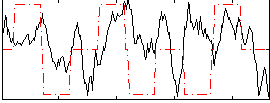  |
Correlation:0.19596
  |
Correlation:0.19596
  |
Correlation:0.14364
  |
Correlation:0.14364
  |
Correlation:0.14364
  |
Correlation:0.14364
  |
Component:059; Jaccard:0.08808
 |
Component:088; Jaccard:0.1059
 |
Component:088; Jaccard:0.12254
 |
Component:307; Jaccard:0.13617
 |
Component:307; Jaccard:0.12644
 |
Component:279; Jaccard:0.11789
 |
Component:279; Jaccard:0.098083
 |
Correlation:0.21538
  |
Correlation:0.14364
  |
Correlation:0.14364
  |
Correlation:0.33558
  |
Correlation:0.33558
  |
Correlation:0.13663
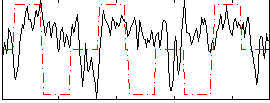  |
Correlation:0.13663
  |
Component:291; Jaccard:0.086962
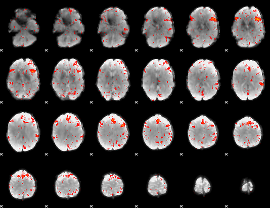 |
Component:280; Jaccard:0.099942
 |
Component:280; Jaccard:0.11335
 |
Component:280; Jaccard:0.12465
 |
Component:280; Jaccard:0.12592
 |
Component:280; Jaccard:0.11493
 |
Component:280; Jaccard:0.093045
 |
Correlation:0.27383
  |
Correlation:-0.16493
  |
Correlation:-0.16493
  |
Correlation:-0.16493
  |
Correlation:-0.16493
  |
Correlation:-0.16493
  |
Correlation:-0.16493
  |
Component:023; Jaccard:0.086332
 |
Component:291; Jaccard:0.098903
 |
Component:153; Jaccard:0.11048
 |
Component:153; Jaccard:0.11918
 |
Component:279; Jaccard:0.12419
 |
Component:307; Jaccard:0.10901
 |
Component:386; Jaccard:0.090491
 |
Correlation:-0.0067473
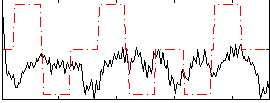  |
Correlation:0.27383
  |
Correlation:0.075578
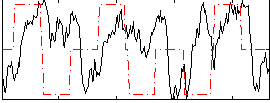  |
Correlation:0.075578
  |
Correlation:0.13663
  |
Correlation:0.33558
  |
Correlation:0.24151
  |
Component:280; Jaccard:0.085083
 |
Component:153; Jaccard:0.097598
 |
Component:291; Jaccard:0.1084
 |
Component:279; Jaccard:0.11695
 |
Component:153; Jaccard:0.11918
 |
Component:153; Jaccard:0.10746
 |
Component:307; Jaccard:0.085169
 |
Correlation:-0.16493
  |
Correlation:0.075578
  |
Correlation:0.27383
  |
Correlation:0.13663
  |
Correlation:0.075578
  |
Correlation:0.075578
  |
Correlation:0.33558
  |
Component:355; Jaccard:0.0845
 |
Component:023; Jaccard:0.095201
 |
Component:386; Jaccard:0.10683
 |
Component:386; Jaccard:0.11385
 |
Component:386; Jaccard:0.11636
 |
Component:386; Jaccard:0.10706
 |
Component:153; Jaccard:0.083948
 |
Correlation:0.046819
 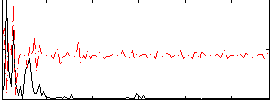 |
Correlation:-0.0067473
  |
Correlation:0.24151
  |
Correlation:0.24151
  |
Correlation:0.24151
  |
Correlation:0.24151
  |
Correlation:0.075578
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
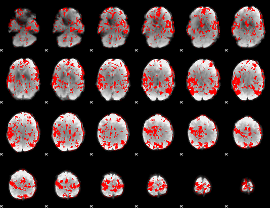 |
GLM binary result thresholded by 2.5
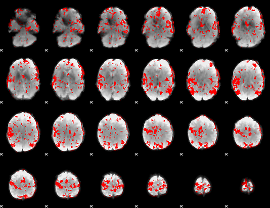 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
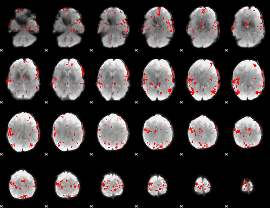 |
GLM binary result thresholded by 4
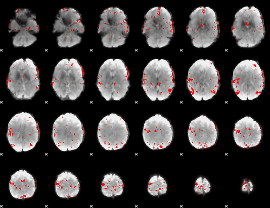 |
GLM Z-score result thresholded by 1
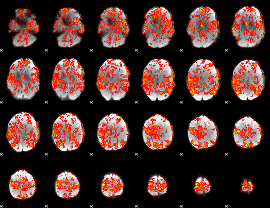 |
GLM Z-score result thresholded by 1.5
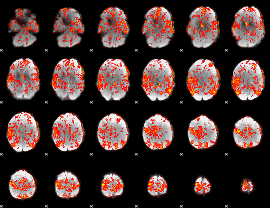 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
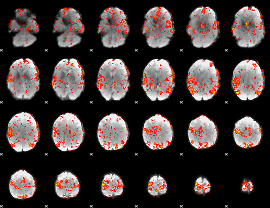 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
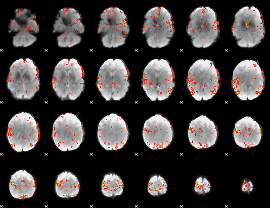 |
GLM Z-score result thresholded by 4
 |
Component:014; Jaccard:0.10639
 |
Component:014; Jaccard:0.12199
 |
Component:014; Jaccard:0.13512
 |
Component:014; Jaccard:0.14565
 |
Component:272; Jaccard:0.15607
 |
Component:295; Jaccard:0.1564
 |
Component:348; Jaccard:0.15354
 |
Correlation:-0.014144
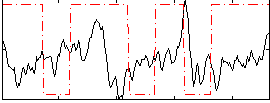  |
Correlation:-0.014144
  |
Correlation:-0.014144
  |
Correlation:-0.014144
  |
Correlation:0.2572
  |
Correlation:0.23541
 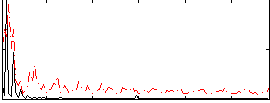 |
Correlation:0.34366
  |
Component:272; Jaccard:0.088444
 |
Component:272; Jaccard:0.10598
 |
Component:272; Jaccard:0.12695
 |
Component:272; Jaccard:0.14309
 |
Component:295; Jaccard:0.14873
 |
Component:272; Jaccard:0.15551
 |
Component:295; Jaccard:0.14934
 |
Correlation:0.2572
  |
Correlation:0.2572
  |
Correlation:0.2572
  |
Correlation:0.2572
  |
Correlation:0.23541
  |
Correlation:0.2572
  |
Correlation:0.23541
  |
Component:335; Jaccard:0.078656
 |
Component:295; Jaccard:0.092211
 |
Component:295; Jaccard:0.11178
 |
Component:295; Jaccard:0.13168
 |
Component:014; Jaccard:0.14741
 |
Component:348; Jaccard:0.15141
 |
Component:272; Jaccard:0.14069
 |
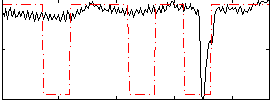
Correlation:0.15038
 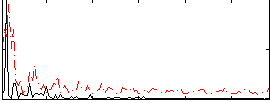 |
Correlation:0.23541
  |
Correlation:0.23541
  |
Correlation:0.23541
  |
Correlation:-0.014144
  |
Correlation:0.34366
  |
Correlation:0.2572
  |
Component:175; Jaccard:0.077063
 |
Component:175; Jaccard:0.091369
 |
Component:175; Jaccard:0.10544
 |
Component:175; Jaccard:0.11871
 |
Component:348; Jaccard:0.13735
 |
Component:014; Jaccard:0.13987
 |
Component:014; Jaccard:0.11574
 |
Correlation:0.25024
  |
Correlation:0.25024
  |
Correlation:0.25024
  |
Correlation:0.25024
  |
Correlation:0.34366
  |
Correlation:-0.014144
  |
Correlation:-0.014144
  |
Component:295; Jaccard:0.07561
 |
Component:068; Jaccard:0.085369
 |
Component:068; Jaccard:0.096865
 |
Component:348; Jaccard:0.11612
 |
Component:175; Jaccard:0.12795
 |
Component:175; Jaccard:0.12432
 |
Component:175; Jaccard:0.11308
 |
Correlation:0.23541
  |
Correlation:0.14288
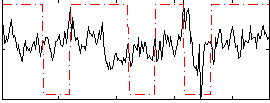  |
Correlation:0.14288
  |
Correlation:0.34366
  |
Correlation:0.25024
  |
Correlation:0.25024
  |
Correlation:0.25024
  |
Component:116; Jaccard:0.075356
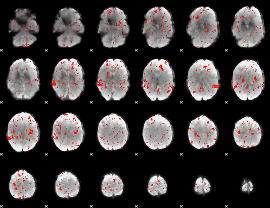 |
Component:057; Jaccard:0.084099
 |
Component:348; Jaccard:0.094804
 |
Component:068; Jaccard:0.10555
 |
Component:068; Jaccard:0.10769
 |
Component:237; Jaccard:0.10323
 |
Component:237; Jaccard:0.09215
 |
Correlation:0.12525
  |
Correlation:0.0088414
  |
Correlation:0.34366
  |
Correlation:0.14288
  |
Correlation:0.14288
  |
Correlation:0.12835
  |
Correlation:0.12835
  |
Component:149; Jaccard:0.075198
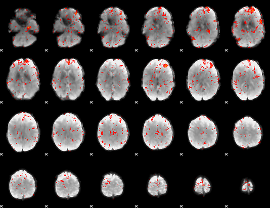 |
Component:335; Jaccard:0.083588
 |
Component:237; Jaccard:0.092981
 |
Component:237; Jaccard:0.1031
 |
Component:237; Jaccard:0.10638
 |
Component:068; Jaccard:0.10321
 |
Component:068; Jaccard:0.087846
 |
Correlation:-0.0022025
 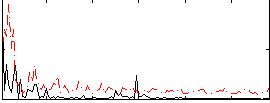 |
Correlation:0.15038
  |
Correlation:0.12835
  |
Correlation:0.12835
  |
Correlation:0.12835
  |
Correlation:0.14288
  |
Correlation:0.14288
  |
Component:057; Jaccard:0.075088
 |
Component:237; Jaccard:0.07985
 |
Component:057; Jaccard:0.092704
 |
Component:057; Jaccard:0.098831
 |
Component:057; Jaccard:0.098659
 |
Component:003; Jaccard:0.093629
 |
Component:378; Jaccard:0.082866
 |
Correlation:0.0088414
  |
Correlation:0.12835
  |
Correlation:0.0088414
  |
Correlation:0.0088414
  |
Correlation:0.0088414
  |
Correlation:0.1396
  |
Correlation:0.3163
  |
Component:068; Jaccard:0.074352
 |
Component:133; Jaccard:0.079773
 |
Component:335; Jaccard:0.087756
 |
Component:003; Jaccard:0.094096
 |
Component:003; Jaccard:0.09583
 |
Component:378; Jaccard:0.092338
 |
Component:003; Jaccard:0.0813
 |
Correlation:0.14288
  |
Correlation:0.43818
  |
Correlation:0.15038
  |
Correlation:0.1396
  |
Correlation:0.1396
  |
Correlation:0.3163
  |
Correlation:0.1396
  |
Component:162; Jaccard:0.073647
 |
Component:116; Jaccard:0.079304
 |
Component:003; Jaccard:0.08576
 |
Component:378; Jaccard:0.091315
 |
Component:378; Jaccard:0.09527
 |
Component:057; Jaccard:0.09093
 |
Component:057; Jaccard:0.075535
 |
Correlation:0.12439
  |
Correlation:0.12525
  |
Correlation:0.1396
  |
Correlation:0.3163
  |
Correlation:0.3163
  |
Correlation:0.0088414
  |
Correlation:0.0088414
  |
Cope: 06:neg_REL BACK
|
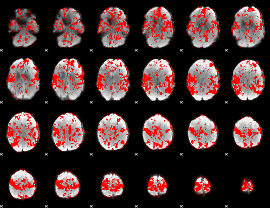
GLM result group-wise
 |
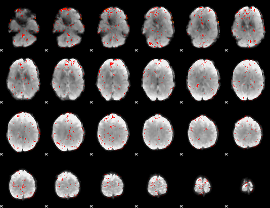
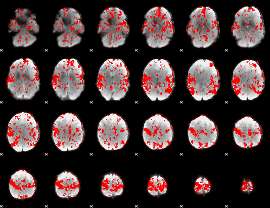
GLM binary result thresholded by 1
 |
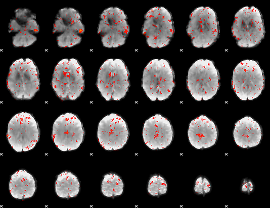
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
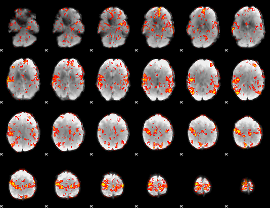
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
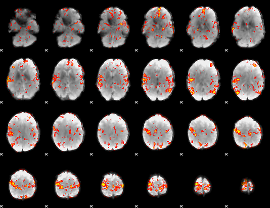
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:014; Jaccard:0.12073
 |
Component:014; Jaccard:0.142
 |
Component:014; Jaccard:0.16794
 |
Component:014; Jaccard:0.19024
 |
Component:014; Jaccard:0.20302
 |
Component:295; Jaccard:0.21026
 |
Component:295; Jaccard:0.21164
 |
Correlation:0.3817
  |
Correlation:0.3817
  |
Correlation:0.3817
  |
Correlation:0.3817
  |
Correlation:0.3817
  |
Correlation:0.39358
 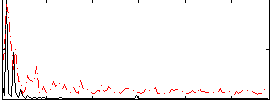 |
Correlation:0.39358
  |
Component:057; Jaccard:0.092111
 |
Component:057; Jaccard:0.10764
 |
Component:295; Jaccard:0.12936
 |
Component:295; Jaccard:0.15884
 |
Component:295; Jaccard:0.18846
 |
Component:014; Jaccard:0.20106
 |
Component:014; Jaccard:0.18572
 |
Correlation:0.34936
  |
Correlation:0.34936
  |
Correlation:0.39358
  |
Correlation:0.39358
  |
Correlation:0.39358
  |
Correlation:0.3817
  |
Correlation:0.3817
  |
Component:272; Jaccard:0.090016
 |
Component:272; Jaccard:0.10583
 |
Component:272; Jaccard:0.1247
 |
Component:272; Jaccard:0.1439
 |
Component:272; Jaccard:0.1567
 |
Component:272; Jaccard:0.16223
 |
Component:272; Jaccard:0.155
 |
Correlation:0.24319
  |
Correlation:0.24319
  |
Correlation:0.24319
  |
Correlation:0.24319
  |
Correlation:0.24319
  |
Correlation:0.24319
  |
Correlation:0.24319
  |
Component:175; Jaccard:0.085692
 |
Component:295; Jaccard:0.10401
 |
Component:057; Jaccard:0.12467
 |
Component:057; Jaccard:0.13982
 |
Component:057; Jaccard:0.15245
 |
Component:057; Jaccard:0.15715
 |
Component:175; Jaccard:0.15132
 |
Correlation:0.31078
  |
Correlation:0.39358
  |
Correlation:0.34936
  |
Correlation:0.34936
  |
Correlation:0.34936
  |
Correlation:0.34936
  |
Correlation:0.31078
  |
Component:295; Jaccard:0.084141
 |
Component:175; Jaccard:0.10201
 |
Component:175; Jaccard:0.12118
 |
Component:175; Jaccard:0.13815
 |
Component:175; Jaccard:0.1522
 |
Component:175; Jaccard:0.15621
 |
Component:057; Jaccard:0.146
 |
Correlation:0.39358
  |
Correlation:0.31078
  |
Correlation:0.31078
  |
Correlation:0.31078
  |
Correlation:0.31078
  |
Correlation:0.31078
  |
Correlation:0.34936
  |
Component:068; Jaccard:0.078478
 |
Component:068; Jaccard:0.088041
 |
Component:237; Jaccard:0.10079
 |
Component:237; Jaccard:0.11422
 |
Component:237; Jaccard:0.12103
 |
Component:237; Jaccard:0.12271
 |
Component:237; Jaccard:0.11585
 |
Correlation:0.19726
  |
Correlation:0.19726
  |
Correlation:0.22334
  |
Correlation:0.22334
  |
Correlation:0.22334
  |
Correlation:0.22334
  |
Correlation:0.22334
  |
Component:378; Jaccard:0.077206
 |
Component:237; Jaccard:0.08734
 |
Component:068; Jaccard:0.097711
 |
Component:068; Jaccard:0.10491
 |
Component:378; Jaccard:0.11163
 |
Component:378; Jaccard:0.10908
 |
Component:378; Jaccard:0.1028
 |
Correlation:0.089832
  |
Correlation:0.22334
  |
Correlation:0.19726
  |
Correlation:0.19726
  |
Correlation:0.089832
  |
Correlation:0.089832
  |
Correlation:0.089832
  |
Component:335; Jaccard:0.07685
 |
Component:378; Jaccard:0.086538
 |
Component:378; Jaccard:0.095444
 |
Component:378; Jaccard:0.10362
 |
Component:068; Jaccard:0.10965
 |
Component:068; Jaccard:0.109
 |
Component:068; Jaccard:0.10051
 |
Correlation:0.052835
  |
Correlation:0.089832
  |
Correlation:0.089832
  |
Correlation:0.089832
  |
Correlation:0.19726
  |
Correlation:0.19726
  |
Correlation:0.19726
  |
Component:149; Jaccard:0.075628
 |
Component:335; Jaccard:0.080818
 |
Component:335; Jaccard:0.08381
 |
Component:335; Jaccard:0.084254
 |
Component:335; Jaccard:0.084475
 |
Component:335; Jaccard:0.078737
 |
Component:259; Jaccard:0.07084
 |
Correlation:0.13281
  |
Correlation:0.052835
  |
Correlation:0.052835
  |
Correlation:0.052835
  |
Correlation:0.052835
  |
Correlation:0.052835
  |
Correlation:0.2023
  |
Component:116; Jaccard:0.07474
 |
Component:149; Jaccard:0.077885
 |
Component:183; Jaccard:0.080324
 |
Component:131; Jaccard:0.083206
 |
Component:131; Jaccard:0.082039
 |
Component:131; Jaccard:0.07719
 |
Component:131; Jaccard:0.069354
 |
Correlation:0.10939
  |
Correlation:0.13281
  |
Correlation:0.040194
  |
Correlation:0.37449
  |
Correlation:0.37449
  |
Correlation:0.37449
  |
Correlation:0.37449
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































