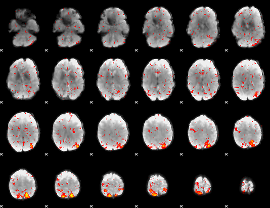
Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
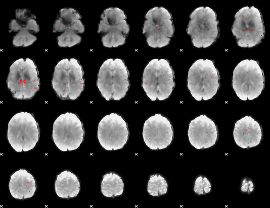
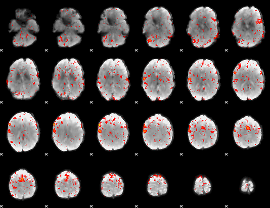
GLM result group-wise
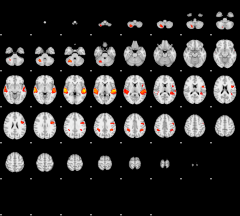 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
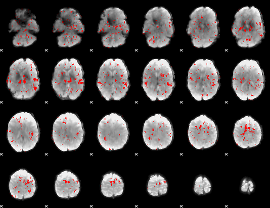 |
GLM binary result thresholded by 2
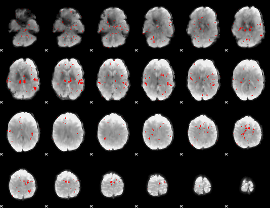 |
GLM binary result thresholded by 2.5
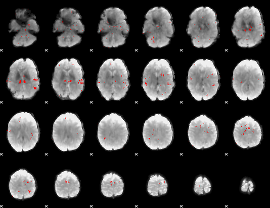 |
GLM binary result thresholded by 3
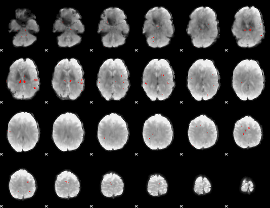 |
GLM binary result thresholded by 3.5
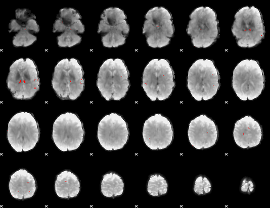 |
GLM binary result thresholded by 4
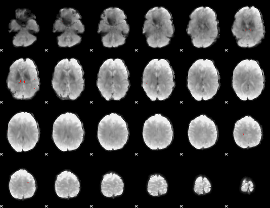 |
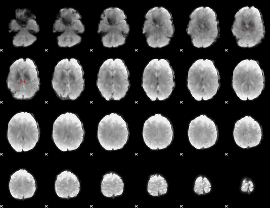
GLM Z-score result thresholded by 1
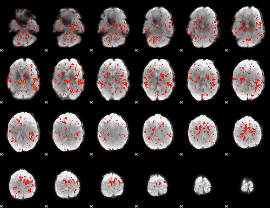 |
GLM Z-score result thresholded by 1.5
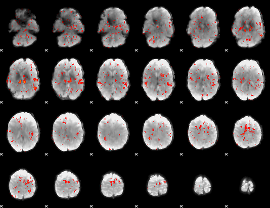 |
GLM Z-score result thresholded by 2
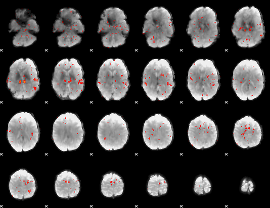 |
GLM Z-score result thresholded by 2.5
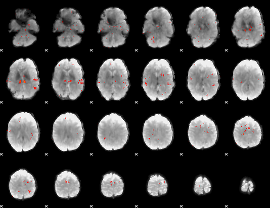 |
GLM Z-score result thresholded by 3
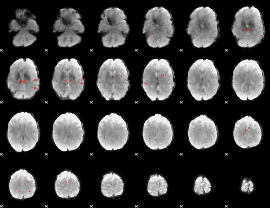 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:243; Jaccard:0.086195
 |
Component:067; Jaccard:0.095358
 |
Component:067; Jaccard:0.08535
 |
Component:067; Jaccard:0.066391
 |
Component:067; Jaccard:0.044815
 |
Component:067; Jaccard:0.025837
 |
Component:067; Jaccard:0.012719
 |
Correlation:-0.0019197
  |
Correlation:-0.21156
 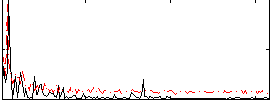 |
Correlation:-0.21156
  |
Correlation:-0.21156
  |
Correlation:-0.21156
  |
Correlation:-0.21156
  |
Correlation:-0.21156
  |
Component:067; Jaccard:0.085765
 |
Component:243; Jaccard:0.081256
 |
Component:243; Jaccard:0.060478
 |
Component:243; Jaccard:0.033836
 |
Component:243; Jaccard:0.013593
 |
Component:243; Jaccard:0.006529
 |
Component:223; Jaccard:0.003073
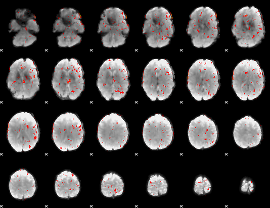 |
Correlation:-0.21156
  |
Correlation:-0.0019197
  |
Correlation:-0.0019197
  |
Correlation:-0.0019197
  |
Correlation:-0.0019197
  |
Correlation:-0.0019197
  |
Correlation:-0.10144
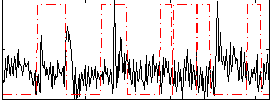 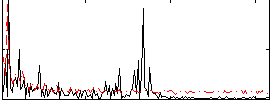 |
Component:176; Jaccard:0.074296
 |
Component:176; Jaccard:0.064212
 |
Component:176; Jaccard:0.045498
 |
Component:180; Jaccard:0.028364
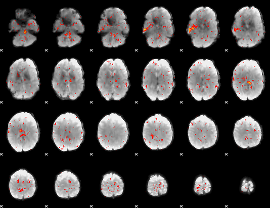 |
Component:180; Jaccard:0.013217
 |
Component:223; Jaccard:0.005848
 |
Component:262; Jaccard:0.002251
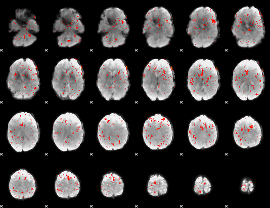 |
Correlation:0.076985
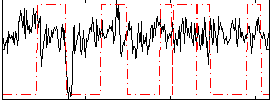 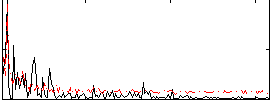 |
Correlation:0.076985
  |
Correlation:0.076985
  |
Correlation:-0.12519
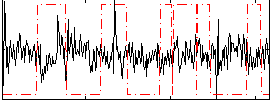  |
Correlation:-0.12519
  |
Correlation:-0.10144
  |
Correlation:0.26393
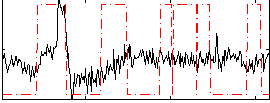  |
Component:073; Jaccard:0.071415
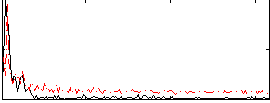
 |
Component:180; Jaccard:0.061412
 |
Component:180; Jaccard:0.044033
 |
Component:176; Jaccard:0.027959
 |
Component:176; Jaccard:0.011991
 |
Component:180; Jaccard:0.005621
 |
Component:180; Jaccard:0.001975
 |
Correlation:0.14112
  |
Correlation:-0.12519
  |
Correlation:-0.12519
  |
Correlation:0.076985
  |
Correlation:0.076985
  |
Correlation:-0.12519
  |
Correlation:-0.12519
  |
Component:353; Jaccard:0.069579
 |
Component:353; Jaccard:0.060078
 |
Component:353; Jaccard:0.043773
 |
Component:022; Jaccard:0.027643
 |
Component:353; Jaccard:0.011762
 |
Component:353; Jaccard:0.004924
 |
Component:243; Jaccard:0.001929
 |
Correlation:-0.27565
 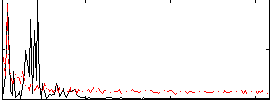 |
Correlation:-0.27565
  |
Correlation:-0.27565
  |
Correlation:0.047537
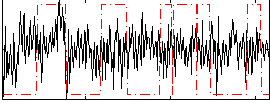 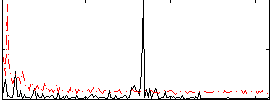 |
Correlation:-0.27565
  |
Correlation:-0.27565
  |
Correlation:-0.0019197
  |
Component:288; Jaccard:0.066899
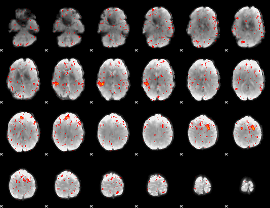 |
Component:288; Jaccard:0.058067
 |
Component:022; Jaccard:0.043474
 |
Component:353; Jaccard:0.024717
 |
Component:012; Jaccard:0.010705
 |
Component:176; Jaccard:0.004784
 |
Component:374; Jaccard:0.001881
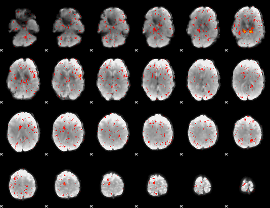 |
Correlation:-0.0018941
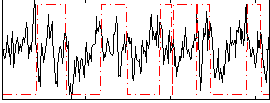 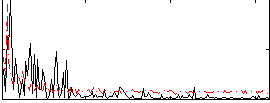 |
Correlation:-0.0018941
  |
Correlation:0.047537
  |
Correlation:-0.27565
  |
Correlation:0.29239
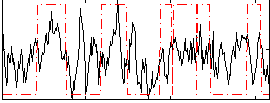 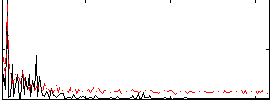 |
Correlation:0.076985
  |
Correlation:-0.030682
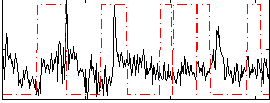 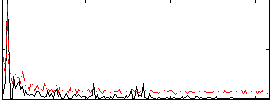 |
Component:022; Jaccard:0.066535
 |
Component:022; Jaccard:0.057963
 |
Component:012; Jaccard:0.04009
 |
Component:012; Jaccard:0.023567
 |
Component:022; Jaccard:0.009962
 |
Component:284; Jaccard:0.004337
 |
Component:176; Jaccard:0.001839
 |
Correlation:0.047537
  |
Correlation:0.047537
  |
Correlation:0.29239
  |
Correlation:0.29239
  |
Correlation:0.047537
  |
Correlation:0.16809
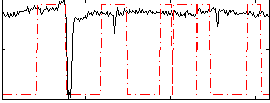  |
Correlation:0.076985
  |
Component:012; Jaccard:0.066046
 |
Component:012; Jaccard:0.05765
 |
Component:284; Jaccard:0.04002
 |
Component:284; Jaccard:0.022328
 |
Component:284; Jaccard:0.009002
 |
Component:262; Jaccard:0.004282
 |
Component:264; Jaccard:0.0018
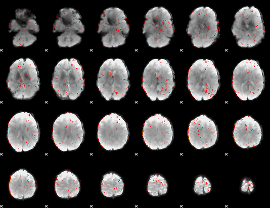 |
Correlation:0.29239
  |
Correlation:0.29239
  |
Correlation:0.16809
  |
Correlation:0.16809
  |
Correlation:0.16809
  |
Correlation:0.26393
  |
Correlation:-0.20148
  |
Component:284; Jaccard:0.065046
 |
Component:284; Jaccard:0.056241
 |
Component:288; Jaccard:0.036913
 |
Component:288; Jaccard:0.018621
 |
Component:006; Jaccard:0.008115
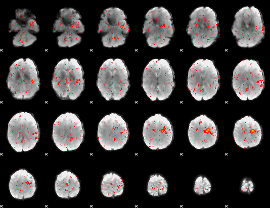 |
Component:022; Jaccard:0.003947
 |
Component:068; Jaccard:0.001576
 |
Correlation:0.16809
  |
Correlation:0.16809
  |
Correlation:-0.0018941
  |
Correlation:-0.0018941
  |
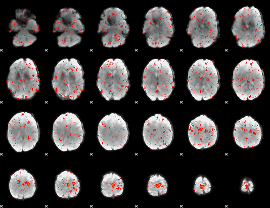
Correlation:-0.19116
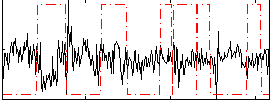  |
Correlation:0.047537
  |
Correlation:-0.1377
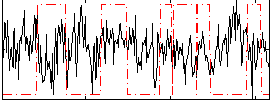 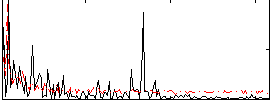 |
Component:180; Jaccard:0.063579
 |
Component:073; Jaccard:0.053911
 |
Component:073; Jaccard:0.033258
 |
Component:086; Jaccard:0.016886
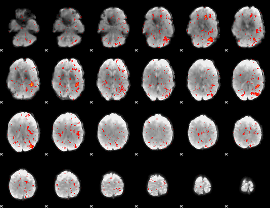 |
Component:288; Jaccard:0.007995
 |
Component:012; Jaccard:0.003604
 |
Component:055; Jaccard:0.001358
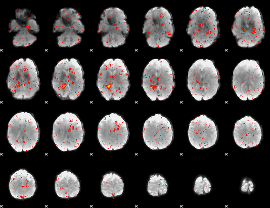 |
Correlation:-0.12519
  |
Correlation:0.14112
  |
Correlation:0.14112
  |
Correlation:0.018849
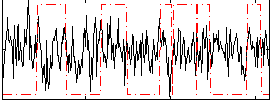  |
Correlation:-0.0018941
  |
Correlation:0.29239
  |
Correlation:-0.096891
  |
Cope: 02:STORY BACK
|
GLM result group-wise
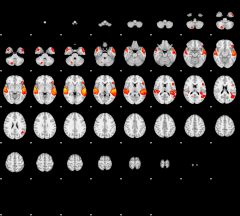 |
GLM binary result thresholded by 1
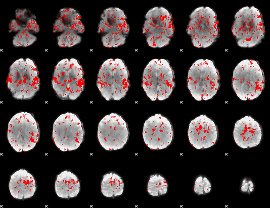 |
GLM binary result thresholded by 1.5
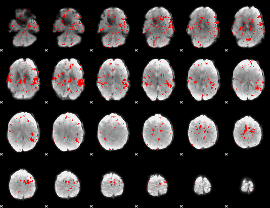 |
GLM binary result thresholded by 2
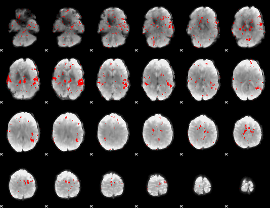 |
GLM binary result thresholded by 2.5
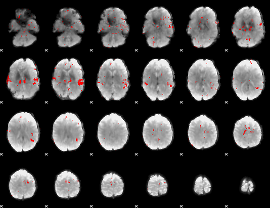 |
GLM binary result thresholded by 3
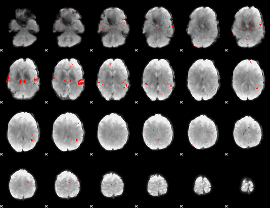 |
GLM binary result thresholded by 3.5
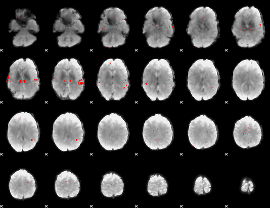 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
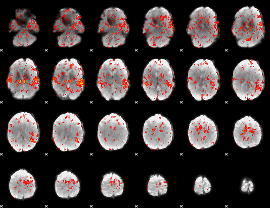 |
GLM Z-score result thresholded by 1.5
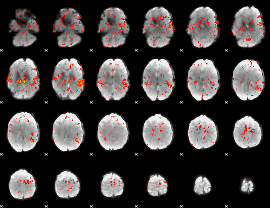 |
GLM Z-score result thresholded by 2
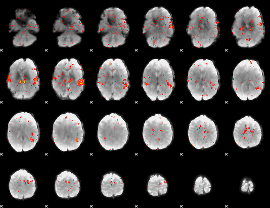 |
GLM Z-score result thresholded by 2.5
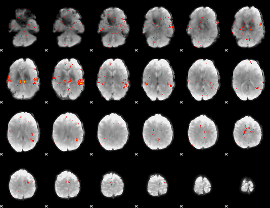 |
GLM Z-score result thresholded by 3
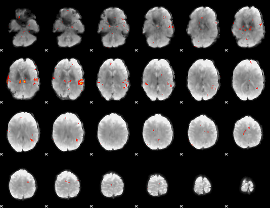 |
GLM Z-score result thresholded by 3.5
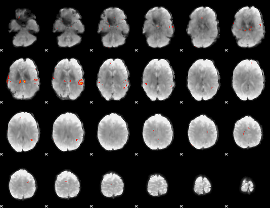 |
GLM Z-score result thresholded by 4
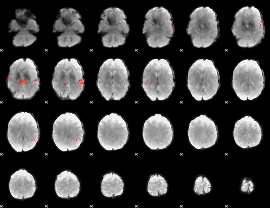 |
Component:353; Jaccard:0.11614
 |
Component:353; Jaccard:0.12476
 |
Component:353; Jaccard:0.11937
 |
Component:067; Jaccard:0.10127
 |
Component:067; Jaccard:0.076885
 |
Component:067; Jaccard:0.054217
 |
Component:067; Jaccard:0.036471
 |
Correlation:0.33227
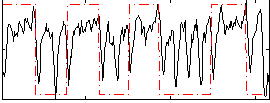 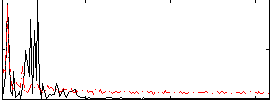 |
Correlation:0.33227
  |
Correlation:0.33227
  |
Correlation:0.22869
 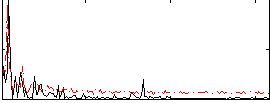 |
Correlation:0.22869
  |
Correlation:0.22869
  |
Correlation:0.22869
  |
Component:092; Jaccard:0.11431
 |
Component:092; Jaccard:0.12248
 |
Component:092; Jaccard:0.11244
 |
Component:353; Jaccard:0.095322
 |
Component:353; Jaccard:0.066895
 |
Component:353; Jaccard:0.042773
 |
Component:353; Jaccard:0.026098
 |
Correlation:0.43143
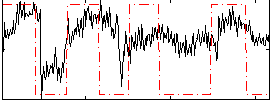 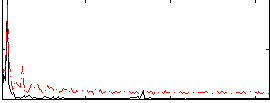 |
Correlation:0.43143
  |
Correlation:0.43143
  |
Correlation:0.33227
  |
Correlation:0.33227
  |
Correlation:0.33227
  |
Correlation:0.33227
  |
Component:192; Jaccard:0.097945
 |
Component:067; Jaccard:0.10835
 |
Component:067; Jaccard:0.11223
 |
Component:092; Jaccard:0.08875
 |
Component:092; Jaccard:0.06
 |
Component:092; Jaccard:0.03807
 |
Component:092; Jaccard:0.022407
 |
Correlation:0.65104
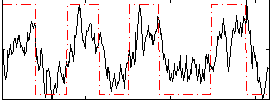 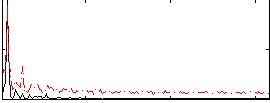 |
Correlation:0.22869
  |
Correlation:0.22869
  |
Correlation:0.43143
  |
Correlation:0.43143
  |
Correlation:0.43143
  |
Correlation:0.43143
  |
Component:067; Jaccard:0.094532
 |
Component:192; Jaccard:0.096377
 |
Component:192; Jaccard:0.084831
 |
Component:192; Jaccard:0.063979
 |
Component:192; Jaccard:0.043322
 |
Component:192; Jaccard:0.025486
 |
Component:192; Jaccard:0.014215
 |
Correlation:0.22869
  |
Correlation:0.65104
  |
Correlation:0.65104
  |
Correlation:0.65104
  |
Correlation:0.65104
  |
Correlation:0.65104
  |
Correlation:0.65104
  |
Component:146; Jaccard:0.088099
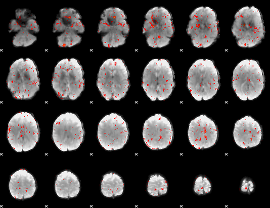 |
Component:243; Jaccard:0.087917
 |
Component:243; Jaccard:0.07821
 |
Component:243; Jaccard:0.051447
 |
Component:379; Jaccard:0.032589
 |
Component:379; Jaccard:0.01994
 |
Component:389; Jaccard:0.011673
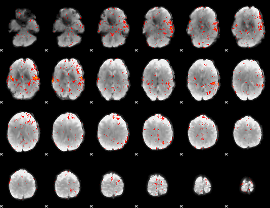 |
Correlation:0.2508
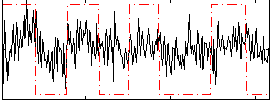 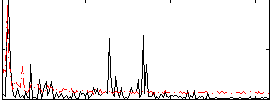 |
Correlation:0.017596
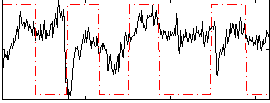  |
Correlation:0.017596
  |
Correlation:0.017596
  |
Correlation:0.47254
  |
Correlation:0.47254
  |
Correlation:0.28417
  |
Component:044; Jaccard:0.08793
 |
Component:146; Jaccard:0.086531
 |
Component:146; Jaccard:0.07086
 |
Component:379; Jaccard:0.050826
 |
Component:146; Jaccard:0.030866
 |
Component:389; Jaccard:0.017611
 |
Component:085; Jaccard:0.010728
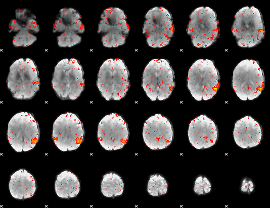 |
Correlation:0.5982
  |
Correlation:0.2508
  |
Correlation:0.2508
  |
Correlation:0.47254
  |
Correlation:0.2508
  |
Correlation:0.28417
  |
Correlation:0.46445
 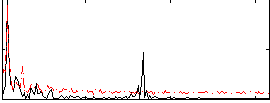 |
Component:243; Jaccard:0.086158
 |
Component:044; Jaccard:0.080509
 |
Component:379; Jaccard:0.065653
 |
Component:146; Jaccard:0.048971
 |
Component:389; Jaccard:0.028936
 |
Component:176; Jaccard:0.016773
 |
Component:365; Jaccard:0.009992
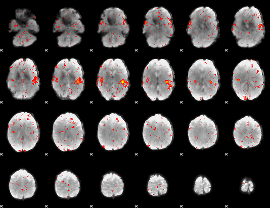 |
Correlation:0.017596
  |
Correlation:0.5982
  |
Correlation:0.47254
  |
Correlation:0.2508
  |
Correlation:0.28417
  |
Correlation:-0.09564
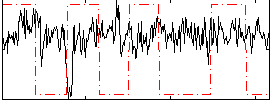 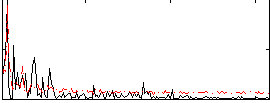 |
Correlation:0.42078
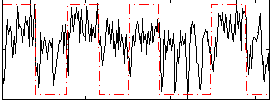 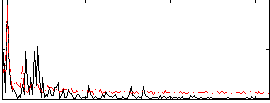 |
Component:176; Jaccard:0.084895
 |
Component:176; Jaccard:0.079389
 |
Component:176; Jaccard:0.065117
 |
Component:176; Jaccard:0.045472
 |
Component:176; Jaccard:0.028666
 |
Component:085; Jaccard:0.016385
 |
Component:379; Jaccard:0.009697
 |
Correlation:-0.09564
  |
Correlation:-0.09564
  |
Correlation:-0.09564
  |
Correlation:-0.09564
  |
Correlation:-0.09564
  |
Correlation:0.46445
  |
Correlation:0.47254
  |
Component:142; Jaccard:0.079006
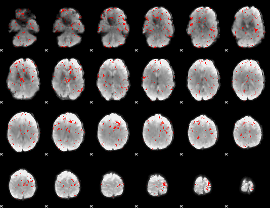 |
Component:379; Jaccard:0.072459
 |
Component:044; Jaccard:0.062924
 |
Component:365; Jaccard:0.043776
 |
Component:243; Jaccard:0.028563
 |
Component:146; Jaccard:0.016082
 |
Component:044; Jaccard:0.007996
 |
Correlation:0.18829
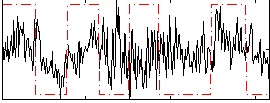 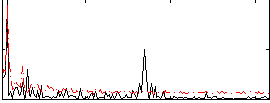 |
Correlation:0.47254
  |
Correlation:0.5982
  |
Correlation:0.42078
  |
Correlation:0.017596
  |
Correlation:0.2508
  |
Correlation:0.5982
  |
Component:085; Jaccard:0.073363
 |
Component:365; Jaccard:0.070005
 |
Component:365; Jaccard:0.061471
 |
Component:044; Jaccard:0.042188
 |
Component:365; Jaccard:0.027522
 |
Component:365; Jaccard:0.015854
 |
Component:146; Jaccard:0.007872
 |
Correlation:0.46445
  |
Correlation:0.42078
  |
Correlation:0.42078
  |
Correlation:0.5982
  |
Correlation:0.42078
  |
Correlation:0.42078
  |
Correlation:0.2508
  |
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
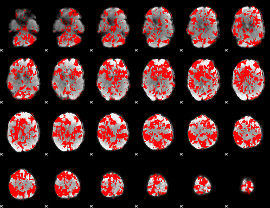 |
GLM binary result thresholded by 1.5
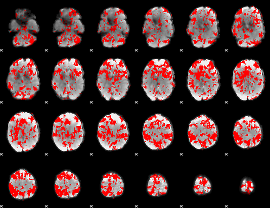 |
GLM binary result thresholded by 2
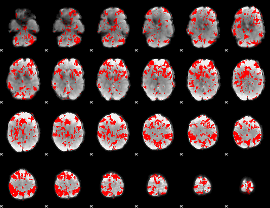 |
GLM binary result thresholded by 2.5
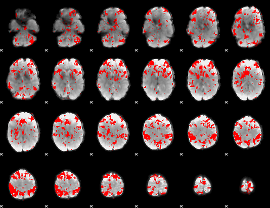 |
GLM binary result thresholded by 3
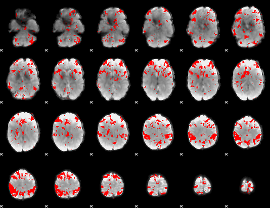 |
GLM binary result thresholded by 3.5
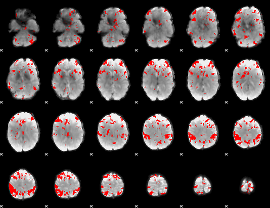 |
GLM binary result thresholded by 4
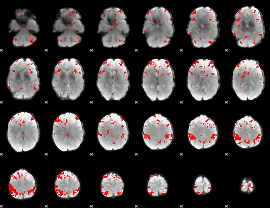 |
GLM Z-score result thresholded by 1
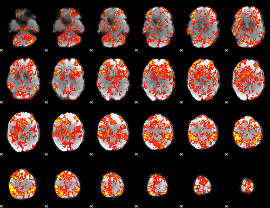 |
GLM Z-score result thresholded by 1.5
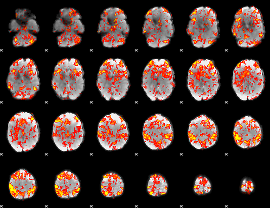 |
GLM Z-score result thresholded by 2
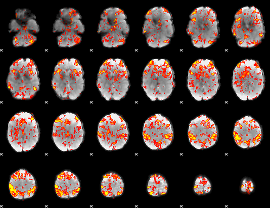 |
GLM Z-score result thresholded by 2.5
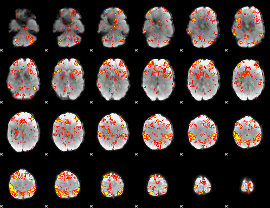 |
GLM Z-score result thresholded by 3
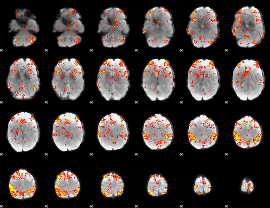 |
GLM Z-score result thresholded by 3.5
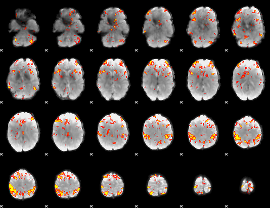 |
GLM Z-score result thresholded by 4
 |
Component:363; Jaccard:0.1402
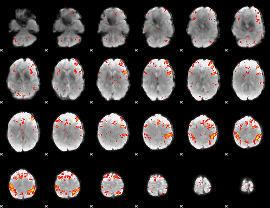 |
Component:363; Jaccard:0.17356
 |
Component:363; Jaccard:0.21285
 |
Component:363; Jaccard:0.25621
 |
Component:363; Jaccard:0.29566
 |
Component:363; Jaccard:0.31878
 |
Component:363; Jaccard:0.32577
 |
Correlation:0.62503
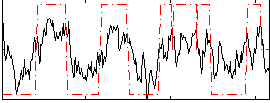 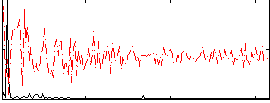 |
Correlation:0.62503
  |
Correlation:0.62503
  |
Correlation:0.62503
  |
Correlation:0.62503
  |
Correlation:0.62503
  |
Correlation:0.62503
  |
Component:095; Jaccard:0.12863
 |
Component:400; Jaccard:0.14998
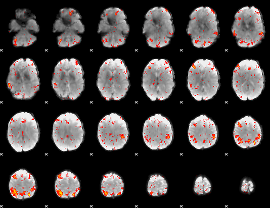 |
Component:400; Jaccard:0.17153
 |
Component:400; Jaccard:0.19302
 |
Component:400; Jaccard:0.20866
 |
Component:400; Jaccard:0.21301
 |
Component:400; Jaccard:0.20765
 |
Correlation:0.41142
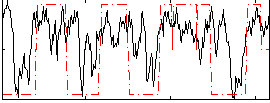 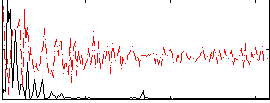 |
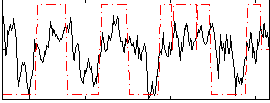
Correlation:0.5597
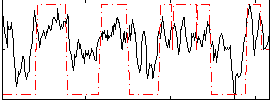  |
Correlation:0.5597
  |
Correlation:0.5597
  |
Correlation:0.5597
  |
Correlation:0.5597
  |
Correlation:0.5597
  |
Component:400; Jaccard:0.12853
 |
Component:095; Jaccard:0.14651
 |
Component:095; Jaccard:0.16507
 |
Component:095; Jaccard:0.17754
 |
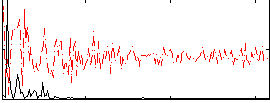
Component:058; Jaccard:0.18882
 |
Component:058; Jaccard:0.20133
 |
Component:058; Jaccard:0.20527
 |
Correlation:0.5597
  |
Correlation:0.41142
  |
Correlation:0.41142
  |
Correlation:0.41142
  |
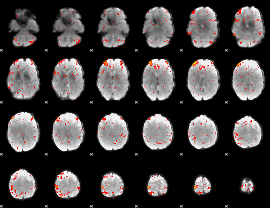
Correlation:0.45514
 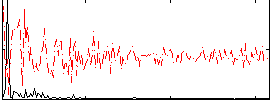 |
Correlation:0.45514
  |
Correlation:0.45514
  |
Component:018; Jaccard:0.11766
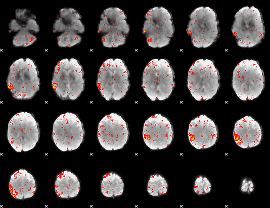 |
Component:018; Jaccard:0.13519
 |
Component:018; Jaccard:0.15381
 |
Component:018; Jaccard:0.17185
 |
Component:018; Jaccard:0.18637
 |
Component:018; Jaccard:0.19231
 |
Component:018; Jaccard:0.18953
 |
Correlation:0.40406
 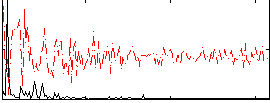 |
Correlation:0.40406
  |
Correlation:0.40406
  |
Correlation:0.40406
  |
Correlation:0.40406
  |
Correlation:0.40406
  |
Correlation:0.40406
  |
Component:218; Jaccard:0.1173
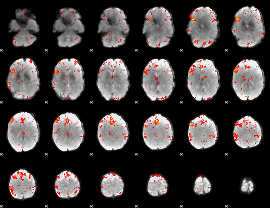 |
Component:218; Jaccard:0.13126
 |
Component:058; Jaccard:0.14605
 |
Component:058; Jaccard:0.16808
 |
Component:095; Jaccard:0.18339
 |
Component:095; Jaccard:0.18172
 |
Component:095; Jaccard:0.17169
 |
Correlation:0.40018
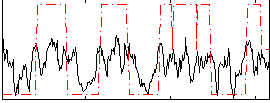 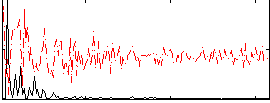 |
Correlation:0.40018
  |
Correlation:0.45514
  |
Correlation:0.45514
  |
Correlation:0.41142
  |
Correlation:0.41142
  |
Correlation:0.41142
  |
Component:316; Jaccard:0.10894
 |
Component:058; Jaccard:0.12433
 |
Component:218; Jaccard:0.146
 |
Component:218; Jaccard:0.15611
 |
Component:224; Jaccard:0.16202
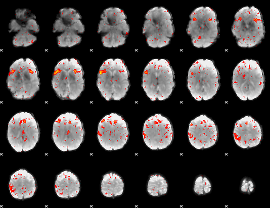 |
Component:224; Jaccard:0.17066
 |
Component:224; Jaccard:0.16898
 |
Correlation:0.33844
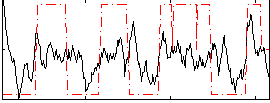 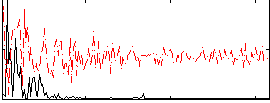 |
Correlation:0.45514
  |
Correlation:0.40018
  |
Correlation:0.40018
  |
Correlation:0.52167
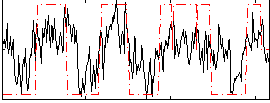 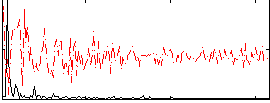 |
Correlation:0.52167
  |
Correlation:0.52167
  |
Component:058; Jaccard:0.10459
 |
Component:316; Jaccard:0.11799
 |
Component:224; Jaccard:0.1305
 |
Component:224; Jaccard:0.14786
 |
Component:218; Jaccard:0.16144
 |
Component:218; Jaccard:0.16079
 |
Component:218; Jaccard:0.15077
 |
Correlation:0.45514
  |
Correlation:0.33844
  |
Correlation:0.52167
  |
Correlation:0.52167
  |
Correlation:0.40018
  |
Correlation:0.40018
  |
Correlation:0.40018
  |
Component:282; Jaccard:0.10336
 |
Component:282; Jaccard:0.11632
 |
Component:282; Jaccard:0.12886
 |
Component:282; Jaccard:0.13977
 |
Component:282; Jaccard:0.14782
 |
Component:282; Jaccard:0.15028
 |
Component:282; Jaccard:0.14546
 |
Correlation:0.45137
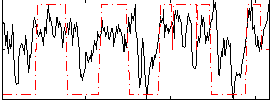 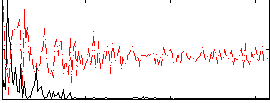 |
Correlation:0.45137
  |
Correlation:0.45137
  |
Correlation:0.45137
  |
Correlation:0.45137
  |
Correlation:0.45137
  |
Correlation:0.45137
  |
Component:219; Jaccard:0.10179
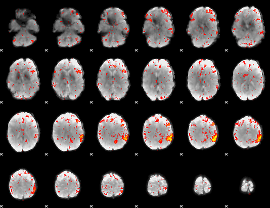 |
Component:224; Jaccard:0.11221
 |
Component:316; Jaccard:0.12694
 |
Component:316; Jaccard:0.13225
 |
Component:251; Jaccard:0.13436
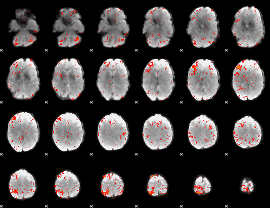 |
Component:251; Jaccard:0.13634
 |
Component:251; Jaccard:0.13069
 |
Correlation:0.25112
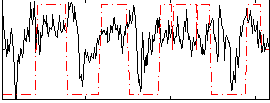 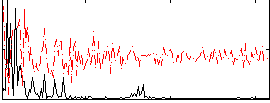 |
Correlation:0.52167
  |
Correlation:0.33844
  |
Correlation:0.33844
  |
Correlation:0.41996
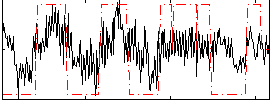 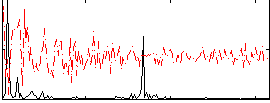 |
Correlation:0.41996
  |
Correlation:0.41996
  |
Component:202; Jaccard:0.098066
 |
Component:219; Jaccard:0.11037
 |
Component:251; Jaccard:0.12023
 |
Component:251; Jaccard:0.13075
 |
Component:316; Jaccard:0.13397
 |
Component:316; Jaccard:0.12929
 |
Component:316; Jaccard:0.12006
 |
Correlation:0.33182
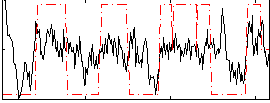  |
Correlation:0.25112
  |
Correlation:0.41996
  |
Correlation:0.41996
  |
Correlation:0.33844
  |
Correlation:0.33844
  |
Correlation:0.33844
  |
Cope: 04:STORY-MATH BACK
|
GLM result group-wise
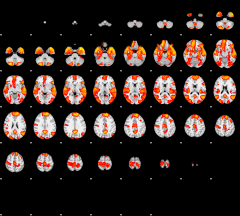 |
GLM binary result thresholded by 1
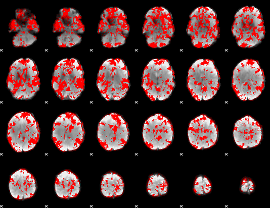 |
GLM binary result thresholded by 1.5
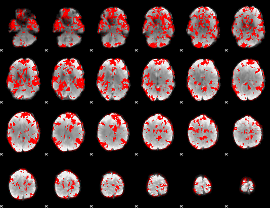 |
GLM binary result thresholded by 2
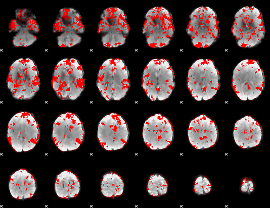 |
GLM binary result thresholded by 2.5
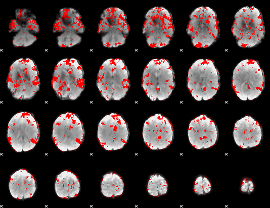 |
GLM binary result thresholded by 3
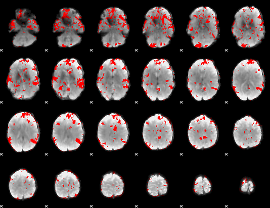 |
GLM binary result thresholded by 3.5
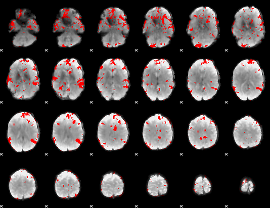 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
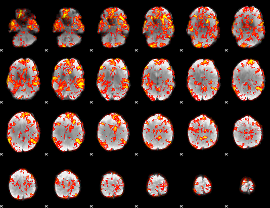 |
GLM Z-score result thresholded by 1.5
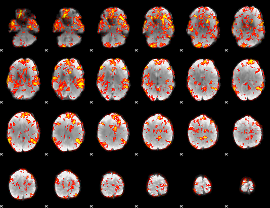 |
GLM Z-score result thresholded by 2
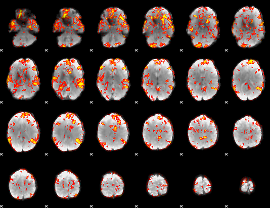 |
GLM Z-score result thresholded by 2.5
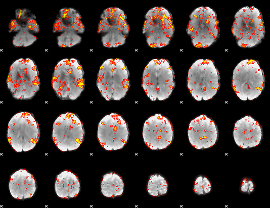 |
GLM Z-score result thresholded by 3
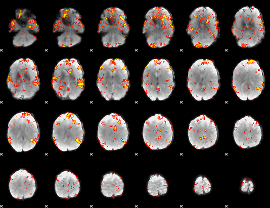 |
GLM Z-score result thresholded by 3.5
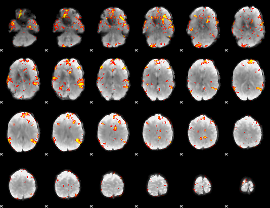 |
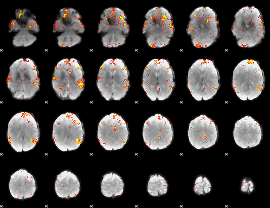
GLM Z-score result thresholded by 4
 |
Component:192; Jaccard:0.13056
 |
Component:192; Jaccard:0.1583
 |
Component:192; Jaccard:0.19017
 |
Component:192; Jaccard:0.22411
 |
Component:192; Jaccard:0.25606
 |
Component:192; Jaccard:0.27727
 |
Component:192; Jaccard:0.28261
 |
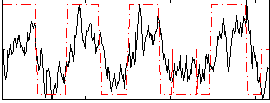
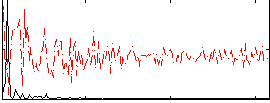
Correlation:0.64794
  |
Correlation:0.64794
  |
Correlation:0.64794
  |
Correlation:0.64794
  |
Correlation:0.64794
  |
Correlation:0.64794
  |
Correlation:0.64794
  |
Component:044; Jaccard:0.11228
 |
Component:044; Jaccard:0.1336
 |
Component:044; Jaccard:0.15845
 |
Component:044; Jaccard:0.18535
 |
Component:044; Jaccard:0.20959
 |
Component:044; Jaccard:0.22648
 |
Component:044; Jaccard:0.23303
 |
Correlation:0.59718
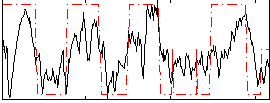  |
Correlation:0.59718
  |
Correlation:0.59718
  |
Correlation:0.59718
  |
Correlation:0.59718
  |
Correlation:0.59718
  |
Correlation:0.59718
  |
Component:032; Jaccard:0.10657
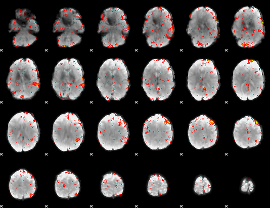 |
Component:032; Jaccard:0.11754
 |
Component:032; Jaccard:0.12732
 |
Component:092; Jaccard:0.13979
 |
Component:092; Jaccard:0.15622
 |
Component:092; Jaccard:0.16741
 |
Component:379; Jaccard:0.17131
 |
Correlation:0.20367
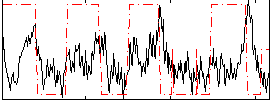 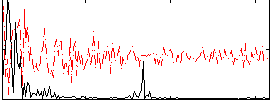 |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.45256
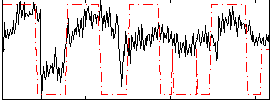 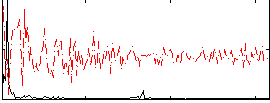 |
Correlation:0.45256
  |
Correlation:0.45256
  |
Correlation:0.47019
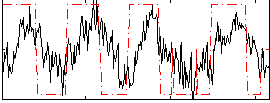 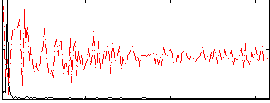 |
Component:085; Jaccard:0.095919
 |
Component:085; Jaccard:0.10677
 |
Component:092; Jaccard:0.12021
 |
Component:379; Jaccard:0.13495
 |
Component:379; Jaccard:0.15277
 |
Component:379; Jaccard:0.16711
 |
Component:092; Jaccard:0.16724
 |
Correlation:0.47756
  |
Correlation:0.47756
  |
Correlation:0.45256
  |
Correlation:0.47019
  |
Correlation:0.47019
  |
Correlation:0.47019
  |
Correlation:0.45256
  |
Component:335; Jaccard:0.095759
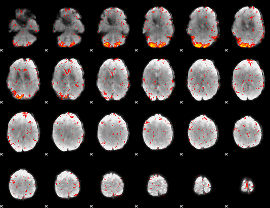 |
Component:335; Jaccard:0.10121
 |
Component:085; Jaccard:0.11813
 |
Component:032; Jaccard:0.13469
 |
Component:032; Jaccard:0.13845
 |
Component:032; Jaccard:0.13746
 |
Component:085; Jaccard:0.13195
 |
Correlation:0.10315
  |
Correlation:0.10315
  |
Correlation:0.47756
  |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.47756
  |
Component:215; Jaccard:0.090776
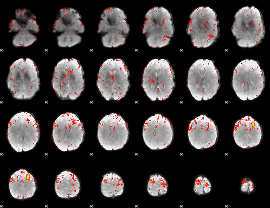 |
Component:092; Jaccard:0.10064
 |
Component:379; Jaccard:0.11619
 |
Component:085; Jaccard:0.12915
 |
Component:085; Jaccard:0.13449
 |
Component:085; Jaccard:0.1352
 |
Component:032; Jaccard:0.13138
 |
Correlation:0.20275
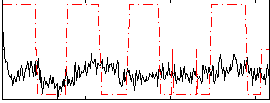 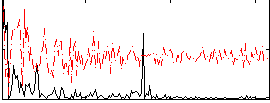 |
Correlation:0.45256
  |
Correlation:0.47019
  |
Correlation:0.47756
  |
Correlation:0.47756
  |
Correlation:0.47756
  |
Correlation:0.20367
  |
Component:169; Jaccard:0.089356
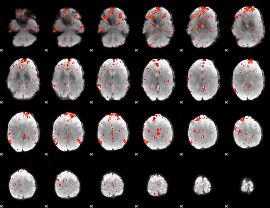 |
Component:379; Jaccard:0.097634
 |
Component:031; Jaccard:0.10579
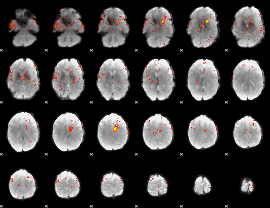 |
Component:031; Jaccard:0.11677
 |
Component:031; Jaccard:0.12535
 |
Component:031; Jaccard:0.12896
 |
Component:031; Jaccard:0.12681
 |
Correlation:0.34867
  |
Correlation:0.47019
  |
Correlation:0.39848
  |
Correlation:0.39848
  |
Correlation:0.39848
  |
Correlation:0.39848
  |
Correlation:0.39848
  |
Component:286; Jaccard:0.086519
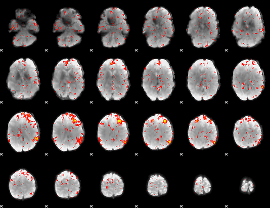 |
Component:169; Jaccard:0.094738
 |
Component:335; Jaccard:0.10483
 |
Component:335; Jaccard:0.10599
 |
Component:037; Jaccard:0.11266
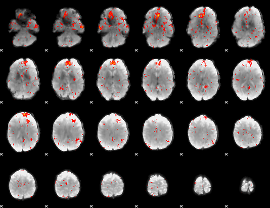 |
Component:037; Jaccard:0.11722
 |
Component:037; Jaccard:0.11604
 |
Correlation:0.35967
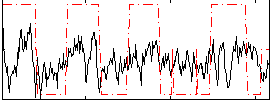  |
Correlation:0.34867
  |
Correlation:0.10315
  |
Correlation:0.10315
  |
Correlation:0.40614
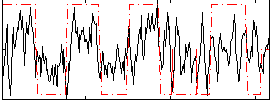 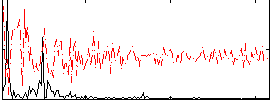 |
Correlation:0.40614
  |
Correlation:0.40614
  |
Component:102; Jaccard:0.08568
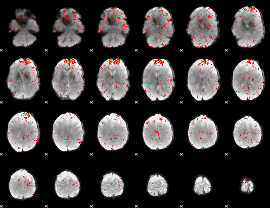 |
Component:215; Jaccard:0.093498
 |
Component:169; Jaccard:0.099087
 |
Component:037; Jaccard:0.10526
 |
Component:077; Jaccard:0.10543
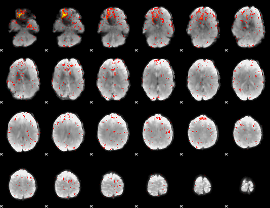 |
Component:077; Jaccard:0.10872
 |
Component:077; Jaccard:0.10709
 |
Correlation:0.2959
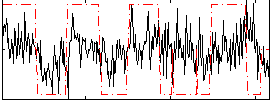 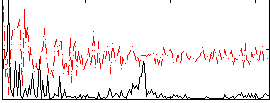 |
Correlation:0.20275
  |
Correlation:0.34867
  |
Correlation:0.40614
  |
Correlation:0.1964
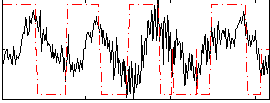 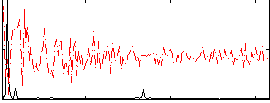 |
Correlation:0.1964
  |
Correlation:0.1964
  |
Component:092; Jaccard:0.084023
 |
Component:031; Jaccard:0.092761
 |
Component:102; Jaccard:0.096224
 |
Component:169; Jaccard:0.1025
 |
Component:335; Jaccard:0.1052
 |
Component:169; Jaccard:0.10145
 |
Component:169; Jaccard:0.0965
 |
Correlation:0.45256
  |
Correlation:0.39848
  |
Correlation:0.2959
  |
Correlation:0.34867
  |
Correlation:0.10315
  |
Correlation:0.34867
  |
Correlation:0.34867
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
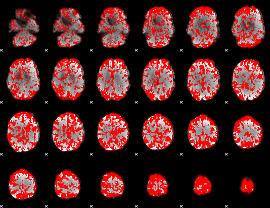 |
GLM binary result thresholded by 1.5
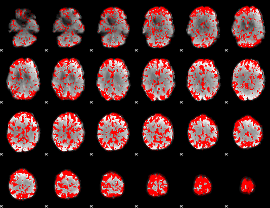 |
GLM binary result thresholded by 2
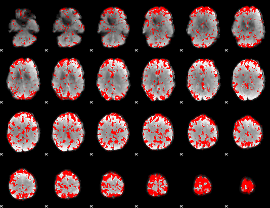 |
GLM binary result thresholded by 2.5
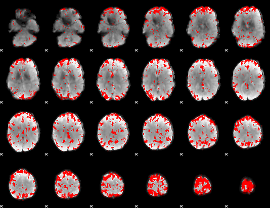 |
GLM binary result thresholded by 3
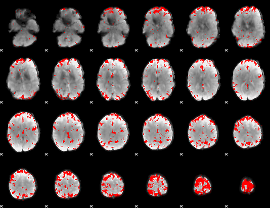 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
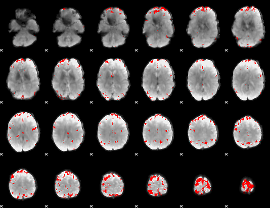 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
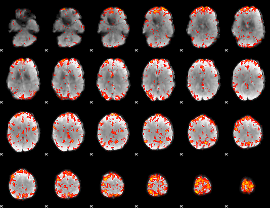 |
GLM Z-score result thresholded by 3
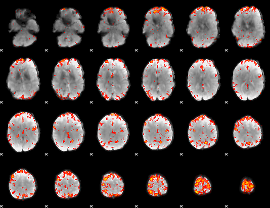 |
GLM Z-score result thresholded by 3.5
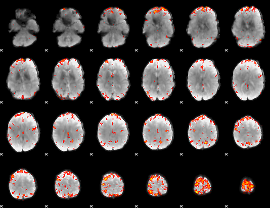 |
GLM Z-score result thresholded by 4
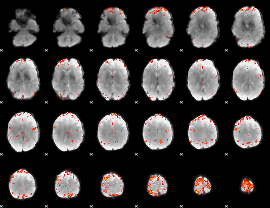 |
Component:088; Jaccard:0.11578
 |
Component:088; Jaccard:0.13821
 |
Component:088; Jaccard:0.16508
 |
Component:088; Jaccard:0.18471
 |
Component:088; Jaccard:0.1948
 |
Component:088; Jaccard:0.18643
 |
Component:088; Jaccard:0.15714
 |
Correlation:-0.069077
  |
Correlation:-0.069077
  |
Correlation:-0.069077
  |
Correlation:-0.069077
  |
Correlation:-0.069077
  |
Correlation:-0.069077
  |
Correlation:-0.069077
  |
Component:254; Jaccard:0.10693
 |
Component:254; Jaccard:0.12564
 |
Component:254; Jaccard:0.14665
 |
Component:254; Jaccard:0.16536
 |
Component:254; Jaccard:0.17522
 |
Component:254; Jaccard:0.17045
 |
Component:254; Jaccard:0.14465
 |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.10502
  |
Component:396; Jaccard:0.10409
 |
Component:396; Jaccard:0.11811
 |
Component:316; Jaccard:0.13333
 |
Component:316; Jaccard:0.14802
 |
Component:316; Jaccard:0.1529
 |
Component:316; Jaccard:0.14081
 |
Component:316; Jaccard:0.11422
 |
Correlation:-0.17967
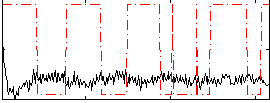 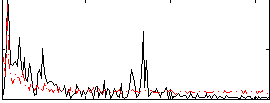 |
Correlation:-0.17967
  |
Correlation:-0.31707
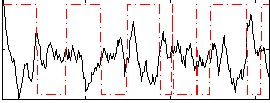 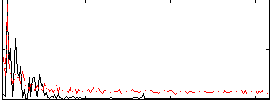 |
Correlation:-0.31707
  |
Correlation:-0.31707
  |
Correlation:-0.31707
  |
Correlation:-0.31707
  |
Component:316; Jaccard:0.099185
 |
Component:316; Jaccard:0.11623
 |
Component:396; Jaccard:0.1304
 |
Component:396; Jaccard:0.1385
 |
Component:396; Jaccard:0.13551
 |
Component:344; Jaccard:0.13042
 |
Component:202; Jaccard:0.10856
 |
Correlation:-0.31707
  |
Correlation:-0.31707
  |
Correlation:-0.17967
  |
Correlation:-0.17967
  |
Correlation:-0.17967
  |
Correlation:-0.16161
  |
Correlation:-0.29434
  |
Component:095; Jaccard:0.092708
 |
Component:095; Jaccard:0.1055
 |
Component:202; Jaccard:0.11698
 |
Component:202; Jaccard:0.13086
 |
Component:344; Jaccard:0.13529
 |
Component:202; Jaccard:0.12661
 |
Component:344; Jaccard:0.10856
 |
Correlation:-0.36762
 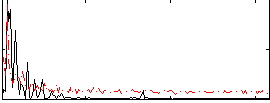 |
Correlation:-0.36762
  |
Correlation:-0.29434
  |
Correlation:-0.29434
  |
Correlation:-0.16161
  |
Correlation:-0.29434
  |
Correlation:-0.16161
  |
Component:087; Jaccard:0.091983
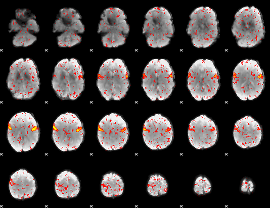 |
Component:202; Jaccard:0.10181
 |
Component:095; Jaccard:0.11671
 |
Component:344; Jaccard:0.12693
 |
Component:202; Jaccard:0.13501
 |
Component:396; Jaccard:0.11984
 |
Component:136; Jaccard:0.099994
 |
Correlation:-0.028504
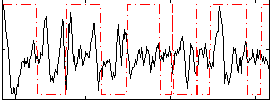 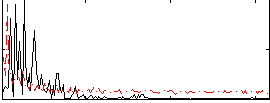 |
Correlation:-0.29434
  |
Correlation:-0.36762
  |
Correlation:-0.16161
  |
Correlation:-0.29434
  |
Correlation:-0.17967
  |
Correlation:-0.12296
  |
Component:277; Jaccard:0.088149
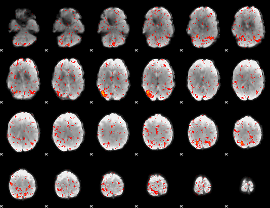 |
Component:087; Jaccard:0.099392
 |
Component:344; Jaccard:0.11209
 |
Component:095; Jaccard:0.12329
 |
Component:136; Jaccard:0.1248
 |
Component:136; Jaccard:0.11939
 |
Component:396; Jaccard:0.095728
 |
Correlation:0.11592
  |
Correlation:-0.028504
  |
Correlation:-0.16161
  |
Correlation:-0.36762
  |
Correlation:-0.12296
  |
Correlation:-0.12296
  |
Correlation:-0.17967
  |
Component:347; Jaccard:0.087834
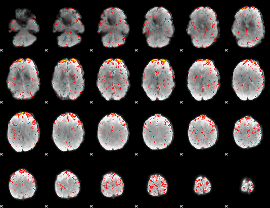 |
Component:347; Jaccard:0.099267
 |
Component:347; Jaccard:0.1119
 |
Component:347; Jaccard:0.12062
 |
Component:347; Jaccard:0.12237
 |
Component:347; Jaccard:0.11234
 |
Component:347; Jaccard:0.09144
 |
Correlation:0.073898
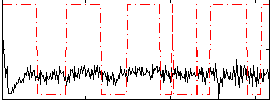 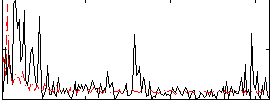 |
Correlation:0.073898
  |
Correlation:0.073898
  |
Correlation:0.073898
  |
Correlation:0.073898
  |
Correlation:0.073898
  |
Correlation:0.073898
  |
Component:202; Jaccard:0.0871
 |
Component:344; Jaccard:0.095935
 |
Component:136; Jaccard:0.10684
 |
Component:136; Jaccard:0.11901
 |
Component:095; Jaccard:0.12079
 |
Component:095; Jaccard:0.10713
 |
Component:095; Jaccard:0.086727
 |
Correlation:-0.29434
  |
Correlation:-0.16161
  |
Correlation:-0.12296
  |
Correlation:-0.12296
  |
Correlation:-0.36762
  |
Correlation:-0.36762
  |
Correlation:-0.36762
  |
Component:247; Jaccard:0.08653
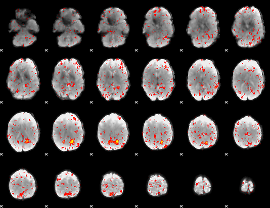 |
Component:277; Jaccard:0.095498
 |
Component:087; Jaccard:0.10382
 |
Component:087; Jaccard:0.1029
 |
Component:247; Jaccard:0.095524
 |
Component:169; Jaccard:0.088678
 |
Component:235; Jaccard:0.078588
 |
Correlation:-0.019159
  |
Correlation:0.11592
  |
Correlation:-0.028504
  |
Correlation:-0.028504
  |
Correlation:-0.019159
  |
Correlation:0.33478
  |
Correlation:0.0019381
  |
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
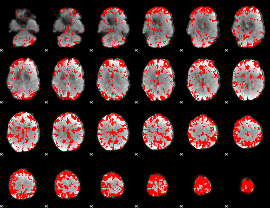 |
GLM binary result thresholded by 2.5
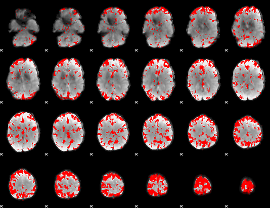 |
GLM binary result thresholded by 3
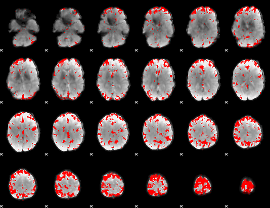 |
GLM binary result thresholded by 3.5
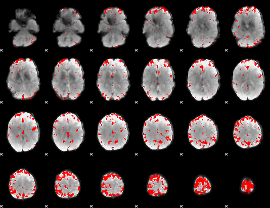 |
GLM binary result thresholded by 4
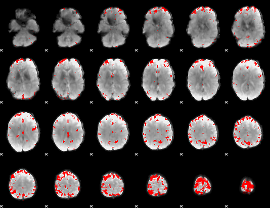 |
GLM Z-score result thresholded by 1
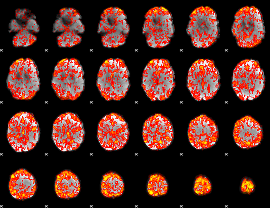 |
GLM Z-score result thresholded by 1.5
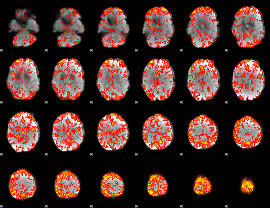 |
GLM Z-score result thresholded by 2
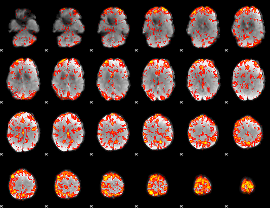 |
GLM Z-score result thresholded by 2.5
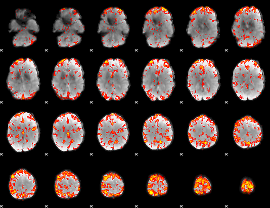 |
GLM Z-score result thresholded by 3
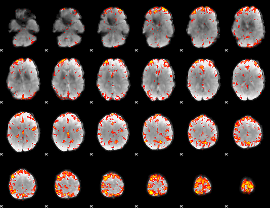 |
GLM Z-score result thresholded by 3.5
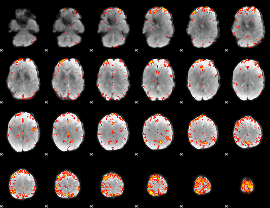 |
GLM Z-score result thresholded by 4
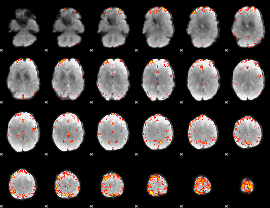 |
Component:088; Jaccard:0.11915
 |
Component:088; Jaccard:0.14264
 |
Component:088; Jaccard:0.16912
 |
Component:088; Jaccard:0.19318
 |
Component:088; Jaccard:0.20977
 |
Component:088; Jaccard:0.21223
 |
Component:088; Jaccard:0.19888
 |
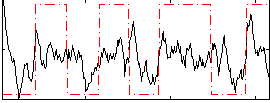
Correlation:0.085515
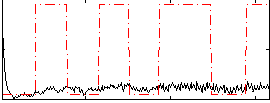  |
Correlation:0.085515
  |
Correlation:0.085515
  |
Correlation:0.085515
  |
Correlation:0.085515
  |
Correlation:0.085515
  |
Correlation:0.085515
  |
Component:095; Jaccard:0.11198
 |
Component:095; Jaccard:0.13655
 |
Component:095; Jaccard:0.16381
 |
Component:095; Jaccard:0.18598
 |
Component:316; Jaccard:0.19986
 |
Component:316; Jaccard:0.20671
 |
Component:316; Jaccard:0.19685
 |
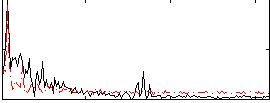
Correlation:0.42674
  |
Correlation:0.42674
  |
Correlation:0.42674
  |
Correlation:0.42674
  |
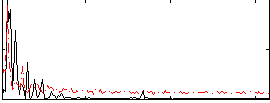
Correlation:0.33638
 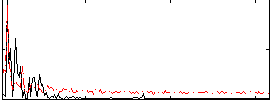 |
Correlation:0.33638
  |
Correlation:0.33638
  |
Component:316; Jaccard:0.1106
 |
Component:316; Jaccard:0.13327
 |
Component:316; Jaccard:0.15838
 |
Component:316; Jaccard:0.18107
 |
Component:095; Jaccard:0.19816
 |
Component:095; Jaccard:0.19398
 |
Component:095; Jaccard:0.17626
 |
Correlation:0.33638
  |
Correlation:0.33638
  |
Correlation:0.33638
  |
Correlation:0.33638
  |
Correlation:0.42674
  |
Correlation:0.42674
  |
Correlation:0.42674
  |
Component:396; Jaccard:0.10813
 |
Component:396; Jaccard:0.12271
 |
Component:202; Jaccard:0.14064
 |
Component:202; Jaccard:0.15938
 |
Component:202; Jaccard:0.17472
 |
Component:202; Jaccard:0.17886
 |
Component:202; Jaccard:0.17263
 |
Correlation:0.15632
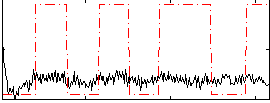 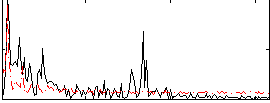 |
Correlation:0.15632
  |
Correlation:0.34633
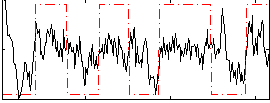  |
Correlation:0.34633
  |
Correlation:0.34633
  |
Correlation:0.34633
  |
Correlation:0.34633
  |
Component:254; Jaccard:0.10245
 |
Component:202; Jaccard:0.11865
 |
Component:396; Jaccard:0.13674
 |
Component:254; Jaccard:0.14685
 |
Component:344; Jaccard:0.15373
 |
Component:344; Jaccard:0.15832
 |
Component:344; Jaccard:0.14749
 |
Correlation:-0.12493
  |
Correlation:0.34633
  |
Correlation:0.15632
  |
Correlation:-0.12493
  |
Correlation:0.15307
  |
Correlation:0.15307
  |
Correlation:0.15307
  |
Component:202; Jaccard:0.098926
 |
Component:254; Jaccard:0.11756
 |
Component:254; Jaccard:0.1338
 |
Component:396; Jaccard:0.14676
 |
Component:254; Jaccard:0.15345
 |
Component:254; Jaccard:0.15099
 |
Component:254; Jaccard:0.13937
 |
Correlation:0.34633
  |
Correlation:-0.12493
  |
Correlation:-0.12493
  |
Correlation:0.15632
  |
Correlation:-0.12493
  |
Correlation:-0.12493
  |
Correlation:-0.12493
  |
Component:087; Jaccard:0.092634
 |
Component:344; Jaccard:0.10439
 |
Component:344; Jaccard:0.12258
 |
Component:344; Jaccard:0.14064
 |
Component:396; Jaccard:0.14983
 |
Component:136; Jaccard:0.14273
 |
Component:136; Jaccard:0.13757
 |
Correlation:-0.020585
  |
Correlation:0.15307
  |
Correlation:0.15307
  |
Correlation:0.15307
  |
Correlation:0.15632
  |
Correlation:0.13508
  |
Correlation:0.13508
  |
Component:104; Jaccard:0.091729
 |
Component:104; Jaccard:0.10277
 |
Component:282; Jaccard:0.11294
 |
Component:136; Jaccard:0.1274
 |
Component:136; Jaccard:0.13707
 |
Component:396; Jaccard:0.14088
 |
Component:396; Jaccard:0.12503
 |
Correlation:0.11764
 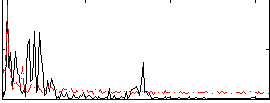 |
Correlation:0.11764
  |
Correlation:0.47612
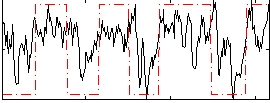 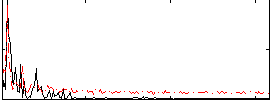 |
Correlation:0.13508
  |
Correlation:0.13508
  |
Correlation:0.15632
  |
Correlation:0.15632
  |
Component:091; Jaccard:0.089371
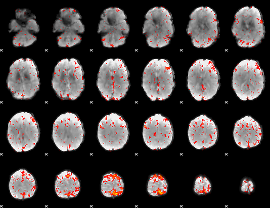 |
Component:087; Jaccard:0.10001
 |
Component:104; Jaccard:0.11294
 |
Component:282; Jaccard:0.125
 |
Component:282; Jaccard:0.131
 |
Component:282; Jaccard:0.13023
 |
Component:282; Jaccard:0.11687
 |
Correlation:0.1799
  |
Correlation:-0.020585
  |
Correlation:0.11764
  |
Correlation:0.47612
  |
Correlation:0.47612
  |
Correlation:0.47612
  |
Correlation:0.47612
  |
Component:344; Jaccard:0.088537
 |
Component:091; Jaccard:0.098113
 |
Component:136; Jaccard:0.11237
 |
Component:104; Jaccard:0.12153
 |
Component:104; Jaccard:0.12263
 |
Component:104; Jaccard:0.11647
 |
Component:104; Jaccard:0.10301
 |
Correlation:0.15307
  |
Correlation:0.1799
  |
Correlation:0.13508
  |
Correlation:0.11764
  |
Correlation:0.11764
  |
Correlation:0.11764
  |
Correlation:0.11764
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































