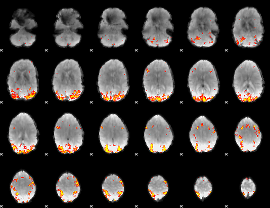
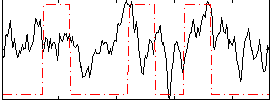
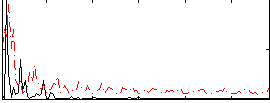
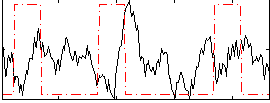
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
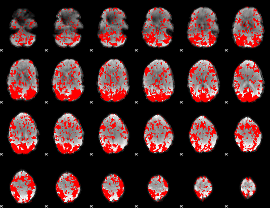 |
GLM binary result thresholded by 1.5
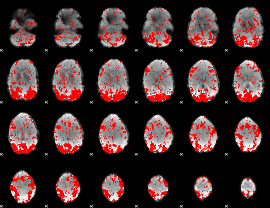 |
GLM binary result thresholded by 2
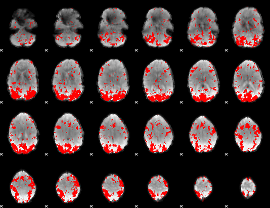 |
GLM binary result thresholded by 2.5
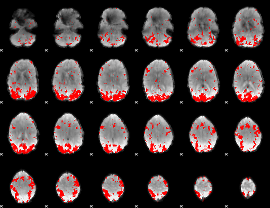 |
GLM binary result thresholded by 3
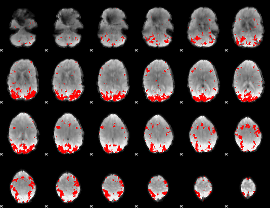 |
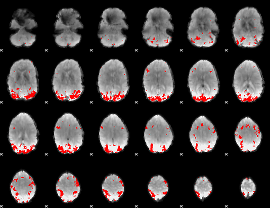
GLM binary result thresholded by 3.5
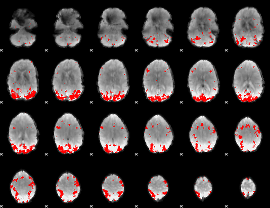 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
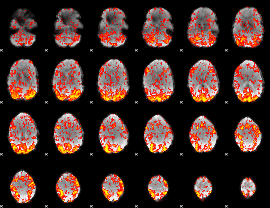 |
GLM Z-score result thresholded by 1.5
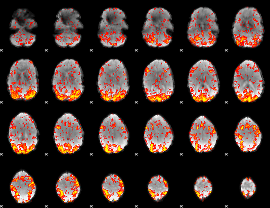 |
GLM Z-score result thresholded by 2
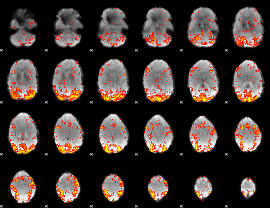 |
GLM Z-score result thresholded by 2.5
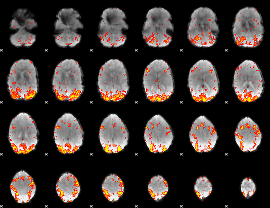 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
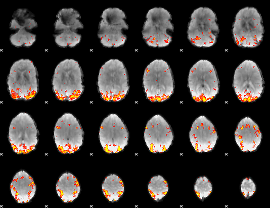 |
GLM Z-score result thresholded by 4
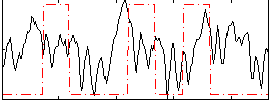
 |
Component:079; Jaccard:0.16272
 |
Component:079; Jaccard:0.20924
 |
Component:079; Jaccard:0.25823
 |
Component:079; Jaccard:0.30415
 |
Component:079; Jaccard:0.33474
 |
Component:079; Jaccard:0.34852
 |
Component:079; Jaccard:0.34596
 |
Correlation:0.10084
 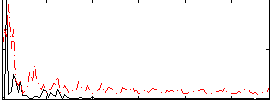 |
Correlation:0.10084
  |
Correlation:0.10084
  |
Correlation:0.10084
  |
Correlation:0.10084
  |
Correlation:0.10084
  |
Correlation:0.10084
  |
Component:218; Jaccard:0.14717
 |
Component:042; Jaccard:0.1794
 |
Component:042; Jaccard:0.21655
 |
Component:042; Jaccard:0.24724
 |
Component:042; Jaccard:0.26495
 |
Component:042; Jaccard:0.26995
 |
Component:163; Jaccard:0.26573
 |
Correlation:0.24169
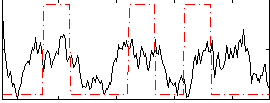 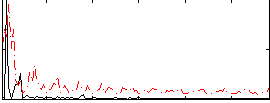 |
Correlation:0.28823
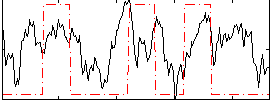 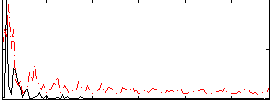 |
Correlation:0.28823
  |
Correlation:0.28823
  |
Correlation:0.28823
  |
Correlation:0.28823
  |
Correlation:0.12046
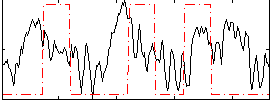 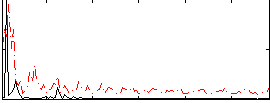 |
Component:042; Jaccard:0.14199
 |
Component:218; Jaccard:0.17634
 |
Component:218; Jaccard:0.20052
 |
Component:163; Jaccard:0.23264
 |
Component:163; Jaccard:0.2546
 |
Component:163; Jaccard:0.26742
 |
Component:042; Jaccard:0.25974
 |
Correlation:0.28823
  |
Correlation:0.24169
  |
Correlation:0.24169
  |
Correlation:0.12046
  |
Correlation:0.12046
  |
Correlation:0.12046
  |
Correlation:0.28823
  |
Component:147; Jaccard:0.13593
 |
Component:163; Jaccard:0.16415
 |
Component:163; Jaccard:0.20028
 |
Component:218; Jaccard:0.21368
 |
Component:015; Jaccard:0.22345
 |
Component:015; Jaccard:0.23553
 |
Component:015; Jaccard:0.23713
 |
Correlation:0.14858
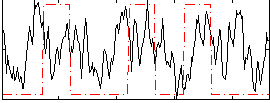 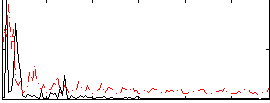 |
Correlation:0.12046
  |
Correlation:0.12046
  |
Correlation:0.24169
  |
Correlation:0.17197
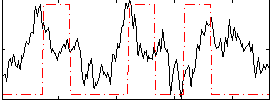 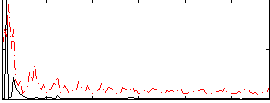 |
Correlation:0.17197
  |
Correlation:0.17197
  |
Component:163; Jaccard:0.12972
 |
Component:147; Jaccard:0.16011
 |
Component:015; Jaccard:0.17683
 |
Component:015; Jaccard:0.203
 |
Component:218; Jaccard:0.21637
 |
Component:218; Jaccard:0.20763
 |
Component:218; Jaccard:0.19155
 |
Correlation:0.12046
  |
Correlation:0.14858
  |
Correlation:0.17197
  |
Correlation:0.17197
  |
Correlation:0.24169
  |
Correlation:0.24169
  |
Correlation:0.24169
  |
Component:386; Jaccard:0.1205
 |
Component:015; Jaccard:0.14666
 |
Component:147; Jaccard:0.1764
 |
Component:147; Jaccard:0.18286
 |
Component:147; Jaccard:0.17832
 |
Component:153; Jaccard:0.17517
 |
Component:153; Jaccard:0.16752
 |
Correlation:-0.039908
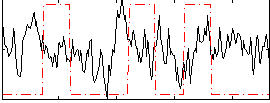 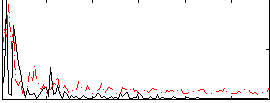 |
Correlation:0.17197
  |
Correlation:0.14858
  |
Correlation:0.14858
  |
Correlation:0.14858
  |
Correlation:0.27496
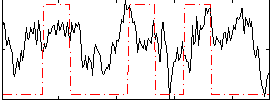  |
Correlation:0.27496
  |
Component:015; Jaccard:0.11761
 |
Component:386; Jaccard:0.13876
 |
Component:207; Jaccard:0.15635
 |
Component:207; Jaccard:0.17128
 |
Component:207; Jaccard:0.17595
 |
Component:315; Jaccard:0.1731
 |
Component:315; Jaccard:0.16631
 |
Correlation:0.17197
  |
Correlation:-0.039908
  |
Correlation:0.027163
  |
Correlation:0.027163
  |
Correlation:0.027163
  |
Correlation:0.13866
 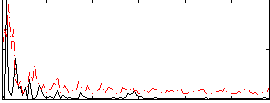 |
Correlation:0.13866
  |
Component:153; Jaccard:0.1145
 |
Component:207; Jaccard:0.13693
 |
Component:153; Jaccard:0.15517
 |
Component:153; Jaccard:0.17011
 |
Component:153; Jaccard:0.17591
 |
Component:207; Jaccard:0.16948
 |
Component:147; Jaccard:0.15407
 |
Correlation:0.27496
  |
Correlation:0.027163
  |
Correlation:0.27496
  |
Correlation:0.27496
  |
Correlation:0.27496
  |
Correlation:0.027163
  |
Correlation:0.14858
  |
Component:276; Jaccard:0.11446
 |
Component:153; Jaccard:0.13576
 |
Component:315; Jaccard:0.15203
 |
Component:315; Jaccard:0.16603
 |
Component:315; Jaccard:0.17375
 |
Component:147; Jaccard:0.16883
 |
Component:207; Jaccard:0.15266
 |
Correlation:-0.054498
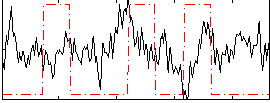 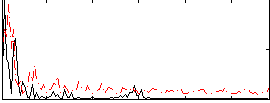 |
Correlation:0.27496
  |
Correlation:0.13866
  |
Correlation:0.13866
  |
Correlation:0.13866
  |
Correlation:0.14858
  |
Correlation:0.027163
  |
Component:207; Jaccard:0.11432
 |
Component:315; Jaccard:0.13157
 |
Component:386; Jaccard:0.149
 |
Component:386; Jaccard:0.15322
 |
Component:091; Jaccard:0.15316
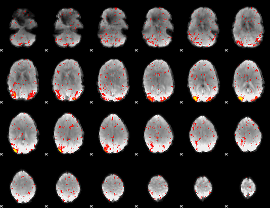 |
Component:091; Jaccard:0.15038
 |
Component:091; Jaccard:0.14518
 |
Correlation:0.027163
  |
Correlation:0.13866
  |
Correlation:-0.039908
  |
Correlation:-0.039908
  |
Correlation:0.23137
  |
Correlation:0.23137
  |
Correlation:0.23137
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
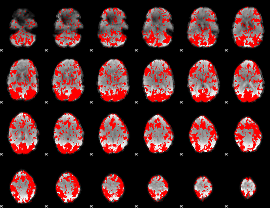 |
GLM binary result thresholded by 1.5
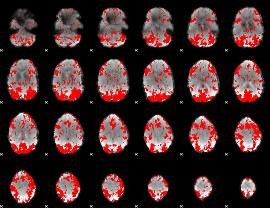 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
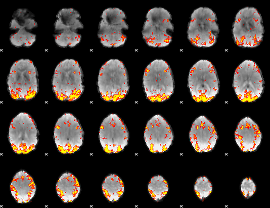 |
GLM Z-score result thresholded by 3.5
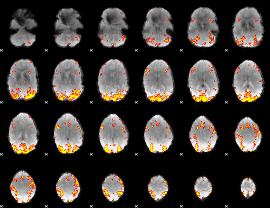 |
GLM Z-score result thresholded by 4
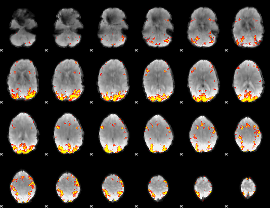 |
Component:079; Jaccard:0.15165
 |
Component:079; Jaccard:0.18622
 |
Component:079; Jaccard:0.22473
 |
Component:079; Jaccard:0.26323
 |
Component:079; Jaccard:0.29787
 |
Component:079; Jaccard:0.32746
 |
Component:079; Jaccard:0.34707
 |
Correlation:0.23358
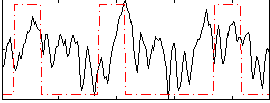 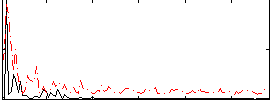 |
Correlation:0.23358
  |
Correlation:0.23358
  |
Correlation:0.23358
  |
Correlation:0.23358
  |
Correlation:0.23358
  |
Correlation:0.23358
  |
Component:218; Jaccard:0.13483
 |
Component:163; Jaccard:0.16216
 |
Component:163; Jaccard:0.19753
 |
Component:163; Jaccard:0.23254
 |
Component:163; Jaccard:0.26472
 |
Component:163; Jaccard:0.29328
 |
Component:163; Jaccard:0.31241
 |
Correlation:-0.016741
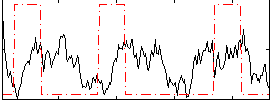 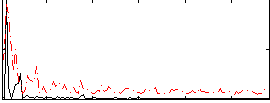 |
Correlation:0.38812
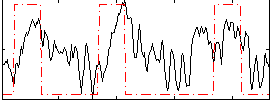 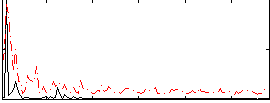 |
Correlation:0.38812
  |
Correlation:0.38812
  |
Correlation:0.38812
  |
Correlation:0.38812
  |
Correlation:0.38812
  |
Component:147; Jaccard:0.13207
 |
Component:218; Jaccard:0.1576
 |
Component:042; Jaccard:0.1859
 |
Component:042; Jaccard:0.21385
 |
Component:042; Jaccard:0.23647
 |
Component:042; Jaccard:0.2526
 |
Component:042; Jaccard:0.25673
 |
Correlation:0.086089
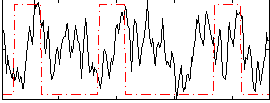 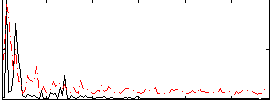 |
Correlation:-0.016741
  |
Correlation:0.04633
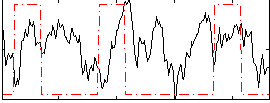  |
Correlation:0.04633
  |
Correlation:0.04633
  |
Correlation:0.04633
  |
Correlation:0.04633
  |
Component:163; Jaccard:0.13066
 |
Component:042; Jaccard:0.15543
 |
Component:218; Jaccard:0.17744
 |
Component:015; Jaccard:0.19631
 |
Component:015; Jaccard:0.22008
 |
Component:015; Jaccard:0.23746
 |
Component:015; Jaccard:0.25136
 |
Correlation:0.38812
  |
Correlation:0.04633
  |
Correlation:-0.016741
  |
Correlation:0.16073
  |
Correlation:0.16073
  |
Correlation:0.16073
  |
Correlation:0.16073
  |
Component:042; Jaccard:0.12772
 |
Component:147; Jaccard:0.15186
 |
Component:015; Jaccard:0.16992
 |
Component:218; Jaccard:0.19516
 |
Component:218; Jaccard:0.20634
 |
Component:218; Jaccard:0.20715
 |
Component:218; Jaccard:0.20182
 |
Correlation:0.04633
  |
Correlation:0.086089
  |
Correlation:0.16073
  |
Correlation:-0.016741
  |
Correlation:-0.016741
  |
Correlation:-0.016741
  |
Correlation:-0.016741
  |
Component:386; Jaccard:0.12054
 |
Component:015; Jaccard:0.14247
 |
Component:147; Jaccard:0.16861
 |
Component:147; Jaccard:0.17996
 |
Component:207; Jaccard:0.19135
 |
Component:207; Jaccard:0.19791
 |
Component:207; Jaccard:0.19742
 |
Correlation:0.2227
 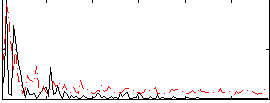 |
Correlation:0.16073
  |
Correlation:0.086089
  |
Correlation:0.086089
  |
Correlation:0.23006
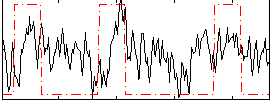 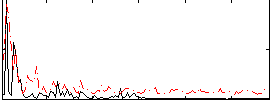 |
Correlation:0.23006
  |
Correlation:0.23006
  |
Component:015; Jaccard:0.11686
 |
Component:386; Jaccard:0.13693
 |
Component:207; Jaccard:0.15828
 |
Component:207; Jaccard:0.1775
 |
Component:147; Jaccard:0.18476
 |
Component:147; Jaccard:0.18282
 |
Component:147; Jaccard:0.17699
 |
Correlation:0.16073
  |
Correlation:0.2227
  |
Correlation:0.23006
  |
Correlation:0.23006
  |
Correlation:0.086089
  |
Correlation:0.086089
  |
Correlation:0.086089
  |
Component:367; Jaccard:0.11508
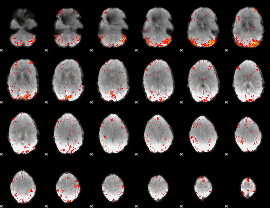 |
Component:207; Jaccard:0.13576
 |
Component:386; Jaccard:0.15054
 |
Component:386; Jaccard:0.16007
 |
Component:153; Jaccard:0.16627
 |
Component:153; Jaccard:0.17068
 |
Component:153; Jaccard:0.16927
 |
Correlation:-0.1728
 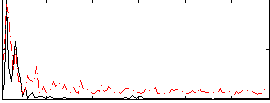 |
Correlation:0.23006
  |
Correlation:0.2227
  |
Correlation:0.2227
  |
Correlation:-0.030283
 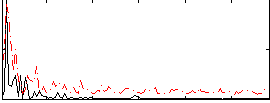 |
Correlation:-0.030283
  |
Correlation:-0.030283
  |
Component:207; Jaccard:0.11455
 |
Component:367; Jaccard:0.12848
 |
Component:153; Jaccard:0.14337
 |
Component:153; Jaccard:0.15706
 |
Component:386; Jaccard:0.16282
 |
Component:315; Jaccard:0.16653
 |
Component:315; Jaccard:0.16893
 |
Correlation:0.23006
  |
Correlation:-0.1728
  |
Correlation:-0.030283
  |
Correlation:-0.030283
  |
Correlation:0.2227
  |
Correlation:0.25501
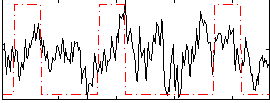 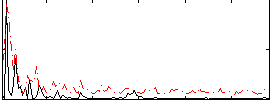 |
Correlation:0.25501
  |
Component:276; Jaccard:0.10723
 |
Component:153; Jaccard:0.12563
 |
Component:367; Jaccard:0.13963
 |
Component:315; Jaccard:0.14948
 |
Component:315; Jaccard:0.15945
 |
Component:386; Jaccard:0.15852
 |
Component:002; Jaccard:0.156
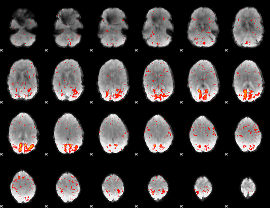 |
Correlation:0.034645
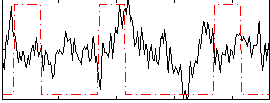 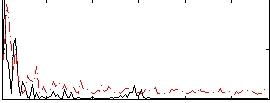 |
Correlation:-0.030283
  |
Correlation:-0.1728
  |
Correlation:0.25501
  |
Correlation:0.25501
  |
Correlation:0.2227
  |
Correlation:0.24231
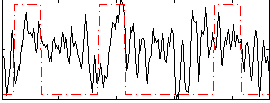 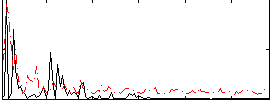 |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
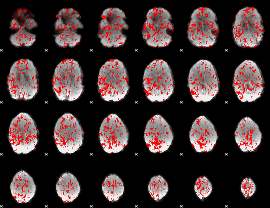 |
GLM binary result thresholded by 1.5
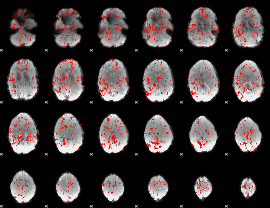 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
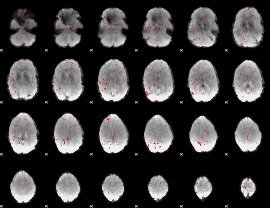 |
GLM binary result thresholded by 3
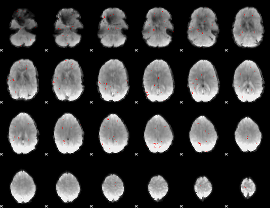 |
GLM binary result thresholded by 3.5
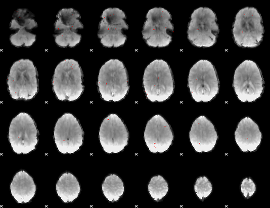 |
GLM binary result thresholded by 4
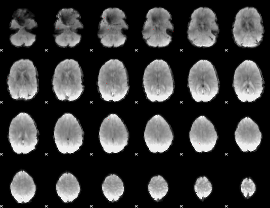 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
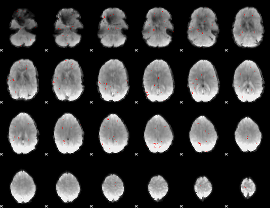 |
GLM Z-score result thresholded by 3.5
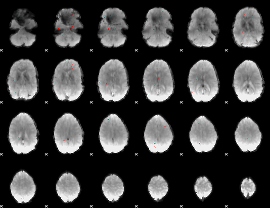 |
GLM Z-score result thresholded by 4
 |
Component:243; Jaccard:0.090423
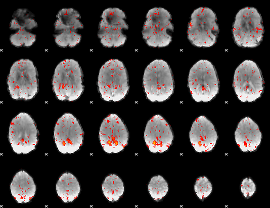 |
Component:243; Jaccard:0.092724
 |
Component:166; Jaccard:0.081603
 |
Component:166; Jaccard:0.05547
 |
Component:166; Jaccard:0.024242
 |
Component:166; Jaccard:0.007833
 |
Component:191; Jaccard:0.001938
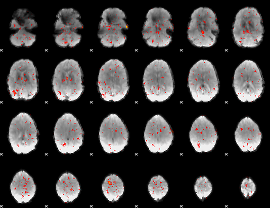 |
Correlation:0.032213
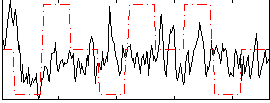 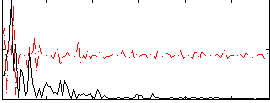 |
Correlation:0.032213
  |
Correlation:0.28632
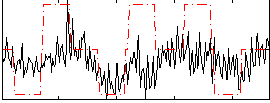 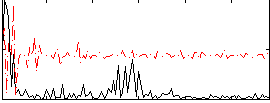 |
Correlation:0.28632
  |
Correlation:0.28632
  |
Correlation:0.28632
  |
Correlation:0.059692
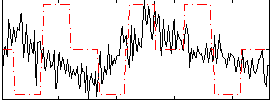 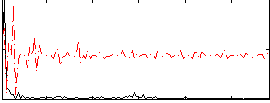 |
Component:260; Jaccard:0.084812
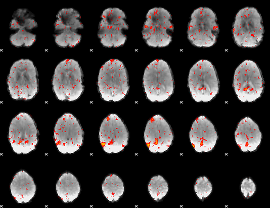 |
Component:166; Jaccard:0.091956
 |
Component:243; Jaccard:0.080211
 |
Component:233; Jaccard:0.046644
 |
Component:233; Jaccard:0.020573
 |
Component:341; Jaccard:0.005583
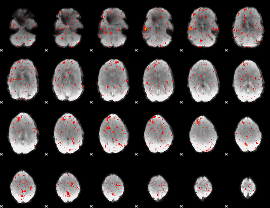 |
Component:351; Jaccard:0.00165
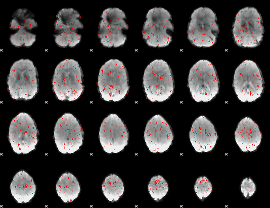 |
Correlation:-0.093405
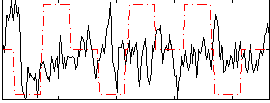 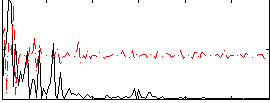 |
Correlation:0.28632
  |
Correlation:0.032213
  |
Correlation:-0.048994
 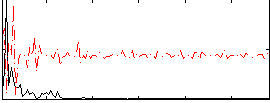 |
Correlation:-0.048994
  |
Correlation:0.23842
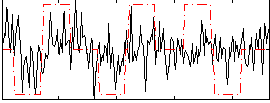 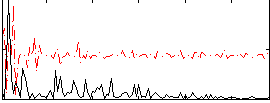 |
Correlation:0.22177
 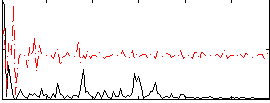 |
Component:341; Jaccard:0.083929
 |
Component:233; Jaccard:0.084646
 |
Component:233; Jaccard:0.076037
 |
Component:243; Jaccard:0.04481
 |
Component:243; Jaccard:0.019369
 |
Component:233; Jaccard:0.005412
 |
Component:144; Jaccard:0.001295
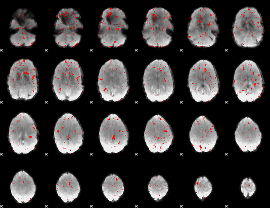 |
Correlation:0.23842
  |
Correlation:-0.048994
  |
Correlation:-0.048994
  |
Correlation:0.032213
  |
Correlation:0.032213
  |
Correlation:-0.048994
  |
Correlation:0.10446
  |
Component:166; Jaccard:0.08228
 |
Component:341; Jaccard:0.083784
 |
Component:260; Jaccard:0.070825
 |
Component:144; Jaccard:0.041258
 |
Component:144; Jaccard:0.018037
 |
Component:265; Jaccard:0.005286
 |
Component:260; Jaccard:0.001142
 |
Correlation:0.28632
  |
Correlation:0.23842
  |
Correlation:-0.093405
  |
Correlation:0.10446
  |
Correlation:0.10446
  |
Correlation:-0.013721
  |
Correlation:-0.093405
  |
Component:233; Jaccard:0.081446
 |
Component:260; Jaccard:0.083452
 |
Component:341; Jaccard:0.068765
 |
Component:341; Jaccard:0.040879
 |
Component:260; Jaccard:0.017266
 |
Component:144; Jaccard:0.005213
 |
Component:033; Jaccard:0.001095
 |
Correlation:-0.048994
  |
Correlation:-0.093405
  |
Correlation:0.23842
  |
Correlation:0.23842
  |
Correlation:-0.093405
  |
Correlation:0.10446
  |
Correlation:0.039436
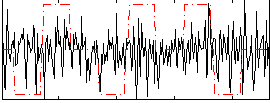 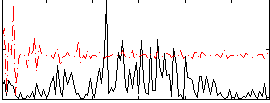 |
Component:294; Jaccard:0.08064
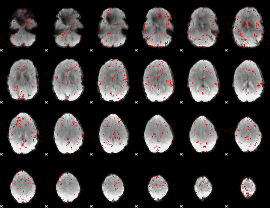 |
Component:294; Jaccard:0.078777
 |
Component:144; Jaccard:0.063613
 |
Component:265; Jaccard:0.040387
 |
Component:265; Jaccard:0.017086
 |
Component:260; Jaccard:0.004609
 |
Component:027; Jaccard:0.001063
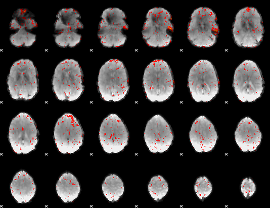 |
Correlation:0.17856
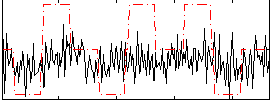 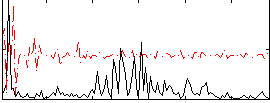 |
Correlation:0.17856
  |
Correlation:0.10446
  |
Correlation:-0.013721
  |
Correlation:-0.013721
  |
Correlation:-0.093405
  |
Correlation:0.024652
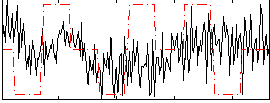 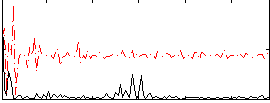 |
Component:265; Jaccard:0.074648
 |
Component:265; Jaccard:0.076005
 |
Component:265; Jaccard:0.062727
 |
Component:260; Jaccard:0.039941
 |
Component:341; Jaccard:0.016503
 |
Component:191; Jaccard:0.00451
 |
Component:334; Jaccard:0.001018
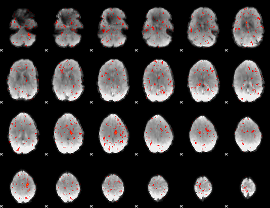 |
Correlation:-0.013721
  |
Correlation:-0.013721
  |
Correlation:-0.013721
  |
Correlation:-0.093405
  |
Correlation:0.23842
  |
Correlation:0.059692
  |
Correlation:0.11837
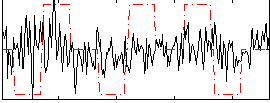 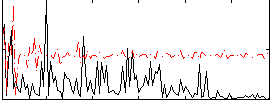 |
Component:264; Jaccard:0.073582
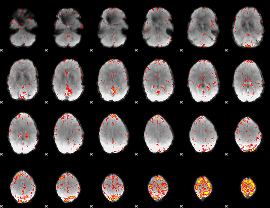 |
Component:143; Jaccard:0.072387
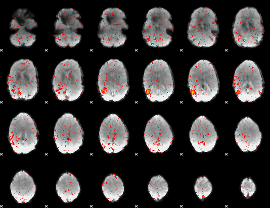 |
Component:143; Jaccard:0.061329
 |
Component:294; Jaccard:0.036856
 |
Component:143; Jaccard:0.014659
 |
Component:243; Jaccard:0.004398
 |
Component:166; Jaccard:0.00087
 |
Correlation:0.064018
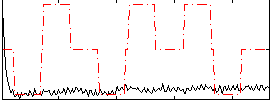 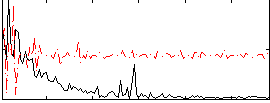 |
Correlation:0.09362
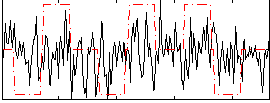 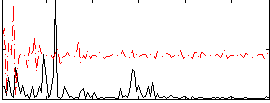 |
Correlation:0.09362
  |
Correlation:0.17856
  |
Correlation:0.09362
  |
Correlation:0.032213
  |
Correlation:0.28632
  |
Component:027; Jaccard:0.07206
 |
Component:144; Jaccard:0.071606
 |
Component:294; Jaccard:0.060194
 |
Component:143; Jaccard:0.036362
 |
Component:191; Jaccard:0.013856
 |
Component:295; Jaccard:0.004381
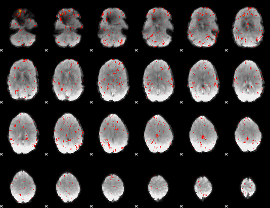 |
Component:243; Jaccard:0.000847
 |
Correlation:0.024652
  |
Correlation:0.10446
  |
Correlation:0.17856
  |
Correlation:0.09362
  |
Correlation:0.059692
  |
Correlation:0.045114
 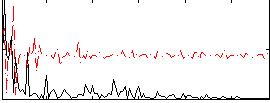 |
Correlation:0.032213
  |
Component:143; Jaccard:0.071881
 |
Component:027; Jaccard:0.068118
 |
Component:027; Jaccard:0.052796
 |
Component:295; Jaccard:0.033149
 |
Component:295; Jaccard:0.013687
 |
Component:351; Jaccard:0.004179
 |
Component:233; Jaccard:0.000828
 |
Correlation:0.09362
  |
Correlation:0.024652
  |
Correlation:0.024652
  |
Correlation:0.045114
  |
Correlation:0.045114
  |
Correlation:0.22177
  |
Correlation:-0.048994
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
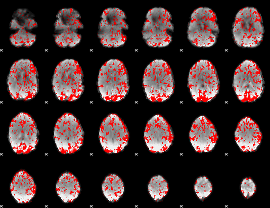 |
GLM binary result thresholded by 1.5
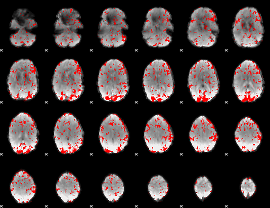 |
GLM binary result thresholded by 2
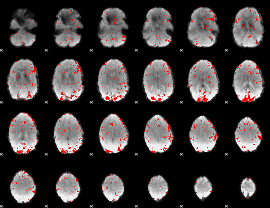 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
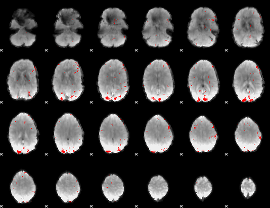 |
GLM binary result thresholded by 3.5
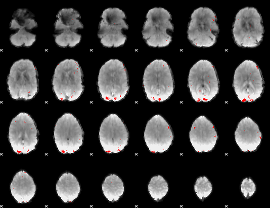 |
GLM binary result thresholded by 4
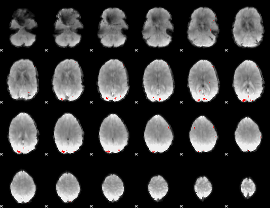 |
GLM Z-score result thresholded by 1
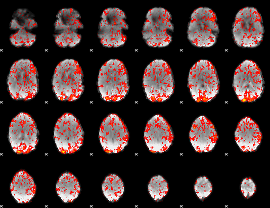 |
GLM Z-score result thresholded by 1.5
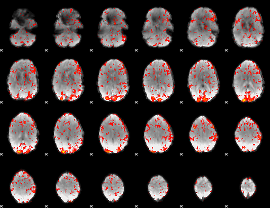 |
GLM Z-score result thresholded by 2
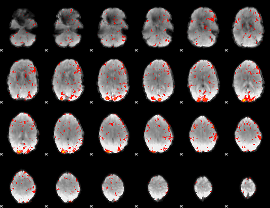 |
GLM Z-score result thresholded by 2.5
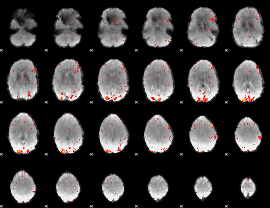 |
GLM Z-score result thresholded by 3
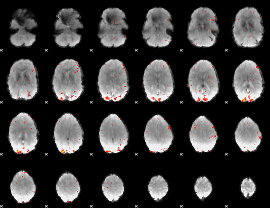 |
GLM Z-score result thresholded by 3.5
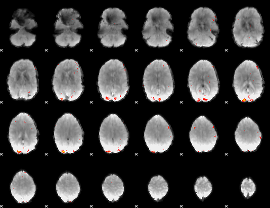 |
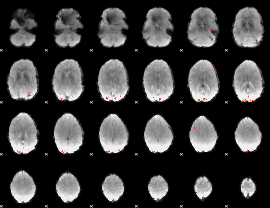
GLM Z-score result thresholded by 4
 |
Component:163; Jaccard:0.12977
 |
Component:163; Jaccard:0.15854
 |
Component:163; Jaccard:0.17179
 |
Component:163; Jaccard:0.15206
 |
Component:163; Jaccard:0.10869
 |
Component:163; Jaccard:0.070414
 |
Component:163; Jaccard:0.043785
 |
Correlation:0.15864
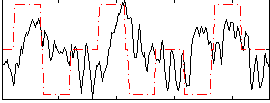 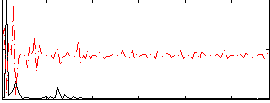 |
Correlation:0.15864
  |
Correlation:0.15864
  |
Correlation:0.15864
  |
Correlation:0.15864
  |
Correlation:0.15864
  |
Correlation:0.15864
  |
Component:079; Jaccard:0.10737
 |
Component:079; Jaccard:0.11754
 |
Component:015; Jaccard:0.11759
 |
Component:015; Jaccard:0.10425
 |
Component:015; Jaccard:0.076432
 |
Component:015; Jaccard:0.051141
 |
Component:015; Jaccard:0.032473
 |
Correlation:0.078673
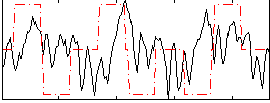 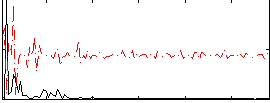 |
Correlation:0.078673
  |
Correlation:-0.0066587
  |
Correlation:-0.0066587
  |
Correlation:-0.0066587
  |
Correlation:-0.0066587
  |
Correlation:-0.0066587
  |
Component:015; Jaccard:0.098956
 |
Component:015; Jaccard:0.11248
 |
Component:079; Jaccard:0.11395
 |
Component:079; Jaccard:0.09439
 |
Component:079; Jaccard:0.065304
 |
Component:079; Jaccard:0.042336
 |
Component:079; Jaccard:0.02583
 |
Correlation:-0.0066587
  |
Correlation:-0.0066587
  |
Correlation:0.078673
  |
Correlation:0.078673
  |
Correlation:0.078673
  |
Correlation:0.078673
  |
Correlation:0.078673
  |
Component:162; Jaccard:0.098274
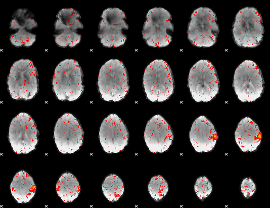 |
Component:207; Jaccard:0.10118
 |
Component:207; Jaccard:0.097902
 |
Component:207; Jaccard:0.075773
 |
Component:207; Jaccard:0.049216
 |
Component:207; Jaccard:0.031764
 |
Component:207; Jaccard:0.020401
 |
Correlation:0.087825
 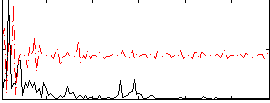 |
Correlation:0.12026
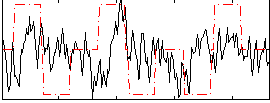 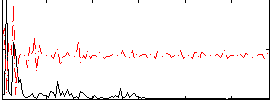 |
Correlation:0.12026
  |
Correlation:0.12026
  |
Correlation:0.12026
  |
Correlation:0.12026
  |
Correlation:0.12026
  |
Component:207; Jaccard:0.091838
 |
Component:162; Jaccard:0.10012
 |
Component:162; Jaccard:0.08669
 |
Component:162; Jaccard:0.062189
 |
Component:380; Jaccard:0.036265
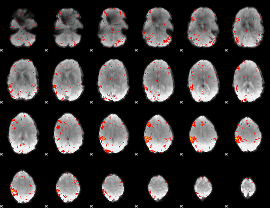 |
Component:380; Jaccard:0.026675
 |
Component:380; Jaccard:0.017475
 |
Correlation:0.12026
  |
Correlation:0.087825
  |
Correlation:0.087825
  |
Correlation:0.087825
  |
Correlation:-0.032431
  |
Correlation:-0.032431
  |
Correlation:-0.032431
  |
Component:386; Jaccard:0.083742
 |
Component:395; Jaccard:0.085476
 |
Component:395; Jaccard:0.081752
 |
Component:017; Jaccard:0.06031
 |
Component:017; Jaccard:0.035011
 |
Component:002; Jaccard:0.022346
 |
Component:002; Jaccard:0.012948
 |
Correlation:0.15565
  |
Correlation:-0.07521
  |
Correlation:-0.07521
  |
Correlation:0.082887
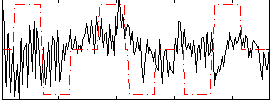 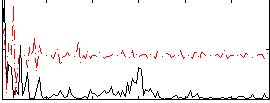 |
Correlation:0.082887
  |
Correlation:0.11522
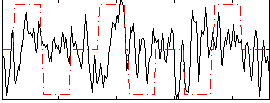 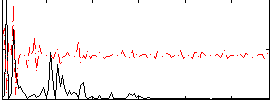 |
Correlation:0.11522
  |
Component:367; Jaccard:0.082922
 |
Component:386; Jaccard:0.085122
 |
Component:017; Jaccard:0.079448
 |
Component:395; Jaccard:0.06004
 |
Component:002; Jaccard:0.035006
 |
Component:386; Jaccard:0.020217
 |
Component:244; Jaccard:0.012653
 |
Correlation:-0.19323
 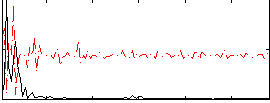 |
Correlation:0.15565
  |
Correlation:0.082887
  |
Correlation:-0.07521
  |
Correlation:0.11522
  |
Correlation:0.15565
  |
Correlation:0.11478
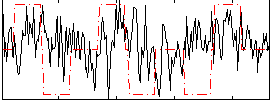 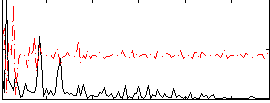 |
Component:380; Jaccard:0.081876
 |
Component:017; Jaccard:0.084828
 |
Component:386; Jaccard:0.07517
 |
Component:386; Jaccard:0.05532
 |
Component:162; Jaccard:0.033305
 |
Component:244; Jaccard:0.018991
 |
Component:386; Jaccard:0.012476
 |
Correlation:-0.032431
  |
Correlation:0.082887
  |
Correlation:0.15565
  |
Correlation:0.15565
  |
Correlation:0.087825
  |
Correlation:0.11478
  |
Correlation:0.15565
  |
Component:117; Jaccard:0.081163
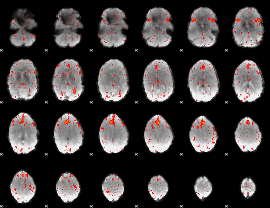 |
Component:380; Jaccard:0.082764
 |
Component:380; Jaccard:0.073249
 |
Component:380; Jaccard:0.053423
 |
Component:386; Jaccard:0.032686
 |
Component:367; Jaccard:0.018591
 |
Component:147; Jaccard:0.011214
 |
Correlation:0.060429
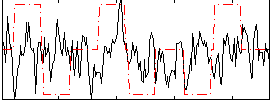  |
Correlation:-0.032431
  |
Correlation:-0.032431
  |
Correlation:-0.032431
  |
Correlation:0.15565
  |
Correlation:-0.19323
  |
Correlation:-0.037036
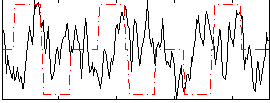 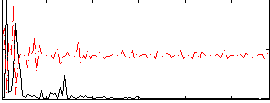 |
Component:017; Jaccard:0.079432
 |
Component:367; Jaccard:0.081286
 |
Component:367; Jaccard:0.070602
 |
Component:002; Jaccard:0.052376
 |
Component:367; Jaccard:0.031796
 |
Component:147; Jaccard:0.017801
 |
Component:099; Jaccard:0.009619
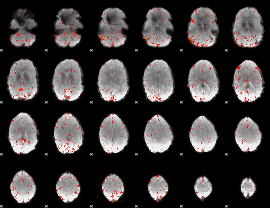 |
Correlation:0.082887
  |
Correlation:-0.19323
  |
Correlation:-0.19323
  |
Correlation:0.11522
  |
Correlation:-0.19323
  |
Correlation:-0.037036
  |
Correlation:0.014606
 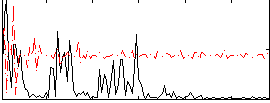 |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
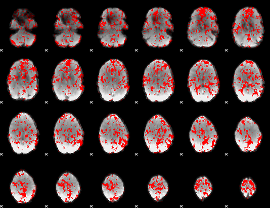 |
GLM binary result thresholded by 1.5
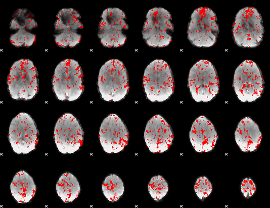 |
GLM binary result thresholded by 2
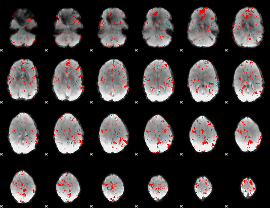 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
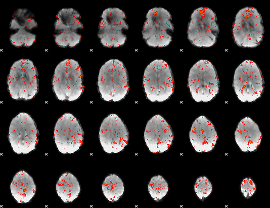 |
GLM Z-score result thresholded by 2.5
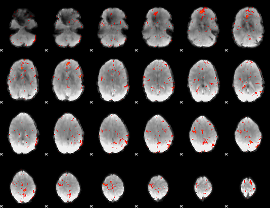 |
GLM Z-score result thresholded by 3
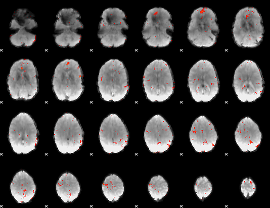 |
GLM Z-score result thresholded by 3.5
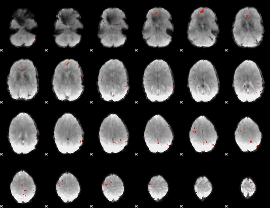 |
GLM Z-score result thresholded by 4
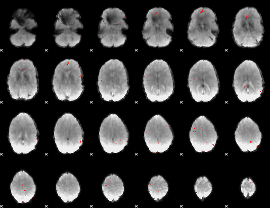 |
Component:264; Jaccard:0.11184
 |
Component:233; Jaccard:0.10543
 |
Component:233; Jaccard:0.10979
 |
Component:233; Jaccard:0.10316
 |
Component:233; Jaccard:0.077798
 |
Component:233; Jaccard:0.047115
 |
Component:233; Jaccard:0.023374
 |
Correlation:0.055692
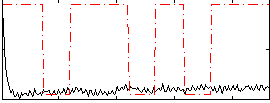 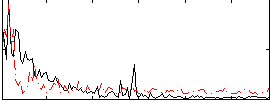 |
Correlation:0.20824
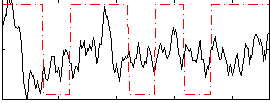 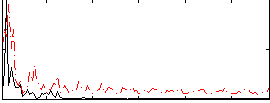 |
Correlation:0.20824
  |
Correlation:0.20824
  |
Correlation:0.20824
  |
Correlation:0.20824
  |
Correlation:0.20824
  |
Component:233; Jaccard:0.092331
 |
Component:264; Jaccard:0.10126
 |
Component:328; Jaccard:0.097103
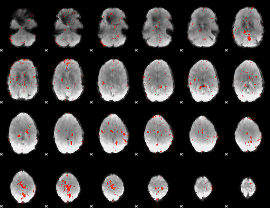 |
Component:328; Jaccard:0.091262
 |
Component:328; Jaccard:0.069153
 |
Component:122; Jaccard:0.041955
 |
Component:190; Jaccard:0.023355
 |
Correlation:0.20824
  |
Correlation:0.055692
  |
Correlation:0.093546
 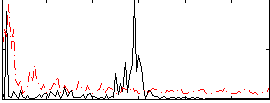 |
Correlation:0.093546
  |
Correlation:0.093546
  |
Correlation:0.29311
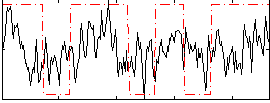 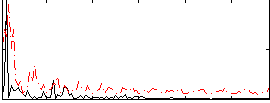 |
Correlation:0.25188
  |
Component:064; Jaccard:0.088247
 |
Component:064; Jaccard:0.093645
 |
Component:122; Jaccard:0.093188
 |
Component:122; Jaccard:0.087901
 |
Component:122; Jaccard:0.067017
 |
Component:190; Jaccard:0.040506
 |
Component:122; Jaccard:0.022921
 |
Correlation:0.10133
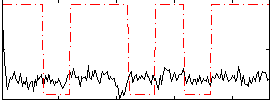 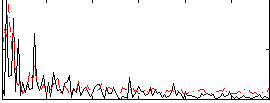 |
Correlation:0.10133
  |
Correlation:0.29311
  |
Correlation:0.29311
  |
Correlation:0.29311
  |
Correlation:0.25188
  |
Correlation:0.29311
  |
Component:036; Jaccard:0.086456
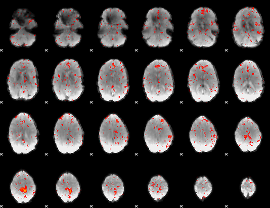 |
Component:036; Jaccard:0.091907
 |
Component:190; Jaccard:0.092839
 |
Component:190; Jaccard:0.084131
 |
Component:190; Jaccard:0.06457
 |
Component:328; Jaccard:0.039405
 |
Component:328; Jaccard:0.018155
 |
Correlation:0.08216
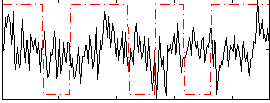 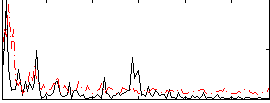 |
Correlation:0.08216
  |
Correlation:0.25188
  |
Correlation:0.25188
  |
Correlation:0.25188
  |
Correlation:0.093546
  |
Correlation:0.093546
  |
Component:213; Jaccard:0.083929
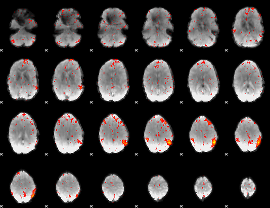 |
Component:213; Jaccard:0.088972
 |
Component:036; Jaccard:0.086913
 |
Component:036; Jaccard:0.068384
 |
Component:340; Jaccard:0.044845
 |
Component:083; Jaccard:0.025281
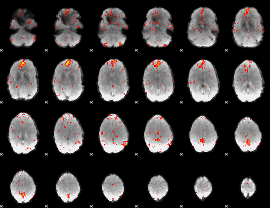 |
Component:083; Jaccard:0.013087
 |
Correlation:0.058509
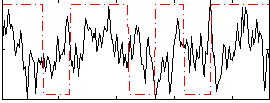  |
Correlation:0.058509
  |
Correlation:0.08216
  |
Correlation:0.08216
  |
Correlation:0.091455
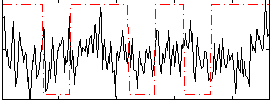 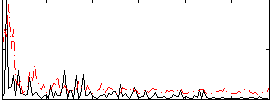 |
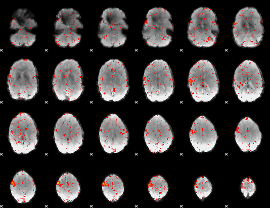
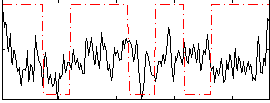
Correlation:0.23315
  |
Correlation:0.23315
  |
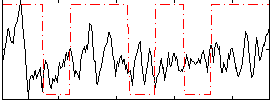
Component:263; Jaccard:0.08139
 |
Component:190; Jaccard:0.087577
 |
Component:064; Jaccard:0.086047
 |
Component:213; Jaccard:0.067969
 |
Component:036; Jaccard:0.044459
 |
Component:036; Jaccard:0.022215
 |
Component:036; Jaccard:0.011167
 |
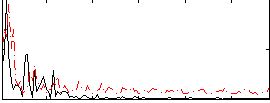
Correlation:0.13792
 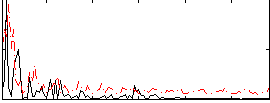 |
Correlation:0.25188
  |
Correlation:0.10133
  |
Correlation:0.058509
  |
Correlation:0.08216
  |
Correlation:0.08216
  |
Correlation:0.08216
  |
Component:342; Jaccard:0.080551
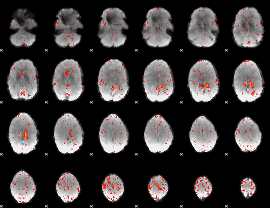 |
Component:328; Jaccard:0.085686
 |
Component:213; Jaccard:0.082084
 |
Component:340; Jaccard:0.066568
 |
Component:083; Jaccard:0.042099
 |
Component:249; Jaccard:0.021725
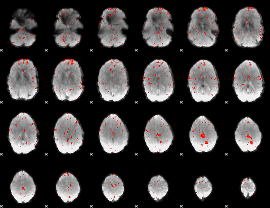 |
Component:249; Jaccard:0.011092
 |
Correlation:0.1197
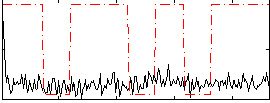  |
Correlation:0.093546
  |
Correlation:0.058509
  |
Correlation:0.091455
  |
Correlation:0.23315
  |
Correlation:0.072225
  |
Correlation:0.072225
  |
Component:340; Jaccard:0.080536
 |
Component:340; Jaccard:0.085636
 |
Component:340; Jaccard:0.079432
 |
Component:064; Jaccard:0.062331
 |
Component:213; Jaccard:0.040319
 |
Component:340; Jaccard:0.020525
 |
Component:350; Jaccard:0.01048
 |
Correlation:0.091455
  |
Correlation:0.091455
  |
Correlation:0.091455
  |
Correlation:0.10133
  |
Correlation:0.058509
  |
Correlation:0.091455
  |
Correlation:0.19619
 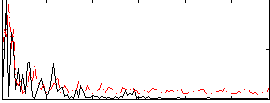 |
Component:190; Jaccard:0.077353
 |
Component:122; Jaccard:0.084219
 |
Component:264; Jaccard:0.074636
 |
Component:083; Jaccard:0.061193
 |
Component:056; Jaccard:0.037935
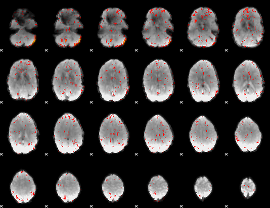 |
Component:296; Jaccard:0.020519
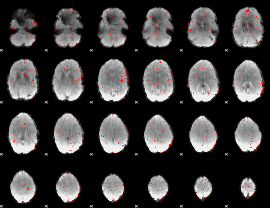 |
Component:296; Jaccard:0.009984
 |
Correlation:0.25188
  |
Correlation:0.29311
  |
Correlation:0.055692
  |
Correlation:0.23315
  |
Correlation:0.22381
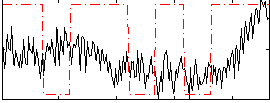 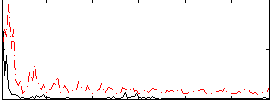 |
Correlation:0.222
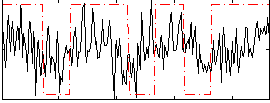  |
Correlation:0.222
  |
Component:350; Jaccard:0.076591
 |
Component:263; Jaccard:0.079742
 |
Component:350; Jaccard:0.074401
 |
Component:350; Jaccard:0.059266
 |
Component:064; Jaccard:0.037363
 |
Component:350; Jaccard:0.020144
 |
Component:340; Jaccard:0.009792
 |
Correlation:0.19619
  |
Correlation:0.13792
  |
Correlation:0.19619
  |
Correlation:0.19619
  |
Correlation:0.10133
  |
Correlation:0.19619
  |
Correlation:0.091455
  |
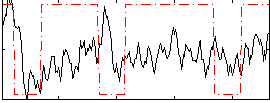
Cope: 06:neg_REL BACK
|
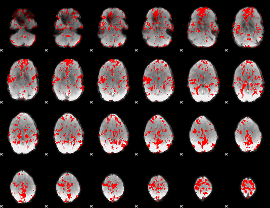
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
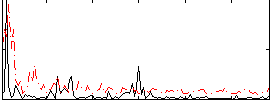
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
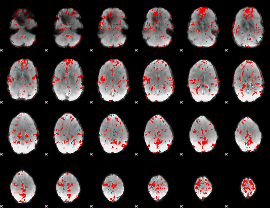 |
GLM binary result thresholded by 2.5
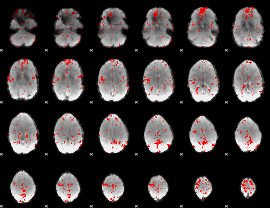 |
GLM binary result thresholded by 3
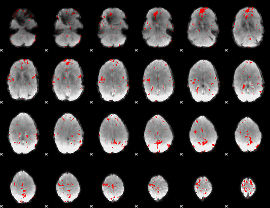 |
GLM binary result thresholded by 3.5
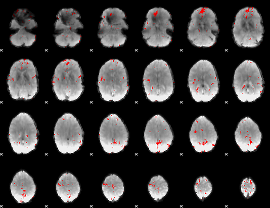 |
GLM binary result thresholded by 4
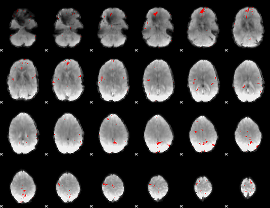 |
GLM Z-score result thresholded by 1
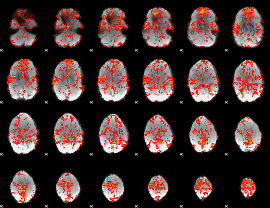 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
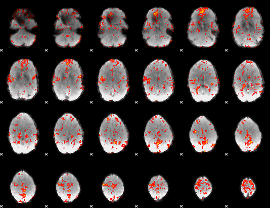 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
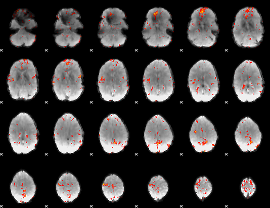 |
GLM Z-score result thresholded by 3.5
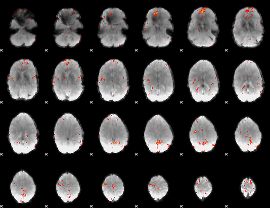 |
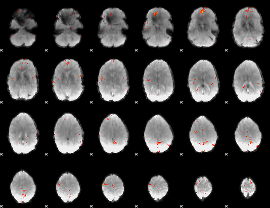
GLM Z-score result thresholded by 4
 |
Component:264; Jaccard:0.13088
 |
Component:233; Jaccard:0.13897
 |
Component:233; Jaccard:0.16127
 |
Component:233; Jaccard:0.17719
 |
Component:233; Jaccard:0.17226
 |
Component:233; Jaccard:0.13906
 |
Component:233; Jaccard:0.10179
 |
Correlation:0.1637
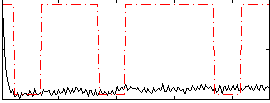 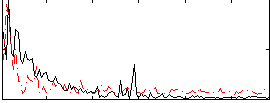 |
Correlation:0.12557
 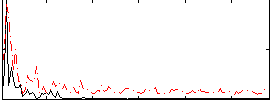 |
Correlation:0.12557
  |
Correlation:0.12557
  |
Correlation:0.12557
  |
Correlation:0.12557
  |
Correlation:0.12557
  |
Component:233; Jaccard:0.11363
 |
Component:264; Jaccard:0.13811
 |
Component:264; Jaccard:0.13496
 |
Component:264; Jaccard:0.11243
 |
Component:190; Jaccard:0.094535
 |
Component:190; Jaccard:0.077884
 |
Component:190; Jaccard:0.059436
 |
Correlation:0.12557
  |
Correlation:0.1637
  |
Correlation:0.1637
  |
Correlation:0.1637
  |
Correlation:-0.067791
  |
Correlation:-0.067791
  |
Correlation:-0.067791
  |
Component:064; Jaccard:0.095652
 |
Component:064; Jaccard:0.10793
 |
Component:064; Jaccard:0.11586
 |
Component:064; Jaccard:0.1118
 |
Component:064; Jaccard:0.091876
 |
Component:122; Jaccard:0.075982
 |
Component:122; Jaccard:0.057513
 |
Correlation:0.15759
 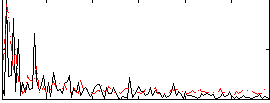 |
Correlation:0.15759
  |
Correlation:0.15759
  |
Correlation:0.15759
  |
Correlation:0.15759
  |
Correlation:0.016931
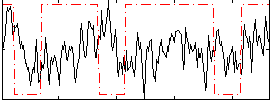  |
Correlation:0.016931
  |
Component:166; Jaccard:0.085295
 |
Component:166; Jaccard:0.096035
 |
Component:166; Jaccard:0.10342
 |
Component:166; Jaccard:0.10074
 |
Component:122; Jaccard:0.091709
 |
Component:328; Jaccard:0.06891
 |
Component:328; Jaccard:0.050594
 |
Correlation:0.37341
  |
Correlation:0.37341
  |
Correlation:0.37341
  |
Correlation:0.37341
  |
Correlation:0.016931
  |
Correlation:-0.052857
  |
Correlation:-0.052857
  |
Component:340; Jaccard:0.082135
 |
Component:190; Jaccard:0.088382
 |
Component:190; Jaccard:0.096736
 |
Component:190; Jaccard:0.099904
 |
Component:166; Jaccard:0.084351
 |
Component:083; Jaccard:0.059117
 |
Component:083; Jaccard:0.042568
 |
Correlation:0.16168
  |
Correlation:-0.067791
  |
Correlation:-0.067791
  |
Correlation:-0.067791
  |
Correlation:0.37341
  |
Correlation:-0.11909
  |
Correlation:-0.11909
  |
Component:342; Jaccard:0.081276
 |
Component:340; Jaccard:0.088265
 |
Component:083; Jaccard:0.090249
 |
Component:122; Jaccard:0.092381
 |
Component:328; Jaccard:0.079709
 |
Component:166; Jaccard:0.058461
 |
Component:249; Jaccard:0.039716
 |
Correlation:0.044832
  |
Correlation:0.16168
  |
Correlation:-0.11909
  |
Correlation:0.016931
  |
Correlation:-0.052857
  |
Correlation:0.37341
  |
Correlation:-0.027246
  |
Component:263; Jaccard:0.080878
 |
Component:083; Jaccard:0.08599
 |
Component:340; Jaccard:0.090103
 |
Component:083; Jaccard:0.087641
 |
Component:264; Jaccard:0.076765
 |
Component:064; Jaccard:0.058444
 |
Component:350; Jaccard:0.038217
 |
Correlation:0.10407
  |
Correlation:-0.11909
  |
Correlation:0.16168
  |
Correlation:-0.11909
  |
Correlation:0.1637
  |
Correlation:0.15759
  |
Correlation:0.14131
  |
Component:389; Jaccard:0.079743
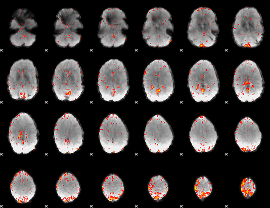 |
Component:350; Jaccard:0.084793
 |
Component:350; Jaccard:0.087636
 |
Component:350; Jaccard:0.084755
 |
Component:083; Jaccard:0.075732
 |
Component:249; Jaccard:0.056714
 |
Component:166; Jaccard:0.033353
 |
Correlation:0.12082
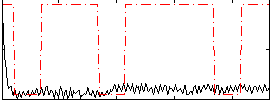 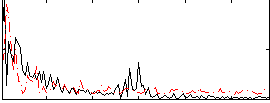 |
Correlation:0.14131
  |
Correlation:0.14131
  |
Correlation:0.14131
  |
Correlation:-0.11909
  |
Correlation:-0.027246
  |
Correlation:0.37341
  |
Component:350; Jaccard:0.079518
 |
Component:342; Jaccard:0.082941
 |
Component:122; Jaccard:0.08694
 |
Component:249; Jaccard:0.08329
 |
Component:249; Jaccard:0.074446
 |
Component:350; Jaccard:0.054262
 |
Component:340; Jaccard:0.032755
 |
Correlation:0.14131
  |
Correlation:0.044832
  |
Correlation:0.016931
  |
Correlation:-0.027246
  |
Correlation:-0.027246
  |
Correlation:0.14131
  |
Correlation:0.16168
  |
Component:265; Jaccard:0.078998
 |
Component:265; Jaccard:0.082503
 |
Component:249; Jaccard:0.085378
 |
Component:328; Jaccard:0.083228
 |
Component:350; Jaccard:0.073008
 |
Component:340; Jaccard:0.051155
 |
Component:064; Jaccard:0.031898
 |
Correlation:0.070846
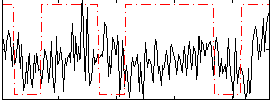  |
Correlation:0.070846
  |
Correlation:-0.027246
  |
Correlation:-0.052857
  |
Correlation:0.14131
  |
Correlation:0.16168
  |
Correlation:0.15759
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































