Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
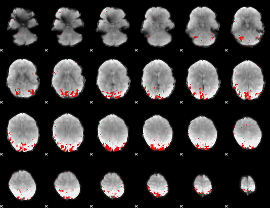
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
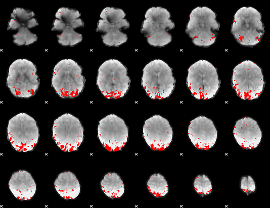 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
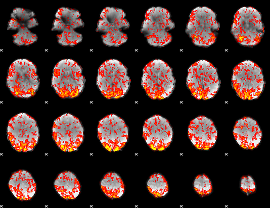 |
GLM Z-score result thresholded by 1.5
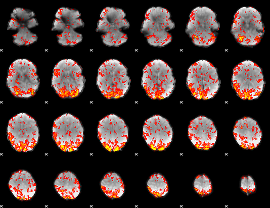 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:327; Jaccard:0.14093
 |
Component:327; Jaccard:0.19303
 |
Component:327; Jaccard:0.25718
 |
Component:327; Jaccard:0.31542
 |
Component:327; Jaccard:0.35269
 |
Component:327; Jaccard:0.34782
 |
Component:327; Jaccard:0.32248
 |
Correlation:0.11945
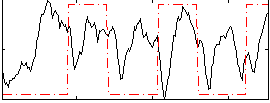 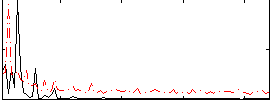 |
Correlation:0.11945
  |
Correlation:0.11945
  |
Correlation:0.11945
  |
Correlation:0.11945
  |
Correlation:0.11945
  |
Correlation:0.11945
  |
Component:206; Jaccard:0.13683
 |
Component:210; Jaccard:0.17597
 |
Component:210; Jaccard:0.22045
 |
Component:210; Jaccard:0.25329
 |
Component:210; Jaccard:0.26492
 |
Component:210; Jaccard:0.25451
 |
Component:210; Jaccard:0.22348
 |
Correlation:0.19549
 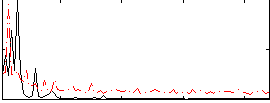 |
Correlation:0.23329
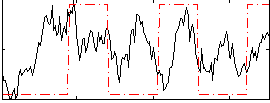  |
Correlation:0.23329
  |
Correlation:0.23329
  |
Correlation:0.23329
  |
Correlation:0.23329
  |
Correlation:0.23329
  |
Component:210; Jaccard:0.13459
 |
Component:206; Jaccard:0.16734
 |
Component:206; Jaccard:0.19143
 |
Component:206; Jaccard:0.19979
 |
Component:131; Jaccard:0.20586
 |
Component:131; Jaccard:0.19531
 |
Component:131; Jaccard:0.16584
 |
Correlation:0.23329
  |
Correlation:0.19549
  |
Correlation:0.19549
  |
Correlation:0.19549
  |
Correlation:-0.098693
  |
Correlation:-0.098693
  |
Correlation:-0.098693
  |
Component:241; Jaccard:0.13241
 |
Component:241; Jaccard:0.15704
 |
Component:131; Jaccard:0.1777
 |
Component:131; Jaccard:0.19978
 |
Component:259; Jaccard:0.1905
 |
Component:259; Jaccard:0.17726
 |
Component:259; Jaccard:0.15496
 |
Correlation:0.030639
 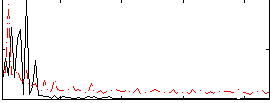 |
Correlation:0.030639
  |
Correlation:-0.098693
  |
Correlation:-0.098693
  |
Correlation:0.13061
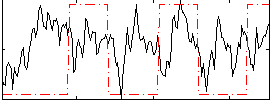  |
Correlation:0.13061
  |
Correlation:0.13061
  |
Component:337; Jaccard:0.12266
 |
Component:131; Jaccard:0.14791
 |
Component:241; Jaccard:0.17407
 |
Component:259; Jaccard:0.18407
 |
Component:206; Jaccard:0.18774
 |
Component:206; Jaccard:0.1653
 |
Component:206; Jaccard:0.13509
 |
Correlation:0.63665
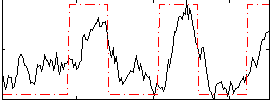 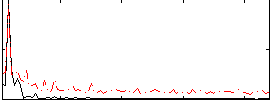 |
Correlation:-0.098693
  |
Correlation:0.030639
  |
Correlation:0.13061
  |
Correlation:0.19549
  |
Correlation:0.19549
  |
Correlation:0.19549
  |
Component:131; Jaccard:0.11775
 |
Component:337; Jaccard:0.14437
 |
Component:259; Jaccard:0.1631
 |
Component:337; Jaccard:0.17335
 |
Component:337; Jaccard:0.17085
 |
Component:337; Jaccard:0.1523
 |
Component:337; Jaccard:0.13179
 |
Correlation:-0.098693
  |
Correlation:0.63665
  |
Correlation:0.13061
  |
Correlation:0.63665
  |
Correlation:0.63665
  |
Correlation:0.63665
  |
Correlation:0.63665
  |
Component:116; Jaccard:0.10898
 |
Component:259; Jaccard:0.13265
 |
Component:337; Jaccard:0.16273
 |
Component:241; Jaccard:0.17082
 |
Component:116; Jaccard:0.1524
 |
Component:116; Jaccard:0.13701
 |
Component:116; Jaccard:0.11376
 |
Correlation:0.26598
  |
Correlation:0.13061
  |
Correlation:0.63665
  |
Correlation:0.030639
  |
Correlation:0.26598
  |
Correlation:0.26598
  |
Correlation:0.26598
  |
Component:262; Jaccard:0.10566
 |
Component:116; Jaccard:0.13164
 |
Component:116; Jaccard:0.15181
 |
Component:116; Jaccard:0.15777
 |
Component:241; Jaccard:0.15045
 |
Component:241; Jaccard:0.12493
 |
Component:241; Jaccard:0.097844
 |
Correlation:0.16516
  |
Correlation:0.26598
  |
Correlation:0.26598
  |
Correlation:0.26598
  |
Correlation:0.030639
  |
Correlation:0.030639
  |
Correlation:0.030639
  |
Component:259; Jaccard:0.10457
 |
Component:262; Jaccard:0.12485
 |
Component:262; Jaccard:0.13844
 |
Component:262; Jaccard:0.14125
 |
Component:262; Jaccard:0.13164
 |
Component:262; Jaccard:0.11359
 |
Component:262; Jaccard:0.094151
 |
Correlation:0.13061
  |
Correlation:0.16516
  |
Correlation:0.16516
  |
Correlation:0.16516
  |
Correlation:0.16516
  |
Correlation:0.16516
  |
Correlation:0.16516
  |
Component:054; Jaccard:0.095178
 |
Component:054; Jaccard:0.10527
 |
Component:052; Jaccard:0.11534
 |
Component:052; Jaccard:0.11708
 |
Component:052; Jaccard:0.10719
 |
Component:052; Jaccard:0.089139
 |
Component:054; Jaccard:0.071798
 |
Correlation:0.39741
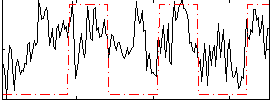 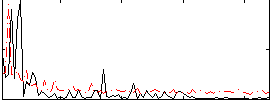 |
Correlation:0.39741
  |
Correlation:0.31643
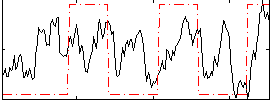 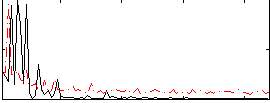 |
Correlation:0.31643
  |
Correlation:0.31643
  |
Correlation:0.31643
  |
Correlation:0.39741
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
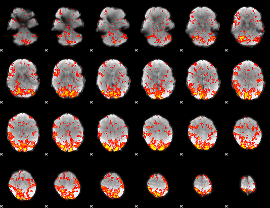 |
GLM Z-score result thresholded by 2
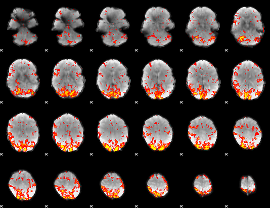 |
GLM Z-score result thresholded by 2.5
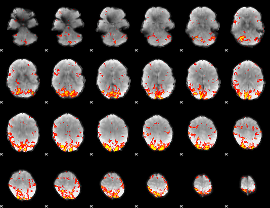 |
GLM Z-score result thresholded by 3
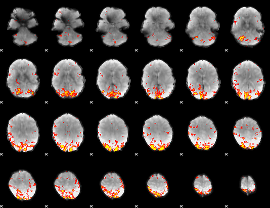 |
GLM Z-score result thresholded by 3.5
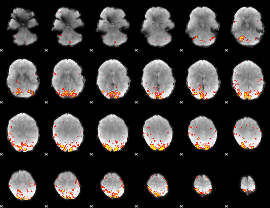 |
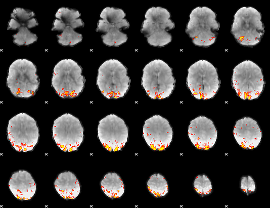
GLM Z-score result thresholded by 4
 |
Component:206; Jaccard:0.14782
 |
Component:327; Jaccard:0.19655
 |
Component:327; Jaccard:0.25315
 |
Component:131; Jaccard:0.30493
 |
Component:131; Jaccard:0.33548
 |
Component:327; Jaccard:0.34433
 |
Component:327; Jaccard:0.32843
 |
Correlation:0.06603
  |
Correlation:0.099476
  |
Correlation:0.099476
  |
Correlation:0.34884
  |
Correlation:0.34884
  |
Correlation:0.099476
  |
Correlation:0.099476
  |
Component:327; Jaccard:0.14715
 |
Component:131; Jaccard:0.19534
 |
Component:131; Jaccard:0.2519
 |
Component:327; Jaccard:0.30344
 |
Component:327; Jaccard:0.33348
 |
Component:131; Jaccard:0.33548
 |
Component:131; Jaccard:0.31477
 |
Correlation:0.099476
  |
Correlation:0.34884
  |
Correlation:0.34884
  |
Correlation:0.099476
  |
Correlation:0.099476
  |
Correlation:0.34884
  |
Correlation:0.34884
  |
Component:241; Jaccard:0.14682
 |
Component:210; Jaccard:0.18775
 |
Component:210; Jaccard:0.23576
 |
Component:210; Jaccard:0.27359
 |
Component:210; Jaccard:0.29286
 |
Component:210; Jaccard:0.28896
 |
Component:210; Jaccard:0.26876
 |
Correlation:0.11369
  |
Correlation:0.064808
  |
Correlation:0.064808
  |
Correlation:0.064808
  |
Correlation:0.064808
  |
Correlation:0.064808
  |
Correlation:0.064808
  |
Component:131; Jaccard:0.14626
 |
Component:206; Jaccard:0.17971
 |
Component:206; Jaccard:0.20538
 |
Component:206; Jaccard:0.21424
 |
Component:206; Jaccard:0.20604
 |
Component:259; Jaccard:0.18763
 |
Component:259; Jaccard:0.17299
 |
Correlation:0.34884
  |
Correlation:0.06603
  |
Correlation:0.06603
  |
Correlation:0.06603
  |
Correlation:0.06603
  |
Correlation:0.074375
  |
Correlation:0.074375
  |
Component:210; Jaccard:0.14492
 |
Component:241; Jaccard:0.17414
 |
Component:241; Jaccard:0.19283
 |
Component:241; Jaccard:0.1927
 |
Component:259; Jaccard:0.19402
 |
Component:206; Jaccard:0.18531
 |
Component:206; Jaccard:0.15574
 |
Correlation:0.064808
  |
Correlation:0.11369
  |
Correlation:0.11369
  |
Correlation:0.11369
  |
Correlation:0.074375
  |
Correlation:0.06603
  |
Correlation:0.06603
  |
Component:262; Jaccard:0.11275
 |
Component:259; Jaccard:0.13741
 |
Component:259; Jaccard:0.16388
 |
Component:259; Jaccard:0.18527
 |
Component:241; Jaccard:0.17765
 |
Component:241; Jaccard:0.15402
 |
Component:241; Jaccard:0.1277
 |
Correlation:0.084101
  |
Correlation:0.074375
  |
Correlation:0.074375
  |
Correlation:0.074375
  |
Correlation:0.11369
  |
Correlation:0.11369
  |
Correlation:0.11369
  |
Component:259; Jaccard:0.11006
 |
Component:262; Jaccard:0.13271
 |
Component:262; Jaccard:0.14828
 |
Component:116; Jaccard:0.15318
 |
Component:116; Jaccard:0.14877
 |
Component:116; Jaccard:0.13689
 |
Component:116; Jaccard:0.11906
 |
Correlation:0.074375
  |
Correlation:0.084101
  |
Correlation:0.084101
  |
Correlation:-0.12777
  |
Correlation:-0.12777
  |
Correlation:-0.12777
  |
Correlation:-0.12777
  |
Component:116; Jaccard:0.10754
 |
Component:116; Jaccard:0.1286
 |
Component:116; Jaccard:0.14533
 |
Component:262; Jaccard:0.15156
 |
Component:262; Jaccard:0.14559
 |
Component:262; Jaccard:0.13255
 |
Component:262; Jaccard:0.11471
 |
Correlation:-0.12777
  |
Correlation:-0.12777
  |
Correlation:-0.12777
  |
Correlation:0.084101
  |
Correlation:0.084101
  |
Correlation:0.084101
  |
Correlation:0.084101
  |
Component:227; Jaccard:0.10162
 |
Component:227; Jaccard:0.11688
 |
Component:227; Jaccard:0.12615
 |
Component:227; Jaccard:0.12742
 |
Component:207; Jaccard:0.11553
 |
Component:207; Jaccard:0.10077
 |
Component:207; Jaccard:0.084736
 |
Correlation:0.20679
  |
Correlation:0.20679
  |
Correlation:0.20679
  |
Correlation:0.20679
  |
Correlation:0.19067
  |
Correlation:0.19067
  |
Correlation:0.19067
  |
Component:207; Jaccard:0.093032
 |
Component:207; Jaccard:0.10888
 |
Component:207; Jaccard:0.1185
 |
Component:207; Jaccard:0.12105
 |
Component:227; Jaccard:0.11511
 |
Component:227; Jaccard:0.09835
 |
Component:227; Jaccard:0.081844
 |
Correlation:0.19067
  |
Correlation:0.19067
  |
Correlation:0.19067
  |
Correlation:0.19067
  |
Correlation:0.20679
  |
Correlation:0.20679
  |
Correlation:0.20679
  |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
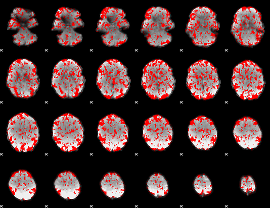 |
GLM binary result thresholded by 1.5
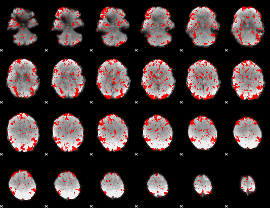 |
GLM binary result thresholded by 2
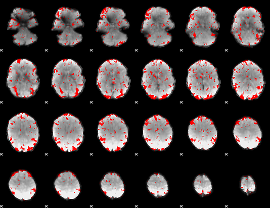 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
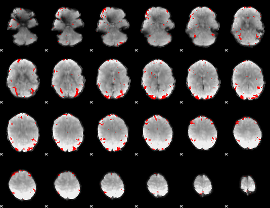 |
GLM binary result thresholded by 3.5
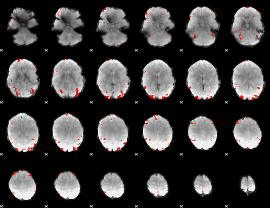 |
GLM binary result thresholded by 4
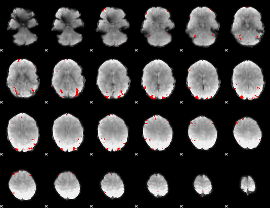 |
GLM Z-score result thresholded by 1
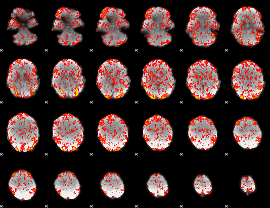 |
GLM Z-score result thresholded by 1.5
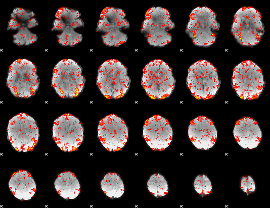 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:337; Jaccard:0.15249
 |
Component:337; Jaccard:0.20379
 |
Component:337; Jaccard:0.24961
 |
Component:337; Jaccard:0.26631
 |
Component:337; Jaccard:0.23954
 |
Component:337; Jaccard:0.1958
 |
Component:337; Jaccard:0.15071
 |
Correlation:0.60881
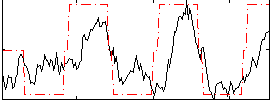  |
Correlation:0.60881
  |
Correlation:0.60881
  |
Correlation:0.60881
  |
Correlation:0.60881
  |
Correlation:0.60881
  |
Correlation:0.60881
  |
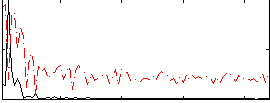
Component:135; Jaccard:0.11267
 |
Component:135; Jaccard:0.13729
 |
Component:135; Jaccard:0.15143
 |
Component:135; Jaccard:0.15384
 |
Component:135; Jaccard:0.144
 |
Component:135; Jaccard:0.12224
 |
Component:135; Jaccard:0.098146
 |
Correlation:0.44838
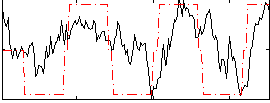  |
Correlation:0.44838
  |
Correlation:0.44838
  |
Correlation:0.44838
  |
Correlation:0.44838
  |
Correlation:0.44838
  |
Correlation:0.44838
  |
Component:319; Jaccard:0.10436
 |
Component:319; Jaccard:0.12831
 |
Component:319; Jaccard:0.14879
 |
Component:319; Jaccard:0.15067
 |
Component:319; Jaccard:0.13008
 |
Component:319; Jaccard:0.09481
 |
Component:319; Jaccard:0.062153
 |
Correlation:0.42799
  |
Correlation:0.42799
  |
Correlation:0.42799
  |
Correlation:0.42799
  |
Correlation:0.42799
  |
Correlation:0.42799
  |
Correlation:0.42799
  |
Component:349; Jaccard:0.10249
 |
Component:349; Jaccard:0.11909
 |
Component:349; Jaccard:0.12921
 |
Component:349; Jaccard:0.12343
 |
Component:349; Jaccard:0.10289
 |
Component:349; Jaccard:0.07804
 |
Component:326; Jaccard:0.054782
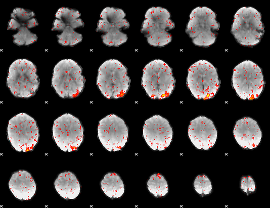 |
Correlation:0.32316
 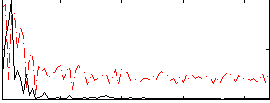 |
Correlation:0.32316
  |
Correlation:0.32316
  |
Correlation:0.32316
  |
Correlation:0.32316
  |
Correlation:0.32316
  |
Correlation:0.34971
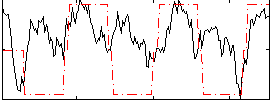 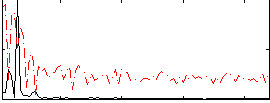 |
Component:017; Jaccard:0.096817
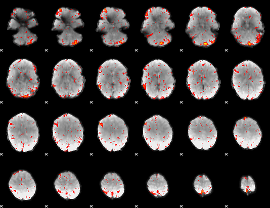 |
Component:017; Jaccard:0.10777
 |
Component:017; Jaccard:0.10927
 |
Component:017; Jaccard:0.10061
 |
Component:017; Jaccard:0.083407
 |
Component:326; Jaccard:0.067247
 |
Component:349; Jaccard:0.054217
 |
Correlation:0.41174
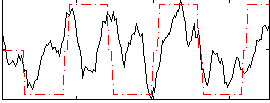 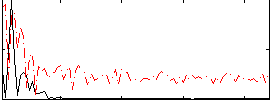 |
Correlation:0.41174
  |
Correlation:0.41174
  |
Correlation:0.41174
  |
Correlation:0.41174
  |
Correlation:0.34971
  |
Correlation:0.32316
  |
Component:298; Jaccard:0.085102
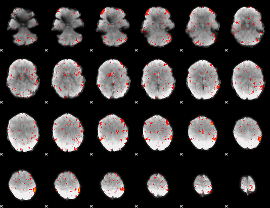 |
Component:024; Jaccard:0.094101
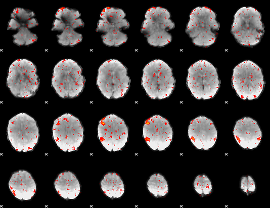 |
Component:024; Jaccard:0.099247
 |
Component:197; Jaccard:0.09396
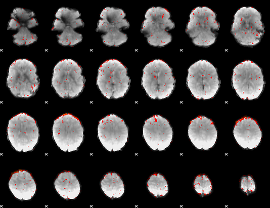 |
Component:326; Jaccard:0.081789
 |
Component:024; Jaccard:0.063079
 |
Component:024; Jaccard:0.048563
 |
Correlation:0.14819
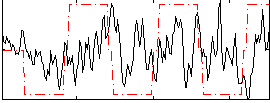 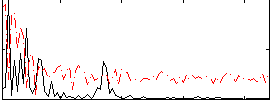 |
Correlation:0.42682
  |
Correlation:0.42682
  |
Correlation:0.12426
  |
Correlation:0.34971
  |
Correlation:0.42682
  |
Correlation:0.42682
  |
Component:024; Jaccard:0.082727
 |
Component:298; Jaccard:0.091601
 |
Component:197; Jaccard:0.098839
 |
Component:024; Jaccard:0.093633
 |
Component:024; Jaccard:0.079672
 |
Component:017; Jaccard:0.061401
 |
Component:017; Jaccard:0.043523
 |
Correlation:0.42682
  |
Correlation:0.14819
  |
Correlation:0.12426
  |
Correlation:0.42682
  |
Correlation:0.42682
  |
Correlation:0.41174
  |
Correlation:0.41174
  |
Component:197; Jaccard:0.07749
 |
Component:197; Jaccard:0.09105
 |
Component:298; Jaccard:0.090392
 |
Component:326; Jaccard:0.08547
 |
Component:197; Jaccard:0.075636
 |
Component:197; Jaccard:0.055311
 |
Component:327; Jaccard:0.036759
 |
Correlation:0.12426
  |
Correlation:0.12426
  |
Correlation:0.14819
  |
Correlation:0.34971
  |
Correlation:0.12426
  |
Correlation:0.12426
  |
Correlation:0.010835
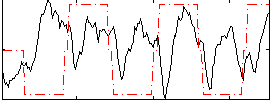  |
Component:326; Jaccard:0.074251
 |
Component:305; Jaccard:0.081224
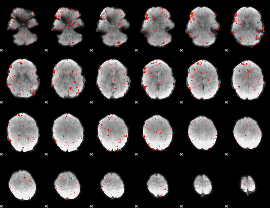 |
Component:305; Jaccard:0.084128
 |
Component:298; Jaccard:0.081296
 |
Component:298; Jaccard:0.067163
 |
Component:342; Jaccard:0.049652
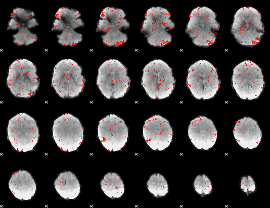 |
Component:342; Jaccard:0.035819
 |
Correlation:0.34971
  |
Correlation:0.32389
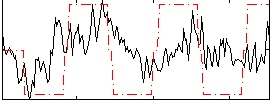  |
Correlation:0.32389
  |
Correlation:0.14819
  |
Correlation:0.14819
  |
Correlation:0.19091
  |
Correlation:0.19091
  |
Component:305; Jaccard:0.072439
 |
Component:326; Jaccard:0.080543
 |
Component:326; Jaccard:0.083973
 |
Component:305; Jaccard:0.077228
 |
Component:305; Jaccard:0.065214
 |
Component:305; Jaccard:0.049127
 |
Component:305; Jaccard:0.034563
 |
Correlation:0.32389
  |
Correlation:0.34971
  |
Correlation:0.34971
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Cope: 04:neg_FACES BACK
|
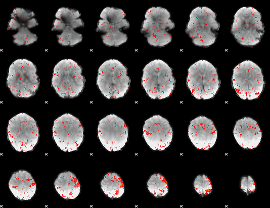
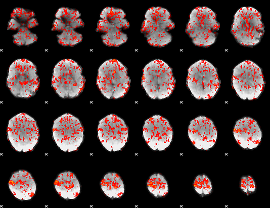
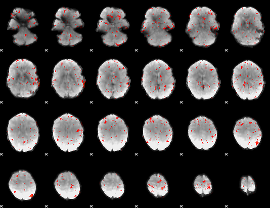
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
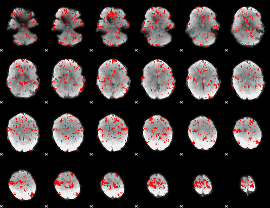 |
GLM binary result thresholded by 2
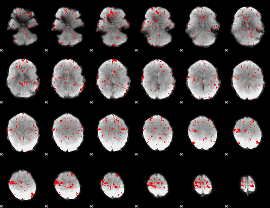 |
GLM binary result thresholded by 2.5
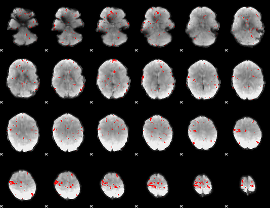 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
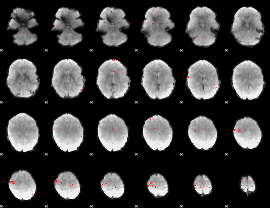 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
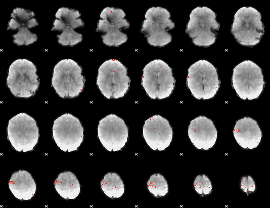 |
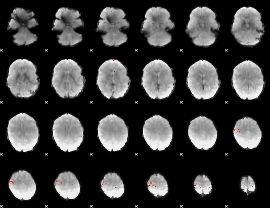
GLM Z-score result thresholded by 4
 |
Component:164; Jaccard:0.1045
 |
Component:164; Jaccard:0.11085
 |
Component:138; Jaccard:0.11869
 |
Component:138; Jaccard:0.11312
 |
Component:138; Jaccard:0.083756
 |
Component:138; Jaccard:0.050881
 |
Component:138; Jaccard:0.029421
 |
Correlation:0.11282
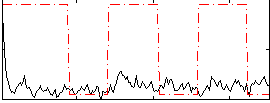 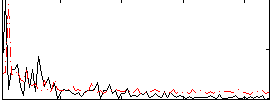 |
Correlation:0.11282
  |
Correlation:0.27979
  |
Correlation:0.27979
  |
Correlation:0.27979
  |
Correlation:0.27979
  |
Correlation:0.27979
  |
Component:138; Jaccard:0.090157
 |
Component:138; Jaccard:0.10679
 |
Component:164; Jaccard:0.10667
 |
Component:164; Jaccard:0.087327
 |
Component:164; Jaccard:0.057187
 |
Component:164; Jaccard:0.030535
 |
Component:164; Jaccard:0.015335
 |
Correlation:0.27979
  |
Correlation:0.27979
  |
Correlation:0.11282
  |
Correlation:0.11282
  |
Correlation:0.11282
  |
Correlation:0.11282
  |
Correlation:0.11282
  |
Component:053; Jaccard:0.080602
 |
Component:205; Jaccard:0.087821
 |
Component:205; Jaccard:0.083653
 |
Component:205; Jaccard:0.066028
 |
Component:205; Jaccard:0.040484
 |
Component:205; Jaccard:0.022095
 |
Component:274; Jaccard:0.010473
 |
Correlation:0.12773
  |
Correlation:-0.011129
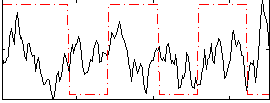 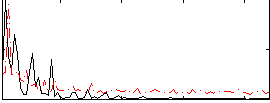 |
Correlation:-0.011129
  |
Correlation:-0.011129
  |
Correlation:-0.011129
  |
Correlation:-0.011129
  |
Correlation:0.16717
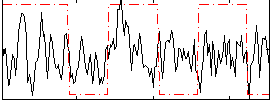 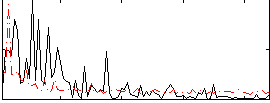 |
Component:074; Jaccard:0.078257
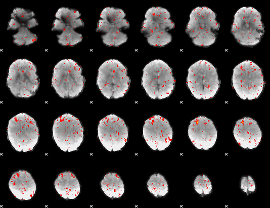 |
Component:053; Jaccard:0.086806
 |
Component:053; Jaccard:0.082965
 |
Component:053; Jaccard:0.062488
 |
Component:053; Jaccard:0.039136
 |
Component:274; Jaccard:0.020496
 |
Component:205; Jaccard:0.010093
 |
Correlation:0.11936
  |
Correlation:0.12773
  |
Correlation:0.12773
  |
Correlation:0.12773
  |
Correlation:0.12773
  |
Correlation:0.16717
  |
Correlation:-0.011129
  |
Component:205; Jaccard:0.077802
 |
Component:074; Jaccard:0.08377
 |
Component:252; Jaccard:0.075328
 |
Component:252; Jaccard:0.055809
 |
Component:274; Jaccard:0.03462
 |
Component:117; Jaccard:0.017815
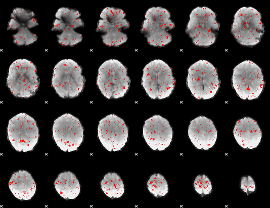 |
Component:117; Jaccard:0.009912
 |
Correlation:-0.011129
  |
Correlation:0.11936
  |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.16717
  |
Correlation:0.02982
  |
Correlation:0.02982
  |
Component:252; Jaccard:0.07569
 |
Component:252; Jaccard:0.082858
 |
Component:074; Jaccard:0.074633
 |
Component:074; Jaccard:0.051998
 |
Component:117; Jaccard:0.032414
 |
Component:053; Jaccard:0.016992
 |
Component:280; Jaccard:0.009211
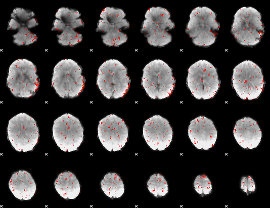 |
Correlation:0.10502
  |
Correlation:0.10502
  |
Correlation:0.11936
  |
Correlation:0.11936
  |
Correlation:0.02982
  |
Correlation:0.12773
  |
Correlation:0.35953
  |
Component:199; Jaccard:0.074623
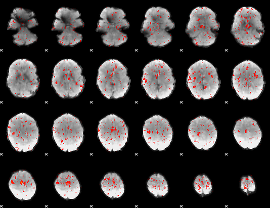 |
Component:140; Jaccard:0.075201
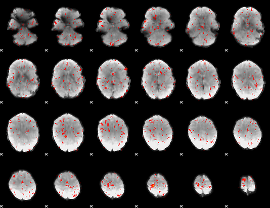 |
Component:248; Jaccard:0.068263
 |
Component:199; Jaccard:0.050617
 |
Component:252; Jaccard:0.032289
 |
Component:199; Jaccard:0.01646
 |
Component:053; Jaccard:0.008951
 |
Correlation:0.15176
  |
Correlation:0.069141
  |
Correlation:0.18065
  |
Correlation:0.15176
  |
Correlation:0.10502
  |
Correlation:0.15176
  |
Correlation:0.12773
  |
Component:140; Jaccard:0.074147
 |
Component:199; Jaccard:0.073231
 |
Component:199; Jaccard:0.067156
 |
Component:274; Jaccard:0.050412
 |
Component:199; Jaccard:0.031083
 |
Component:252; Jaccard:0.016441
 |
Component:199; Jaccard:0.008845
 |
Correlation:0.069141
  |
Correlation:0.15176
  |
Correlation:0.15176
  |
Correlation:0.16717
  |
Correlation:0.15176
  |
Correlation:0.10502
  |
Correlation:0.15176
  |
Component:397; Jaccard:0.069383
 |
Component:397; Jaccard:0.072279
 |
Component:140; Jaccard:0.066709
 |
Component:117; Jaccard:0.049654
 |
Component:280; Jaccard:0.02928
 |
Component:329; Jaccard:0.016129
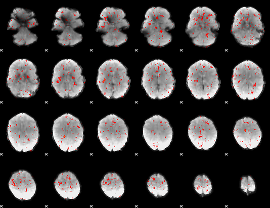 |
Component:329; Jaccard:0.008603
 |
Correlation:0.10491
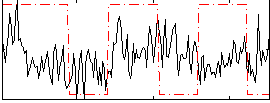 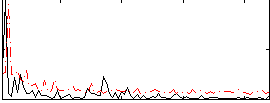 |
Correlation:0.10491
  |
Correlation:0.069141
  |
Correlation:0.02982
  |
Correlation:0.35953
  |
Correlation:0.15994
 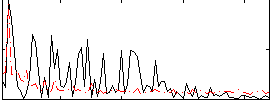 |
Correlation:0.15994
  |
Component:117; Jaccard:0.065902
 |
Component:248; Jaccard:0.069824
 |
Component:397; Jaccard:0.066399
 |
Component:280; Jaccard:0.048759
 |
Component:329; Jaccard:0.028854
 |
Component:280; Jaccard:0.015489
 |
Component:252; Jaccard:0.008476
 |
Correlation:0.02982
  |
Correlation:0.18065
  |
Correlation:0.10491
  |
Correlation:0.35953
  |
Correlation:0.15994
  |
Correlation:0.35953
  |
Correlation:0.10502
  |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
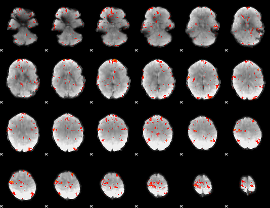 |
GLM Z-score result thresholded by 3
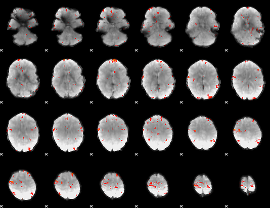 |
GLM Z-score result thresholded by 3.5
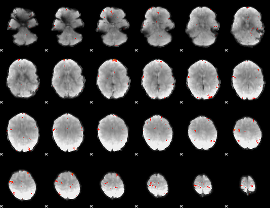 |
GLM Z-score result thresholded by 4
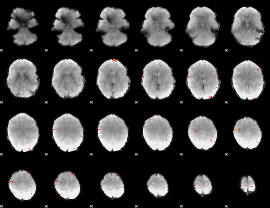 |
Component:164; Jaccard:0.096493
 |
Component:164; Jaccard:0.10367
 |
Component:394; Jaccard:0.11186
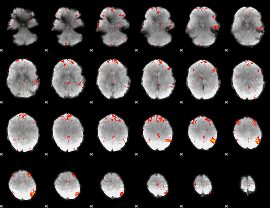 |
Component:394; Jaccard:0.10751
 |
Component:394; Jaccard:0.085435
 |
Component:156; Jaccard:0.053714
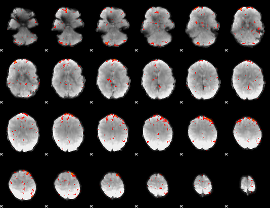 |
Component:156; Jaccard:0.032634
 |
Correlation:0.009073
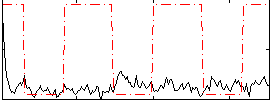 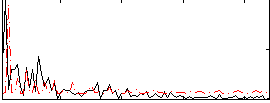 |
Correlation:0.009073
  |
Correlation:0.25671
  |
Correlation:0.25671
  |
Correlation:0.25671
  |
Correlation:0.13659
  |
Correlation:0.13659
  |
Component:349; Jaccard:0.092721
 |
Component:394; Jaccard:0.10118
 |
Component:205; Jaccard:0.10954
 |
Component:205; Jaccard:0.105
 |
Component:205; Jaccard:0.081363
 |
Component:394; Jaccard:0.052905
 |
Component:205; Jaccard:0.029803
 |
Correlation:0.28454
  |
Correlation:0.25671
  |
Correlation:0.1353
  |
Correlation:0.1353
  |
Correlation:0.1353
  |
Correlation:0.25671
  |
Correlation:0.1353
  |
Component:394; Jaccard:0.083938
 |
Component:349; Jaccard:0.10108
 |
Component:164; Jaccard:0.10412
 |
Component:161; Jaccard:0.10259
 |
Component:161; Jaccard:0.079182
 |
Component:161; Jaccard:0.051895
 |
Component:394; Jaccard:0.027574
 |
Correlation:0.25671
  |
Correlation:0.28454
  |
Correlation:0.009073
  |
Correlation:0.15264
  |
Correlation:0.15264
  |
Correlation:0.15264
  |
Correlation:0.25671
  |
Component:205; Jaccard:0.082683
 |
Component:205; Jaccard:0.097825
 |
Component:161; Jaccard:0.10375
 |
Component:164; Jaccard:0.094206
 |
Component:156; Jaccard:0.078332
 |
Component:205; Jaccard:0.051375
 |
Component:161; Jaccard:0.02752
 |
Correlation:0.1353
  |
Correlation:0.1353
  |
Correlation:0.15264
  |
Correlation:0.009073
  |
Correlation:0.13659
  |
Correlation:0.1353
  |
Correlation:0.15264
  |
Component:024; Jaccard:0.08111
 |
Component:161; Jaccard:0.093095
 |
Component:349; Jaccard:0.10256
 |
Component:156; Jaccard:0.093943
 |
Component:164; Jaccard:0.068516
 |
Component:138; Jaccard:0.0416
 |
Component:138; Jaccard:0.025292
 |
Correlation:0.43484
  |
Correlation:0.15264
  |
Correlation:0.28454
  |
Correlation:0.13659
  |
Correlation:0.009073
  |
Correlation:-0.19237
  |
Correlation:-0.19237
  |
Component:294; Jaccard:0.079263
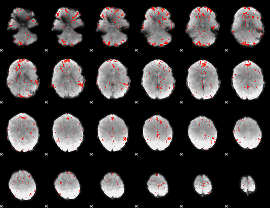 |
Component:252; Jaccard:0.089994
 |
Component:252; Jaccard:0.097429
 |
Component:349; Jaccard:0.089373
 |
Component:349; Jaccard:0.066403
 |
Component:252; Jaccard:0.041531
 |
Component:252; Jaccard:0.02168
 |
Correlation:0.31476
  |
Correlation:0.033649
 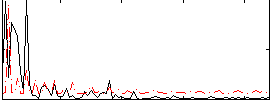 |
Correlation:0.033649
  |
Correlation:0.28454
  |
Correlation:0.28454
  |
Correlation:0.033649
  |
Correlation:0.033649
  |
Component:161; Jaccard:0.0774
 |
Component:024; Jaccard:0.087137
 |
Component:156; Jaccard:0.091637
 |
Component:252; Jaccard:0.089018
 |
Component:209; Jaccard:0.065533
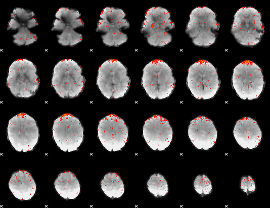 |
Component:349; Jaccard:0.039802
 |
Component:349; Jaccard:0.020207
 |
Correlation:0.15264
  |
Correlation:0.43484
  |
Correlation:0.13659
  |
Correlation:0.033649
  |
Correlation:0.27443
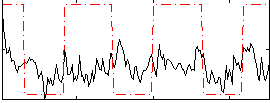 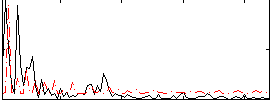 |
Correlation:0.28454
  |
Correlation:0.28454
  |
Component:252; Jaccard:0.076364
 |
Component:053; Jaccard:0.08493
 |
Component:209; Jaccard:0.088098
 |
Component:209; Jaccard:0.084513
 |
Component:252; Jaccard:0.065447
 |
Component:164; Jaccard:0.039457
 |
Component:209; Jaccard:0.020045
 |
Correlation:0.033649
  |
Correlation:0.034976
  |
Correlation:0.27443
  |
Correlation:0.27443
  |
Correlation:0.033649
  |
Correlation:0.009073
  |
Correlation:0.27443
  |
Component:053; Jaccard:0.075768
 |
Component:156; Jaccard:0.084359
 |
Component:053; Jaccard:0.087679
 |
Component:138; Jaccard:0.080487
 |
Component:138; Jaccard:0.065443
 |
Component:209; Jaccard:0.038848
 |
Component:164; Jaccard:0.019952
 |
Correlation:0.034976
  |
Correlation:0.13659
  |
Correlation:0.034976
  |
Correlation:-0.19237
  |
Correlation:-0.19237
  |
Correlation:0.27443
  |
Correlation:0.009073
  |
Component:209; Jaccard:0.074256
 |
Component:209; Jaccard:0.082446
 |
Component:024; Jaccard:0.086707
 |
Component:053; Jaccard:0.077745
 |
Component:053; Jaccard:0.053789
 |
Component:053; Jaccard:0.034605
 |
Component:053; Jaccard:0.017439
 |
Correlation:0.27443
  |
Correlation:0.27443
  |
Correlation:0.43484
  |
Correlation:0.034976
  |
Correlation:0.034976
  |
Correlation:0.034976
  |
Correlation:0.034976
  |
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
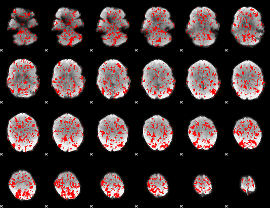
GLM binary result thresholded by 1
 |
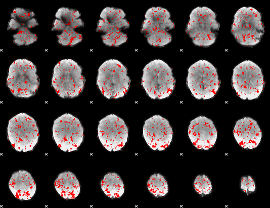
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
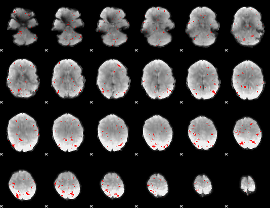
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
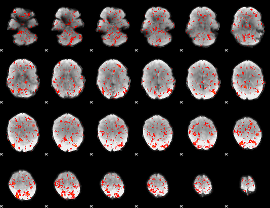 |
GLM Z-score result thresholded by 2
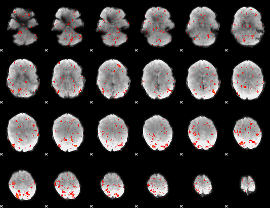 |
GLM Z-score result thresholded by 2.5
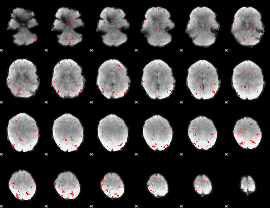 |
GLM Z-score result thresholded by 3
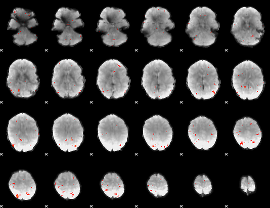 |
GLM Z-score result thresholded by 3.5
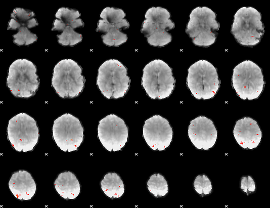 |
GLM Z-score result thresholded by 4
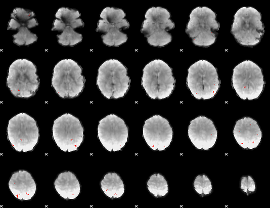 |
Component:131; Jaccard:0.12283
 |
Component:131; Jaccard:0.13757
 |
Component:131; Jaccard:0.12816
 |
Component:131; Jaccard:0.095801
 |
Component:131; Jaccard:0.056836
 |
Component:131; Jaccard:0.029265
 |
Component:131; Jaccard:0.014105
 |
Correlation:0.24275
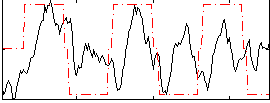 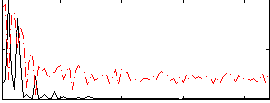 |
Correlation:0.24275
  |
Correlation:0.24275
  |
Correlation:0.24275
  |
Correlation:0.24275
  |
Correlation:0.24275
  |
Correlation:0.24275
  |
Component:227; Jaccard:0.12118
 |
Component:227; Jaccard:0.13193
 |
Component:227; Jaccard:0.12182
 |
Component:227; Jaccard:0.090318
 |
Component:227; Jaccard:0.051975
 |
Component:227; Jaccard:0.02793
 |
Component:227; Jaccard:0.012023
 |
Correlation:0.14546
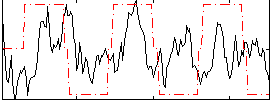  |
Correlation:0.14546
  |
Correlation:0.14546
  |
Correlation:0.14546
  |
Correlation:0.14546
  |
Correlation:0.14546
  |
Correlation:0.14546
  |
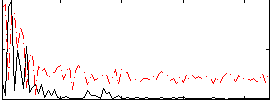
Component:273; Jaccard:0.098386
 |
Component:273; Jaccard:0.10841
 |
Component:273; Jaccard:0.097447
 |
Component:273; Jaccard:0.06829
 |
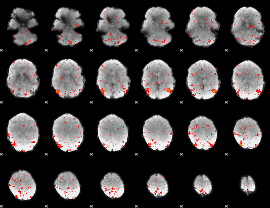
Component:236; Jaccard:0.039646
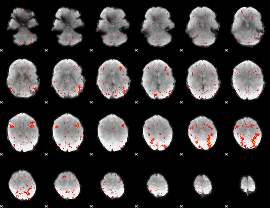 |
Component:236; Jaccard:0.019585
 |
Component:273; Jaccard:0.008875
 |
Correlation:0.12801
  |
Correlation:0.12801
  |
Correlation:0.12801
  |
Correlation:0.12801
  |
Correlation:0.10314
  |
Correlation:0.10314
  |
Correlation:0.12801
  |
Component:043; Jaccard:0.083713
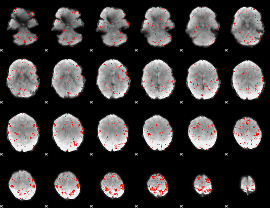 |
Component:143; Jaccard:0.088889
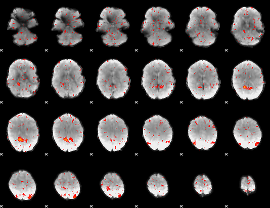 |
Component:236; Jaccard:0.080791
 |
Component:236; Jaccard:0.063672
 |
Component:273; Jaccard:0.038918
 |
Component:273; Jaccard:0.019437
 |
Component:236; Jaccard:0.007722
 |
Correlation:0.25167
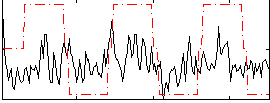 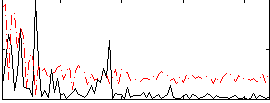 |
Correlation:0.079951
  |
Correlation:0.10314
  |
Correlation:0.10314
  |
Correlation:0.12801
  |
Correlation:0.12801
  |
Correlation:0.10314
  |
Component:143; Jaccard:0.079607
 |
Component:236; Jaccard:0.083996
 |
Component:143; Jaccard:0.078738
 |
Component:143; Jaccard:0.057465
 |
Component:143; Jaccard:0.030706
 |
Component:143; Jaccard:0.016694
 |
Component:143; Jaccard:0.007067
 |
Correlation:0.079951
  |
Correlation:0.10314
  |
Correlation:0.079951
  |
Correlation:0.079951
  |
Correlation:0.079951
  |
Correlation:0.079951
  |
Correlation:0.079951
  |
Component:236; Jaccard:0.077842
 |
Component:043; Jaccard:0.080888
 |
Component:043; Jaccard:0.063964
 |
Component:043; Jaccard:0.040252
 |
Component:330; Jaccard:0.023572
 |
Component:330; Jaccard:0.011984
 |
Component:330; Jaccard:0.004513
 |
Correlation:0.10314
  |
Correlation:0.25167
  |
Correlation:0.25167
  |
Correlation:0.25167
  |
Correlation:0.14704
  |
Correlation:0.14704
  |
Correlation:0.14704
  |
Component:330; Jaccard:0.075286
 |
Component:330; Jaccard:0.072907
 |
Component:330; Jaccard:0.058536
 |
Component:330; Jaccard:0.038436
 |
Component:043; Jaccard:0.022551
 |
Component:043; Jaccard:0.01068
 |
Component:115; Jaccard:0.004185
 |
Correlation:0.14704
  |
Correlation:0.14704
  |
Correlation:0.14704
  |
Correlation:0.14704
  |
Correlation:0.25167
  |
Correlation:0.25167
  |
Correlation:0.16205
  |
Component:076; Jaccard:0.075086
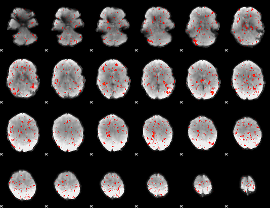 |
Component:076; Jaccard:0.06944
 |
Component:138; Jaccard:0.057319
 |
Component:138; Jaccard:0.036354
 |
Component:115; Jaccard:0.017423
 |
Component:055; Jaccard:0.009075
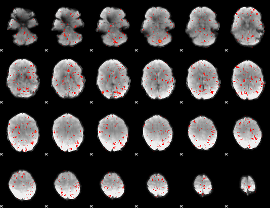 |
Component:055; Jaccard:0.003816
 |
Correlation:0.026134
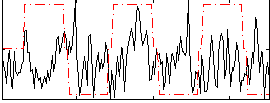 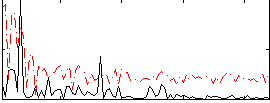 |
Correlation:0.026134
  |
Correlation:0.2561
  |
Correlation:0.2561
  |
Correlation:0.16205
  |
Correlation:0.11337
  |
Correlation:0.11337
  |
Component:138; Jaccard:0.063687
 |
Component:138; Jaccard:0.064379
 |
Component:076; Jaccard:0.055022
 |
Component:076; Jaccard:0.033831
 |
Component:055; Jaccard:0.016594
 |
Component:115; Jaccard:0.008276
 |
Component:043; Jaccard:0.003367
 |
Correlation:0.2561
  |
Correlation:0.2561
  |
Correlation:0.026134
  |
Correlation:0.026134
  |
Correlation:0.11337
  |
Correlation:0.16205
  |
Correlation:0.25167
  |
Component:312; Jaccard:0.06363
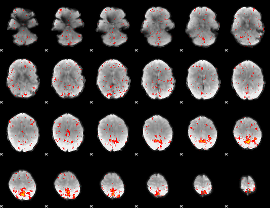 |
Component:074; Jaccard:0.060179
 |
Component:055; Jaccard:0.048953
 |
Component:115; Jaccard:0.033597
 |
Component:362; Jaccard:0.016314
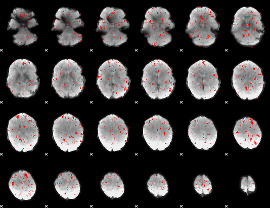 |
Component:312; Jaccard:0.008017
 |
Component:003; Jaccard:0.003226
 |
Correlation:-0.21226
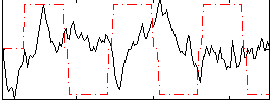  |
Correlation:0.10484
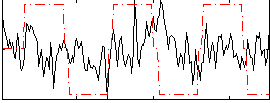  |
Correlation:0.11337
  |
Correlation:0.16205
  |
Correlation:0.080787
  |
Correlation:-0.21226
  |
Correlation:0.26499
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































