Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
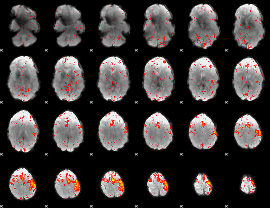
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
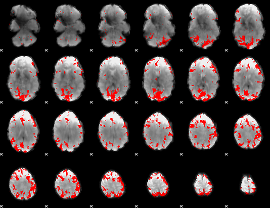 |
GLM binary result thresholded by 4
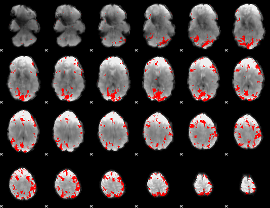 |
GLM Z-score result thresholded by 1
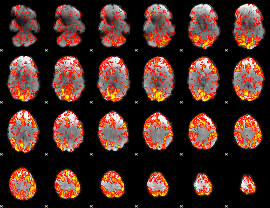 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
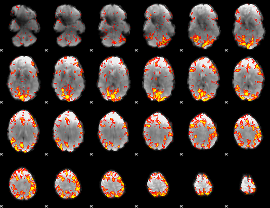 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
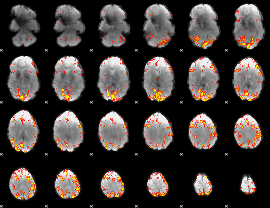 |
GLM Z-score result thresholded by 4
 |
Component:079; Jaccard:0.13697
 |
Component:079; Jaccard:0.16593
 |
Component:079; Jaccard:0.19867
 |
Component:079; Jaccard:0.2314
 |
Component:079; Jaccard:0.2589
 |
Component:079; Jaccard:0.27403
 |
Component:327; Jaccard:0.2928
 |
Correlation:0.077835
  |
Correlation:0.077835
  |
Correlation:0.077835
  |
Correlation:0.077835
  |
Correlation:0.077835
  |
Correlation:0.077835
  |
Correlation:0.18412
  |
Component:159; Jaccard:0.12716
 |
Component:159; Jaccard:0.14965
 |
Component:042; Jaccard:0.17496
 |
Component:042; Jaccard:0.2069
 |
Component:327; Jaccard:0.23847
 |
Component:327; Jaccard:0.27123
 |
Component:079; Jaccard:0.27947
 |
Correlation:0.074153
  |
Correlation:0.074153
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.18412
  |
Correlation:0.18412
  |
Correlation:0.077835
  |
Component:334; Jaccard:0.1201
 |
Component:042; Jaccard:0.14311
 |
Component:159; Jaccard:0.16911
 |
Component:327; Jaccard:0.20208
 |
Component:042; Jaccard:0.2327
 |
Component:042; Jaccard:0.24777
 |
Component:042; Jaccard:0.25288
 |
Correlation:0.052492
  |
Correlation:0.14653
  |
Correlation:0.074153
  |
Correlation:0.18412
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Correlation:0.14653
  |
Component:197; Jaccard:0.11646
 |
Component:015; Jaccard:0.13842
 |
Component:327; Jaccard:0.16572
 |
Component:015; Jaccard:0.19129
 |
Component:015; Jaccard:0.21184
 |
Component:015; Jaccard:0.22549
 |
Component:015; Jaccard:0.22577
 |
Correlation:-0.036903
  |
Correlation:0.17802
  |
Correlation:0.18412
  |
Correlation:0.17802
  |
Correlation:0.17802
  |
Correlation:0.17802
  |
Correlation:0.17802
  |
Component:042; Jaccard:0.11611
 |
Component:334; Jaccard:0.13515
 |
Component:015; Jaccard:0.16498
 |
Component:159; Jaccard:0.18324
 |
Component:159; Jaccard:0.18827
 |
Component:159; Jaccard:0.18662
 |
Component:222; Jaccard:0.17889
 |
Correlation:0.14653
  |
Correlation:0.052492
  |
Correlation:0.17802
  |
Correlation:0.074153
  |
Correlation:0.074153
  |
Correlation:0.074153
  |
Correlation:0.13362
  |
Component:015; Jaccard:0.11415
 |
Component:327; Jaccard:0.13307
 |
Component:334; Jaccard:0.14616
 |
Component:334; Jaccard:0.15382
 |
Component:222; Jaccard:0.1664
 |
Component:222; Jaccard:0.17549
 |
Component:159; Jaccard:0.17855
 |
Correlation:0.17802
  |
Correlation:0.18412
  |
Correlation:0.052492
  |
Correlation:0.052492
  |
Correlation:0.13362
  |
Correlation:0.13362
  |
Correlation:0.074153
  |
Component:145; Jaccard:0.10587
 |
Component:197; Jaccard:0.13063
 |
Component:197; Jaccard:0.14317
 |
Component:222; Jaccard:0.1522
 |
Component:285; Jaccard:0.16091
 |
Component:285; Jaccard:0.16664
 |
Component:285; Jaccard:0.16464
 |
Correlation:0.002046
  |
Correlation:-0.036903
  |
Correlation:-0.036903
  |
Correlation:0.13362
  |
Correlation:0.044636
  |
Correlation:0.044636
  |
Correlation:0.044636
  |
Component:327; Jaccard:0.1056
 |
Component:185; Jaccard:0.11898
 |
Component:285; Jaccard:0.13581
 |
Component:197; Jaccard:0.1514
 |
Component:334; Jaccard:0.15414
 |
Component:372; Jaccard:0.1493
 |
Component:372; Jaccard:0.14435
 |
Correlation:0.18412
  |
Correlation:0.23755
  |
Correlation:0.044636
  |
Correlation:-0.036903
  |
Correlation:0.052492
  |
Correlation:0.28552
  |
Correlation:0.28552
  |
Component:185; Jaccard:0.10504
 |
Component:285; Jaccard:0.11846
 |
Component:222; Jaccard:0.1341
 |
Component:285; Jaccard:0.15115
 |
Component:197; Jaccard:0.15175
 |
Component:154; Jaccard:0.14865
 |
Component:154; Jaccard:0.14404
 |
Correlation:0.23755
  |
Correlation:0.044636
  |
Correlation:0.13362
  |
Correlation:0.044636
  |
Correlation:-0.036903
  |
Correlation:-0.003456
  |
Correlation:-0.003456
  |
Component:207; Jaccard:0.10328
 |
Component:145; Jaccard:0.11805
 |
Component:154; Jaccard:0.13067
 |
Component:154; Jaccard:0.14297
 |
Component:154; Jaccard:0.14929
 |
Component:334; Jaccard:0.1477
 |
Component:364; Jaccard:0.14109
 |
Correlation:-0.022867
  |
Correlation:0.002046
  |
Correlation:-0.003456
  |
Correlation:-0.003456
  |
Correlation:-0.003456
  |
Correlation:0.052492
  |
Correlation:0.089653
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
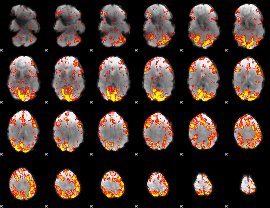 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
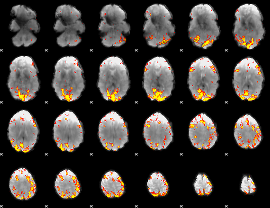 |
Component:079; Jaccard:0.14873
 |
Component:079; Jaccard:0.18265
 |
Component:079; Jaccard:0.21972
 |
Component:079; Jaccard:0.25681
 |
Component:079; Jaccard:0.28802
 |
Component:079; Jaccard:0.31039
 |
Component:079; Jaccard:0.32081
 |
Correlation:0.20297
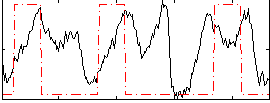 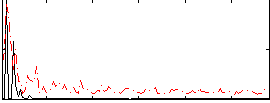 |
Correlation:0.20297
  |
Correlation:0.20297
  |
Correlation:0.20297
  |
Correlation:0.20297
  |
Correlation:0.20297
  |
Correlation:0.20297
  |
Component:042; Jaccard:0.12346
 |
Component:042; Jaccard:0.15125
 |
Component:042; Jaccard:0.18391
 |
Component:042; Jaccard:0.21722
 |
Component:042; Jaccard:0.24638
 |
Component:042; Jaccard:0.26893
 |
Component:042; Jaccard:0.2832
 |
Correlation:0.23275
  |
Correlation:0.23275
  |
Correlation:0.23275
  |
Correlation:0.23275
  |
Correlation:0.23275
  |
Correlation:0.23275
  |
Correlation:0.23275
  |
Component:334; Jaccard:0.1221
 |
Component:015; Jaccard:0.14678
 |
Component:015; Jaccard:0.17414
 |
Component:327; Jaccard:0.20111
 |
Component:327; Jaccard:0.23296
 |
Component:327; Jaccard:0.26019
 |
Component:327; Jaccard:0.283
 |
Correlation:0.065928
  |
Correlation:0.28668
  |
Correlation:0.28668
  |
Correlation:0.31615
  |
Correlation:0.31615
  |
Correlation:0.31615
  |
Correlation:0.31615
  |
Component:015; Jaccard:0.12101
 |
Component:334; Jaccard:0.13641
 |
Component:327; Jaccard:0.16693
 |
Component:015; Jaccard:0.19974
 |
Component:015; Jaccard:0.22281
 |
Component:015; Jaccard:0.23655
 |
Component:015; Jaccard:0.24127
 |
Correlation:0.28668
  |
Correlation:0.065928
  |
Correlation:0.31615
  |
Correlation:0.28668
  |
Correlation:0.28668
  |
Correlation:0.28668
  |
Correlation:0.28668
  |
Component:197; Jaccard:0.11863
 |
Component:285; Jaccard:0.13634
 |
Component:285; Jaccard:0.15711
 |
Component:285; Jaccard:0.17702
 |
Component:285; Jaccard:0.19391
 |
Component:285; Jaccard:0.20497
 |
Component:285; Jaccard:0.21126
 |
Correlation:0.21063
  |
Correlation:0.23429
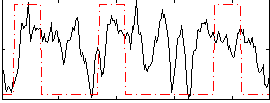  |
Correlation:0.23429
  |
Correlation:0.23429
  |
Correlation:0.23429
  |
Correlation:0.23429
  |
Correlation:0.23429
  |
Component:285; Jaccard:0.11587
 |
Component:327; Jaccard:0.13605
 |
Component:334; Jaccard:0.14746
 |
Component:364; Jaccard:0.16431
 |
Component:364; Jaccard:0.18341
 |
Component:364; Jaccard:0.19822
 |
Component:364; Jaccard:0.21012
 |
Correlation:0.23429
  |
Correlation:0.31615
  |
Correlation:0.065928
  |
Correlation:0.45549
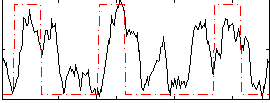 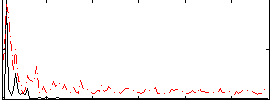 |
Correlation:0.45549
  |
Correlation:0.45549
  |
Correlation:0.45549
  |
Component:207; Jaccard:0.11076
 |
Component:197; Jaccard:0.13214
 |
Component:364; Jaccard:0.14388
 |
Component:222; Jaccard:0.15858
 |
Component:222; Jaccard:0.17332
 |
Component:222; Jaccard:0.18367
 |
Component:222; Jaccard:0.18948
 |
Correlation:0.095009
 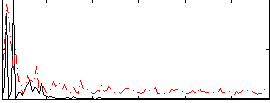 |
Correlation:0.21063
  |
Correlation:0.45549
  |
Correlation:0.22293
  |
Correlation:0.22293
  |
Correlation:0.22293
  |
Correlation:0.22293
  |
Component:154; Jaccard:0.11047
 |
Component:154; Jaccard:0.12779
 |
Component:154; Jaccard:0.14339
 |
Component:154; Jaccard:0.1567
 |
Component:154; Jaccard:0.16166
 |
Component:154; Jaccard:0.16128
 |
Component:188; Jaccard:0.16307
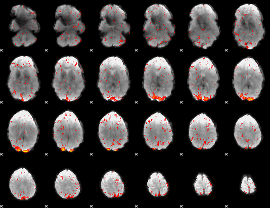 |
Correlation:0.23599
  |
Correlation:0.23599
  |
Correlation:0.23599
  |
Correlation:0.23599
  |
Correlation:0.23599
  |
Correlation:0.23599
  |
Correlation:0.3856
  |
Component:327; Jaccard:0.10927
 |
Component:364; Jaccard:0.12284
 |
Component:197; Jaccard:0.14151
 |
Component:334; Jaccard:0.1538
 |
Component:188; Jaccard:0.15538
 |
Component:188; Jaccard:0.15969
 |
Component:154; Jaccard:0.15667
 |
Correlation:0.31615
  |
Correlation:0.45549
  |
Correlation:0.21063
  |
Correlation:0.065928
  |
Correlation:0.3856
  |
Correlation:0.3856
  |
Correlation:0.23599
  |
Component:145; Jaccard:0.10868
 |
Component:207; Jaccard:0.11967
 |
Component:222; Jaccard:0.14011
 |
Component:197; Jaccard:0.14697
 |
Component:334; Jaccard:0.15397
 |
Component:334; Jaccard:0.14686
 |
Component:372; Jaccard:0.14362
 |
Correlation:0.15675
  |
Correlation:0.095009
  |
Correlation:0.22293
  |
Correlation:0.21063
  |
Correlation:0.065928
  |
Correlation:0.065928
  |
Correlation:0.097573
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
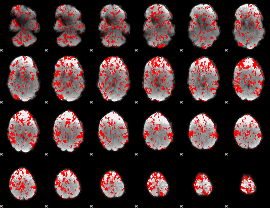 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
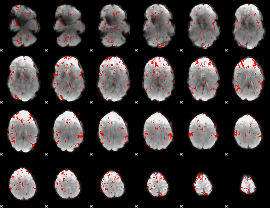 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
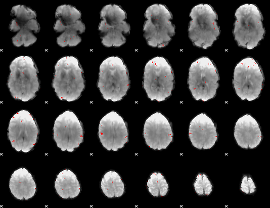 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
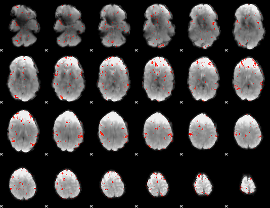 |
GLM Z-score result thresholded by 3
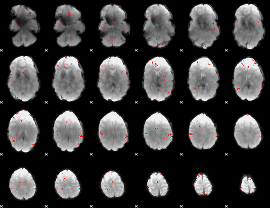 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:351; Jaccard:0.12026
 |
Component:351; Jaccard:0.12993
 |
Component:351; Jaccard:0.11854
 |
Component:351; Jaccard:0.088741
 |
Component:193; Jaccard:0.050243
 |
Component:066; Jaccard:0.022556
 |
Component:226; Jaccard:0.007167
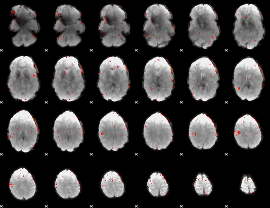 |
Correlation:0.083986
  |
Correlation:0.083986
  |
Correlation:0.083986
  |
Correlation:0.083986
  |
Correlation:0.060456
  |
Correlation:0.20045
 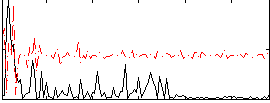 |
Correlation:0.066683
  |
Component:159; Jaccard:0.10643
 |
Component:159; Jaccard:0.1091
 |
Component:193; Jaccard:0.10626
 |
Component:193; Jaccard:0.086883
 |
Component:066; Jaccard:0.050021
 |
Component:193; Jaccard:0.021722
 |
Component:066; Jaccard:0.006278
 |
Correlation:0.058069
  |
Correlation:0.058069
  |
Correlation:0.060456
  |
Correlation:0.060456
  |
Correlation:0.20045
  |
Correlation:0.060456
  |
Correlation:0.20045
  |
Component:400; Jaccard:0.098439
 |
Component:193; Jaccard:0.10492
 |
Component:159; Jaccard:0.099991
 |
Component:244; Jaccard:0.071188
 |
Component:351; Jaccard:0.046139
 |
Component:351; Jaccard:0.02031
 |
Component:244; Jaccard:0.006151
 |
Correlation:0.03888
  |
Correlation:0.060456
  |
Correlation:0.058069
  |
Correlation:0.26091
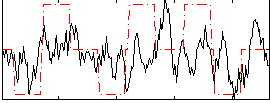  |
Correlation:0.083986
  |
Correlation:0.083986
  |
Correlation:0.26091
  |
Component:244; Jaccard:0.098135
 |
Component:244; Jaccard:0.1049
 |
Component:244; Jaccard:0.095851
 |
Component:159; Jaccard:0.068855
 |
Component:244; Jaccard:0.037994
 |
Component:226; Jaccard:0.018365
 |
Component:193; Jaccard:0.006056
 |
Correlation:0.26091
  |
Correlation:0.26091
  |
Correlation:0.26091
  |
Correlation:0.058069
  |
Correlation:0.26091
  |
Correlation:0.066683
  |
Correlation:0.060456
  |
Component:193; Jaccard:0.091786
 |
Component:400; Jaccard:0.10048
 |
Component:400; Jaccard:0.087689
 |
Component:066; Jaccard:0.068787
 |
Component:159; Jaccard:0.034767
 |
Component:237; Jaccard:0.016351
 |
Component:351; Jaccard:0.00566
 |
Correlation:0.060456
  |
Correlation:0.03888
  |
Correlation:0.03888
  |
Correlation:0.20045
  |
Correlation:0.058069
  |
Correlation:0.11755
  |
Correlation:0.083986
  |
Component:128; Jaccard:0.084688
 |
Component:236; Jaccard:0.089435
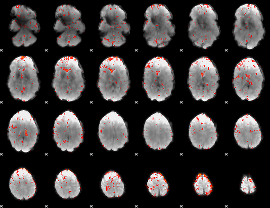 |
Component:236; Jaccard:0.083942
 |
Component:400; Jaccard:0.063083
 |
Component:400; Jaccard:0.034285
 |
Component:244; Jaccard:0.016202
 |
Component:237; Jaccard:0.005261
 |
Correlation:0.041491
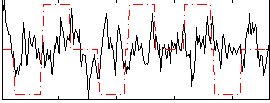  |
Correlation:0.043676
 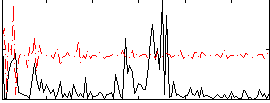 |
Correlation:0.043676
  |
Correlation:0.03888
  |
Correlation:0.03888
  |
Correlation:0.26091
  |
Correlation:0.11755
  |
Component:073; Jaccard:0.084457
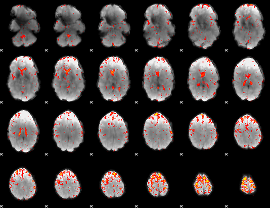 |
Component:128; Jaccard:0.085989
 |
Component:237; Jaccard:0.078315
 |
Component:236; Jaccard:0.061544
 |
Component:226; Jaccard:0.033397
 |
Component:159; Jaccard:0.015318
 |
Component:159; Jaccard:0.004381
 |
Correlation:0.074536
  |
Correlation:0.041491
  |
Correlation:0.11755
  |
Correlation:0.043676
  |
Correlation:0.066683
  |
Correlation:0.058069
  |
Correlation:0.058069
  |
Component:236; Jaccard:0.083185
 |
Component:146; Jaccard:0.081995
 |
Component:066; Jaccard:0.076237
 |
Component:237; Jaccard:0.059658
 |
Component:237; Jaccard:0.032859
 |
Component:146; Jaccard:0.01482
 |
Component:400; Jaccard:0.004366
 |
Correlation:0.043676
  |
Correlation:0.10833
  |
Correlation:0.20045
  |
Correlation:0.11755
  |
Correlation:0.11755
  |
Correlation:0.10833
  |
Correlation:0.03888
  |
Component:295; Jaccard:0.08243
 |
Component:237; Jaccard:0.08179
 |
Component:146; Jaccard:0.075178
 |
Component:146; Jaccard:0.055909
 |
Component:236; Jaccard:0.031219
 |
Component:400; Jaccard:0.01446
 |
Component:376; Jaccard:0.004132
 |
Correlation:0.12176
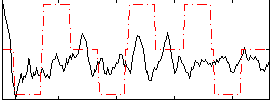  |
Correlation:0.11755
  |
Correlation:0.10833
  |
Correlation:0.10833
  |
Correlation:0.043676
  |
Correlation:0.03888
  |
Correlation:0.015009
 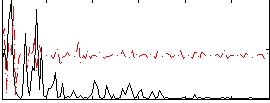 |
Component:146; Jaccard:0.080285
 |
Component:376; Jaccard:0.077916
 |
Component:128; Jaccard:0.074128
 |
Component:376; Jaccard:0.052665
 |
Component:291; Jaccard:0.030703
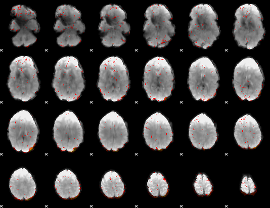 |
Component:376; Jaccard:0.014182
 |
Component:282; Jaccard:0.003911
 |
Correlation:0.10833
  |
Correlation:0.015009
  |
Correlation:0.041491
  |
Correlation:0.015009
  |
Correlation:0.0050844
  |
Correlation:0.015009
  |
Correlation:0.1764
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
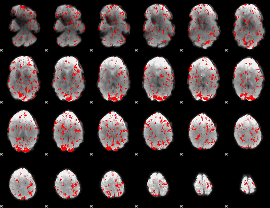 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
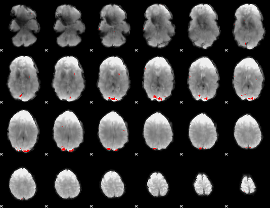 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
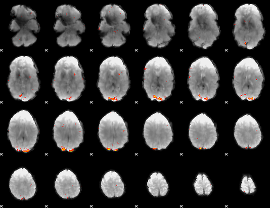 |
GLM Z-score result thresholded by 4
 |
Component:188; Jaccard:0.11982
 |
Component:188; Jaccard:0.1464
 |
Component:188; Jaccard:0.1618
 |
Component:188; Jaccard:0.1581
 |
Component:188; Jaccard:0.13387
 |
Component:188; Jaccard:0.10413
 |
Component:188; Jaccard:0.081336
 |
Correlation:0.33982
  |
Correlation:0.33982
  |
Correlation:0.33982
  |
Correlation:0.33982
  |
Correlation:0.33982
  |
Correlation:0.33982
  |
Correlation:0.33982
  |
Component:364; Jaccard:0.11416
 |
Component:364; Jaccard:0.1389
 |
Component:364; Jaccard:0.15208
 |
Component:364; Jaccard:0.14318
 |
Component:364; Jaccard:0.11803
 |
Component:364; Jaccard:0.091805
 |
Component:364; Jaccard:0.068878
 |
Correlation:0.21683
  |
Correlation:0.21683
  |
Correlation:0.21683
  |
Correlation:0.21683
  |
Correlation:0.21683
  |
Correlation:0.21683
  |
Correlation:0.21683
  |
Component:172; Jaccard:0.094863
 |
Component:172; Jaccard:0.10084
 |
Component:172; Jaccard:0.099224
 |
Component:172; Jaccard:0.088058
 |
Component:172; Jaccard:0.067328
 |
Component:172; Jaccard:0.051653
 |
Component:172; Jaccard:0.038099
 |
Correlation:0.31173
  |
Correlation:0.31173
  |
Correlation:0.31173
  |
Correlation:0.31173
  |
Correlation:0.31173
  |
Correlation:0.31173
  |
Correlation:0.31173
  |
Component:075; Jaccard:0.085359
 |
Component:075; Jaccard:0.08993
 |
Component:075; Jaccard:0.08081
 |
Component:049; Jaccard:0.06697
 |
Component:049; Jaccard:0.047137
 |
Component:042; Jaccard:0.035481
 |
Component:042; Jaccard:0.027001
 |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.11231
  |
Correlation:0.41379
  |
Correlation:0.41379
  |
Correlation:0.051106
  |
Correlation:0.051106
  |
Component:285; Jaccard:0.085303
 |
Component:285; Jaccard:0.086321
 |
Component:285; Jaccard:0.079879
 |
Component:285; Jaccard:0.064004
 |
Component:285; Jaccard:0.045072
 |
Component:285; Jaccard:0.032272
 |
Component:285; Jaccard:0.022139
 |
Correlation:0.11241
  |
Correlation:0.11241
  |
Correlation:0.11241
  |
Correlation:0.11241
  |
Correlation:0.11241
  |
Correlation:0.11241
  |
Correlation:0.11241
  |
Component:302; Jaccard:0.084248
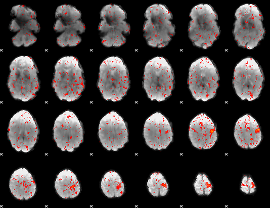 |
Component:208; Jaccard:0.084846
 |
Component:049; Jaccard:0.07899
 |
Component:075; Jaccard:0.055641
 |
Component:042; Jaccard:0.044026
 |
Component:049; Jaccard:0.03015
 |
Component:049; Jaccard:0.020884
 |
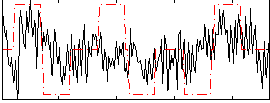
Correlation:0.26054
  |
Correlation:0.27366
  |
Correlation:0.41379
  |
Correlation:0.11231
  |
Correlation:0.051106
  |
Correlation:0.41379
  |
Correlation:0.41379
  |
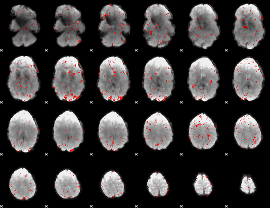
Component:079; Jaccard:0.083398
 |
Component:302; Jaccard:0.082367
 |
Component:208; Jaccard:0.07236
 |
Component:042; Jaccard:0.053719
 |
Component:327; Jaccard:0.038677
 |
Component:327; Jaccard:0.028211
 |
Component:029; Jaccard:0.018952
 |
Correlation:0.07417
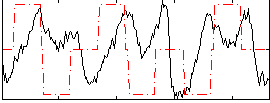  |
Correlation:0.26054
  |
Correlation:0.27366
  |
Correlation:0.051106
  |
Correlation:0.078258
  |
Correlation:0.078258
  |
Correlation:0.23473
  |
Component:208; Jaccard:0.082804
 |
Component:049; Jaccard:0.079665
 |
Component:302; Jaccard:0.066963
 |
Component:029; Jaccard:0.051933
 |
Component:029; Jaccard:0.0371
 |
Component:029; Jaccard:0.026827
 |
Component:327; Jaccard:0.018414
 |
Correlation:0.27366
  |
Correlation:0.41379
  |
Correlation:0.26054
  |
Correlation:0.23473
  |
Correlation:0.23473
  |
Correlation:0.23473
  |
Correlation:0.078258
  |
Component:277; Jaccard:0.074723
 |
Component:079; Jaccard:0.079124
 |
Component:079; Jaccard:0.066962
 |
Component:327; Jaccard:0.051603
 |
Component:369; Jaccard:0.036501
 |
Component:369; Jaccard:0.025378
 |
Component:369; Jaccard:0.018404
 |
Correlation:0.15395
  |
Correlation:0.07417
  |
Correlation:0.07417
  |
Correlation:0.078258
  |
Correlation:0.10568
  |
Correlation:0.10568
  |
Correlation:0.10568
  |
Component:049; Jaccard:0.073733
 |
Component:277; Jaccard:0.069052
 |
Component:029; Jaccard:0.061232
 |
Component:369; Jaccard:0.050495
 |
Component:079; Jaccard:0.034227
 |
Component:079; Jaccard:0.024554
 |
Component:079; Jaccard:0.017843
 |
Correlation:0.41379
  |
Correlation:0.15395
  |
Correlation:0.23473
  |
Correlation:0.10568
  |
Correlation:0.07417
  |
Correlation:0.07417
  |
Correlation:0.07417
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
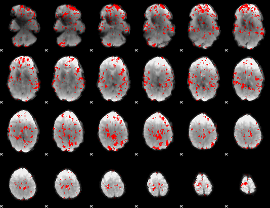 |
GLM binary result thresholded by 2.5
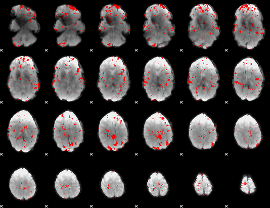 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
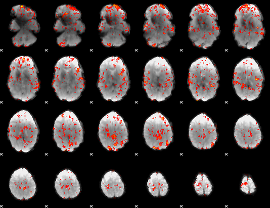 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
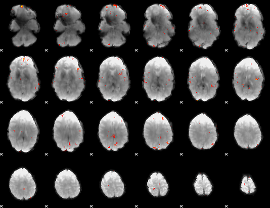 |
Component:161; Jaccard:0.10723
 |
Component:161; Jaccard:0.12764
 |
Component:161; Jaccard:0.14668
 |
Component:161; Jaccard:0.15588
 |
Component:161; Jaccard:0.14574
 |
Component:161; Jaccard:0.1196
 |
Component:161; Jaccard:0.080558
 |
Correlation:0.25459
  |
Correlation:0.25459
  |
Correlation:0.25459
  |
Correlation:0.25459
  |
Correlation:0.25459
  |
Correlation:0.25459
  |
Correlation:0.25459
  |
Component:005; Jaccard:0.10273
 |
Component:005; Jaccard:0.11199
 |
Component:005; Jaccard:0.11424
 |
Component:005; Jaccard:0.10775
 |
Component:005; Jaccard:0.088637
 |
Component:005; Jaccard:0.064195
 |
Component:012; Jaccard:0.041737
 |
Correlation:0.32382
  |
Correlation:0.32382
  |
Correlation:0.32382
  |
Correlation:0.32382
  |
Correlation:0.32382
  |
Correlation:0.32382
  |
Correlation:0.15832
  |
Component:341; Jaccard:0.098526
 |
Component:341; Jaccard:0.1017
 |
Component:012; Jaccard:0.10264
 |
Component:012; Jaccard:0.097084
 |
Component:012; Jaccard:0.082352
 |
Component:012; Jaccard:0.063654
 |
Component:005; Jaccard:0.039714
 |
Correlation:0.18643
  |
Correlation:0.18643
  |
Correlation:0.15832
  |
Correlation:0.15832
  |
Correlation:0.15832
  |
Correlation:0.15832
  |
Correlation:0.32382
  |
Component:012; Jaccard:0.093982
 |
Component:230; Jaccard:0.10013
 |
Component:230; Jaccard:0.10037
 |
Component:230; Jaccard:0.092574
 |
Component:230; Jaccard:0.075755
 |
Component:060; Jaccard:0.056134
 |
Component:060; Jaccard:0.036753
 |
Correlation:0.15832
  |
Correlation:-0.045543
  |
Correlation:-0.045543
  |
Correlation:-0.045543
  |
Correlation:-0.045543
  |
Correlation:0.08509
  |
Correlation:0.08509
  |
Component:230; Jaccard:0.093411
 |
Component:012; Jaccard:0.099975
 |
Component:341; Jaccard:0.095965
 |
Component:014; Jaccard:0.08722
 |
Component:014; Jaccard:0.071564
 |
Component:014; Jaccard:0.052811
 |
Component:014; Jaccard:0.033453
 |
Correlation:-0.045543
  |
Correlation:0.15832
  |
Correlation:0.18643
  |
Correlation:0.019198
  |
Correlation:0.019198
  |
Correlation:0.019198
  |
Correlation:0.019198
  |
Component:219; Jaccard:0.082742
 |
Component:014; Jaccard:0.088265
 |
Component:014; Jaccard:0.089695
 |
Component:341; Jaccard:0.083297
 |
Component:060; Jaccard:0.069867
 |
Component:230; Jaccard:0.052451
 |
Component:053; Jaccard:0.030691
 |
Correlation:-0.029522
  |
Correlation:0.019198
  |
Correlation:0.019198
  |
Correlation:0.18643
  |
Correlation:0.08509
  |
Correlation:-0.045543
  |
Correlation:0.052499
  |
Component:014; Jaccard:0.080856
 |
Component:075; Jaccard:0.086607
 |
Component:075; Jaccard:0.088268
 |
Component:075; Jaccard:0.082236
 |
Component:075; Jaccard:0.068045
 |
Component:075; Jaccard:0.04905
 |
Component:230; Jaccard:0.030568
 |
Correlation:0.019198
  |
Correlation:0.25681
  |
Correlation:0.25681
  |
Correlation:0.25681
  |
Correlation:0.25681
  |
Correlation:0.25681
  |
Correlation:-0.045543
  |
Component:053; Jaccard:0.080273
 |
Component:219; Jaccard:0.085125
 |
Component:053; Jaccard:0.086076
 |
Component:053; Jaccard:0.081314
 |
Component:053; Jaccard:0.064897
 |
Component:053; Jaccard:0.046462
 |
Component:008; Jaccard:0.028514
 |
Correlation:0.052499
  |
Correlation:-0.029522
  |
Correlation:0.052499
  |
Correlation:0.052499
  |
Correlation:0.052499
  |
Correlation:0.052499
  |
Correlation:0.10634
  |
Component:384; Jaccard:0.079971
 |
Component:053; Jaccard:0.08435
 |
Component:219; Jaccard:0.081105
 |
Component:201; Jaccard:0.07208
 |
Component:341; Jaccard:0.060744
 |
Component:008; Jaccard:0.044841
 |
Component:075; Jaccard:0.027897
 |
Correlation:0.14349
  |
Correlation:0.052499
  |
Correlation:-0.029522
  |
Correlation:0.25483
  |
Correlation:0.18643
  |
Correlation:0.10634
  |
Correlation:0.25681
  |
Component:346; Jaccard:0.07911
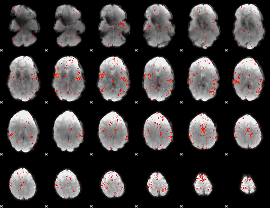 |
Component:346; Jaccard:0.07929
 |
Component:201; Jaccard:0.077105
 |
Component:060; Jaccard:0.070985
 |
Component:008; Jaccard:0.055988
 |
Component:057; Jaccard:0.041258
 |
Component:087; Jaccard:0.025285
 |
Correlation:0.2089
  |
Correlation:0.2089
  |
Correlation:0.25483
  |
Correlation:0.08509
  |
Correlation:0.10634
  |
Correlation:0.0057002
  |
Correlation:0.086711
  |
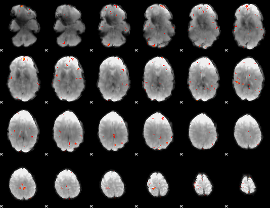
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
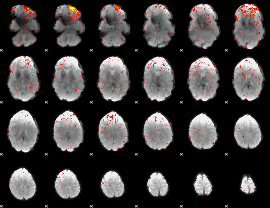
GLM binary result thresholded by 1
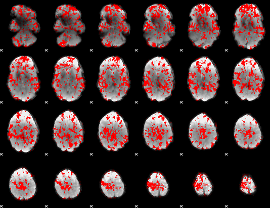 |
GLM binary result thresholded by 1.5
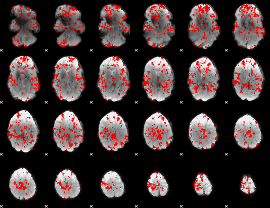 |
GLM binary result thresholded by 2
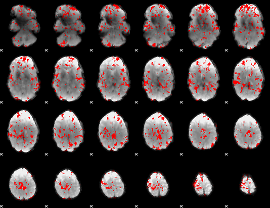 |
GLM binary result thresholded by 2.5
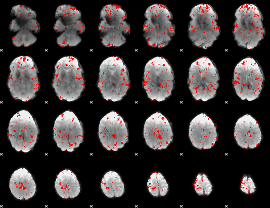 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
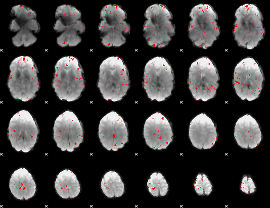 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:161; Jaccard:0.11792
 |
Component:161; Jaccard:0.14684
 |
Component:161; Jaccard:0.17773
 |
Component:161; Jaccard:0.19594
 |
Component:161; Jaccard:0.18881
 |
Component:161; Jaccard:0.15153
 |
Component:161; Jaccard:0.10483
 |
Correlation:0.28095
  |
Correlation:0.28095
  |
Correlation:0.28095
  |
Correlation:0.28095
  |
Correlation:0.28095
  |
Correlation:0.28095
  |
Correlation:0.28095
  |
Component:341; Jaccard:0.10724
 |
Component:230; Jaccard:0.11461
 |
Component:230; Jaccard:0.12295
 |
Component:230; Jaccard:0.12268
 |
Component:230; Jaccard:0.10634
 |
Component:230; Jaccard:0.078989
 |
Component:230; Jaccard:0.050396
 |
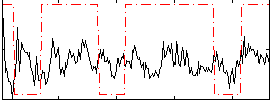
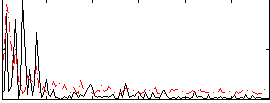
Correlation:0.10308
  |
Correlation:0.058957
  |
Correlation:0.058957
  |
Correlation:0.058957
  |
Correlation:0.058957
  |
Correlation:0.058957
  |
Correlation:0.058957
  |
Component:230; Jaccard:0.10335
 |
Component:341; Jaccard:0.11153
 |
Component:341; Jaccard:0.11175
 |
Component:012; Jaccard:0.10299
 |
Component:012; Jaccard:0.088141
 |
Component:012; Jaccard:0.066425
 |
Component:012; Jaccard:0.045612
 |
Correlation:0.058957
  |
Correlation:0.10308
  |
Correlation:0.10308
  |
Correlation:0.041251
  |
Correlation:0.041251
  |
Correlation:0.041251
  |
Correlation:0.041251
  |
Component:351; Jaccard:0.09777
 |
Component:351; Jaccard:0.10498
 |
Component:012; Jaccard:0.1059
 |
Component:351; Jaccard:0.10146
 |
Component:351; Jaccard:0.082671
 |
Component:351; Jaccard:0.06019
 |
Component:351; Jaccard:0.038166
 |
Correlation:0.19631
  |
Correlation:0.19631
  |
Correlation:0.041251
  |
Correlation:0.19631
  |
Correlation:0.19631
  |
Correlation:0.19631
  |
Correlation:0.19631
  |
Component:012; Jaccard:0.093864
 |
Component:012; Jaccard:0.10319
 |
Component:351; Jaccard:0.10552
 |
Component:341; Jaccard:0.10127
 |
Component:341; Jaccard:0.081501
 |
Component:341; Jaccard:0.058609
 |
Component:259; Jaccard:0.037558
 |
Correlation:0.041251
  |
Correlation:0.041251
  |
Correlation:0.19631
  |
Correlation:0.10308
  |
Correlation:0.10308
  |
Correlation:0.10308
  |
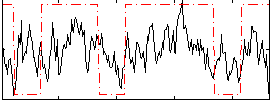
Correlation:0.28494
  |
Component:259; Jaccard:0.089967
 |
Component:346; Jaccard:0.095412
 |
Component:346; Jaccard:0.095056
 |
Component:346; Jaccard:0.090237
 |
Component:259; Jaccard:0.074874
 |
Component:259; Jaccard:0.055977
 |
Component:008; Jaccard:0.0358
 |
Correlation:0.28494
  |
Correlation:0.12858
  |
Correlation:0.12858
  |
Correlation:0.12858
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.10199
  |
Component:346; Jaccard:0.089499
 |
Component:259; Jaccard:0.094477
 |
Component:259; Jaccard:0.094956
 |
Component:259; Jaccard:0.089284
 |
Component:346; Jaccard:0.073585
 |
Component:346; Jaccard:0.055511
 |
Component:346; Jaccard:0.035334
 |
Correlation:0.12858
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.28494
  |
Correlation:0.12858
  |
Correlation:0.12858
  |
Correlation:0.12858
  |
Component:005; Jaccard:0.079378
 |
Component:005; Jaccard:0.083325
 |
Component:053; Jaccard:0.082602
 |
Component:053; Jaccard:0.076746
 |
Component:053; Jaccard:0.064131
 |
Component:008; Jaccard:0.047384
 |
Component:087; Jaccard:0.035264
 |
Correlation:-0.16455
  |
Correlation:-0.16455
  |
Correlation:0.15331
  |
Correlation:0.15331
  |
Correlation:0.15331
  |
Correlation:0.10199
  |
Correlation:0.0040255
  |
Component:053; Jaccard:0.077957
 |
Component:053; Jaccard:0.082295
 |
Component:014; Jaccard:0.081642
 |
Component:005; Jaccard:0.075186
 |
Component:005; Jaccard:0.060667
 |
Component:053; Jaccard:0.045899
 |
Component:341; Jaccard:0.03522
 |
Correlation:0.15331
  |
Correlation:0.15331
  |
Correlation:0.018404
  |
Correlation:-0.16455
  |
Correlation:-0.16455
  |
Correlation:0.15331
  |
Correlation:0.10308
  |
Component:244; Jaccard:0.076148
 |
Component:178; Jaccard:0.079369
 |
Component:005; Jaccard:0.081307
 |
Component:014; Jaccard:0.075078
 |
Component:014; Jaccard:0.059059
 |
Component:005; Jaccard:0.04426
 |
Component:053; Jaccard:0.032008
 |
Correlation:0.34982
  |
Correlation:0.30526
  |
Correlation:-0.16455
  |
Correlation:0.018404
  |
Correlation:0.018404
  |
Correlation:-0.16455
  |
Correlation:0.15331
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































