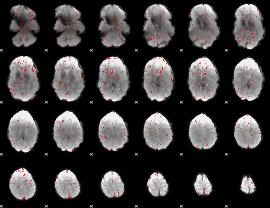
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
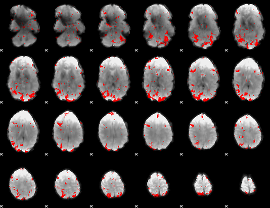 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
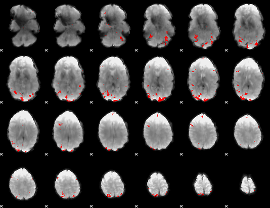 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
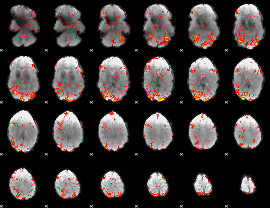 |
GLM Z-score result thresholded by 2.5
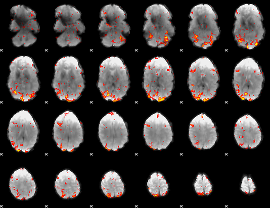 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
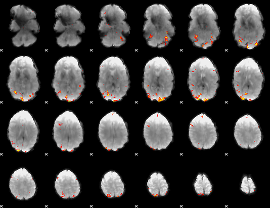 |
Component:191; Jaccard:0.142
 |
Component:191; Jaccard:0.1801
 |
Component:191; Jaccard:0.2112
 |
Component:191; Jaccard:0.22699
 |
Component:191; Jaccard:0.21598
 |
Component:191; Jaccard:0.18208
 |
Component:191; Jaccard:0.14377
 |
Correlation:0.41238
  |
Correlation:0.41238
  |
Correlation:0.41238
  |
Correlation:0.41238
  |
Correlation:0.41238
  |
Correlation:0.41238
  |
Correlation:0.41238
  |
Component:236; Jaccard:0.13811
 |
Component:236; Jaccard:0.16393
 |
Component:236; Jaccard:0.18509
 |
Component:236; Jaccard:0.19183
 |
Component:236; Jaccard:0.17823
 |
Component:236; Jaccard:0.15864
 |
Component:236; Jaccard:0.12843
 |
Correlation:0.28962
  |
Correlation:0.28962
  |
Correlation:0.28962
  |
Correlation:0.28962
  |
Correlation:0.28962
  |
Correlation:0.28962
  |
Correlation:0.28962
  |
Component:400; Jaccard:0.12279
 |
Component:400; Jaccard:0.14104
 |
Component:182; Jaccard:0.15596
 |
Component:182; Jaccard:0.16692
 |
Component:182; Jaccard:0.16564
 |
Component:182; Jaccard:0.1493
 |
Component:182; Jaccard:0.12381
 |
Correlation:0.53062
  |
Correlation:0.53062
  |
Correlation:0.031704
  |
Correlation:0.031704
  |
Correlation:0.031704
  |
Correlation:0.031704
  |
Correlation:0.031704
  |
Component:003; Jaccard:0.11658
 |
Component:182; Jaccard:0.13306
 |
Component:400; Jaccard:0.15309
 |
Component:400; Jaccard:0.15628
 |
Component:305; Jaccard:0.14648
 |
Component:305; Jaccard:0.13744
 |
Component:305; Jaccard:0.12154
 |
Correlation:0.11948
  |
Correlation:0.031704
  |
Correlation:0.53062
  |
Correlation:0.53062
  |
Correlation:0.42484
  |
Correlation:0.42484
  |
Correlation:0.42484
  |
Component:143; Jaccard:0.10926
 |
Component:188; Jaccard:0.12644
 |
Component:188; Jaccard:0.13998
 |
Component:305; Jaccard:0.14283
 |
Component:400; Jaccard:0.14635
 |
Component:400; Jaccard:0.12528
 |
Component:188; Jaccard:0.10514
 |
Correlation:0.6249
  |
Correlation:0.46585
  |
Correlation:0.46585
  |
Correlation:0.42484
  |
Correlation:0.53062
  |
Correlation:0.53062
  |
Correlation:0.46585
  |
Component:182; Jaccard:0.10716
 |
Component:143; Jaccard:0.12551
 |
Component:143; Jaccard:0.13366
 |
Component:188; Jaccard:0.1417
 |
Component:188; Jaccard:0.13683
 |
Component:188; Jaccard:0.12291
 |
Component:400; Jaccard:0.10102
 |
Correlation:0.031704
  |
Correlation:0.6249
  |
Correlation:0.6249
  |
Correlation:0.46585
  |
Correlation:0.46585
  |
Correlation:0.46585
  |
Correlation:0.53062
  |
Component:188; Jaccard:0.10546
 |
Component:003; Jaccard:0.12348
 |
Component:305; Jaccard:0.12821
 |
Component:143; Jaccard:0.13031
 |
Component:243; Jaccard:0.12159
 |
Component:243; Jaccard:0.11218
 |
Component:243; Jaccard:0.098933
 |
Correlation:0.46585
  |
Correlation:0.11948
  |
Correlation:0.42484
  |
Correlation:0.6249
  |
Correlation:0.52816
  |
Correlation:0.52816
  |
Correlation:0.52816
  |
Component:091; Jaccard:0.098633
 |
Component:305; Jaccard:0.10832
 |
Component:003; Jaccard:0.11844
 |
Component:243; Jaccard:0.11896
 |
Component:143; Jaccard:0.11628
 |
Component:143; Jaccard:0.095466
 |
Component:143; Jaccard:0.077254
 |
Correlation:0.29589
  |
Correlation:0.42484
  |
Correlation:0.11948
  |
Correlation:0.52816
  |
Correlation:0.6249
  |
Correlation:0.6249
  |
Correlation:0.6249
  |
Component:192; Jaccard:0.09565
 |
Component:197; Jaccard:0.10668
 |
Component:243; Jaccard:0.11361
 |
Component:192; Jaccard:0.10965
 |
Component:197; Jaccard:0.099741
 |
Component:192; Jaccard:0.085781
 |
Component:192; Jaccard:0.075812
 |
Correlation:0.27304
  |
Correlation:0.39916
  |
Correlation:0.52816
  |
Correlation:0.27304
  |
Correlation:0.39916
  |
Correlation:0.27304
  |
Correlation:0.27304
  |
Component:197; Jaccard:0.094085
 |
Component:192; Jaccard:0.10589
 |
Component:197; Jaccard:0.1134
 |
Component:197; Jaccard:0.10956
 |
Component:192; Jaccard:0.099285
 |
Component:197; Jaccard:0.082922
 |
Component:197; Jaccard:0.064398
 |
Correlation:0.39916
  |
Correlation:0.27304
  |
Correlation:0.39916
  |
Correlation:0.39916
  |
Correlation:0.27304
  |
Correlation:0.39916
  |
Correlation:0.39916
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
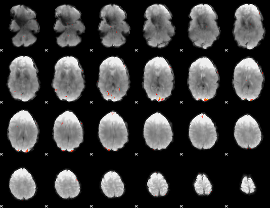 |
Component:182; Jaccard:0.1433
 |
Component:182; Jaccard:0.17014
 |
Component:182; Jaccard:0.18652
 |
Component:182; Jaccard:0.17261
 |
Component:182; Jaccard:0.14119
 |
Component:182; Jaccard:0.10267
 |
Component:182; Jaccard:0.068865
 |
Correlation:0.16619
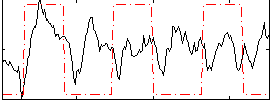 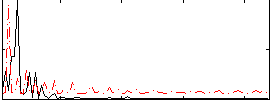 |
Correlation:0.16619
  |
Correlation:0.16619
  |
Correlation:0.16619
  |
Correlation:0.16619
  |
Correlation:0.16619
  |
Correlation:0.16619
  |
Component:208; Jaccard:0.08826
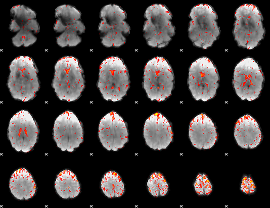 |
Component:389; Jaccard:0.078497
 |
Component:318; Jaccard:0.07577
 |
Component:318; Jaccard:0.067252
 |
Component:318; Jaccard:0.047119
 |
Component:099; Jaccard:0.027665
 |
Component:099; Jaccard:0.016697
 |
Correlation:-0.031431
  |
Correlation:-0.14661
  |
Correlation:-0.041191
  |
Correlation:-0.041191
  |
Correlation:-0.041191
  |
Correlation:0.03003
  |
Correlation:0.03003
  |
Component:039; Jaccard:0.081945
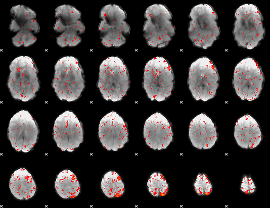 |
Component:208; Jaccard:0.077879
 |
Component:389; Jaccard:0.069878
 |
Component:307; Jaccard:0.058994
 |
Component:307; Jaccard:0.042304
 |
Component:069; Jaccard:0.025555
 |
Component:389; Jaccard:0.015145
 |
Correlation:0.14082
  |
Correlation:-0.031431
  |
Correlation:-0.14661
  |
Correlation:0.030526
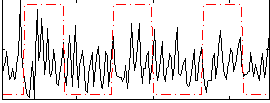  |
Correlation:0.030526
  |
Correlation:0.18978
  |
Correlation:-0.14661
  |
Component:261; Jaccard:0.077717
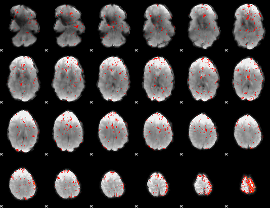 |
Component:261; Jaccard:0.076063
 |
Component:307; Jaccard:0.068589
 |
Component:389; Jaccard:0.054166
 |
Component:099; Jaccard:0.039309
 |
Component:389; Jaccard:0.025035
 |
Component:192; Jaccard:0.013868
 |
Correlation:0.23344
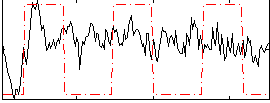  |
Correlation:0.23344
  |
Correlation:0.030526
  |
Correlation:-0.14661
  |
Correlation:0.03003
  |
Correlation:-0.14661
  |
Correlation:-0.27931
  |
Component:389; Jaccard:0.075611
 |
Component:039; Jaccard:0.075323
 |
Component:261; Jaccard:0.066832
 |
Component:069; Jaccard:0.04965
 |
Component:389; Jaccard:0.037627
 |
Component:307; Jaccard:0.024264
 |
Component:305; Jaccard:0.013792
 |
Correlation:-0.14661
  |
Correlation:0.14082
  |
Correlation:0.23344
  |
Correlation:0.18978
  |
Correlation:-0.14661
  |
Correlation:0.030526
  |
Correlation:-0.38015
  |
Component:345; Jaccard:0.064788
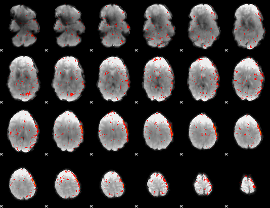 |
Component:318; Jaccard:0.068801
 |
Component:039; Jaccard:0.062192
 |
Component:099; Jaccard:0.049623
 |
Component:069; Jaccard:0.035835
 |
Component:318; Jaccard:0.023995
 |
Component:069; Jaccard:0.013485
 |
Correlation:0.005278
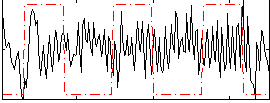 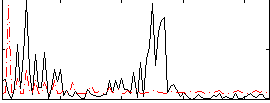 |
Correlation:-0.041191
  |
Correlation:0.14082
  |
Correlation:0.03003
  |
Correlation:0.18978
  |
Correlation:-0.041191
  |
Correlation:0.18978
  |
Component:221; Jaccard:0.064787
 |
Component:221; Jaccard:0.066659
 |
Component:099; Jaccard:0.058996
 |
Component:261; Jaccard:0.0479
 |
Component:261; Jaccard:0.029477
 |
Component:107; Jaccard:0.019561
 |
Component:107; Jaccard:0.013074
 |
Correlation:0.18652
  |
Correlation:0.18652
  |
Correlation:0.03003
  |
Correlation:0.23344
  |
Correlation:0.23344
  |
Correlation:0.55149
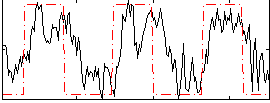  |
Correlation:0.55149
  |
Component:191; Jaccard:0.063726
 |
Component:307; Jaccard:0.064997
 |
Component:069; Jaccard:0.058679
 |
Component:039; Jaccard:0.043546
 |
Component:192; Jaccard:0.028659
 |
Component:236; Jaccard:0.018779
 |
Component:236; Jaccard:0.012711
 |
Correlation:-0.46222
  |
Correlation:0.030526
  |
Correlation:0.18978
  |
Correlation:0.14082
  |
Correlation:-0.27931
  |
Correlation:-0.27391
  |
Correlation:-0.27391
  |
Component:091; Jaccard:0.060931
 |
Component:099; Jaccard:0.062926
 |
Component:208; Jaccard:0.057392
 |
Component:221; Jaccard:0.041146
 |
Component:107; Jaccard:0.027592
 |
Component:192; Jaccard:0.018722
 |
Component:307; Jaccard:0.012238
 |
Correlation:-0.22382
  |
Correlation:0.03003
  |
Correlation:-0.031431
  |
Correlation:0.18652
  |
Correlation:0.55149
  |
Correlation:-0.27931
  |
Correlation:0.030526
  |
Component:099; Jaccard:0.060028
 |
Component:069; Jaccard:0.061033
 |
Component:221; Jaccard:0.056132
 |
Component:319; Jaccard:0.037857
 |
Component:221; Jaccard:0.027447
 |
Component:305; Jaccard:0.017601
 |
Component:319; Jaccard:0.011561
 |
Correlation:0.03003
  |
Correlation:0.18978
  |
Correlation:0.18652
  |
Correlation:-0.077432
  |
Correlation:0.18652
  |
Correlation:-0.38015
  |
Correlation:-0.077432
  |
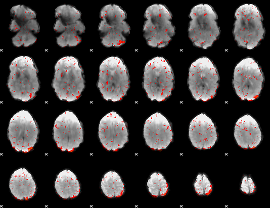
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
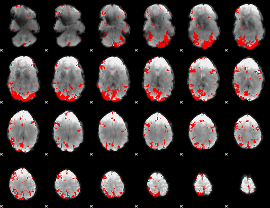 |
GLM binary result thresholded by 4
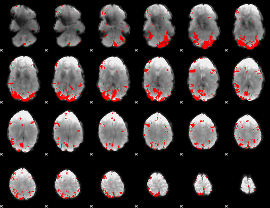 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
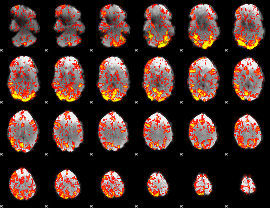 |
GLM Z-score result thresholded by 2
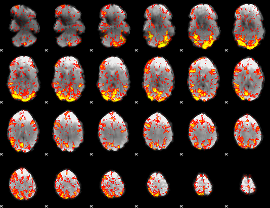 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
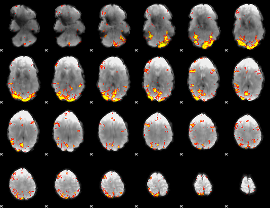 |
Component:400; Jaccard:0.12054
 |
Component:400; Jaccard:0.15293
 |
Component:400; Jaccard:0.19277
 |
Component:400; Jaccard:0.23245
 |
Component:400; Jaccard:0.26807
 |
Component:400; Jaccard:0.29174
 |
Component:377; Jaccard:0.30216
 |
Correlation:0.50741
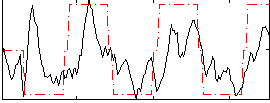 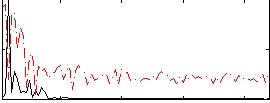 |
Correlation:0.50741
  |
Correlation:0.50741
  |
Correlation:0.50741
  |
Correlation:0.50741
  |
Correlation:0.50741
  |
Correlation:0.62017
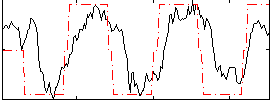 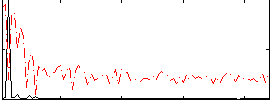 |
Component:377; Jaccard:0.11425
 |
Component:377; Jaccard:0.1469
 |
Component:377; Jaccard:0.18713
 |
Component:377; Jaccard:0.22871
 |
Component:377; Jaccard:0.26518
 |
Component:377; Jaccard:0.28917
 |
Component:400; Jaccard:0.29904
 |
Correlation:0.62017
  |
Correlation:0.62017
  |
Correlation:0.62017
  |
Correlation:0.62017
  |
Correlation:0.62017
  |
Correlation:0.62017
  |
Correlation:0.50741
  |
Component:303; Jaccard:0.10116
 |
Component:236; Jaccard:0.12469
 |
Component:236; Jaccard:0.15165
 |
Component:236; Jaccard:0.17874
 |
Component:236; Jaccard:0.20138
 |
Component:236; Jaccard:0.21482
 |
Component:236; Jaccard:0.2189
 |
Correlation:0.43019
  |
Correlation:0.30567
  |
Correlation:0.30567
  |
Correlation:0.30567
  |
Correlation:0.30567
  |
Correlation:0.30567
  |
Correlation:0.30567
  |
Component:236; Jaccard:0.10115
 |
Component:303; Jaccard:0.1171
 |
Component:191; Jaccard:0.14052
 |
Component:191; Jaccard:0.16817
 |
Component:191; Jaccard:0.19228
 |
Component:191; Jaccard:0.20606
 |
Component:191; Jaccard:0.20615
 |
Correlation:0.30567
  |
Correlation:0.43019
  |
Correlation:0.4744
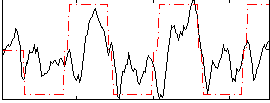  |
Correlation:0.4744
  |
Correlation:0.4744
  |
Correlation:0.4744
  |
Correlation:0.4744
  |
Component:003; Jaccard:0.097533
 |
Component:003; Jaccard:0.1134
 |
Component:303; Jaccard:0.12977
 |
Component:143; Jaccard:0.14127
 |
Component:143; Jaccard:0.1571
 |
Component:143; Jaccard:0.16465
 |
Component:305; Jaccard:0.1652
 |
Correlation:0.16629
  |
Correlation:0.16629
  |
Correlation:0.43019
  |
Correlation:0.58841
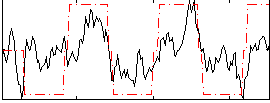  |
Correlation:0.58841
  |
Correlation:0.58841
  |
Correlation:0.43664
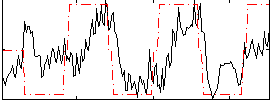  |
Component:332; Jaccard:0.094143
 |
Component:191; Jaccard:0.11335
 |
Component:003; Jaccard:0.12688
 |
Component:303; Jaccard:0.1394
 |
Component:128; Jaccard:0.15379
 |
Component:128; Jaccard:0.16198
 |
Component:128; Jaccard:0.16234
 |
Correlation:0.22811
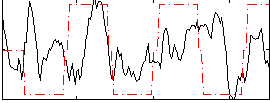  |
Correlation:0.4744
  |
Correlation:0.16629
  |
Correlation:0.43019
  |
Correlation:0.49626
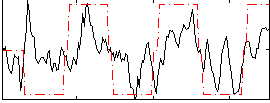  |
Correlation:0.49626
  |
Correlation:0.49626
  |
Component:060; Jaccard:0.092515
 |
Component:332; Jaccard:0.10817
 |
Component:332; Jaccard:0.12175
 |
Component:128; Jaccard:0.13823
 |
Component:188; Jaccard:0.14226
 |
Component:305; Jaccard:0.15458
 |
Component:143; Jaccard:0.16179
 |
Correlation:0.050427
  |
Correlation:0.22811
  |
Correlation:0.22811
  |
Correlation:0.49626
  |
Correlation:0.45197
  |
Correlation:0.43664
  |
Correlation:0.58841
  |
Component:376; Jaccard:0.090834
 |
Component:060; Jaccard:0.10533
 |
Component:143; Jaccard:0.12091
 |
Component:003; Jaccard:0.13466
 |
Component:303; Jaccard:0.14219
 |
Component:188; Jaccard:0.15312
 |
Component:188; Jaccard:0.1571
 |
Correlation:0.37019
  |
Correlation:0.050427
  |
Correlation:0.58841
  |
Correlation:0.16629
  |
Correlation:0.43019
  |
Correlation:0.45197
  |
Correlation:0.45197
  |
Component:191; Jaccard:0.090115
 |
Component:376; Jaccard:0.10115
 |
Component:128; Jaccard:0.11835
 |
Component:332; Jaccard:0.12816
 |
Component:305; Jaccard:0.14177
 |
Component:243; Jaccard:0.13689
 |
Component:243; Jaccard:0.14272
 |
Correlation:0.4744
  |
Correlation:0.37019
  |
Correlation:0.49626
  |
Correlation:0.22811
  |
Correlation:0.43664
  |
Correlation:0.55616
  |
Correlation:0.55616
  |
Component:323; Jaccard:0.087251
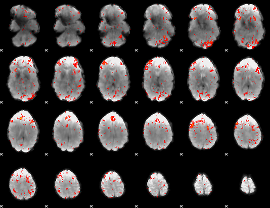 |
Component:323; Jaccard:0.10107
 |
Component:060; Jaccard:0.11631
 |
Component:323; Jaccard:0.12614
 |
Component:003; Jaccard:0.13572
 |
Component:303; Jaccard:0.13663
 |
Component:323; Jaccard:0.12531
 |
Correlation:0.39025
  |
Correlation:0.39025
  |
Correlation:0.050427
  |
Correlation:0.39025
  |
Correlation:0.16629
  |
Correlation:0.43019
  |
Correlation:0.39025
  |
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
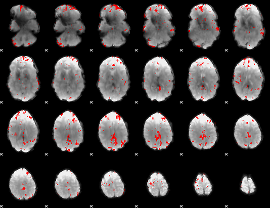 |
GLM binary result thresholded by 4
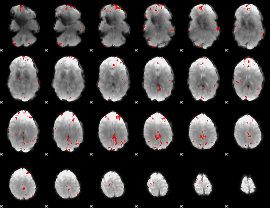 |
GLM Z-score result thresholded by 1
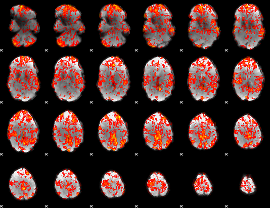 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
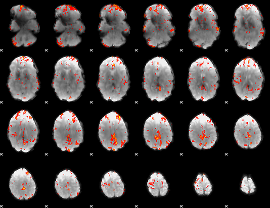 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:072; Jaccard:0.099982
 |
Component:072; Jaccard:0.12656
 |
Component:072; Jaccard:0.15743
 |
Component:037; Jaccard:0.18424
 |
Component:037; Jaccard:0.19979
 |
Component:037; Jaccard:0.18882
 |
Component:037; Jaccard:0.1556
 |
Correlation:0.11549
  |
Correlation:0.11549
  |
Correlation:0.11549
  |
Correlation:0.23185
  |
Correlation:0.23185
  |
Correlation:0.23185
  |
Correlation:0.23185
  |
Component:228; Jaccard:0.098423
 |
Component:037; Jaccard:0.12517
 |
Component:037; Jaccard:0.15686
 |
Component:072; Jaccard:0.18362
 |
Component:072; Jaccard:0.19666
 |
Component:072; Jaccard:0.18546
 |
Component:072; Jaccard:0.14885
 |
Correlation:0.19776
  |
Correlation:0.23185
  |
Correlation:0.23185
  |
Correlation:0.11549
  |
Correlation:0.11549
  |
Correlation:0.11549
  |
Correlation:0.11549
  |
Component:037; Jaccard:0.098108
 |
Component:228; Jaccard:0.11862
 |
Component:228; Jaccard:0.1395
 |
Component:228; Jaccard:0.15374
 |
Component:228; Jaccard:0.15688
 |
Component:228; Jaccard:0.1417
 |
Component:228; Jaccard:0.11361
 |
Correlation:0.23185
  |
Correlation:0.19776
  |
Correlation:0.19776
  |
Correlation:0.19776
  |
Correlation:0.19776
  |
Correlation:0.19776
  |
Correlation:0.19776
  |
Component:174; Jaccard:0.097782
 |
Component:014; Jaccard:0.11413
 |
Component:014; Jaccard:0.1309
 |
Component:014; Jaccard:0.14094
 |
Component:123; Jaccard:0.14273
 |
Component:123; Jaccard:0.13002
 |
Component:123; Jaccard:0.1004
 |
Correlation:-0.1379
  |
Correlation:0.12333
  |
Correlation:0.12333
  |
Correlation:0.12333
  |
Correlation:0.24845
  |
Correlation:0.24845
  |
Correlation:0.24845
  |
Component:014; Jaccard:0.096969
 |
Component:174; Jaccard:0.11341
 |
Component:174; Jaccard:0.12388
 |
Component:123; Jaccard:0.13711
 |
Component:014; Jaccard:0.13924
 |
Component:014; Jaccard:0.12089
 |
Component:014; Jaccard:0.093244
 |
Correlation:0.12333
  |
Correlation:-0.1379
  |
Correlation:-0.1379
  |
Correlation:0.24845
  |
Correlation:0.12333
  |
Correlation:0.12333
  |
Correlation:0.12333
  |
Component:343; Jaccard:0.093943
 |
Component:343; Jaccard:0.10628
 |
Component:123; Jaccard:0.1201
 |
Component:174; Jaccard:0.12633
 |
Component:174; Jaccard:0.11664
 |
Component:284; Jaccard:0.10741
 |
Component:284; Jaccard:0.090885
 |
Correlation:0.088731
  |
Correlation:0.088731
  |
Correlation:0.24845
  |
Correlation:-0.1379
  |
Correlation:-0.1379
  |
Correlation:0.091765
  |
Correlation:0.091765
  |
Component:097; Jaccard:0.086848
 |
Component:123; Jaccard:0.098574
 |
Component:343; Jaccard:0.11531
 |
Component:343; Jaccard:0.11838
 |
Component:284; Jaccard:0.11642
 |
Component:174; Jaccard:0.09342
 |
Component:153; Jaccard:0.07477
 |
Correlation:-0.071503
  |
Correlation:0.24845
  |
Correlation:0.088731
  |
Correlation:0.088731
  |
Correlation:0.091765
  |
Correlation:-0.1379
  |
Correlation:0.067148
  |
Component:352; Jaccard:0.086102
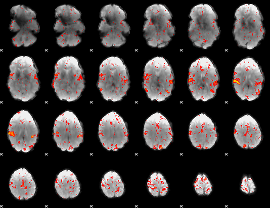 |
Component:284; Jaccard:0.093721
 |
Component:284; Jaccard:0.10588
 |
Component:284; Jaccard:0.11338
 |
Component:343; Jaccard:0.10937
 |
Component:153; Jaccard:0.092506
 |
Component:343; Jaccard:0.068699
 |
Correlation:0.030798
  |
Correlation:0.091765
  |
Correlation:0.091765
  |
Correlation:0.091765
  |
Correlation:0.088731
  |
Correlation:0.067148
  |
Correlation:0.088731
  |
Component:257; Jaccard:0.081635
 |
Component:097; Jaccard:0.091935
 |
Component:153; Jaccard:0.10085
 |
Component:153; Jaccard:0.10596
 |
Component:153; Jaccard:0.10371
 |
Component:343; Jaccard:0.09102
 |
Component:174; Jaccard:0.068472
 |
Correlation:0.04453
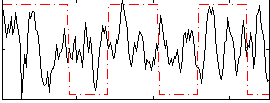  |
Correlation:-0.071503
  |
Correlation:0.067148
  |
Correlation:0.067148
  |
Correlation:0.067148
  |
Correlation:0.088731
  |
Correlation:-0.1379
  |
Component:284; Jaccard:0.080177
 |
Component:153; Jaccard:0.091197
 |
Component:097; Jaccard:0.093671
 |
Component:097; Jaccard:0.092404
 |
Component:097; Jaccard:0.084602
 |
Component:097; Jaccard:0.071888
 |
Component:097; Jaccard:0.055489
 |
Correlation:0.091765
  |
Correlation:0.067148
  |
Correlation:-0.071503
  |
Correlation:-0.071503
  |
Correlation:-0.071503
  |
Correlation:-0.071503
  |
Correlation:-0.071503
  |
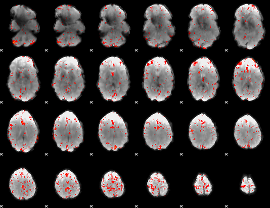
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
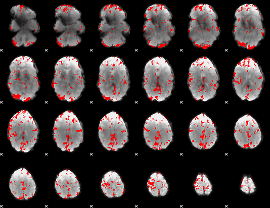 |
GLM binary result thresholded by 3.5
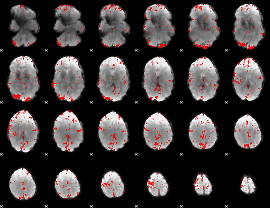 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
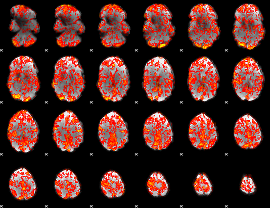 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
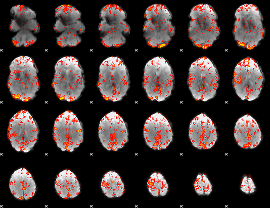 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:174; Jaccard:0.10309
 |
Component:174; Jaccard:0.12914
 |
Component:174; Jaccard:0.16019
 |
Component:174; Jaccard:0.19333
 |
Component:174; Jaccard:0.21207
 |
Component:174; Jaccard:0.20728
 |
Component:174; Jaccard:0.17592
 |
Correlation:0.13292
  |
Correlation:0.13292
  |
Correlation:0.13292
  |
Correlation:0.13292
  |
Correlation:0.13292
  |
Correlation:0.13292
  |
Correlation:0.13292
  |
Component:044; Jaccard:0.091792
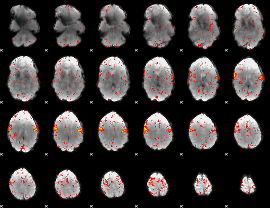 |
Component:044; Jaccard:0.10506
 |
Component:044; Jaccard:0.11703
 |
Component:377; Jaccard:0.12965
 |
Component:377; Jaccard:0.14028
 |
Component:377; Jaccard:0.14032
 |
Component:377; Jaccard:0.13143
 |
Correlation:0.13444
  |
Correlation:0.13444
  |
Correlation:0.13444
  |
Correlation:0.58921
  |
Correlation:0.58921
  |
Correlation:0.58921
  |
Correlation:0.58921
  |
Component:377; Jaccard:0.086202
 |
Component:377; Jaccard:0.099081
 |
Component:377; Jaccard:0.11456
 |
Component:044; Jaccard:0.12569
 |
Component:044; Jaccard:0.12509
 |
Component:044; Jaccard:0.11526
 |
Component:044; Jaccard:0.093558
 |
Correlation:0.58921
  |
Correlation:0.58921
  |
Correlation:0.58921
  |
Correlation:0.13444
  |
Correlation:0.13444
  |
Correlation:0.13444
  |
Correlation:0.13444
  |
Component:227; Jaccard:0.082942
 |
Component:227; Jaccard:0.093031
 |
Component:343; Jaccard:0.10531
 |
Component:343; Jaccard:0.11613
 |
Component:343; Jaccard:0.11762
 |
Component:343; Jaccard:0.1105
 |
Component:343; Jaccard:0.090264
 |
Correlation:0.11221
  |
Correlation:0.11221
  |
Correlation:-0.16697
  |
Correlation:-0.16697
  |
Correlation:-0.16697
  |
Correlation:-0.16697
  |
Correlation:-0.16697
  |
Component:352; Jaccard:0.080307
 |
Component:343; Jaccard:0.092477
 |
Component:227; Jaccard:0.10134
 |
Component:227; Jaccard:0.10509
 |
Component:037; Jaccard:0.10835
 |
Component:037; Jaccard:0.10136
 |
Component:112; Jaccard:0.085757
 |
Correlation:-0.074148
  |
Correlation:-0.16697
  |
Correlation:0.11221
  |
Correlation:0.11221
  |
Correlation:-0.16232
 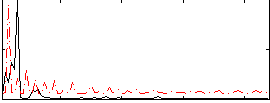 |
Correlation:-0.16232
  |
Correlation:0.25587
 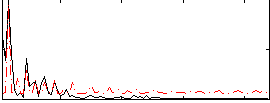 |
Component:343; Jaccard:0.080101
 |
Component:352; Jaccard:0.086883
 |
Component:112; Jaccard:0.096247
 |
Component:112; Jaccard:0.10299
 |
Component:227; Jaccard:0.10639
 |
Component:112; Jaccard:0.099369
 |
Component:037; Jaccard:0.081738
 |
Correlation:-0.16697
  |
Correlation:-0.074148
  |
Correlation:0.25587
  |
Correlation:0.25587
  |
Correlation:0.11221
  |
Correlation:0.25587
  |
Correlation:-0.16232
  |
Component:112; Jaccard:0.076828
 |
Component:112; Jaccard:0.08685
 |
Component:072; Jaccard:0.093941
 |
Component:072; Jaccard:0.10281
 |
Component:112; Jaccard:0.10438
 |
Component:227; Jaccard:0.096885
 |
Component:227; Jaccard:0.080388
 |
Correlation:0.25587
  |
Correlation:0.25587
  |
Correlation:-0.0088346
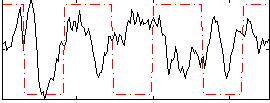  |
Correlation:-0.0088346
  |
Correlation:0.25587
  |
Correlation:0.11221
  |
Correlation:0.11221
  |
Component:097; Jaccard:0.076804
 |
Component:297; Jaccard:0.085763
 |
Component:037; Jaccard:0.092022
 |
Component:037; Jaccard:0.10264
 |
Component:072; Jaccard:0.10433
 |
Component:072; Jaccard:0.096352
 |
Component:072; Jaccard:0.078551
 |
Correlation:0.042102
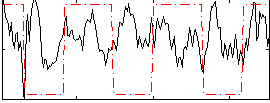  |
Correlation:0.18857
  |
Correlation:-0.16232
  |
Correlation:-0.16232
  |
Correlation:-0.0088346
  |
Correlation:-0.0088346
  |
Correlation:-0.0088346
  |
Component:368; Jaccard:0.076635
 |
Component:097; Jaccard:0.083171
 |
Component:297; Jaccard:0.091793
 |
Component:297; Jaccard:0.094784
 |
Component:228; Jaccard:0.095489
 |
Component:153; Jaccard:0.087345
 |
Component:153; Jaccard:0.077908
 |
Correlation:0.12759
  |
Correlation:0.042102
  |
Correlation:0.18857
  |
Correlation:0.18857
  |
Correlation:-0.20717
 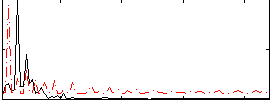 |
Correlation:0.06239
  |
Correlation:0.06239
  |
Component:297; Jaccard:0.076512
 |
Component:201; Jaccard:0.083099
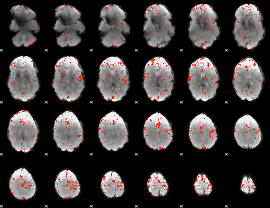 |
Component:352; Jaccard:0.090912
 |
Component:228; Jaccard:0.093261
 |
Component:201; Jaccard:0.090617
 |
Component:228; Jaccard:0.087079
 |
Component:228; Jaccard:0.070321
 |
Correlation:0.18857
  |
Correlation:-0.13665
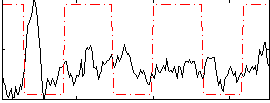 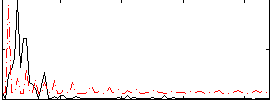 |
Correlation:-0.074148
  |
Correlation:-0.20717
  |
Correlation:-0.13665
  |
Correlation:-0.20717
  |
Correlation:-0.20717
  |
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
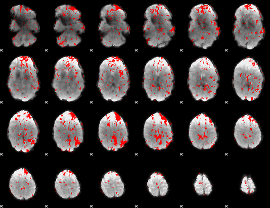 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
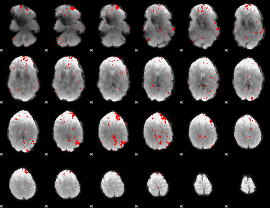 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
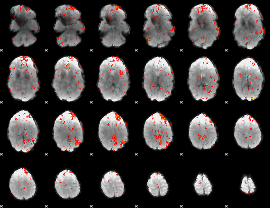 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
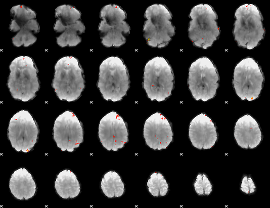 |
Component:107; Jaccard:0.11823
 |
Component:107; Jaccard:0.15242
 |
Component:107; Jaccard:0.17522
 |
Component:107; Jaccard:0.16997
 |
Component:107; Jaccard:0.1399
 |
Component:107; Jaccard:0.095161
 |
Component:107; Jaccard:0.054795
 |
Correlation:0.55223
  |
Correlation:0.55223
  |
Correlation:0.55223
  |
Correlation:0.55223
  |
Correlation:0.55223
  |
Correlation:0.55223
  |
Correlation:0.55223
  |
Component:037; Jaccard:0.11372
 |
Component:072; Jaccard:0.12672
 |
Component:072; Jaccard:0.13045
 |
Component:072; Jaccard:0.11641
 |
Component:072; Jaccard:0.089455
 |
Component:123; Jaccard:0.055372
 |
Component:072; Jaccard:0.027285
 |
Correlation:0.2138
  |
Correlation:0.067436
  |
Correlation:0.067436
  |
Correlation:0.067436
  |
Correlation:0.067436
  |
Correlation:0.18051
  |
Correlation:0.067436
  |
Component:072; Jaccard:0.11224
 |
Component:037; Jaccard:0.12669
 |
Component:037; Jaccard:0.12706
 |
Component:123; Jaccard:0.11601
 |
Component:123; Jaccard:0.08924
 |
Component:072; Jaccard:0.054081
 |
Component:123; Jaccard:0.026638
 |
Correlation:0.067436
  |
Correlation:0.2138
  |
Correlation:0.2138
  |
Correlation:0.18051
  |
Correlation:0.18051
  |
Correlation:0.067436
  |
Correlation:0.18051
  |
Component:123; Jaccard:0.10565
 |
Component:123; Jaccard:0.12059
 |
Component:123; Jaccard:0.12584
 |
Component:154; Jaccard:0.1113
 |
Component:154; Jaccard:0.084354
 |
Component:154; Jaccard:0.053659
 |
Component:154; Jaccard:0.025448
 |
Correlation:0.18051
  |
Correlation:0.18051
  |
Correlation:0.18051
  |
Correlation:0.30573
  |
Correlation:0.30573
  |
Correlation:0.30573
  |
Correlation:0.30573
  |
Component:014; Jaccard:0.10152
 |
Component:154; Jaccard:0.11324
 |
Component:154; Jaccard:0.12063
 |
Component:037; Jaccard:0.10719
 |
Component:037; Jaccard:0.075402
 |
Component:037; Jaccard:0.044991
 |
Component:037; Jaccard:0.022749
 |
Correlation:0.16255
  |
Correlation:0.30573
  |
Correlation:0.30573
  |
Correlation:0.2138
  |
Correlation:0.2138
  |
Correlation:0.2138
  |
Correlation:0.2138
  |
Component:228; Jaccard:0.09977
 |
Component:014; Jaccard:0.10395
 |
Component:228; Jaccard:0.09678
 |
Component:228; Jaccard:0.081666
 |
Component:228; Jaccard:0.057359
 |
Component:360; Jaccard:0.036538
 |
Component:360; Jaccard:0.019126
 |
Correlation:0.21964
  |
Correlation:0.16255
  |
Correlation:0.21964
  |
Correlation:0.21964
  |
Correlation:0.21964
  |
Correlation:0.074959
  |
Correlation:0.074959
  |
Component:154; Jaccard:0.098482
 |
Component:228; Jaccard:0.10166
 |
Component:014; Jaccard:0.096666
 |
Component:014; Jaccard:0.076788
 |
Component:360; Jaccard:0.055696
 |
Component:228; Jaccard:0.034294
 |
Component:228; Jaccard:0.016568
 |
Correlation:0.30573
  |
Correlation:0.21964
  |
Correlation:0.16255
  |
Correlation:0.16255
  |
Correlation:0.074959
  |
Correlation:0.21964
  |
Correlation:0.21964
  |
Component:310; Jaccard:0.083908
 |
Component:310; Jaccard:0.091972
 |
Component:310; Jaccard:0.090155
 |
Component:310; Jaccard:0.075709
 |
Component:310; Jaccard:0.054292
 |
Component:310; Jaccard:0.033748
 |
Component:225; Jaccard:0.01642
 |
Correlation:0.16968
  |
Correlation:0.16968
  |
Correlation:0.16968
  |
Correlation:0.16968
  |
Correlation:0.16968
  |
Correlation:0.16968
  |
Correlation:0.12181
  |
Component:284; Jaccard:0.079252
 |
Component:284; Jaccard:0.082291
 |
Component:360; Jaccard:0.07732
 |
Component:360; Jaccard:0.073021
 |
Component:014; Jaccard:0.050468
 |
Component:014; Jaccard:0.030614
 |
Component:014; Jaccard:0.015518
 |
Correlation:0.027304
  |
Correlation:0.027304
  |
Correlation:0.074959
  |
Correlation:0.074959
  |
Correlation:0.16255
  |
Correlation:0.16255
  |
Correlation:0.16255
  |
Component:068; Jaccard:0.072656
 |
Component:225; Jaccard:0.077563
 |
Component:284; Jaccard:0.076714
 |
Component:225; Jaccard:0.064988
 |
Component:284; Jaccard:0.045795
 |
Component:225; Jaccard:0.030009
 |
Component:310; Jaccard:0.014791
 |
Correlation:0.17413
  |
Correlation:0.12181
  |
Correlation:0.027304
  |
Correlation:0.12181
  |
Correlation:0.027304
  |
Correlation:0.12181
  |
Correlation:0.16968
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































