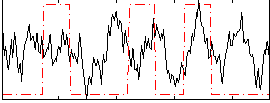
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
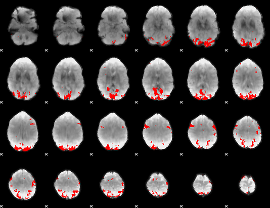
GLM binary result thresholded by 1
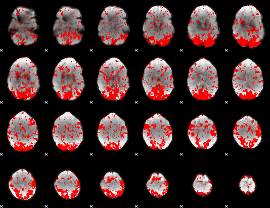 |
GLM binary result thresholded by 1.5
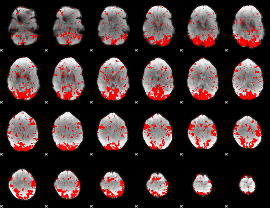 |
GLM binary result thresholded by 2
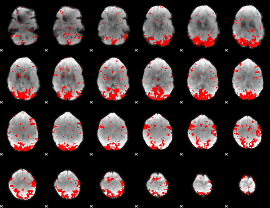 |
GLM binary result thresholded by 2.5
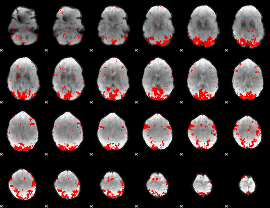 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
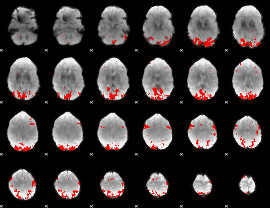 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
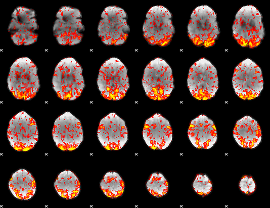 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
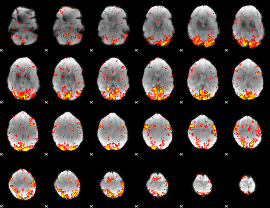 |
GLM Z-score result thresholded by 2.5
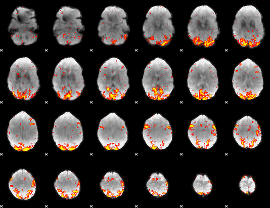 |
GLM Z-score result thresholded by 3
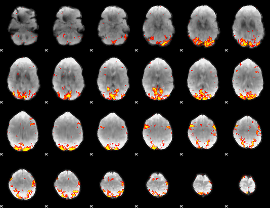 |
GLM Z-score result thresholded by 3.5
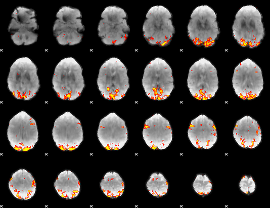 |
GLM Z-score result thresholded by 4
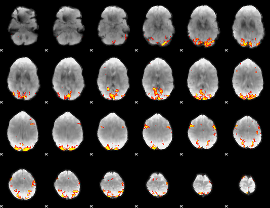 |
Component:111; Jaccard:0.18018
 |
Component:111; Jaccard:0.24205
 |
Component:111; Jaccard:0.31507
 |
Component:111; Jaccard:0.37923
 |
Component:111; Jaccard:0.42245
 |
Component:111; Jaccard:0.42986
 |
Component:111; Jaccard:0.41336
 |
Correlation:0.28292
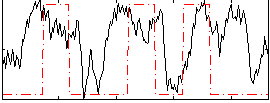 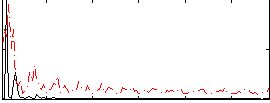 |
Correlation:0.28292
  |
Correlation:0.28292
  |
Correlation:0.28292
  |
Correlation:0.28292
  |
Correlation:0.28292
  |
Correlation:0.28292
  |
Component:213; Jaccard:0.14302
 |
Component:213; Jaccard:0.18668
 |
Component:213; Jaccard:0.23152
 |
Component:213; Jaccard:0.26632
 |
Component:213; Jaccard:0.28579
 |
Component:213; Jaccard:0.2858
 |
Component:213; Jaccard:0.27117
 |
Correlation:0.23971
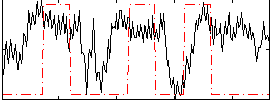 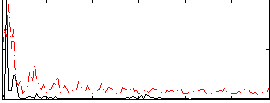 |
Correlation:0.23971
  |
Correlation:0.23971
  |
Correlation:0.23971
  |
Correlation:0.23971
  |
Correlation:0.23971
  |
Correlation:0.23971
  |
Component:391; Jaccard:0.13242
 |
Component:284; Jaccard:0.15937
 |
Component:284; Jaccard:0.19142
 |
Component:284; Jaccard:0.21593
 |
Component:284; Jaccard:0.22889
 |
Component:284; Jaccard:0.22831
 |
Component:284; Jaccard:0.22105
 |
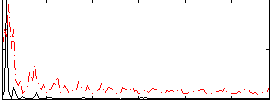
Correlation:0.14008
  |
Correlation:0.28809
  |
Correlation:0.28809
  |
Correlation:0.28809
  |
Correlation:0.28809
  |
Correlation:0.28809
  |
Correlation:0.28809
  |
Component:284; Jaccard:0.12779
 |
Component:059; Jaccard:0.15266
 |
Component:059; Jaccard:0.18516
 |
Component:059; Jaccard:0.20837
 |
Component:059; Jaccard:0.22028
 |
Component:059; Jaccard:0.22002
 |
Component:059; Jaccard:0.2142
 |
Correlation:0.28809
  |
Correlation:0.31541
  |
Correlation:0.31541
  |
Correlation:0.31541
  |
Correlation:0.31541
  |
Correlation:0.31541
  |
Correlation:0.31541
  |
Component:059; Jaccard:0.12215
 |
Component:391; Jaccard:0.15238
 |
Component:391; Jaccard:0.16845
 |
Component:069; Jaccard:0.17693
 |
Component:069; Jaccard:0.18913
 |
Component:069; Jaccard:0.19172
 |
Component:069; Jaccard:0.18303
 |
Correlation:0.31541
  |
Correlation:0.14008
  |
Correlation:0.14008
  |
Correlation:0.26375
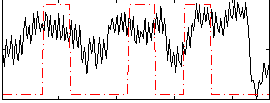 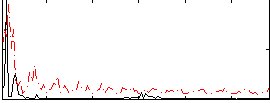 |
Correlation:0.26375
  |
Correlation:0.26375
  |
Correlation:0.26375
  |
Component:230; Jaccard:0.11707
 |
Component:230; Jaccard:0.14018
 |
Component:230; Jaccard:0.16202
 |
Component:230; Jaccard:0.17541
 |
Component:230; Jaccard:0.17882
 |
Component:230; Jaccard:0.17354
 |
Component:230; Jaccard:0.16226
 |
Correlation:0.27645
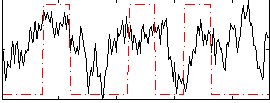 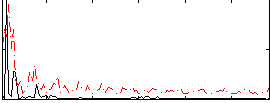 |
Correlation:0.27645
  |
Correlation:0.27645
  |
Correlation:0.27645
  |
Correlation:0.27645
  |
Correlation:0.27645
  |
Correlation:0.27645
  |
Component:069; Jaccard:0.11222
 |
Component:069; Jaccard:0.13739
 |
Component:069; Jaccard:0.15981
 |
Component:391; Jaccard:0.17006
 |
Component:391; Jaccard:0.16307
 |
Component:289; Jaccard:0.15705
 |
Component:289; Jaccard:0.14921
 |
Correlation:0.26375
  |
Correlation:0.26375
  |
Correlation:0.26375
  |
Correlation:0.14008
  |
Correlation:0.14008
  |
Correlation:0.32526
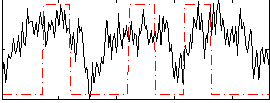  |
Correlation:0.32526
  |
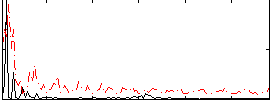
Component:150; Jaccard:0.11053
 |
Component:289; Jaccard:0.13094
 |
Component:289; Jaccard:0.14972
 |
Component:289; Jaccard:0.16178
 |
Component:289; Jaccard:0.16278
 |
Component:391; Jaccard:0.14888
 |
Component:391; Jaccard:0.13181
 |
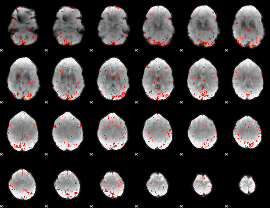
Correlation:0.1253
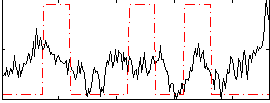 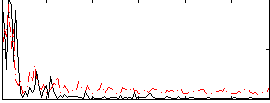 |
Correlation:0.32526
  |
Correlation:0.32526
  |
Correlation:0.32526
  |
Correlation:0.32526
  |
Correlation:0.14008
  |
Correlation:0.14008
  |
Component:289; Jaccard:0.10903
 |
Component:150; Jaccard:0.12168
 |
Component:187; Jaccard:0.12945
 |
Component:187; Jaccard:0.13832
 |
Component:187; Jaccard:0.13947
 |
Component:187; Jaccard:0.13281
 |
Component:187; Jaccard:0.12312
 |
Correlation:0.32526
  |
Correlation:0.1253
  |
Correlation:0.35102
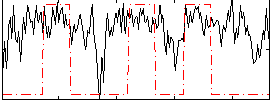 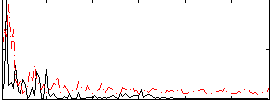 |
Correlation:0.35102
  |
Correlation:0.35102
  |
Correlation:0.35102
  |
Correlation:0.35102
  |
Component:389; Jaccard:0.099695
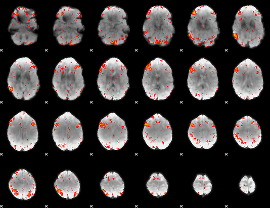 |
Component:187; Jaccard:0.11597
 |
Component:150; Jaccard:0.12558
 |
Component:150; Jaccard:0.12057
 |
Component:389; Jaccard:0.10832
 |
Component:389; Jaccard:0.098913
 |
Component:138; Jaccard:0.092133
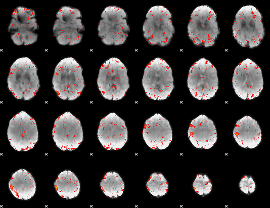 |
Correlation:0.40774
  |
Correlation:0.35102
  |
Correlation:0.1253
  |
Correlation:0.1253
  |
Correlation:0.40774
  |
Correlation:0.40774
  |
Correlation:0.25361
  |
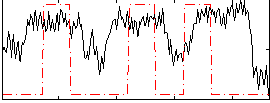
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
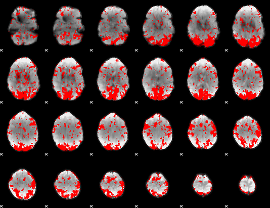 |
GLM binary result thresholded by 2
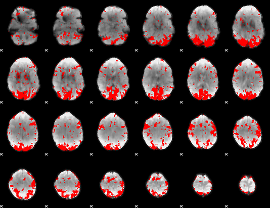 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
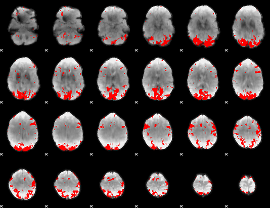 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
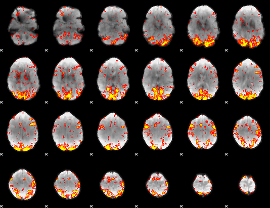
GLM Z-score result thresholded by 2
 |
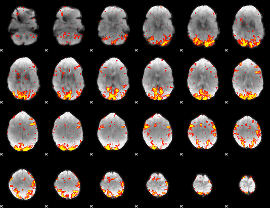
GLM Z-score result thresholded by 2.5
 |
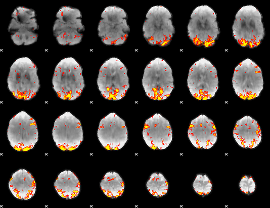
GLM Z-score result thresholded by 3
 |
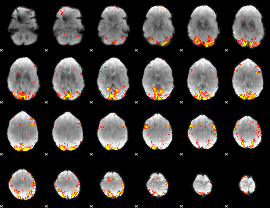
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
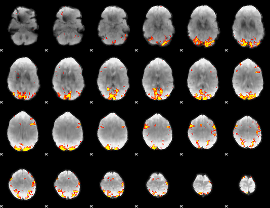 |
Component:111; Jaccard:0.16901
 |
Component:111; Jaccard:0.22563
 |
Component:111; Jaccard:0.29326
 |
Component:111; Jaccard:0.36126
 |
Component:111; Jaccard:0.41707
 |
Component:111; Jaccard:0.45332
 |
Component:111; Jaccard:0.46014
 |
Correlation:0.2433
 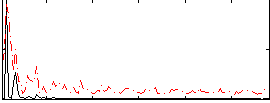 |
Correlation:0.2433
  |
Correlation:0.2433
  |
Correlation:0.2433
  |
Correlation:0.2433
  |
Correlation:0.2433
  |
Correlation:0.2433
  |
Component:391; Jaccard:0.13642
 |
Component:213; Jaccard:0.16745
 |
Component:213; Jaccard:0.20439
 |
Component:213; Jaccard:0.23558
 |
Component:213; Jaccard:0.25734
 |
Component:213; Jaccard:0.26838
 |
Component:213; Jaccard:0.26689
 |
Correlation:0.18082
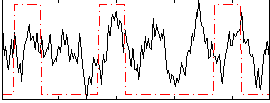 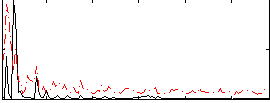 |
Correlation:0.091612
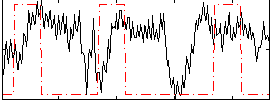 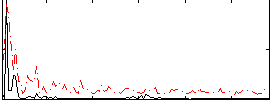 |
Correlation:0.091612
  |
Correlation:0.091612
  |
Correlation:0.091612
  |
Correlation:0.091612
  |
Correlation:0.091612
  |
Component:213; Jaccard:0.13168
 |
Component:284; Jaccard:0.16366
 |
Component:284; Jaccard:0.19974
 |
Component:284; Jaccard:0.22963
 |
Component:284; Jaccard:0.24785
 |
Component:284; Jaccard:0.25413
 |
Component:284; Jaccard:0.24969
 |
Correlation:0.091612
  |
Correlation:0.15218
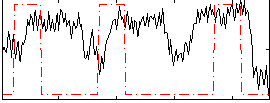 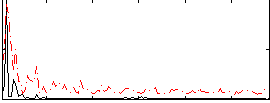 |
Correlation:0.15218
  |
Correlation:0.15218
  |
Correlation:0.15218
  |
Correlation:0.15218
  |
Correlation:0.15218
  |
Component:284; Jaccard:0.1298
 |
Component:391; Jaccard:0.15977
 |
Component:059; Jaccard:0.18203
 |
Component:059; Jaccard:0.21004
 |
Component:059; Jaccard:0.22586
 |
Component:059; Jaccard:0.23265
 |
Component:059; Jaccard:0.22564
 |
Correlation:0.15218
  |
Correlation:0.18082
  |
Correlation:0.06909
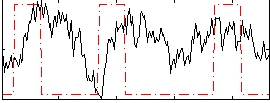 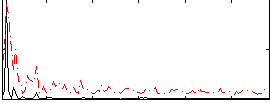 |
Correlation:0.06909
  |
Correlation:0.06909
  |
Correlation:0.06909
  |
Correlation:0.06909
  |
Component:069; Jaccard:0.12109
 |
Component:069; Jaccard:0.15039
 |
Component:391; Jaccard:0.17769
 |
Component:069; Jaccard:0.19866
 |
Component:069; Jaccard:0.21123
 |
Component:069; Jaccard:0.21829
 |
Component:069; Jaccard:0.21675
 |
Correlation:0.24842
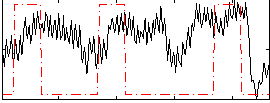 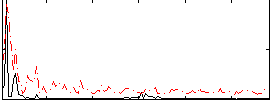 |
Correlation:0.24842
  |
Correlation:0.18082
  |
Correlation:0.24842
  |
Correlation:0.24842
  |
Correlation:0.24842
  |
Correlation:0.24842
  |
Component:059; Jaccard:0.11761
 |
Component:059; Jaccard:0.14905
 |
Component:069; Jaccard:0.1766
 |
Component:391; Jaccard:0.18399
 |
Component:391; Jaccard:0.18057
 |
Component:391; Jaccard:0.16764
 |
Component:230; Jaccard:0.15787
 |
Correlation:0.06909
  |
Correlation:0.06909
  |
Correlation:0.24842
  |
Correlation:0.18082
  |
Correlation:0.18082
  |
Correlation:0.18082
  |
Correlation:-0.093999
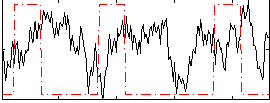 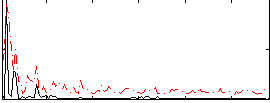 |
Component:389; Jaccard:0.10771
 |
Component:230; Jaccard:0.12692
 |
Component:230; Jaccard:0.14618
 |
Component:230; Jaccard:0.15699
 |
Component:230; Jaccard:0.16487
 |
Component:230; Jaccard:0.16306
 |
Component:391; Jaccard:0.15229
 |
Correlation:0.034878
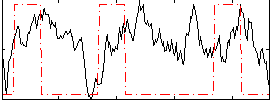 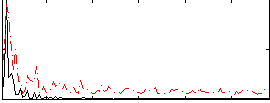 |
Correlation:-0.093999
  |
Correlation:-0.093999
  |
Correlation:-0.093999
  |
Correlation:-0.093999
  |
Correlation:-0.093999
  |
Correlation:0.18082
  |
Component:230; Jaccard:0.10694
 |
Component:389; Jaccard:0.12191
 |
Component:289; Jaccard:0.13672
 |
Component:289; Jaccard:0.15109
 |
Component:289; Jaccard:0.15541
 |
Component:289; Jaccard:0.15253
 |
Component:289; Jaccard:0.14945
 |
Correlation:-0.093999
  |
Correlation:0.034878
  |
Correlation:0.16417
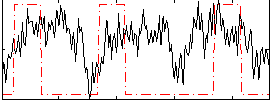 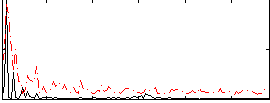 |
Correlation:0.16417
  |
Correlation:0.16417
  |
Correlation:0.16417
  |
Correlation:0.16417
  |
Component:121; Jaccard:0.10419
 |
Component:289; Jaccard:0.11917
 |
Component:389; Jaccard:0.13335
 |
Component:389; Jaccard:0.13667
 |
Component:187; Jaccard:0.13991
 |
Component:187; Jaccard:0.13825
 |
Component:187; Jaccard:0.13508
 |
Correlation:0.23308
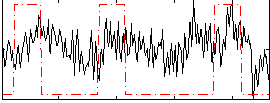  |
Correlation:0.16417
  |
Correlation:0.034878
  |
Correlation:0.034878
  |
Correlation:-0.0036184
  |
Correlation:-0.0036184
  |
Correlation:-0.0036184
  |
Component:289; Jaccard:0.10019
 |
Component:121; Jaccard:0.11607
 |
Component:187; Jaccard:0.12517
 |
Component:187; Jaccard:0.13523
 |
Component:389; Jaccard:0.13084
 |
Component:389; Jaccard:0.12186
 |
Component:389; Jaccard:0.10712
 |
Correlation:0.16417
  |
Correlation:0.23308
  |
Correlation:-0.0036184
  |
Correlation:-0.0036184
  |
Correlation:0.034878
  |
Correlation:0.034878
  |
Correlation:0.034878
  |
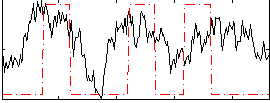
Cope: 03:MATCH-REL BACK
|
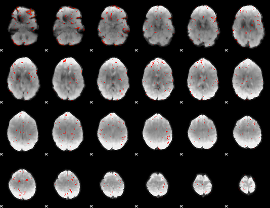
GLM result group-wise
 |
GLM binary result thresholded by 1
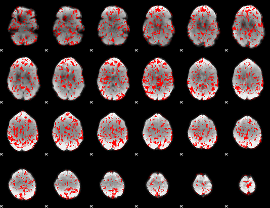 |
GLM binary result thresholded by 1.5
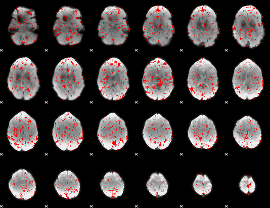 |
GLM binary result thresholded by 2
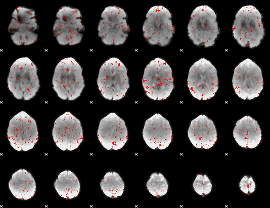 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
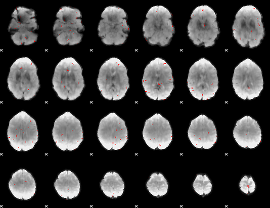 |
GLM binary result thresholded by 3.5
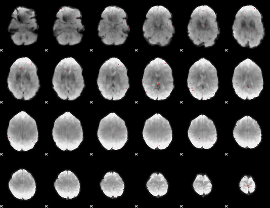 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
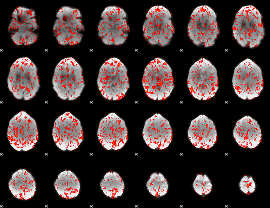 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
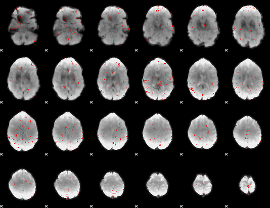 |
GLM Z-score result thresholded by 3
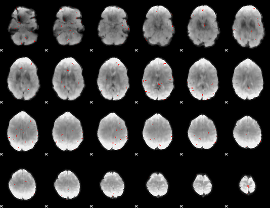 |
GLM Z-score result thresholded by 3.5
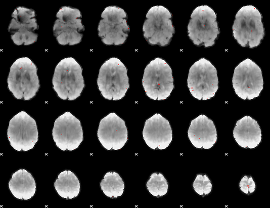 |
GLM Z-score result thresholded by 4
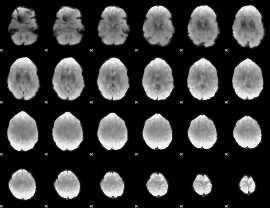 |
Component:380; Jaccard:0.099669
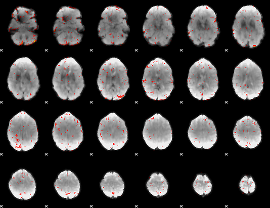 |
Component:380; Jaccard:0.11457
 |
Component:380; Jaccard:0.10957
 |
Component:380; Jaccard:0.080434
 |
Component:380; Jaccard:0.039383
 |
Component:380; Jaccard:0.014024
 |
Component:232; Jaccard:0.00348
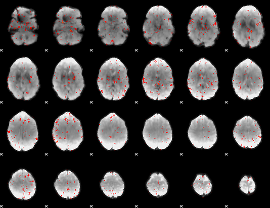 |
Correlation:0.17921
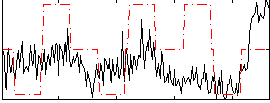 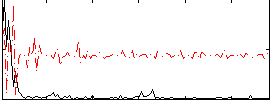 |
Correlation:0.17921
  |
Correlation:0.17921
  |
Correlation:0.17921
  |
Correlation:0.17921
  |
Correlation:0.17921
  |
Correlation:0.28463
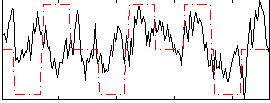 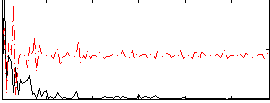 |
Component:047; Jaccard:0.080254
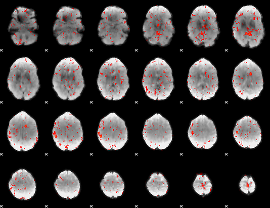 |
Component:397; Jaccard:0.085142
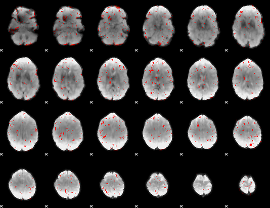 |
Component:265; Jaccard:0.079533
 |
Component:265; Jaccard:0.056045
 |
Component:174; Jaccard:0.027556
 |
Component:232; Jaccard:0.01024
 |
Component:380; Jaccard:0.003455
 |
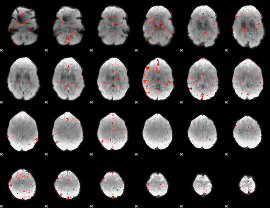
Correlation:0.31044
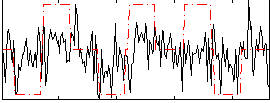 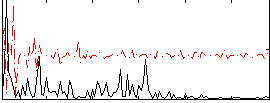 |
Correlation:0.26656
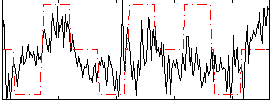 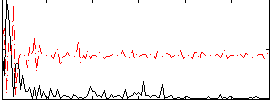 |
Correlation:0.1566
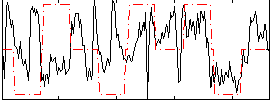 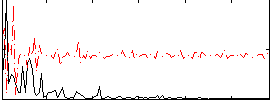 |
Correlation:0.1566
  |
Correlation:0.17952
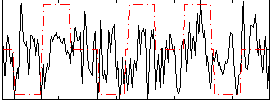  |
Correlation:0.28463
  |
Correlation:0.17921
  |
Component:397; Jaccard:0.079724
 |
Component:047; Jaccard:0.085013
 |
Component:397; Jaccard:0.078845
 |
Component:397; Jaccard:0.055721
 |
Component:265; Jaccard:0.027008
 |
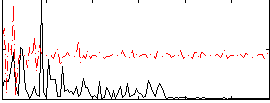
Component:119; Jaccard:0.009425
 |
Component:047; Jaccard:0.002551
 |
Correlation:0.26656
  |
Correlation:0.31044
  |
Correlation:0.26656
  |
Correlation:0.26656
  |
Correlation:0.1566
  |
Correlation:0.16288
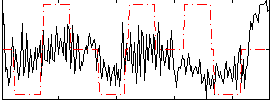 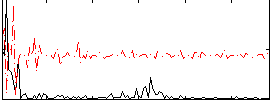 |
Correlation:0.31044
  |
Component:308; Jaccard:0.077892
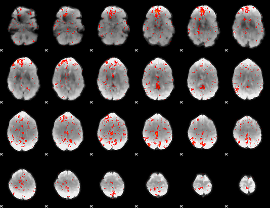 |
Component:265; Jaccard:0.080582
 |
Component:174; Jaccard:0.0781
 |
Component:174; Jaccard:0.054183
 |
Component:397; Jaccard:0.026668
 |
Component:174; Jaccard:0.009415
 |
Component:265; Jaccard:0.002256
 |
Correlation:0.12704
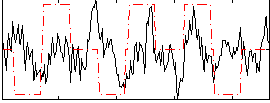 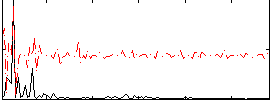 |
Correlation:0.1566
  |
Correlation:0.17952
  |
Correlation:0.17952
  |
Correlation:0.26656
  |
Correlation:0.17952
  |
Correlation:0.1566
  |
Component:364; Jaccard:0.073675
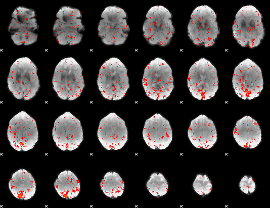 |
Component:174; Jaccard:0.080066
 |
Component:047; Jaccard:0.074333
 |
Component:047; Jaccard:0.051237
 |
Component:232; Jaccard:0.026031
 |
Component:265; Jaccard:0.00872
 |
Component:397; Jaccard:0.002137
 |
Correlation:0.16887
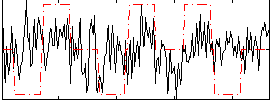 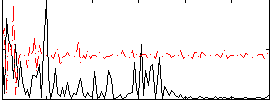 |
Correlation:0.17952
  |
Correlation:0.31044
  |
Correlation:0.31044
  |
Correlation:0.28463
  |
Correlation:0.1566
  |
Correlation:0.26656
  |
Component:174; Jaccard:0.072946
 |
Component:119; Jaccard:0.078743
 |
Component:119; Jaccard:0.072082
 |
Component:119; Jaccard:0.049458
 |
Component:119; Jaccard:0.02507
 |
Component:397; Jaccard:0.008575
 |
Component:048; Jaccard:0.002
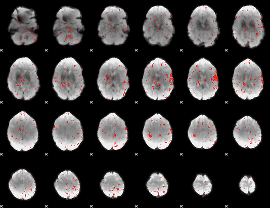 |
Correlation:0.17952
  |
Correlation:0.16288
  |
Correlation:0.16288
  |
Correlation:0.16288
  |
Correlation:0.16288
  |
Correlation:0.26656
  |
Correlation:0.090462
  |
Component:119; Jaccard:0.071776
 |
Component:308; Jaccard:0.071415
 |
Component:232; Jaccard:0.065276
 |
Component:232; Jaccard:0.047661
 |
Component:047; Jaccard:0.024188
 |
Component:047; Jaccard:0.00807
 |
Component:174; Jaccard:0.001941
 |
Correlation:0.16288
  |
Correlation:0.12704
  |
Correlation:0.28463
  |
Correlation:0.28463
  |
Correlation:0.31044
  |
Correlation:0.31044
  |
Correlation:0.17952
  |
Component:265; Jaccard:0.06958
 |
Component:232; Jaccard:0.07038
 |
Component:166; Jaccard:0.058503
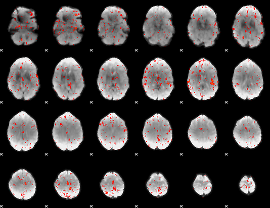 |
Component:166; Jaccard:0.037641
 |
Component:195; Jaccard:0.019152
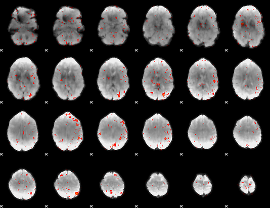 |
Component:195; Jaccard:0.006841
 |
Component:256; Jaccard:0.001915
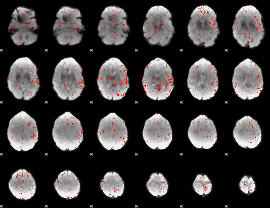 |
Correlation:0.1566
  |
Correlation:0.28463
  |
Correlation:0.43498
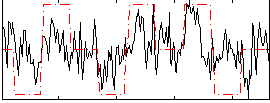 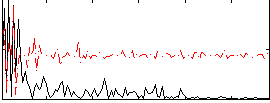 |
Correlation:0.43498
  |
Correlation:0.19181
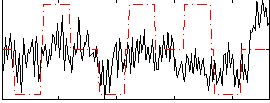 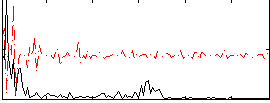 |
Correlation:0.19181
  |
Correlation:0.16434
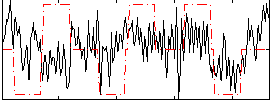 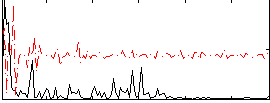 |
Component:194; Jaccard:0.069214
 |
Component:166; Jaccard:0.069078
 |
Component:274; Jaccard:0.055544
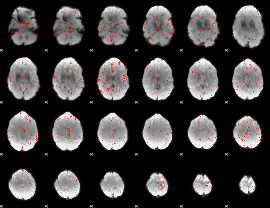 |
Component:195; Jaccard:0.036188
 |
Component:166; Jaccard:0.01827
 |
Component:194; Jaccard:0.006355
 |
Component:205; Jaccard:0.001891
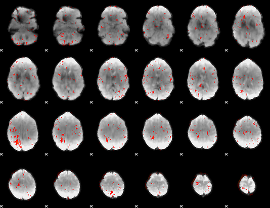 |
Correlation:0.27157
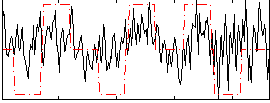  |
Correlation:0.43498
  |
Correlation:0.21657
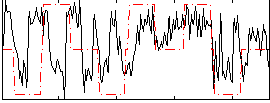 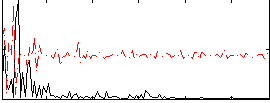 |
Correlation:0.19181
  |
Correlation:0.43498
  |
Correlation:0.27157
  |
Correlation:0.13607
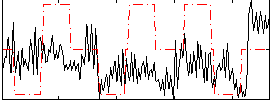 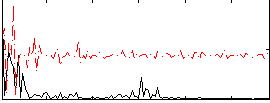 |
Component:166; Jaccard:0.069121
 |
Component:194; Jaccard:0.067096
 |
Component:048; Jaccard:0.055251
 |
Component:274; Jaccard:0.035968
 |
Component:243; Jaccard:0.017458
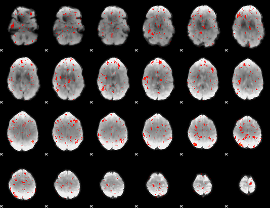 |
Component:166; Jaccard:0.005697
 |
Component:194; Jaccard:0.001828
 |
Correlation:0.43498
  |
Correlation:0.27157
  |
Correlation:0.090462
  |
Correlation:0.21657
  |
Correlation:0.15366
  |
Correlation:0.43498
  |
Correlation:0.27157
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
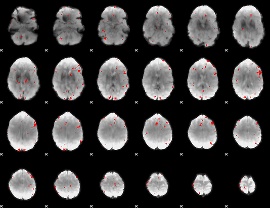 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
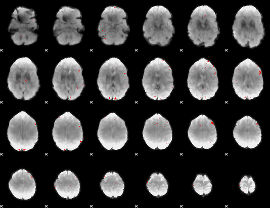 |
GLM Z-score result thresholded by 1
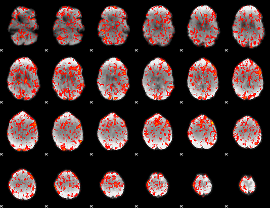 |
GLM Z-score result thresholded by 1.5
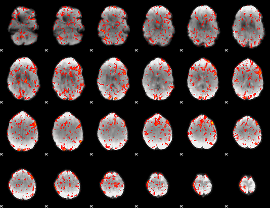 |
GLM Z-score result thresholded by 2
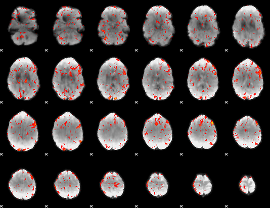 |
GLM Z-score result thresholded by 2.5
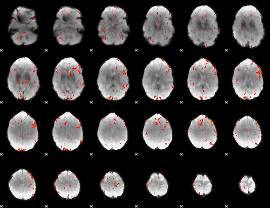 |
GLM Z-score result thresholded by 3
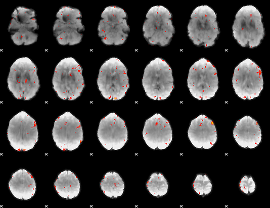 |
GLM Z-score result thresholded by 3.5
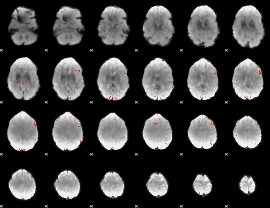 |
GLM Z-score result thresholded by 4
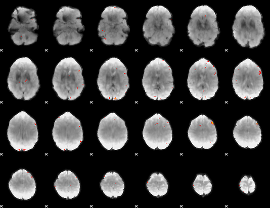 |
Component:306; Jaccard:0.12796
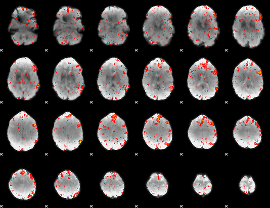 |
Component:306; Jaccard:0.14176
 |
Component:306; Jaccard:0.14036
 |
Component:306; Jaccard:0.11265
 |
Component:306; Jaccard:0.076262
 |
Component:306; Jaccard:0.043863
 |
Component:306; Jaccard:0.022226
 |
Correlation:0.10063
 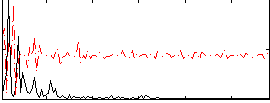 |
Correlation:0.10063
  |
Correlation:0.10063
  |
Correlation:0.10063
  |
Correlation:0.10063
  |
Correlation:0.10063
  |
Correlation:0.10063
  |
Component:393; Jaccard:0.082231
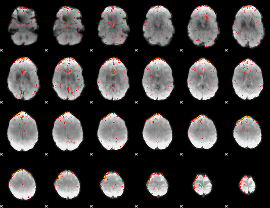 |
Component:393; Jaccard:0.092313
 |
Component:106; Jaccard:0.096092
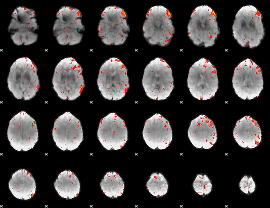 |
Component:106; Jaccard:0.084505
 |
Component:106; Jaccard:0.060143
 |
Component:106; Jaccard:0.033476
 |
Component:106; Jaccard:0.018459
 |
Correlation:0.13568
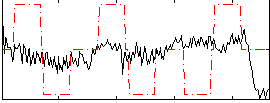 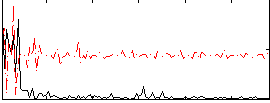 |
Correlation:0.13568
  |
Correlation:-0.054467
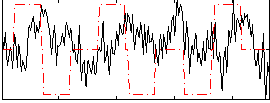 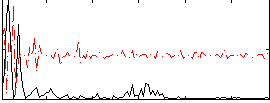 |
Correlation:-0.054467
  |
Correlation:-0.054467
  |
Correlation:-0.054467
  |
Correlation:-0.054467
  |
Component:068; Jaccard:0.082152
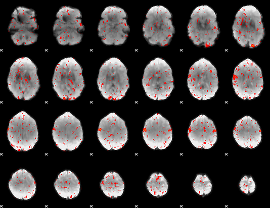 |
Component:106; Jaccard:0.091544
 |
Component:393; Jaccard:0.092504
 |
Component:393; Jaccard:0.072252
 |
Component:227; Jaccard:0.04658
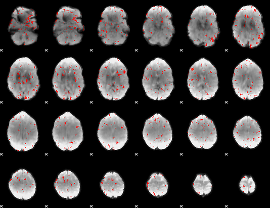 |
Component:329; Jaccard:0.029008
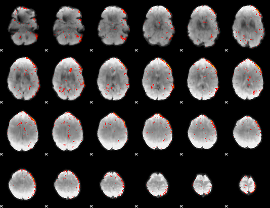 |
Component:329; Jaccard:0.017168
 |
Correlation:0.19713
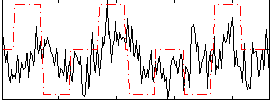 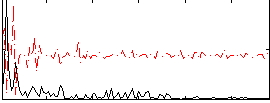 |
Correlation:-0.054467
  |
Correlation:0.13568
  |
Correlation:0.13568
  |
Correlation:0.036302
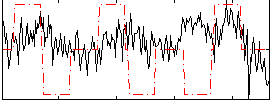  |
Correlation:0.072931
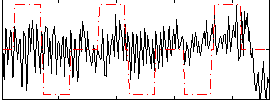 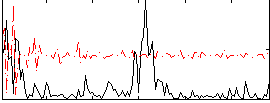 |
Correlation:0.072931
  |
Component:121; Jaccard:0.08128
 |
Component:068; Jaccard:0.086296
 |
Component:227; Jaccard:0.083007
 |
Component:227; Jaccard:0.068804
 |
Component:393; Jaccard:0.045287
 |
Component:377; Jaccard:0.027278
 |
Component:377; Jaccard:0.014908
 |
Correlation:0.00083091
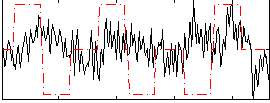 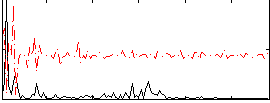 |
Correlation:0.19713
  |
Correlation:0.036302
  |
Correlation:0.036302
  |
Correlation:0.13568
  |
Correlation:0.052714
 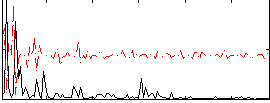 |
Correlation:0.052714
  |
Component:106; Jaccard:0.078277
 |
Component:227; Jaccard:0.085358
 |
Component:068; Jaccard:0.080802
 |
Component:068; Jaccard:0.065186
 |
Component:329; Jaccard:0.044724
 |
Component:227; Jaccard:0.025641
 |
Component:069; Jaccard:0.013369
 |
Correlation:-0.054467
  |
Correlation:0.036302
  |
Correlation:0.19713
  |
Correlation:0.19713
  |
Correlation:0.072931
  |
Correlation:0.036302
  |
Correlation:-0.0090854
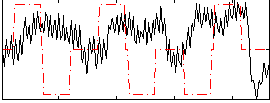  |
Component:069; Jaccard:0.077169
 |
Component:121; Jaccard:0.08317
 |
Component:377; Jaccard:0.075381
 |
Component:377; Jaccard:0.062817
 |
Component:377; Jaccard:0.044366
 |
Component:393; Jaccard:0.024423
 |
Component:198; Jaccard:0.012621
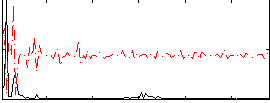
 |
Correlation:-0.0090854
  |
Correlation:0.00083091
  |
Correlation:0.052714
  |
Correlation:0.052714
  |
Correlation:0.052714
  |
Correlation:0.13568
  |
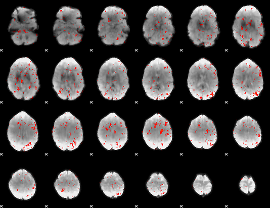
Correlation:0.19163
 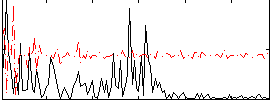 |
Component:227; Jaccard:0.075359
 |
Component:069; Jaccard:0.080439
 |
Component:108; Jaccard:0.074811
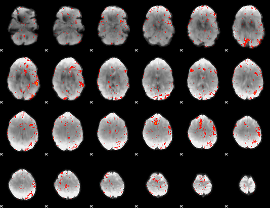 |
Component:329; Jaccard:0.062267
 |
Component:068; Jaccard:0.043284
 |
Component:198; Jaccard:0.023819
 |
Component:158; Jaccard:0.012562
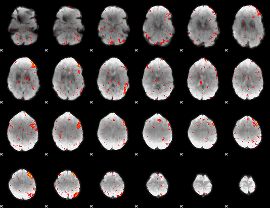 |
Correlation:0.036302
  |
Correlation:-0.0090854
  |
Correlation:0.10304
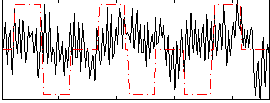  |
Correlation:0.072931
  |
Correlation:0.19713
  |
Correlation:0.19163
  |
Correlation:0.0065139
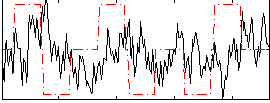 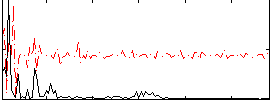 |
Component:377; Jaccard:0.074881
 |
Component:377; Jaccard:0.077971
 |
Component:121; Jaccard:0.074626
 |
Component:158; Jaccard:0.061203
 |
Component:158; Jaccard:0.042892
 |
Component:068; Jaccard:0.023645
 |
Component:227; Jaccard:0.012529
 |
Correlation:0.052714
  |
Correlation:0.052714
  |
Correlation:0.00083091
  |
Correlation:0.0065139
  |
Correlation:0.0065139
  |
Correlation:0.19713
  |
Correlation:0.036302
  |
Component:111; Jaccard:0.074215
 |
Component:108; Jaccard:0.075696
 |
Component:329; Jaccard:0.074216
 |
Component:108; Jaccard:0.059911
 |
Component:198; Jaccard:0.041046
 |
Component:158; Jaccard:0.023369
 |
Component:393; Jaccard:0.011816
 |
Correlation:-0.023485
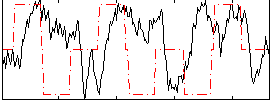 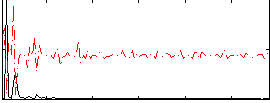 |
Correlation:0.10304
  |
Correlation:0.072931
  |
Correlation:0.10304
  |
Correlation:0.19163
  |
Correlation:0.0065139
  |
Correlation:0.13568
  |
Component:335; Jaccard:0.073542
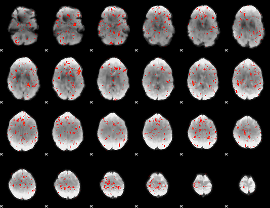 |
Component:111; Jaccard:0.075069
 |
Component:069; Jaccard:0.072433
 |
Component:069; Jaccard:0.057245
 |
Component:069; Jaccard:0.03903
 |
Component:069; Jaccard:0.022696
 |
Component:121; Jaccard:0.011408
 |
Correlation:0.34268
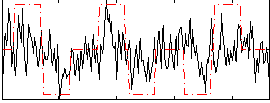 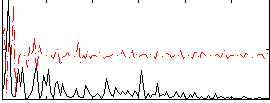 |
Correlation:-0.023485
  |
Correlation:-0.0090854
  |
Correlation:-0.0090854
  |
Correlation:-0.0090854
  |
Correlation:-0.0090854
  |
Correlation:0.00083091
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
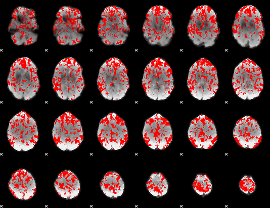 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
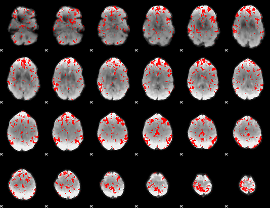 |
GLM binary result thresholded by 2.5
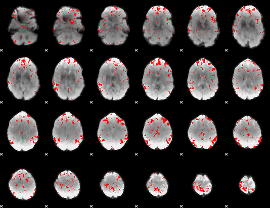 |
GLM binary result thresholded by 3
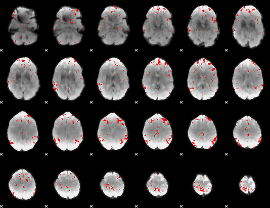 |
GLM binary result thresholded by 3.5
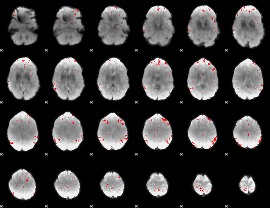 |
GLM binary result thresholded by 4
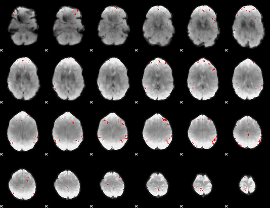 |
GLM Z-score result thresholded by 1
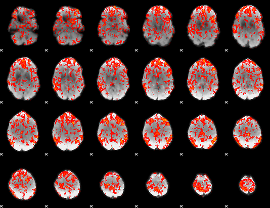 |
GLM Z-score result thresholded by 1.5
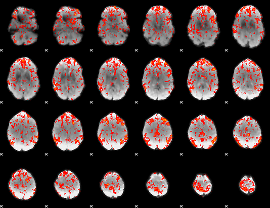 |
GLM Z-score result thresholded by 2
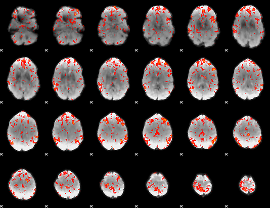 |
GLM Z-score result thresholded by 2.5
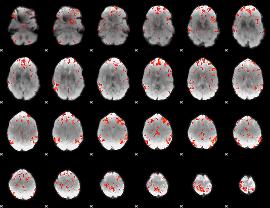 |
GLM Z-score result thresholded by 3
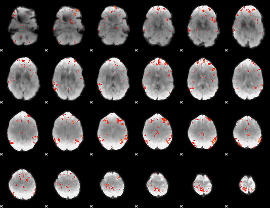 |
GLM Z-score result thresholded by 3.5
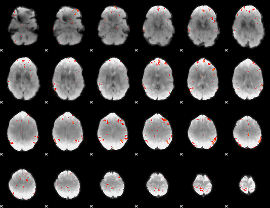 |
GLM Z-score result thresholded by 4
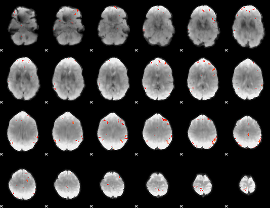 |
Component:346; Jaccard:0.11911
 |
Component:346; Jaccard:0.14293
 |
Component:346; Jaccard:0.15937
 |
Component:346; Jaccard:0.15405
 |
Component:321; Jaccard:0.13719
 |
Component:321; Jaccard:0.11023
 |
Component:321; Jaccard:0.070125
 |
Correlation:0.0072872
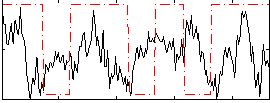 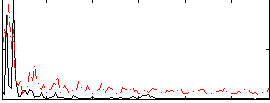 |
Correlation:0.0072872
  |
Correlation:0.0072872
  |
Correlation:0.0072872
  |
Correlation:0.2464
 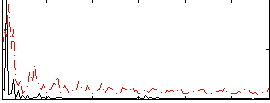 |
Correlation:0.2464
  |
Correlation:0.2464
  |
Component:347; Jaccard:0.11472
 |
Component:347; Jaccard:0.13637
 |
Component:347; Jaccard:0.15215
 |
Component:347; Jaccard:0.15035
 |
Component:124; Jaccard:0.12655
 |
Component:298; Jaccard:0.085662
 |
Component:298; Jaccard:0.051528
 |
Correlation:0.11727
 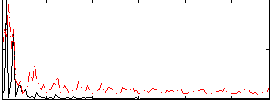 |
Correlation:0.11727
  |
Correlation:0.11727
  |
Correlation:0.11727
  |
Correlation:0.10159
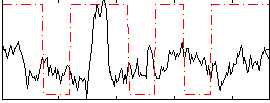 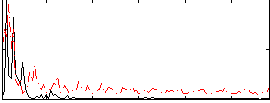 |
Correlation:0.2942
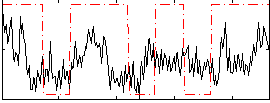 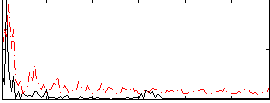 |
Correlation:0.2942
  |
Component:124; Jaccard:0.1085
 |
Component:124; Jaccard:0.13121
 |
Component:124; Jaccard:0.14772
 |
Component:124; Jaccard:0.14978
 |
Component:346; Jaccard:0.12244
 |
Component:124; Jaccard:0.085541
 |
Component:124; Jaccard:0.04954
 |
Correlation:0.10159
  |
Correlation:0.10159
  |
Correlation:0.10159
  |
Correlation:0.10159
  |
Correlation:0.0072872
  |
Correlation:0.10159
  |
Correlation:0.10159
  |
Component:298; Jaccard:0.095001
 |
Component:298; Jaccard:0.11399
 |
Component:298; Jaccard:0.13164
 |
Component:321; Jaccard:0.14538
 |
Component:347; Jaccard:0.12132
 |
Component:347; Jaccard:0.083088
 |
Component:347; Jaccard:0.046007
 |
Correlation:0.2942
  |
Correlation:0.2942
  |
Correlation:0.2942
  |
Correlation:0.2464
  |
Correlation:0.11727
  |
Correlation:0.11727
  |
Correlation:0.11727
  |
Component:361; Jaccard:0.092607
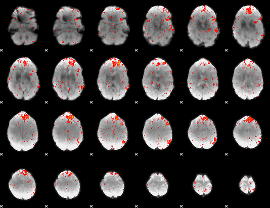 |
Component:321; Jaccard:0.1063
 |
Component:321; Jaccard:0.12981
 |
Component:298; Jaccard:0.13311
 |
Component:298; Jaccard:0.11639
 |
Component:346; Jaccard:0.082261
 |
Component:346; Jaccard:0.045779
 |
Correlation:0.022688
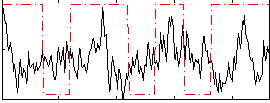 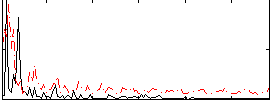 |
Correlation:0.2464
  |
Correlation:0.2464
  |
Correlation:0.2942
  |
Correlation:0.2942
  |
Correlation:0.0072872
  |
Correlation:0.0072872
  |
Component:350; Jaccard:0.090104
 |
Component:361; Jaccard:0.10219
 |
Component:102; Jaccard:0.10431
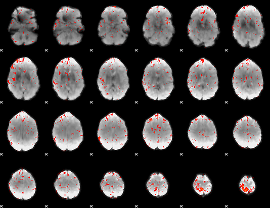 |
Component:288; Jaccard:0.10384
 |
Component:288; Jaccard:0.092689
 |
Component:288; Jaccard:0.063331
 |
Component:288; Jaccard:0.040468
 |
Correlation:-0.13384
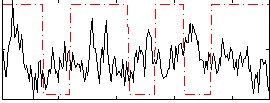 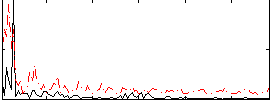 |
Correlation:0.022688
  |
Correlation:0.24321
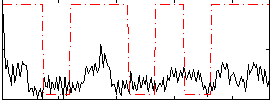 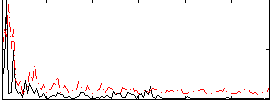 |
Correlation:0.21003
  |
Correlation:0.21003
  |
Correlation:0.21003
  |
Correlation:0.21003
  |
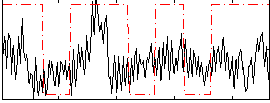
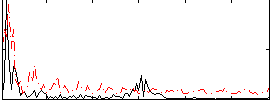
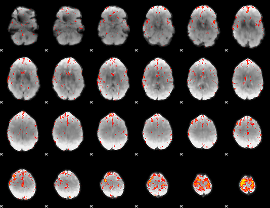
Component:110; Jaccard:0.085964
 |
Component:350; Jaccard:0.098418
 |
Component:361; Jaccard:0.10232
 |
Component:102; Jaccard:0.1021
 |
Component:102; Jaccard:0.083313
 |
Component:102; Jaccard:0.056266
 |
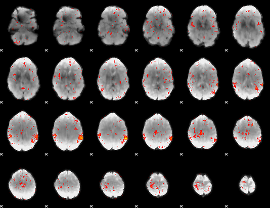
Component:185; Jaccard:0.032685
 |
Correlation:0.09867
 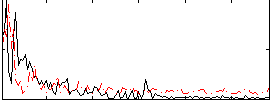 |
Correlation:-0.13384
  |
Correlation:0.022688
  |
Correlation:0.24321
  |
Correlation:0.24321
  |
Correlation:0.24321
  |
Correlation:0.22043
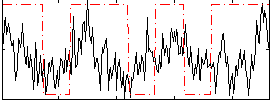 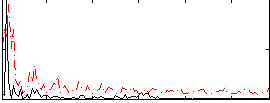 |
Component:321; Jaccard:0.085252
 |
Component:102; Jaccard:0.094976
 |
Component:350; Jaccard:0.099809
 |
Component:361; Jaccard:0.092269
 |
Component:350; Jaccard:0.072356
 |
Component:350; Jaccard:0.049831
 |
Component:102; Jaccard:0.03043
 |
Correlation:0.2464
  |
Correlation:0.24321
  |
Correlation:-0.13384
  |
Correlation:0.022688
  |
Correlation:-0.13384
  |
Correlation:-0.13384
  |
Correlation:0.24321
  |
Component:267; Jaccard:0.082121
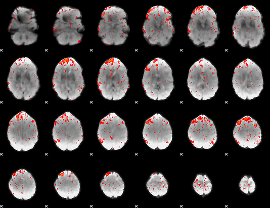 |
Component:110; Jaccard:0.089505
 |
Component:288; Jaccard:0.099216
 |
Component:350; Jaccard:0.091522
 |
Component:361; Jaccard:0.069242
 |
Component:185; Jaccard:0.048512
 |
Component:350; Jaccard:0.026961
 |
Correlation:-0.079181
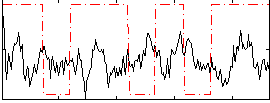  |
Correlation:0.09867
  |
Correlation:0.21003
  |
Correlation:-0.13384
  |
Correlation:0.022688
  |
Correlation:0.22043
  |
Correlation:-0.13384
  |
Component:102; Jaccard:0.082005
 |
Component:288; Jaccard:0.086403
 |
Component:110; Jaccard:0.085012
 |
Component:077; Jaccard:0.076418
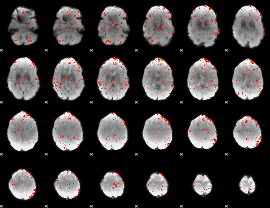 |
Component:185; Jaccard:0.065378
 |
Component:361; Jaccard:0.045093
 |
Component:056; Jaccard:0.025052
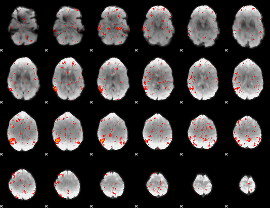 |
Correlation:0.24321
  |
Correlation:0.21003
  |
Correlation:0.09867
  |
Correlation:0.099875
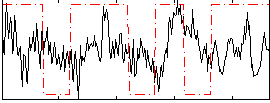  |
Correlation:0.22043
  |
Correlation:0.022688
  |
Correlation:0.041675
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
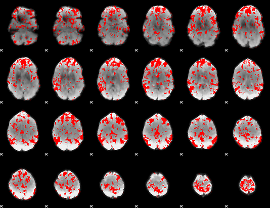 |
GLM binary result thresholded by 2
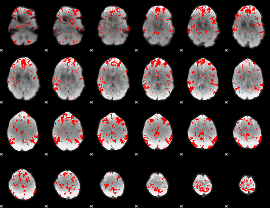 |
GLM binary result thresholded by 2.5
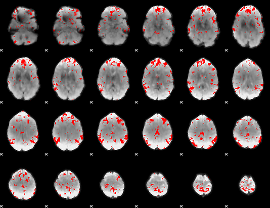 |
GLM binary result thresholded by 3
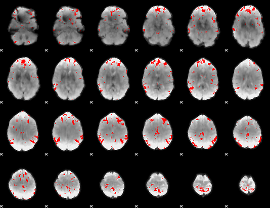 |
GLM binary result thresholded by 3.5
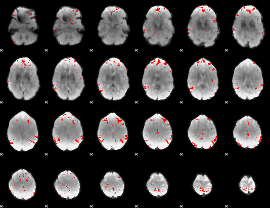 |
GLM binary result thresholded by 4
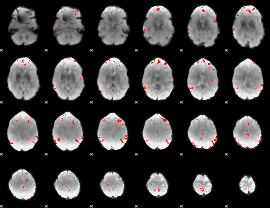 |
GLM Z-score result thresholded by 1
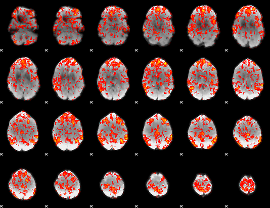 |
GLM Z-score result thresholded by 1.5
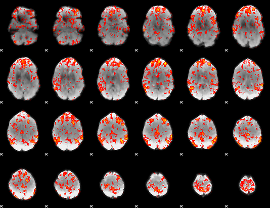 |
GLM Z-score result thresholded by 2
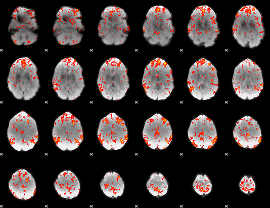 |
GLM Z-score result thresholded by 2.5
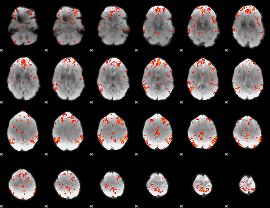 |
GLM Z-score result thresholded by 3
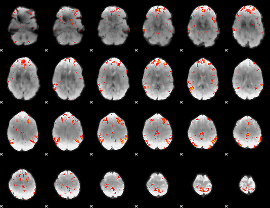 |
GLM Z-score result thresholded by 3.5
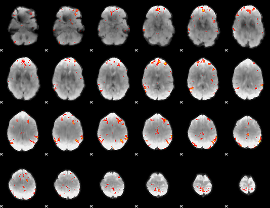 |
GLM Z-score result thresholded by 4
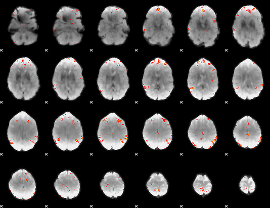 |
Component:346; Jaccard:0.12388
 |
Component:346; Jaccard:0.15111
 |
Component:346; Jaccard:0.17462
 |
Component:346; Jaccard:0.17968
 |
Component:321; Jaccard:0.17692
 |
Component:321; Jaccard:0.16153
 |
Component:321; Jaccard:0.13078
 |
Correlation:0.23597
  |
Correlation:0.23597
  |
Correlation:0.23597
  |
Correlation:0.23597
  |
Correlation:0.16563
  |
Correlation:0.16563
  |
Correlation:0.16563
  |
Component:347; Jaccard:0.12006
 |
Component:347; Jaccard:0.14453
 |
Component:347; Jaccard:0.16529
 |
Component:321; Jaccard:0.1717
 |
Component:346; Jaccard:0.16225
 |
Component:347; Jaccard:0.13084
 |
Component:298; Jaccard:0.099087
 |
Correlation:0.054191
  |
Correlation:0.054191
  |
Correlation:0.054191
  |
Correlation:0.16563
  |
Correlation:0.23597
  |
Correlation:0.054191
  |
Correlation:0.12831
  |
Component:298; Jaccard:0.10553
 |
Component:298; Jaccard:0.12717
 |
Component:321; Jaccard:0.15141
 |
Component:347; Jaccard:0.17114
 |
Component:347; Jaccard:0.1579
 |
Component:346; Jaccard:0.13076
 |
Component:347; Jaccard:0.097345
 |
Correlation:0.12831
  |
Correlation:0.12831
  |
Correlation:0.16563
  |
Correlation:0.054191
  |
Correlation:0.054191
  |
Correlation:0.23597
  |
Correlation:0.054191
  |
Component:124; Jaccard:0.10407
 |
Component:124; Jaccard:0.12334
 |
Component:298; Jaccard:0.14626
 |
Component:298; Jaccard:0.15644
 |
Component:298; Jaccard:0.1487
 |
Component:298; Jaccard:0.1282
 |
Component:346; Jaccard:0.09463
 |
Correlation:0.0088856
  |
Correlation:0.0088856
  |
Correlation:0.12831
  |
Correlation:0.12831
  |
Correlation:0.12831
  |
Correlation:0.12831
  |
Correlation:0.23597
  |
Component:350; Jaccard:0.10341
 |
Component:321; Jaccard:0.12182
 |
Component:124; Jaccard:0.14053
 |
Component:124; Jaccard:0.14778
 |
Component:124; Jaccard:0.13623
 |
Component:124; Jaccard:0.11166
 |
Component:124; Jaccard:0.083681
 |
Correlation:0.052192
  |
Correlation:0.16563
  |
Correlation:0.0088856
  |
Correlation:0.0088856
  |
Correlation:0.0088856
  |
Correlation:0.0088856
  |
Correlation:0.0088856
  |
Component:321; Jaccard:0.096456
 |
Component:350; Jaccard:0.11622
 |
Component:350; Jaccard:0.12474
 |
Component:350; Jaccard:0.12435
 |
Component:350; Jaccard:0.11344
 |
Component:288; Jaccard:0.094564
 |
Component:288; Jaccard:0.072165
 |
Correlation:0.16563
  |
Correlation:0.052192
  |
Correlation:0.052192
  |
Correlation:0.052192
  |
Correlation:0.052192
  |
Correlation:0.015924
  |
Correlation:0.015924
  |
Component:308; Jaccard:0.095119
 |
Component:308; Jaccard:0.1049
 |
Component:308; Jaccard:0.11203
 |
Component:288; Jaccard:0.11668
 |
Component:288; Jaccard:0.11286
 |
Component:350; Jaccard:0.090595
 |
Component:185; Jaccard:0.064943
 |
Correlation:0.050968
  |
Correlation:0.050968
  |
Correlation:0.050968
  |
Correlation:0.015924
  |
Correlation:0.015924
  |
Correlation:0.052192
  |
Correlation:0.23339
  |
Component:361; Jaccard:0.092314
 |
Component:361; Jaccard:0.10078
 |
Component:288; Jaccard:0.10514
 |
Component:308; Jaccard:0.10865
 |
Component:185; Jaccard:0.09811
 |
Component:185; Jaccard:0.082934
 |
Component:350; Jaccard:0.064191
 |
Correlation:0.34743
  |
Correlation:0.34743
  |
Correlation:0.015924
  |
Correlation:0.050968
  |
Correlation:0.23339
  |
Correlation:0.23339
  |
Correlation:0.052192
  |
Component:156; Jaccard:0.083973
 |
Component:056; Jaccard:0.091444
 |
Component:361; Jaccard:0.1044
 |
Component:185; Jaccard:0.10138
 |
Component:308; Jaccard:0.093142
 |
Component:056; Jaccard:0.075542
 |
Component:056; Jaccard:0.055043
 |
Correlation:0.18882
  |
Correlation:0.10631
  |
Correlation:0.34743
  |
Correlation:0.23339
  |
Correlation:0.050968
  |
Correlation:0.10631
  |
Correlation:0.10631
  |
Component:267; Jaccard:0.082958
 |
Component:156; Jaccard:0.090491
 |
Component:056; Jaccard:0.097732
 |
Component:361; Jaccard:0.099233
 |
Component:056; Jaccard:0.089446
 |
Component:334; Jaccard:0.073796
 |
Component:334; Jaccard:0.054698
 |
Correlation:0.22319
  |
Correlation:0.18882
  |
Correlation:0.10631
  |
Correlation:0.34743
  |
Correlation:0.10631
  |
Correlation:-0.041396
  |
Correlation:-0.041396
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































