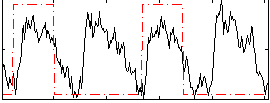
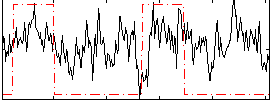
Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:099; Jaccard:0.13712
 |
Component:099; Jaccard:0.17562
 |
Component:099; Jaccard:0.21934
 |
Component:099; Jaccard:0.26281
 |
Component:099; Jaccard:0.30287
 |
Component:099; Jaccard:0.3292
 |
Component:099; Jaccard:0.34036
 |
Correlation:0.29866
  |
Correlation:0.29866
  |
Correlation:0.29866
  |
Correlation:0.29866
  |
Correlation:0.29866
  |
Correlation:0.29866
  |
Correlation:0.29866
  |
Component:049; Jaccard:0.13131
 |
Component:329; Jaccard:0.16248
 |
Component:329; Jaccard:0.19687
 |
Component:329; Jaccard:0.228
 |
Component:329; Jaccard:0.25185
 |
Component:329; Jaccard:0.26313
 |
Component:329; Jaccard:0.26149
 |
Correlation:0.27457
  |
Correlation:0.27063
  |
Correlation:0.27063
  |
Correlation:0.27063
  |
Correlation:0.27063
  |
Correlation:0.27063
  |
Correlation:0.27063
  |
Component:329; Jaccard:0.1293
 |
Component:049; Jaccard:0.16119
 |
Component:049; Jaccard:0.19123
 |
Component:086; Jaccard:0.21938
 |
Component:086; Jaccard:0.24202
 |
Component:086; Jaccard:0.25367
 |
Component:086; Jaccard:0.25379
 |
Correlation:0.27063
  |
Correlation:0.27457
  |
Correlation:0.27457
  |
Correlation:0.2999
  |
Correlation:0.2999
  |
Correlation:0.2999
  |
Correlation:0.2999
  |
Component:286; Jaccard:0.12704
 |
Component:086; Jaccard:0.15613
 |
Component:086; Jaccard:0.18885
 |
Component:049; Jaccard:0.21379
 |
Component:049; Jaccard:0.22482
 |
Component:049; Jaccard:0.22756
 |
Component:049; Jaccard:0.21741
 |
Correlation:0.2603
  |
Correlation:0.2999
  |
Correlation:0.2999
  |
Correlation:0.27457
  |
Correlation:0.27457
  |
Correlation:0.27457
  |
Correlation:0.27457
  |
Component:160; Jaccard:0.12504
 |
Component:160; Jaccard:0.1466
 |
Component:133; Jaccard:0.16727
 |
Component:133; Jaccard:0.18317
 |
Component:133; Jaccard:0.18976
 |
Component:133; Jaccard:0.18486
 |
Component:133; Jaccard:0.17479
 |
Correlation:0.3937
  |
Correlation:0.3937
  |
Correlation:0.27436
  |
Correlation:0.27436
  |
Correlation:0.27436
  |
Correlation:0.27436
  |
Correlation:0.27436
  |
Component:086; Jaccard:0.12457
 |
Component:133; Jaccard:0.14598
 |
Component:160; Jaccard:0.16597
 |
Component:160; Jaccard:0.17748
 |
Component:160; Jaccard:0.1777
 |
Component:006; Jaccard:0.17363
 |
Component:006; Jaccard:0.16596
 |
Correlation:0.2999
  |
Correlation:0.27436
  |
Correlation:0.3937
  |
Correlation:0.3937
  |
Correlation:0.3937
  |
Correlation:0.30213
  |
Correlation:0.30213
  |
Component:133; Jaccard:0.12182
 |
Component:286; Jaccard:0.14205
 |
Component:006; Jaccard:0.15793
 |
Component:006; Jaccard:0.17086
 |
Component:006; Jaccard:0.17644
 |
Component:160; Jaccard:0.17055
 |
Component:160; Jaccard:0.15387
 |
Correlation:0.27436
  |
Correlation:0.2603
  |
Correlation:0.30213
  |
Correlation:0.30213
  |
Correlation:0.30213
  |
Correlation:0.3937
  |
Correlation:0.3937
  |
Component:006; Jaccard:0.11734
 |
Component:006; Jaccard:0.13939
 |
Component:286; Jaccard:0.15254
 |
Component:286; Jaccard:0.15585
 |
Component:286; Jaccard:0.15293
 |
Component:286; Jaccard:0.14275
 |
Component:286; Jaccard:0.13039
 |
Correlation:0.30213
  |
Correlation:0.30213
  |
Correlation:0.2603
  |
Correlation:0.2603
  |
Correlation:0.2603
  |
Correlation:0.2603
  |
Correlation:0.2603
  |
Component:278; Jaccard:0.11361
 |
Component:278; Jaccard:0.12418
 |
Component:278; Jaccard:0.12658
 |
Component:024; Jaccard:0.12927
 |
Component:024; Jaccard:0.13192
 |
Component:024; Jaccard:0.13223
 |
Component:024; Jaccard:0.12856
 |
Correlation:0.27698
 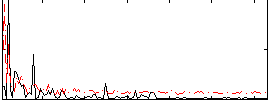 |
Correlation:0.27698
  |
Correlation:0.27698
  |
Correlation:0.14805
 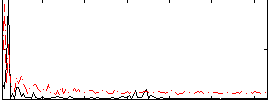 |
Correlation:0.14805
  |
Correlation:0.14805
  |
Correlation:0.14805
  |
Component:013; Jaccard:0.10857
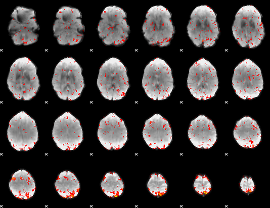 |
Component:104; Jaccard:0.11476
 |
Component:024; Jaccard:0.12102
 |
Component:165; Jaccard:0.12453
 |
Component:262; Jaccard:0.12499
 |
Component:262; Jaccard:0.12051
 |
Component:262; Jaccard:0.11227
 |
Correlation:0.22193
  |
Correlation:0.17965
  |
Correlation:0.14805
  |
Correlation:0.17117
  |
Correlation:0.14469
  |
Correlation:0.14469
  |
Correlation:0.14469
  |
Cope: 02:REWARD BACK
|
GLM result group-wise
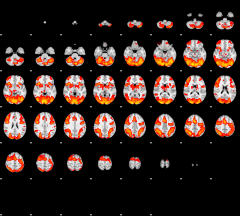 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
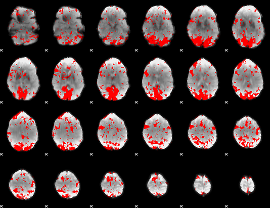 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
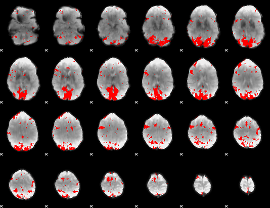 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:099; Jaccard:0.1274
 |
Component:099; Jaccard:0.17331
 |
Component:099; Jaccard:0.23063
 |
Component:099; Jaccard:0.29089
 |
Component:099; Jaccard:0.34619
 |
Component:099; Jaccard:0.37727
 |
Component:099; Jaccard:0.38815
 |
Correlation:0.3653
  |
Correlation:0.3653
  |
Correlation:0.3653
  |
Correlation:0.3653
  |
Correlation:0.3653
  |
Correlation:0.3653
  |
Correlation:0.3653
  |
Component:104; Jaccard:0.11811
 |
Component:329; Jaccard:0.15119
 |
Component:329; Jaccard:0.19332
 |
Component:329; Jaccard:0.22996
 |
Component:329; Jaccard:0.25996
 |
Component:329; Jaccard:0.27115
 |
Component:329; Jaccard:0.26786
 |
Correlation:0.033214
  |
Correlation:0.36743
  |
Correlation:0.36743
  |
Correlation:0.36743
  |
Correlation:0.36743
  |
Correlation:0.36743
  |
Correlation:0.36743
  |
Component:329; Jaccard:0.11494
 |
Component:086; Jaccard:0.141
 |
Component:086; Jaccard:0.17575
 |
Component:086; Jaccard:0.20923
 |
Component:086; Jaccard:0.23709
 |
Component:086; Jaccard:0.25132
 |
Component:086; Jaccard:0.25105
 |
Correlation:0.36743
  |
Correlation:0.20352
  |
Correlation:0.20352
  |
Correlation:0.20352
  |
Correlation:0.20352
  |
Correlation:0.20352
  |
Correlation:0.20352
  |
Component:024; Jaccard:0.11409
 |
Component:024; Jaccard:0.13643
 |
Component:024; Jaccard:0.15595
 |
Component:049; Jaccard:0.1738
 |
Component:049; Jaccard:0.18185
 |
Component:049; Jaccard:0.17849
 |
Component:049; Jaccard:0.16528
 |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:-0.11978
  |
Correlation:-0.11978
  |
Correlation:-0.11978
  |
Correlation:-0.11978
  |
Component:086; Jaccard:0.10861
 |
Component:104; Jaccard:0.1308
 |
Component:049; Jaccard:0.15384
 |
Component:024; Jaccard:0.16728
 |
Component:024; Jaccard:0.17161
 |
Component:024; Jaccard:0.16869
 |
Component:024; Jaccard:0.15873
 |
Correlation:0.20352
  |
Correlation:0.033214
  |
Correlation:-0.11978
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Correlation:0.368
  |
Component:160; Jaccard:0.106
 |
Component:049; Jaccard:0.12995
 |
Component:160; Jaccard:0.14614
 |
Component:133; Jaccard:0.16374
 |
Component:133; Jaccard:0.17016
 |
Component:133; Jaccard:0.16616
 |
Component:133; Jaccard:0.15796
 |
Correlation:-0.027216
  |
Correlation:-0.11978
  |
Correlation:-0.027216
  |
Correlation:0.052513
  |
Correlation:0.052513
  |
Correlation:0.052513
  |
Correlation:0.052513
  |
Component:049; Jaccard:0.10545
 |
Component:160; Jaccard:0.12663
 |
Component:133; Jaccard:0.14539
 |
Component:160; Jaccard:0.1581
 |
Component:160; Jaccard:0.1583
 |
Component:160; Jaccard:0.14771
 |
Component:160; Jaccard:0.13213
 |
Correlation:-0.11978
  |
Correlation:-0.027216
  |
Correlation:0.052513
  |
Correlation:-0.027216
  |
Correlation:-0.027216
  |
Correlation:-0.027216
  |
Correlation:-0.027216
  |
Component:286; Jaccard:0.10528
 |
Component:133; Jaccard:0.12461
 |
Component:104; Jaccard:0.13661
 |
Component:262; Jaccard:0.1413
 |
Component:262; Jaccard:0.14471
 |
Component:262; Jaccard:0.14048
 |
Component:262; Jaccard:0.13158
 |
Correlation:-0.02234
  |
Correlation:0.052513
  |
Correlation:0.033214
  |
Correlation:0.12478
  |
Correlation:0.12478
  |
Correlation:0.12478
  |
Correlation:0.12478
  |
Component:133; Jaccard:0.10324
 |
Component:286; Jaccard:0.11946
 |
Component:286; Jaccard:0.13092
 |
Component:104; Jaccard:0.13703
 |
Component:286; Jaccard:0.1325
 |
Component:006; Jaccard:0.12597
 |
Component:006; Jaccard:0.1202
 |
Correlation:0.052513
  |
Correlation:-0.02234
  |
Correlation:-0.02234
  |
Correlation:0.033214
  |
Correlation:-0.02234
  |
Correlation:0.14988
  |
Correlation:0.14988
  |
Component:294; Jaccard:0.10229
 |
Component:262; Jaccard:0.11221
 |
Component:262; Jaccard:0.12984
 |
Component:286; Jaccard:0.13647
 |
Component:104; Jaccard:0.1287
 |
Component:286; Jaccard:0.12311
 |
Component:286; Jaccard:0.11022
 |
Correlation:0.19783
  |
Correlation:0.12478
  |
Correlation:0.12478
  |
Correlation:-0.02234
  |
Correlation:0.033214
  |
Correlation:-0.02234
  |
Correlation:-0.02234
  |
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
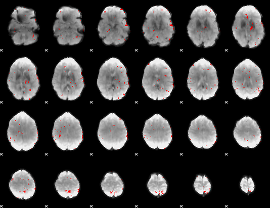 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
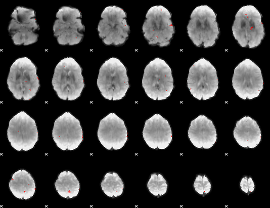 |
GLM Z-score result thresholded by 1
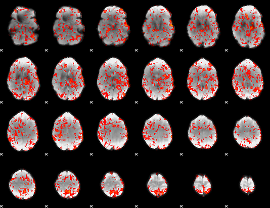 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:013; Jaccard:0.10477
 |
Component:013; Jaccard:0.1037
 |
Component:013; Jaccard:0.091644
 |
Component:013; Jaccard:0.067012
 |
Component:017; Jaccard:0.042969
 |
Component:139; Jaccard:0.026773
 |
Component:139; Jaccard:0.013469
 |
Correlation:0.18843
 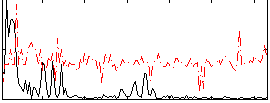 |
Correlation:0.18843
  |
Correlation:0.18843
  |
Correlation:0.18843
  |
Correlation:0.076689
  |
Correlation:0.053568
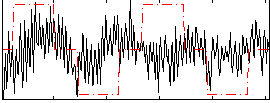  |
Correlation:0.053568
  |
Component:373; Jaccard:0.10291
 |
Component:373; Jaccard:0.098827
 |
Component:373; Jaccard:0.083318
 |
Component:017; Jaccard:0.062961
 |
Component:139; Jaccard:0.040879
 |
Component:017; Jaccard:0.024488
 |
Component:017; Jaccard:0.010809
 |
Correlation:0.16459
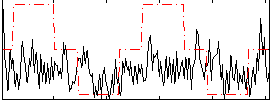  |
Correlation:0.16459
  |
Correlation:0.16459
  |
Correlation:0.076689
  |
Correlation:0.053568
  |
Correlation:0.076689
  |
Correlation:0.076689
  |
Component:012; Jaccard:0.083982
 |
Component:012; Jaccard:0.084194
 |
Component:012; Jaccard:0.078402
 |
Component:176; Jaccard:0.058926
 |
Component:324; Jaccard:0.039027
 |
Component:324; Jaccard:0.022327
 |
Component:324; Jaccard:0.010561
 |
Correlation:0.23791
  |
Correlation:0.23791
  |
Correlation:0.23791
  |
Correlation:0.10198
  |
Correlation:0.081836
  |
Correlation:0.081836
  |
Correlation:0.081836
  |
Component:309; Jaccard:0.075706
 |
Component:017; Jaccard:0.075437
 |
Component:017; Jaccard:0.074757
 |
Component:324; Jaccard:0.057897
 |
Component:013; Jaccard:0.038762
 |
Component:176; Jaccard:0.020606
 |
Component:032; Jaccard:0.00996
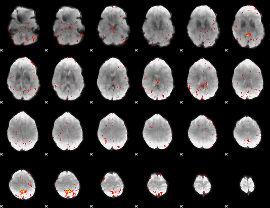 |
Correlation:0.20057
  |
Correlation:0.076689
  |
Correlation:0.076689
  |
Correlation:0.081836
  |
Correlation:0.18843
  |
Correlation:0.10198
  |
Correlation:-0.010355
  |
Component:307; Jaccard:0.074725
 |
Component:176; Jaccard:0.075378
 |
Component:176; Jaccard:0.072833
 |
Component:012; Jaccard:0.05764
 |
Component:176; Jaccard:0.038174
 |
Component:032; Jaccard:0.019248
 |
Component:373; Jaccard:0.009815
 |
Correlation:0.14406
  |
Correlation:0.10198
  |
Correlation:0.10198
  |
Correlation:0.23791
  |
Correlation:0.10198
  |
Correlation:-0.010355
  |
Correlation:0.16459
  |
Component:001; Jaccard:0.074593
 |
Component:309; Jaccard:0.074978
 |
Component:324; Jaccard:0.070613
 |
Component:373; Jaccard:0.057352
 |
Component:373; Jaccard:0.034946
 |
Component:013; Jaccard:0.01913
 |
Component:070; Jaccard:0.009311
 |
Correlation:0.26174
  |
Correlation:0.20057
  |
Correlation:0.081836
  |
Correlation:0.16459
  |
Correlation:0.16459
  |
Correlation:0.18843
  |
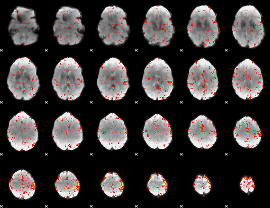
Correlation:0.28143
  |
Component:006; Jaccard:0.074152
 |
Component:307; Jaccard:0.073546
 |
Component:309; Jaccard:0.065593
 |
Component:139; Jaccard:0.053304
 |
Component:012; Jaccard:0.034149
 |
Component:373; Jaccard:0.018325
 |
Component:155; Jaccard:0.00818
 |
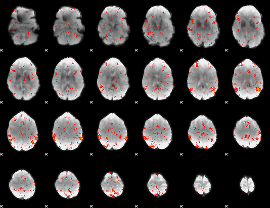
Correlation:0.089537
  |
Correlation:0.14406
  |
Correlation:0.20057
  |
Correlation:0.053568
  |
Correlation:0.23791
  |
Correlation:0.16459
  |
Correlation:0.014276
  |
Component:259; Jaccard:0.073977
 |
Component:006; Jaccard:0.073226
 |
Component:006; Jaccard:0.065109
 |
Component:154; Jaccard:0.049881
 |
Component:032; Jaccard:0.033052
 |
Component:012; Jaccard:0.017645
 |
Component:265; Jaccard:0.00803
 |
Correlation:0.12
  |
Correlation:0.089537
  |
Correlation:0.089537
  |
Correlation:0.26544
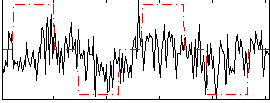  |
Correlation:-0.010355
  |
Correlation:0.23791
  |
Correlation:0.40311
  |
Component:089; Jaccard:0.073887
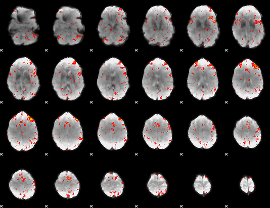 |
Component:089; Jaccard:0.073044
 |
Component:089; Jaccard:0.064863
 |
Component:309; Jaccard:0.049616
 |
Component:154; Jaccard:0.032181
 |
Component:154; Jaccard:0.017366
 |
Component:154; Jaccard:0.007898
 |
Correlation:0.29845
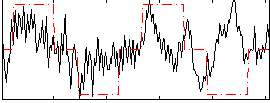  |
Correlation:0.29845
  |
Correlation:0.29845
  |
Correlation:0.20057
  |
Correlation:0.26544
  |
Correlation:0.26544
  |
Correlation:0.26544
  |
Component:278; Jaccard:0.073209
 |
Component:026; Jaccard:0.071126
 |
Component:026; Jaccard:0.063126
 |
Component:265; Jaccard:0.048471
 |
Component:070; Jaccard:0.03105
 |
Component:309; Jaccard:0.016331
 |
Component:013; Jaccard:0.007894
 |
Correlation:0.20523
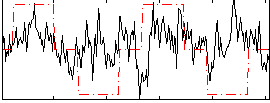  |
Correlation:0.23649
  |
Correlation:0.23649
  |
Correlation:0.40311
  |
Correlation:0.28143
  |
Correlation:0.20057
  |
Correlation:0.18843
  |
Cope: 04:neg_PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
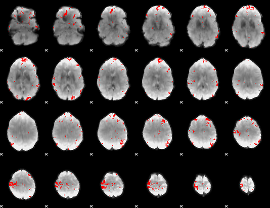 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
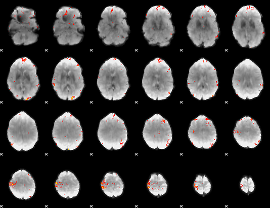 |
Component:150; Jaccard:0.13629
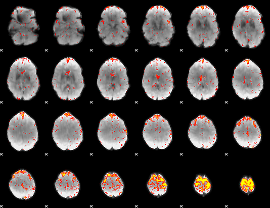 |
Component:169; Jaccard:0.1539
 |
Component:169; Jaccard:0.17345
 |
Component:169; Jaccard:0.17519
 |
Component:169; Jaccard:0.15792
 |
Component:281; Jaccard:0.13117
 |
Component:281; Jaccard:0.095539
 |
Correlation:0.16972
  |
Correlation:0.28953
  |
Correlation:0.28953
  |
Correlation:0.28953
  |
Correlation:0.28953
  |
Correlation:0.1055
 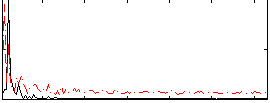 |
Correlation:0.1055
  |
Component:169; Jaccard:0.12997
 |
Component:150; Jaccard:0.1497
 |
Component:150; Jaccard:0.14767
 |
Component:281; Jaccard:0.15737
 |
Component:281; Jaccard:0.15464
 |
Component:169; Jaccard:0.12105
 |
Component:169; Jaccard:0.079845
 |
Correlation:0.28953
  |
Correlation:0.16972
  |
Correlation:0.16972
  |
Correlation:0.1055
  |
Correlation:0.1055
  |
Correlation:0.28953
  |
Correlation:0.28953
  |
Component:330; Jaccard:0.11228
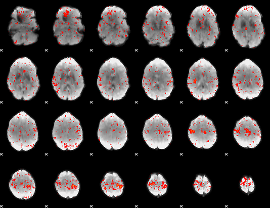 |
Component:281; Jaccard:0.12448
 |
Component:281; Jaccard:0.14382
 |
Component:283; Jaccard:0.12909
 |
Component:283; Jaccard:0.11278
 |
Component:283; Jaccard:0.083595
 |
Component:283; Jaccard:0.053865
 |
Correlation:0.20043
  |
Correlation:0.1055
  |
Correlation:0.1055
  |
Correlation:0.37078
  |
Correlation:0.37078
  |
Correlation:0.37078
  |
Correlation:0.37078
  |
Component:098; Jaccard:0.10926
 |
Component:330; Jaccard:0.12397
 |
Component:098; Jaccard:0.13191
 |
Component:330; Jaccard:0.12721
 |
Component:330; Jaccard:0.10768
 |
Component:098; Jaccard:0.081656
 |
Component:098; Jaccard:0.052985
 |
Correlation:0.301
  |
Correlation:0.20043
  |
Correlation:0.301
  |
Correlation:0.20043
  |
Correlation:0.20043
  |
Correlation:0.301
  |
Correlation:0.301
  |
Component:223; Jaccard:0.10609
 |
Component:098; Jaccard:0.12332
 |
Component:283; Jaccard:0.13123
 |
Component:098; Jaccard:0.12567
 |
Component:098; Jaccard:0.10708
 |
Component:292; Jaccard:0.078956
 |
Component:143; Jaccard:0.052387
 |
Correlation:0.37422
  |
Correlation:0.301
  |
Correlation:0.37078
  |
Correlation:0.301
  |
Correlation:0.301
  |
Correlation:0.21668
  |
Correlation:0.14974
  |
Component:283; Jaccard:0.10505
 |
Component:223; Jaccard:0.12037
 |
Component:330; Jaccard:0.13001
 |
Component:150; Jaccard:0.12323
 |
Component:292; Jaccard:0.1018
 |
Component:330; Jaccard:0.078441
 |
Component:292; Jaccard:0.052345
 |
Correlation:0.37078
  |
Correlation:0.37422
  |
Correlation:0.20043
  |
Correlation:0.16972
  |
Correlation:0.21668
  |
Correlation:0.20043
  |
Correlation:0.21668
  |
Component:281; Jaccard:0.10496
 |
Component:283; Jaccard:0.1186
 |
Component:223; Jaccard:0.12986
 |
Component:223; Jaccard:0.12263
 |
Component:363; Jaccard:0.098692
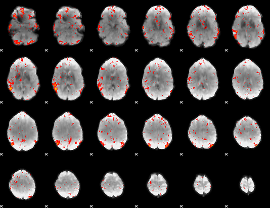 |
Component:363; Jaccard:0.073439
 |
Component:330; Jaccard:0.051428
 |
Correlation:0.1055
  |
Correlation:0.37078
  |
Correlation:0.37422
  |
Correlation:0.37422
  |
Correlation:0.35144
  |
Correlation:0.35144
  |
Correlation:0.20043
  |
Component:363; Jaccard:0.098148
 |
Component:363; Jaccard:0.11008
 |
Component:363; Jaccard:0.11765
 |
Component:292; Jaccard:0.11523
 |
Component:223; Jaccard:0.097866
 |
Component:143; Jaccard:0.073019
 |
Component:183; Jaccard:0.047869
 |
Correlation:0.35144
  |
Correlation:0.35144
  |
Correlation:0.35144
  |
Correlation:0.21668
  |
Correlation:0.37422
  |
Correlation:0.14974
  |
Correlation:0.08983
  |
Component:342; Jaccard:0.096234
 |
Component:292; Jaccard:0.10824
 |
Component:292; Jaccard:0.11638
 |
Component:363; Jaccard:0.11314
 |
Component:143; Jaccard:0.092028
 |
Component:223; Jaccard:0.069558
 |
Component:363; Jaccard:0.047198
 |
Correlation:0.10111
  |
Correlation:0.21668
  |
Correlation:0.21668
  |
Correlation:0.35144
  |
Correlation:0.14974
  |
Correlation:0.37422
  |
Correlation:0.35144
  |
Component:292; Jaccard:0.094605
 |
Component:342; Jaccard:0.10659
 |
Component:342; Jaccard:0.11133
 |
Component:342; Jaccard:0.10765
 |
Component:342; Jaccard:0.089067
 |
Component:342; Jaccard:0.066074
 |
Component:342; Jaccard:0.045458
 |
Correlation:0.21668
  |
Correlation:0.10111
  |
Correlation:0.10111
  |
Correlation:0.10111
  |
Correlation:0.10111
  |
Correlation:0.10111
  |
Correlation:0.10111
  |
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
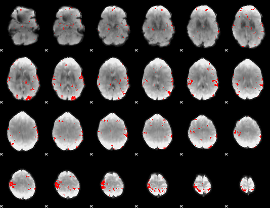 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
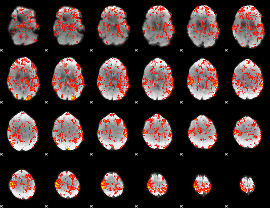 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
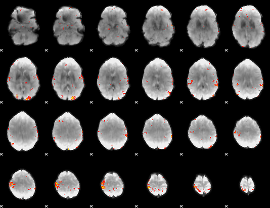 |
GLM Z-score result thresholded by 4
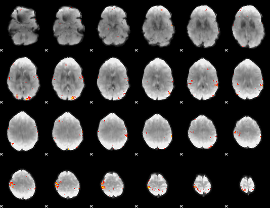 |
Component:281; Jaccard:0.14109
 |
Component:281; Jaccard:0.17215
 |
Component:281; Jaccard:0.19633
 |
Component:281; Jaccard:0.20062
 |
Component:281; Jaccard:0.18093
 |
Component:281; Jaccard:0.14027
 |
Component:281; Jaccard:0.10021
 |
Correlation:-0.021567
 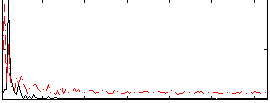 |
Correlation:-0.021567
  |
Correlation:-0.021567
  |
Correlation:-0.021567
  |
Correlation:-0.021567
  |
Correlation:-0.021567
  |
Correlation:-0.021567
  |
Component:143; Jaccard:0.12777
 |
Component:143; Jaccard:0.15076
 |
Component:143; Jaccard:0.16197
 |
Component:143; Jaccard:0.15763
 |
Component:143; Jaccard:0.13693
 |
Component:143; Jaccard:0.10615
 |
Component:143; Jaccard:0.073474
 |
Correlation:0.28762
  |
Correlation:0.28762
  |
Correlation:0.28762
  |
Correlation:0.28762
  |
Correlation:0.28762
  |
Correlation:0.28762
  |
Correlation:0.28762
  |
Component:373; Jaccard:0.12445
 |
Component:373; Jaccard:0.13245
 |
Component:373; Jaccard:0.12895
 |
Component:373; Jaccard:0.11307
 |
Component:183; Jaccard:0.098888
 |
Component:183; Jaccard:0.078003
 |
Component:183; Jaccard:0.056787
 |
Correlation:0.19955
 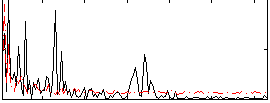 |
Correlation:0.19955
  |
Correlation:0.19955
  |
Correlation:0.19955
  |
Correlation:0.26313
  |
Correlation:0.26313
  |
Correlation:0.26313
  |
Component:157; Jaccard:0.098612
 |
Component:183; Jaccard:0.10772
 |
Component:183; Jaccard:0.11552
 |
Component:183; Jaccard:0.11217
 |
Component:373; Jaccard:0.089212
 |
Component:036; Jaccard:0.065862
 |
Component:036; Jaccard:0.049302
 |
Correlation:0.27253
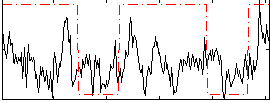  |
Correlation:0.26313
  |
Correlation:0.26313
  |
Correlation:0.26313
  |
Correlation:0.19955
  |
Correlation:0.28891
  |
Correlation:0.28891
  |
Component:361; Jaccard:0.093514
 |
Component:157; Jaccard:0.10742
 |
Component:157; Jaccard:0.10608
 |
Component:036; Jaccard:0.094187
 |
Component:036; Jaccard:0.082392
 |
Component:373; Jaccard:0.060464
 |
Component:373; Jaccard:0.040505
 |
Correlation:0.05772
  |
Correlation:0.27253
  |
Correlation:0.27253
  |
Correlation:0.28891
  |
Correlation:0.28891
  |
Correlation:0.19955
  |
Correlation:0.19955
  |
Component:183; Jaccard:0.092749
 |
Component:361; Jaccard:0.10186
 |
Component:361; Jaccard:0.10174
 |
Component:361; Jaccard:0.092088
 |
Component:361; Jaccard:0.073286
 |
Component:361; Jaccard:0.052511
 |
Component:361; Jaccard:0.035264
 |
Correlation:0.26313
  |
Correlation:0.05772
  |
Correlation:0.05772
  |
Correlation:0.05772
  |
Correlation:0.05772
  |
Correlation:0.05772
  |
Correlation:0.05772
  |
Component:298; Jaccard:0.090009
 |
Component:298; Jaccard:0.097653
 |
Component:298; Jaccard:0.097482
 |
Component:157; Jaccard:0.091662
 |
Component:157; Jaccard:0.069817
 |
Component:157; Jaccard:0.047871
 |
Component:070; Jaccard:0.032431
 |
Correlation:0.26446
  |
Correlation:0.26446
  |
Correlation:0.26446
  |
Correlation:0.27253
  |
Correlation:0.27253
  |
Correlation:0.27253
  |
Correlation:0.33013
  |
Component:397; Jaccard:0.083241
 |
Component:036; Jaccard:0.090777
 |
Component:036; Jaccard:0.096367
 |
Component:298; Jaccard:0.086986
 |
Component:298; Jaccard:0.068741
 |
Component:070; Jaccard:0.047014
 |
Component:298; Jaccard:0.032238
 |
Correlation:0.0030789
  |
Correlation:0.28891
  |
Correlation:0.28891
  |
Correlation:0.26446
  |
Correlation:0.26446
  |
Correlation:0.33013
  |
Correlation:0.26446
  |
Component:257; Jaccard:0.081807
 |
Component:324; Jaccard:0.086492
 |
Component:070; Jaccard:0.088553
 |
Component:070; Jaccard:0.081151
 |
Component:070; Jaccard:0.063354
 |
Component:298; Jaccard:0.047004
 |
Component:157; Jaccard:0.030342
 |
Correlation:0.19832
  |
Correlation:0.12037
  |
Correlation:0.33013
  |
Correlation:0.33013
  |
Correlation:0.33013
  |
Correlation:0.26446
  |
Correlation:0.27253
  |
Component:036; Jaccard:0.081521
 |
Component:070; Jaccard:0.084752
 |
Component:324; Jaccard:0.085457
 |
Component:324; Jaccard:0.076669
 |
Component:324; Jaccard:0.058147
 |
Component:309; Jaccard:0.039683
 |
Component:342; Jaccard:0.029337
 |
Correlation:0.28891
  |
Correlation:0.33013
  |
Correlation:0.12037
  |
Correlation:0.12037
  |
Correlation:0.12037
  |
Correlation:0.1727
  |
Correlation:0.0073054
  |
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
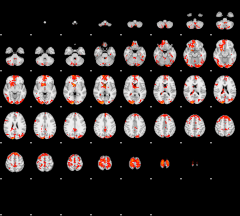 |
GLM binary result thresholded by 1
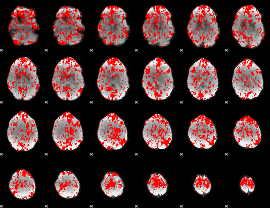 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
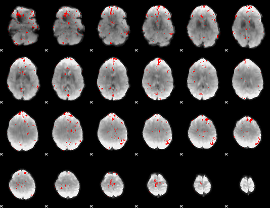 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
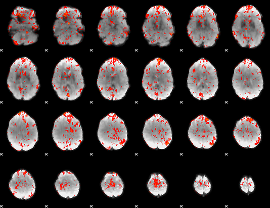 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
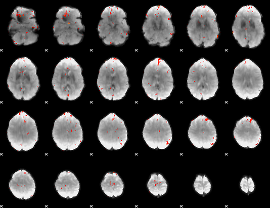 |
GLM Z-score result thresholded by 4
 |
Component:169; Jaccard:0.12153
 |
Component:169; Jaccard:0.14096
 |
Component:169; Jaccard:0.15296
 |
Component:169; Jaccard:0.14597
 |
Component:159; Jaccard:0.12301
 |
Component:159; Jaccard:0.091558
 |
Component:159; Jaccard:0.057718
 |
Correlation:0.38849
  |
Correlation:0.38849
  |
Correlation:0.38849
  |
Correlation:0.38849
  |
Correlation:0.52819
  |
Correlation:0.52819
  |
Correlation:0.52819
  |
Component:395; Jaccard:0.11012
 |
Component:395; Jaccard:0.12322
 |
Component:159; Jaccard:0.13155
 |
Component:159; Jaccard:0.13762
 |
Component:169; Jaccard:0.11955
 |
Component:169; Jaccard:0.079935
 |
Component:169; Jaccard:0.04606
 |
Correlation:0.34266
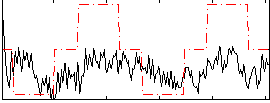  |
Correlation:0.34266
  |
Correlation:0.52819
  |
Correlation:0.52819
  |
Correlation:0.38849
  |
Correlation:0.38849
  |
Correlation:0.38849
  |
Component:223; Jaccard:0.10563
 |
Component:223; Jaccard:0.12204
 |
Component:223; Jaccard:0.12975
 |
Component:223; Jaccard:0.11846
 |
Component:395; Jaccard:0.092724
 |
Component:223; Jaccard:0.058607
 |
Component:209; Jaccard:0.036095
 |
Correlation:0.36583
 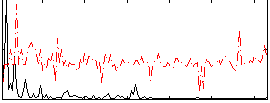 |
Correlation:0.36583
  |
Correlation:0.36583
  |
Correlation:0.36583
  |
Correlation:0.34266
  |
Correlation:0.36583
  |
Correlation:0.32454
  |
Component:150; Jaccard:0.0979
 |
Component:159; Jaccard:0.1097
 |
Component:395; Jaccard:0.12692
 |
Component:395; Jaccard:0.11749
 |
Component:223; Jaccard:0.092657
 |
Component:395; Jaccard:0.057924
 |
Component:223; Jaccard:0.033346
 |
Correlation:0.10301
  |
Correlation:0.52819
  |
Correlation:0.34266
  |
Correlation:0.34266
  |
Correlation:0.36583
  |
Correlation:0.34266
  |
Correlation:0.36583
  |
Component:283; Jaccard:0.095277
 |
Component:283; Jaccard:0.10735
 |
Component:283; Jaccard:0.1145
 |
Component:283; Jaccard:0.109
 |
Component:283; Jaccard:0.085215
 |
Component:209; Jaccard:0.057577
 |
Component:395; Jaccard:0.03293
 |
Correlation:0.39792
  |
Correlation:0.39792
  |
Correlation:0.39792
  |
Correlation:0.39792
  |
Correlation:0.39792
  |
Correlation:0.32454
  |
Correlation:0.34266
  |
Component:098; Jaccard:0.089785
 |
Component:098; Jaccard:0.094867
 |
Component:379; Jaccard:0.096631
 |
Component:379; Jaccard:0.090899
 |
Component:209; Jaccard:0.07625
 |
Component:283; Jaccard:0.055966
 |
Component:283; Jaccard:0.03291
 |
Correlation:0.29953
  |
Correlation:0.29953
  |
Correlation:0.18185
  |
Correlation:0.18185
  |
Correlation:0.32454
  |
Correlation:0.39792
  |
Correlation:0.39792
  |
Component:159; Jaccard:0.089669
 |
Component:379; Jaccard:0.091404
 |
Component:098; Jaccard:0.09094
 |
Component:209; Jaccard:0.085149
 |
Component:379; Jaccard:0.069979
 |
Component:379; Jaccard:0.046007
 |
Component:379; Jaccard:0.026462
 |
Correlation:0.52819
  |
Correlation:0.18185
  |
Correlation:0.29953
  |
Correlation:0.32454
  |
Correlation:0.18185
  |
Correlation:0.18185
  |
Correlation:0.18185
  |
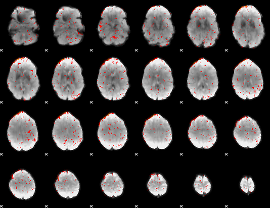
Component:330; Jaccard:0.084383
 |
Component:150; Jaccard:0.090819
 |
Component:279; Jaccard:0.088763
 |
Component:279; Jaccard:0.077926
 |
Component:254; Jaccard:0.062212
 |
Component:254; Jaccard:0.03957
 |
Component:254; Jaccard:0.021833
 |
Correlation:0.25872
  |
Correlation:0.10301
  |
Correlation:0.17
  |
Correlation:0.17
  |
Correlation:0.043549
  |
Correlation:0.043549
  |
Correlation:0.043549
  |
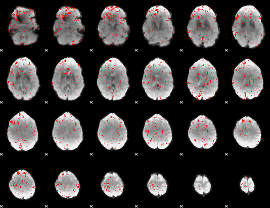
Component:051; Jaccard:0.08199
 |
Component:279; Jaccard:0.086197
 |
Component:209; Jaccard:0.082464
 |
Component:254; Jaccard:0.077505
 |
Component:279; Jaccard:0.061675
 |
Component:279; Jaccard:0.037062
 |
Component:284; Jaccard:0.021087
 |
Correlation:0.2428
  |
Correlation:0.17
  |
Correlation:0.32454
  |
Correlation:0.043549
  |
Correlation:0.17
  |
Correlation:0.17
  |
Correlation:0.076396
  |
Component:379; Jaccard:0.081789
 |
Component:330; Jaccard:0.086063
 |
Component:051; Jaccard:0.081752
 |
Component:098; Jaccard:0.076232
 |
Component:284; Jaccard:0.056725
 |
Component:284; Jaccard:0.035798
 |
Component:277; Jaccard:0.020618
 |
Correlation:0.18185
  |
Correlation:0.25872
  |
Correlation:0.2428
  |
Correlation:0.29953
  |
Correlation:0.076396
  |
Correlation:0.076396
  |
Correlation:0.33057
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































