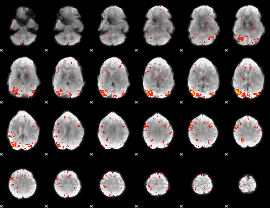
Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
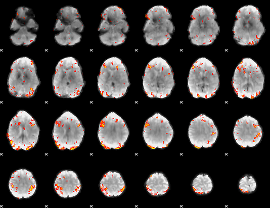
GLM result group-wise
 |
GLM binary result thresholded by 1
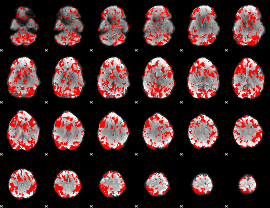 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
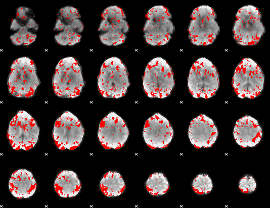 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
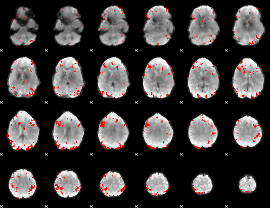 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
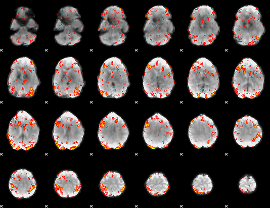 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:158; Jaccard:0.1126
 |
Component:158; Jaccard:0.13606
 |
Component:158; Jaccard:0.16145
 |
Component:003; Jaccard:0.18345
 |
Component:003; Jaccard:0.19956
 |
Component:003; Jaccard:0.20447
 |
Component:003; Jaccard:0.19086
 |
Correlation:0.14085
  |
Correlation:0.14085
  |
Correlation:0.14085
  |
Correlation:0.23907
  |
Correlation:0.23907
  |
Correlation:0.23907
  |
Correlation:0.23907
  |
Component:269; Jaccard:0.11119
 |
Component:003; Jaccard:0.13297
 |
Component:003; Jaccard:0.15969
 |
Component:158; Jaccard:0.18326
 |
Component:158; Jaccard:0.19406
 |
Component:158; Jaccard:0.19213
 |
Component:158; Jaccard:0.17808
 |
Correlation:0.11953
  |
Correlation:0.23907
  |
Correlation:0.23907
  |
Correlation:0.14085
  |
Correlation:0.14085
  |
Correlation:0.14085
  |
Correlation:0.14085
  |
Component:003; Jaccard:0.10805
 |
Component:127; Jaccard:0.12832
 |
Component:127; Jaccard:0.15072
 |
Component:127; Jaccard:0.17166
 |
Component:127; Jaccard:0.18137
 |
Component:127; Jaccard:0.1764
 |
Component:127; Jaccard:0.15738
 |
Correlation:0.23907
  |
Correlation:0.11885
  |
Correlation:0.11885
  |
Correlation:0.11885
  |
Correlation:0.11885
  |
Correlation:0.11885
  |
Correlation:0.11885
  |
Component:127; Jaccard:0.10749
 |
Component:269; Jaccard:0.12512
 |
Component:106; Jaccard:0.13871
 |
Component:106; Jaccard:0.15042
 |
Component:106; Jaccard:0.15276
 |
Component:106; Jaccard:0.14448
 |
Component:106; Jaccard:0.12799
 |
Correlation:0.11885
  |
Correlation:0.11953
  |
Correlation:-0.056976
  |
Correlation:-0.056976
  |
Correlation:-0.056976
  |
Correlation:-0.056976
  |
Correlation:-0.056976
  |
Component:106; Jaccard:0.10736
 |
Component:106; Jaccard:0.1244
 |
Component:269; Jaccard:0.13552
 |
Component:269; Jaccard:0.13978
 |
Component:269; Jaccard:0.13289
 |
Component:257; Jaccard:0.1194
 |
Component:257; Jaccard:0.11238
 |
Correlation:-0.056976
  |
Correlation:-0.056976
  |
Correlation:0.11953
  |
Correlation:0.11953
  |
Correlation:0.11953
  |
Correlation:0.26966
  |
Correlation:0.26966
  |
Component:254; Jaccard:0.10072
 |
Component:254; Jaccard:0.10582
 |
Component:257; Jaccard:0.10923
 |
Component:257; Jaccard:0.11849
 |
Component:257; Jaccard:0.12256
 |
Component:269; Jaccard:0.11834
 |
Component:269; Jaccard:0.097501
 |
Correlation:0.10391
  |
Correlation:0.10391
  |
Correlation:0.26966
  |
Correlation:0.26966
  |
Correlation:0.26966
  |
Correlation:0.11953
  |
Correlation:0.11953
  |
Component:346; Jaccard:0.096132
 |
Component:346; Jaccard:0.10336
 |
Component:254; Jaccard:0.10752
 |
Component:324; Jaccard:0.10519
 |
Component:324; Jaccard:0.10897
 |
Component:324; Jaccard:0.10755
 |
Component:324; Jaccard:0.095892
 |
Correlation:0.13881
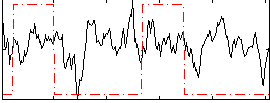  |
Correlation:0.13881
  |
Correlation:0.10391
  |
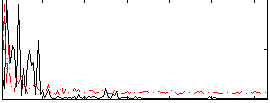
Correlation:0.16088
  |
Correlation:0.16088
  |
Correlation:0.16088
  |
Correlation:0.16088
  |
Component:298; Jaccard:0.090586
 |
Component:257; Jaccard:0.097711
 |
Component:346; Jaccard:0.10675
 |
Component:346; Jaccard:0.10422
 |
Component:298; Jaccard:0.098614
 |
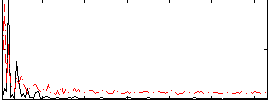
Component:343; Jaccard:0.089081
 |
Component:343; Jaccard:0.080854
 |
Correlation:0.0051033
  |
Correlation:0.26966
  |
Correlation:0.13881
  |
Correlation:0.13881
  |
Correlation:0.0051033
  |
Correlation:0.14813
  |
Correlation:0.14813
  |
Component:257; Jaccard:0.086485
 |
Component:298; Jaccard:0.097061
 |
Component:298; Jaccard:0.10132
 |
Component:254; Jaccard:0.10227
 |
Component:346; Jaccard:0.095558
 |
Component:298; Jaccard:0.088217
 |
Component:019; Jaccard:0.078421
 |
Correlation:0.26966
  |
Correlation:0.0051033
  |
Correlation:0.0051033
  |
Correlation:0.10391
  |
Correlation:0.13881
  |
Correlation:0.0051033
  |
Correlation:0.16762
  |
Component:280; Jaccard:0.085862
 |
Component:019; Jaccard:0.091308
 |
Component:324; Jaccard:0.098095
 |
Component:298; Jaccard:0.10115
 |
Component:019; Jaccard:0.094511
 |
Component:019; Jaccard:0.087855
 |
Component:298; Jaccard:0.074721
 |
Correlation:-0.042023
  |
Correlation:0.16762
  |
Correlation:0.16088
  |
Correlation:0.0051033
  |
Correlation:0.16762
  |
Correlation:0.16762
  |
Correlation:0.0051033
  |
Cope: 02:REWARD BACK
|
GLM result group-wise
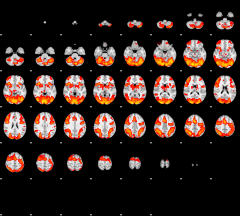 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
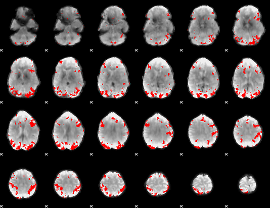 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
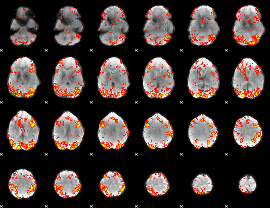 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
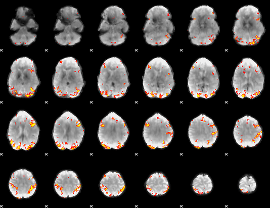
GLM Z-score result thresholded by 3.5
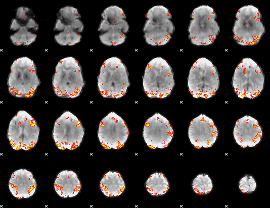 |
GLM Z-score result thresholded by 4
 |
Component:388; Jaccard:0.14217
 |
Component:388; Jaccard:0.16392
 |
Component:388; Jaccard:0.18107
 |
Component:158; Jaccard:0.22088
 |
Component:158; Jaccard:0.2552
 |
Component:158; Jaccard:0.28093
 |
Component:158; Jaccard:0.28964
 |
Correlation:0.32773
  |
Correlation:0.32773
  |
Correlation:0.32773
  |
Correlation:0.24418
  |
Correlation:0.24418
  |
Correlation:0.24418
  |
Correlation:0.24418
  |
Component:106; Jaccard:0.11373
 |
Component:158; Jaccard:0.14308
 |
Component:158; Jaccard:0.18051
 |
Component:388; Jaccard:0.18693
 |
Component:003; Jaccard:0.20725
 |
Component:003; Jaccard:0.22301
 |
Component:003; Jaccard:0.22788
 |
Correlation:0.1156
  |
Correlation:0.24418
  |
Correlation:0.24418
  |
Correlation:0.32773
  |
Correlation:0.28019
  |
Correlation:0.28019
  |
Correlation:0.28019
  |
Component:158; Jaccard:0.11011
 |
Component:106; Jaccard:0.14023
 |
Component:106; Jaccard:0.1661
 |
Component:106; Jaccard:0.18605
 |
Component:106; Jaccard:0.20041
 |
Component:106; Jaccard:0.19889
 |
Component:106; Jaccard:0.18919
 |
Correlation:0.24418
  |
Correlation:0.1156
  |
Correlation:0.1156
  |
Correlation:0.1156
  |
Correlation:0.1156
  |
Correlation:0.1156
  |
Correlation:0.1156
  |
Component:346; Jaccard:0.10617
 |
Component:003; Jaccard:0.1224
 |
Component:003; Jaccard:0.15238
 |
Component:003; Jaccard:0.18263
 |
Component:388; Jaccard:0.18321
 |
Component:388; Jaccard:0.16895
 |
Component:127; Jaccard:0.15994
 |
Correlation:0.14624
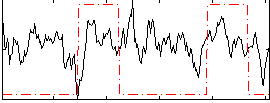  |
Correlation:0.28019
  |
Correlation:0.28019
  |
Correlation:0.28019
  |
Correlation:0.32773
  |
Correlation:0.32773
  |
Correlation:0.17706
  |
Component:269; Jaccard:0.10033
 |
Component:346; Jaccard:0.122
 |
Component:346; Jaccard:0.13771
 |
Component:127; Jaccard:0.15618
 |
Component:127; Jaccard:0.16592
 |
Component:127; Jaccard:0.16838
 |
Component:388; Jaccard:0.15163
 |
Correlation:0.14131
 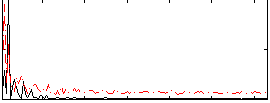 |
Correlation:0.14624
  |
Correlation:0.14624
  |
Correlation:0.17706
  |
Correlation:0.17706
  |
Correlation:0.17706
  |
Correlation:0.32773
  |
Component:298; Jaccard:0.098203
 |
Component:127; Jaccard:0.11584
 |
Component:127; Jaccard:0.13743
 |
Component:346; Jaccard:0.14945
 |
Component:346; Jaccard:0.15361
 |
Component:346; Jaccard:0.1498
 |
Component:343; Jaccard:0.13996
 |
Correlation:0.064267
  |
Correlation:0.17706
  |
Correlation:0.17706
  |
Correlation:0.14624
  |
Correlation:0.14624
  |
Correlation:0.14624
  |
Correlation:0.22285
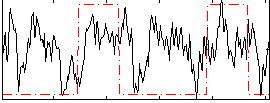 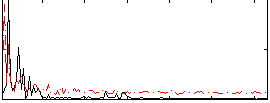 |
Component:003; Jaccard:0.096543
 |
Component:269; Jaccard:0.11313
 |
Component:298; Jaccard:0.12506
 |
Component:298; Jaccard:0.13332
 |
Component:343; Jaccard:0.14272
 |
Component:343; Jaccard:0.14516
 |
Component:346; Jaccard:0.13982
 |
Correlation:0.28019
  |
Correlation:0.14131
  |
Correlation:0.064267
  |
Correlation:0.064267
  |
Correlation:0.22285
  |
Correlation:0.22285
  |
Correlation:0.14624
  |
Component:127; Jaccard:0.094741
 |
Component:298; Jaccard:0.11261
 |
Component:269; Jaccard:0.1217
 |
Component:343; Jaccard:0.13303
 |
Component:298; Jaccard:0.13473
 |
Component:019; Jaccard:0.12815
 |
Component:019; Jaccard:0.12266
 |
Correlation:0.17706
  |
Correlation:0.064267
  |
Correlation:0.14131
  |
Correlation:0.22285
  |
Correlation:0.064267
  |
Correlation:0.086268
  |
Correlation:0.086268
  |
Component:188; Jaccard:0.092999
 |
Component:343; Jaccard:0.1041
 |
Component:343; Jaccard:0.12023
 |
Component:019; Jaccard:0.12583
 |
Component:019; Jaccard:0.13005
 |
Component:298; Jaccard:0.12794
 |
Component:169; Jaccard:0.12062
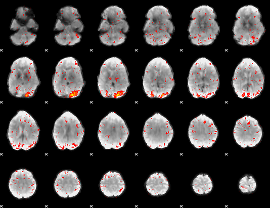 |
Correlation:0.16063
  |
Correlation:0.22285
  |
Correlation:0.22285
  |
Correlation:0.086268
  |
Correlation:0.086268
  |
Correlation:0.064267
  |
Correlation:0.38512
  |
Component:019; Jaccard:0.090621
 |
Component:019; Jaccard:0.10377
 |
Component:019; Jaccard:0.11588
 |
Component:269; Jaccard:0.12559
 |
Component:324; Jaccard:0.12576
 |
Component:324; Jaccard:0.12608
 |
Component:324; Jaccard:0.11891
 |
Correlation:0.086268
  |
Correlation:0.086268
  |
Correlation:0.086268
  |
Correlation:0.14131
  |
Correlation:0.23755
 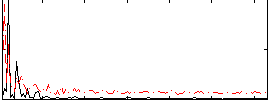 |
Correlation:0.23755
  |
Correlation:0.23755
  |
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
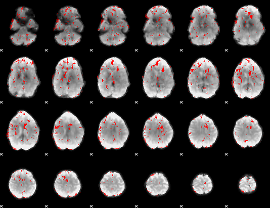 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
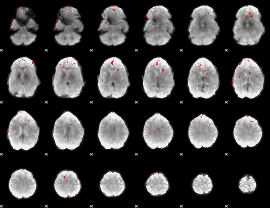 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
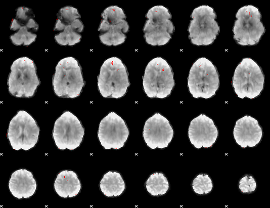 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
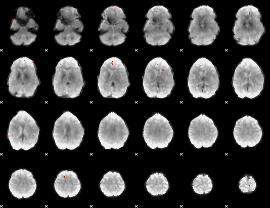 |
Component:074; Jaccard:0.10216
 |
Component:074; Jaccard:0.10009
 |
Component:150; Jaccard:0.089191
 |
Component:233; Jaccard:0.071594
 |
Component:233; Jaccard:0.055107
 |
Component:233; Jaccard:0.032981
 |
Component:052; Jaccard:0.018556
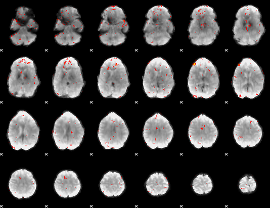 |
Correlation:0.29797
  |
Correlation:0.29797
  |
Correlation:0.11635
  |
Correlation:0.10623
  |
Correlation:0.10623
  |
Correlation:0.10623
  |
Correlation:0.50478
  |
Component:362; Jaccard:0.087614
 |
Component:150; Jaccard:0.093059
 |
Component:074; Jaccard:0.088914
 |
Component:150; Jaccard:0.066793
 |
Component:114; Jaccard:0.048338
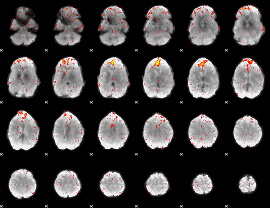 |
Component:114; Jaccard:0.026594
 |
Component:344; Jaccard:0.01517
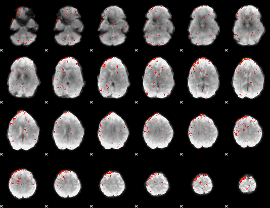 |
Correlation:-0.0026516
 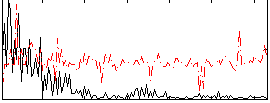 |
Correlation:0.11635
  |
Correlation:0.29797
  |
Correlation:0.11635
  |
Correlation:0.28864
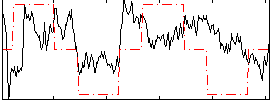  |
Correlation:0.28864
  |
Correlation:0.16084
 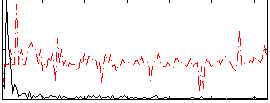 |
Component:150; Jaccard:0.085651
 |
Component:064; Jaccard:0.092749
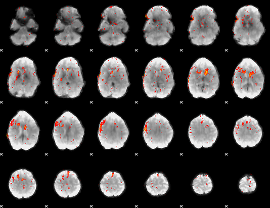 |
Component:064; Jaccard:0.086661
 |
Component:064; Jaccard:0.06676
 |
Component:064; Jaccard:0.042836
 |
Component:344; Jaccard:0.025987
 |
Component:114; Jaccard:0.01467
 |
Correlation:0.11635
  |
Correlation:-0.028076
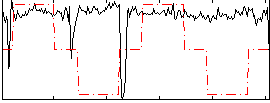 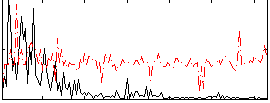 |
Correlation:-0.028076
  |
Correlation:-0.028076
  |
Correlation:-0.028076
  |
Correlation:0.16084
  |
Correlation:0.28864
  |
Component:064; Jaccard:0.083287
 |
Component:362; Jaccard:0.09098
 |
Component:272; Jaccard:0.081101
 |
Component:074; Jaccard:0.065533
 |
Component:344; Jaccard:0.040875
 |
Component:005; Jaccard:0.022594
 |
Component:233; Jaccard:0.014288
 |
Correlation:-0.028076
  |
Correlation:-0.0026516
  |
Correlation:0.041474
 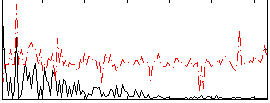 |
Correlation:0.29797
  |
Correlation:0.16084
  |
Correlation:0.32556
  |
Correlation:0.10623
  |
Component:272; Jaccard:0.083201
 |
Component:272; Jaccard:0.087651
 |
Component:264; Jaccard:0.078854
 |
Component:114; Jaccard:0.065391
 |
Component:074; Jaccard:0.040442
 |
Component:064; Jaccard:0.021999
 |
Component:005; Jaccard:0.013314
 |
Correlation:0.041474
  |
Correlation:0.041474
  |
Correlation:0.19318
  |
Correlation:0.28864
  |
Correlation:0.29797
  |
Correlation:-0.028076
  |
Correlation:0.32556
  |
Component:264; Jaccard:0.08082
 |
Component:012; Jaccard:0.084879
 |
Component:233; Jaccard:0.078162
 |
Component:264; Jaccard:0.061524
 |
Component:264; Jaccard:0.04039
 |
Component:074; Jaccard:0.021617
 |
Component:286; Jaccard:0.012297
 |
Correlation:0.19318
  |
Correlation:0.12077
  |
Correlation:0.10623
  |
Correlation:0.19318
  |
Correlation:0.19318
  |
Correlation:0.29797
  |
Correlation:0.20152
  |
Component:012; Jaccard:0.080319
 |
Component:264; Jaccard:0.083107
 |
Component:114; Jaccard:0.077321
 |
Component:272; Jaccard:0.061487
 |
Component:272; Jaccard:0.039663
 |
Component:052; Jaccard:0.021538
 |
Component:246; Jaccard:0.01095
 |
Correlation:0.12077
  |
Correlation:0.19318
  |
Correlation:0.28864
  |
Correlation:0.041474
  |
Correlation:0.041474
  |
Correlation:0.50478
  |
Correlation:0.32139
  |
Component:114; Jaccard:0.076111
 |
Component:114; Jaccard:0.079613
 |
Component:012; Jaccard:0.07652
 |
Component:012; Jaccard:0.056496
 |
Component:150; Jaccard:0.039394
 |
Component:286; Jaccard:0.021305
 |
Component:199; Jaccard:0.010913
 |
Correlation:0.28864
  |
Correlation:0.28864
  |
Correlation:0.12077
  |
Correlation:0.12077
  |
Correlation:0.11635
  |
Correlation:0.20152
  |
Correlation:0.33095
  |
Component:139; Jaccard:0.068126
 |
Component:139; Jaccard:0.076935
 |
Component:362; Jaccard:0.074646
 |
Component:139; Jaccard:0.055444
 |
Component:005; Jaccard:0.03857
 |
Component:199; Jaccard:0.020313
 |
Component:074; Jaccard:0.010862
 |
Correlation:0.20106
  |
Correlation:0.20106
  |
Correlation:-0.0026516
  |
Correlation:0.20106
  |
Correlation:0.32556
  |
Correlation:0.33095
  |
Correlation:0.29797
  |
Component:335; Jaccard:0.066467
 |
Component:233; Jaccard:0.07346
 |
Component:139; Jaccard:0.074086
 |
Component:344; Jaccard:0.050599
 |
Component:052; Jaccard:0.03622
 |
Component:322; Jaccard:0.019482
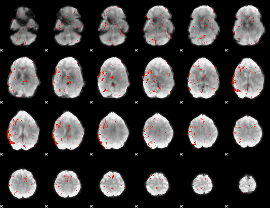 |
Component:322; Jaccard:0.010462
 |
Correlation:0.22538
  |
Correlation:0.10623
  |
Correlation:0.20106
  |
Correlation:0.16084
  |
Correlation:0.50478
  |
Correlation:0.12525
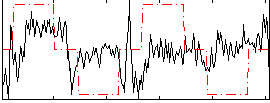  |
Correlation:0.12525
  |
Cope: 04:neg_PUNISH BACK
|
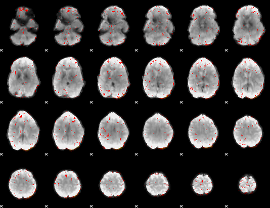
GLM result group-wise
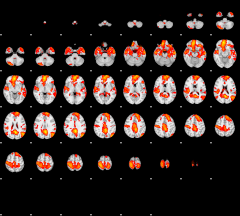 |
GLM binary result thresholded by 1
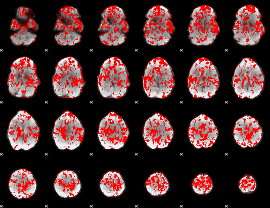 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
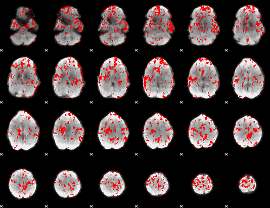 |
GLM binary result thresholded by 2.5
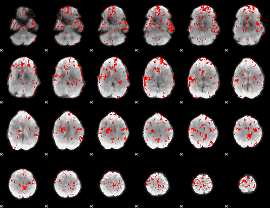 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
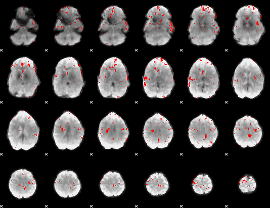 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
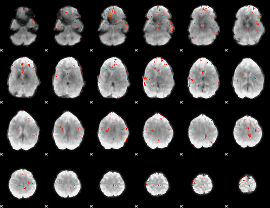 |
Component:035; Jaccard:0.08185
 |
Component:354; Jaccard:0.092401
 |
Component:354; Jaccard:0.10056
 |
Component:035; Jaccard:0.10627
 |
Component:035; Jaccard:0.1038
 |
Component:035; Jaccard:0.092077
 |
Component:035; Jaccard:0.068159
 |
Correlation:0.15482
  |
Correlation:0.24505
  |
Correlation:0.24505
  |
Correlation:0.15482
  |
Correlation:0.15482
  |
Correlation:0.15482
  |
Correlation:0.15482
  |
Component:017; Jaccard:0.081061
 |
Component:035; Jaccard:0.092081
 |
Component:035; Jaccard:0.10019
 |
Component:354; Jaccard:0.10154
 |
Component:178; Jaccard:0.099732
 |
Component:178; Jaccard:0.082174
 |
Component:354; Jaccard:0.058536
 |
Correlation:0.25428
  |
Correlation:0.15482
  |
Correlation:0.15482
  |
Correlation:0.24505
  |
Correlation:0.31779
  |
Correlation:0.31779
  |
Correlation:0.24505
  |
Component:354; Jaccard:0.080156
 |
Component:017; Jaccard:0.09006
 |
Component:017; Jaccard:0.094818
 |
Component:178; Jaccard:0.10092
 |
Component:354; Jaccard:0.093519
 |
Component:354; Jaccard:0.079361
 |
Component:216; Jaccard:0.058252
 |
Correlation:0.24505
  |
Correlation:0.25428
  |
Correlation:0.25428
  |
Correlation:0.31779
  |
Correlation:0.24505
  |
Correlation:0.24505
  |
Correlation:0.082743
  |
Component:129; Jaccard:0.079444
 |
Component:178; Jaccard:0.08453
 |
Component:178; Jaccard:0.093272
 |
Component:017; Jaccard:0.095606
 |
Component:017; Jaccard:0.087398
 |
Component:216; Jaccard:0.076762
 |
Component:389; Jaccard:0.056742
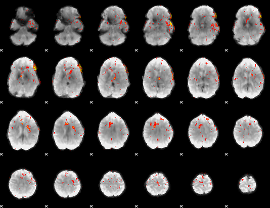 |
Correlation:0.17948
  |
Correlation:0.31779
  |
Correlation:0.31779
  |
Correlation:0.25428
  |
Correlation:0.25428
  |
Correlation:0.082743
  |
Correlation:0.1426
 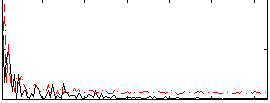 |
Component:270; Jaccard:0.077891
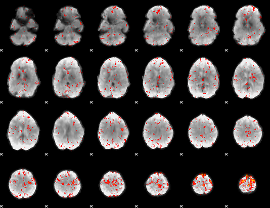 |
Component:129; Jaccard:0.081635
 |
Component:180; Jaccard:0.084295
 |
Component:180; Jaccard:0.084336
 |
Component:216; Jaccard:0.082865
 |
Component:309; Jaccard:0.072853
 |
Component:178; Jaccard:0.056663
 |
Correlation:0.16735
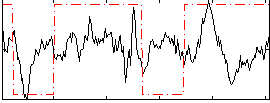  |
Correlation:0.17948
  |
Correlation:0.17761
  |
Correlation:0.17761
  |
Correlation:0.082743
  |
Correlation:0.1083
 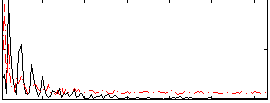 |
Correlation:0.31779
  |
Component:178; Jaccard:0.075935
 |
Component:270; Jaccard:0.081287
 |
Component:270; Jaccard:0.081632
 |
Component:216; Jaccard:0.084325
 |
Component:309; Jaccard:0.081581
 |
Component:017; Jaccard:0.070141
 |
Component:063; Jaccard:0.052098
 |
Correlation:0.31779
  |
Correlation:0.16735
  |
Correlation:0.16735
  |
Correlation:0.082743
  |
Correlation:0.1083
  |
Correlation:0.25428
  |
Correlation:0.30423
  |
Component:260; Jaccard:0.074824
 |
Component:180; Jaccard:0.077858
 |
Component:129; Jaccard:0.080563
 |
Component:309; Jaccard:0.083377
 |
Component:180; Jaccard:0.079401
 |
Component:389; Jaccard:0.069711
 |
Component:017; Jaccard:0.051813
 |
Correlation:0.062604
  |
Correlation:0.17761
  |
Correlation:0.17948
  |
Correlation:0.1083
  |
Correlation:0.17761
  |
Correlation:0.1426
  |
Correlation:0.25428
  |
Component:128; Jaccard:0.069491
 |
Component:260; Jaccard:0.0774
 |
Component:376; Jaccard:0.078595
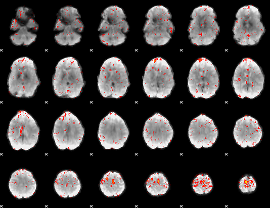 |
Component:313; Jaccard:0.081236
 |
Component:313; Jaccard:0.075844
 |
Component:180; Jaccard:0.066128
 |
Component:309; Jaccard:0.05138
 |
Correlation:0.3072
  |
Correlation:0.062604
  |
Correlation:0.13622
  |
Correlation:0.1896
  |
Correlation:0.1896
  |
Correlation:0.17761
  |
Correlation:0.1083
  |
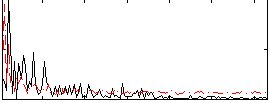
Component:182; Jaccard:0.069402
 |
Component:376; Jaccard:0.074493
 |
Component:313; Jaccard:0.078361
 |
Component:270; Jaccard:0.078843
 |
Component:389; Jaccard:0.075638
 |
Component:063; Jaccard:0.064886
 |
Component:316; Jaccard:0.047092
 |
Correlation:0.10073
  |
Correlation:0.13622
  |
Correlation:0.1896
  |
Correlation:0.16735
  |
Correlation:0.1426
  |
Correlation:0.30423
  |
Correlation:0.23195
  |
Component:313; Jaccard:0.069367
 |
Component:313; Jaccard:0.074425
 |
Component:216; Jaccard:0.078217
 |
Component:077; Jaccard:0.077581
 |
Component:077; Jaccard:0.073297
 |
Component:313; Jaccard:0.063945
 |
Component:180; Jaccard:0.046482
 |
Correlation:0.1896
  |
Correlation:0.1896
  |
Correlation:0.082743
  |
Correlation:0.04033
  |
Correlation:0.04033
  |
Correlation:0.1896
  |
Correlation:0.17761
  |
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
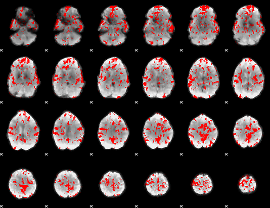 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
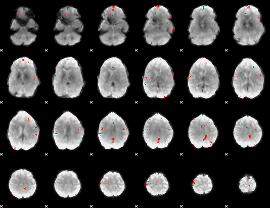 |
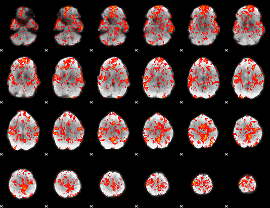
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
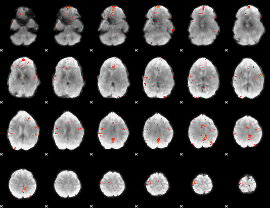
GLM Z-score result thresholded by 3.5
 |
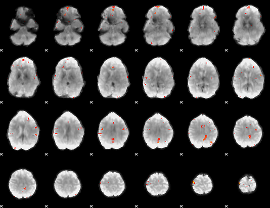
GLM Z-score result thresholded by 4
 |
Component:035; Jaccard:0.13302
 |
Component:035; Jaccard:0.17016
 |
Component:035; Jaccard:0.19363
 |
Component:035; Jaccard:0.19144
 |
Component:035; Jaccard:0.16676
 |
Component:035; Jaccard:0.12595
 |
Component:035; Jaccard:0.083746
 |
Correlation:0.19566
  |
Correlation:0.19566
  |
Correlation:0.19566
  |
Correlation:0.19566
  |
Correlation:0.19566
  |
Correlation:0.19566
  |
Correlation:0.19566
  |
Component:182; Jaccard:0.10136
 |
Component:017; Jaccard:0.11163
 |
Component:017; Jaccard:0.11258
 |
Component:178; Jaccard:0.10499
 |
Component:313; Jaccard:0.086645
 |
Component:313; Jaccard:0.067172
 |
Component:313; Jaccard:0.043517
 |
Correlation:0.043644
  |
Correlation:0.22294
  |
Correlation:0.22294
  |
Correlation:0.095363
  |
Correlation:0.20369
  |
Correlation:0.20369
  |
Correlation:0.20369
  |
Component:017; Jaccard:0.10073
 |
Component:182; Jaccard:0.10882
 |
Component:178; Jaccard:0.11064
 |
Component:017; Jaccard:0.10377
 |
Component:178; Jaccard:0.084827
 |
Component:216; Jaccard:0.062025
 |
Component:216; Jaccard:0.042178
 |
Correlation:0.22294
  |
Correlation:0.043644
  |
Correlation:0.095363
  |
Correlation:0.22294
  |
Correlation:0.095363
  |
Correlation:0.38427
  |
Correlation:0.38427
  |
Component:313; Jaccard:0.095225
 |
Component:178; Jaccard:0.10502
 |
Component:182; Jaccard:0.10714
 |
Component:313; Jaccard:0.10205
 |
Component:017; Jaccard:0.082147
 |
Component:178; Jaccard:0.057596
 |
Component:178; Jaccard:0.035978
 |
Correlation:0.20369
  |
Correlation:0.095363
  |
Correlation:0.043644
  |
Correlation:0.20369
  |
Correlation:0.22294
  |
Correlation:0.095363
  |
Correlation:0.095363
  |
Component:178; Jaccard:0.093794
 |
Component:313; Jaccard:0.10439
 |
Component:313; Jaccard:0.10649
 |
Component:216; Jaccard:0.093641
 |
Component:216; Jaccard:0.081741
 |
Component:017; Jaccard:0.053513
 |
Component:017; Jaccard:0.033548
 |
Correlation:0.095363
  |
Correlation:0.20369
  |
Correlation:0.20369
  |
Correlation:0.38427
  |
Correlation:0.38427
  |
Correlation:0.22294
  |
Correlation:0.22294
  |
Component:260; Jaccard:0.091498
 |
Component:260; Jaccard:0.094941
 |
Component:216; Jaccard:0.09259
 |
Component:182; Jaccard:0.092761
 |
Component:182; Jaccard:0.072237
 |
Component:264; Jaccard:0.053411
 |
Component:052; Jaccard:0.033303
 |
Correlation:0.24815
  |
Correlation:0.24815
  |
Correlation:0.38427
  |
Correlation:0.043644
  |
Correlation:0.043644
  |
Correlation:0.25254
  |
Correlation:0.48165
  |
Component:129; Jaccard:0.088263
 |
Component:129; Jaccard:0.089298
 |
Component:264; Jaccard:0.089698
 |
Component:264; Jaccard:0.084611
 |
Component:264; Jaccard:0.071615
 |
Component:182; Jaccard:0.051562
 |
Component:182; Jaccard:0.033141
 |
Correlation:0.11616
  |
Correlation:0.11616
  |
Correlation:0.25254
  |
Correlation:0.25254
  |
Correlation:0.25254
  |
Correlation:0.043644
  |
Correlation:0.043644
  |
Component:270; Jaccard:0.083675
 |
Component:376; Jaccard:0.087696
 |
Component:260; Jaccard:0.089293
 |
Component:074; Jaccard:0.075749
 |
Component:376; Jaccard:0.061496
 |
Component:052; Jaccard:0.045936
 |
Component:264; Jaccard:0.033072
 |
Correlation:-0.095773
  |
Correlation:0.14499
  |
Correlation:0.24815
  |
Correlation:0.22494
  |
Correlation:0.14499
  |
Correlation:0.48165
  |
Correlation:0.25254
  |
Component:376; Jaccard:0.082523
 |
Component:270; Jaccard:0.085867
 |
Component:376; Jaccard:0.086213
 |
Component:376; Jaccard:0.075728
 |
Component:074; Jaccard:0.061167
 |
Component:074; Jaccard:0.043165
 |
Component:376; Jaccard:0.028466
 |
Correlation:0.14499
  |
Correlation:-0.095773
  |
Correlation:0.14499
  |
Correlation:0.14499
  |
Correlation:0.22494
  |
Correlation:0.22494
  |
Correlation:0.14499
  |
Component:074; Jaccard:0.07849
 |
Component:216; Jaccard:0.084166
 |
Component:074; Jaccard:0.085642
 |
Component:260; Jaccard:0.07555
 |
Component:166; Jaccard:0.057448
 |
Component:376; Jaccard:0.043027
 |
Component:166; Jaccard:0.028404
 |
Correlation:0.22494
  |
Correlation:0.38427
  |
Correlation:0.22494
  |
Correlation:0.24815
  |
Correlation:0.29874
  |
Correlation:0.14499
  |
Correlation:0.29874
  |
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
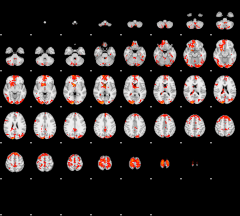 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
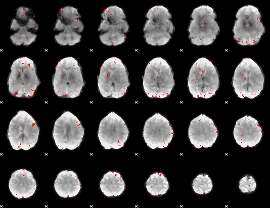 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
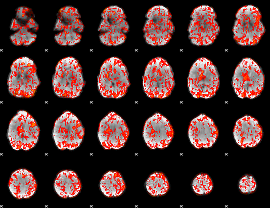 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
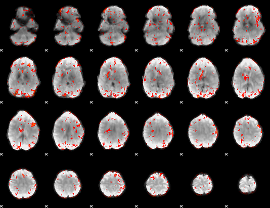 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:388; Jaccard:0.12652
 |
Component:388; Jaccard:0.14842
 |
Component:388; Jaccard:0.16299
 |
Component:388; Jaccard:0.16113
 |
Component:388; Jaccard:0.13124
 |
Component:292; Jaccard:0.086615
 |
Component:292; Jaccard:0.058656
 |
Correlation:0.32937
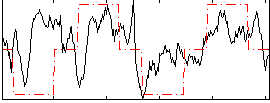  |
Correlation:0.32937
  |
Correlation:0.32937
  |
Correlation:0.32937
  |
Correlation:0.32937
  |
Correlation:0.24332
 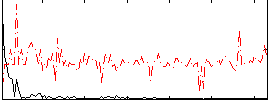 |
Correlation:0.24332
  |
Component:258; Jaccard:0.07399
 |
Component:145; Jaccard:0.087533
 |
Component:145; Jaccard:0.102
 |
Component:292; Jaccard:0.10905
 |
Component:292; Jaccard:0.10633
 |
Component:388; Jaccard:0.086032
 |
Component:111; Jaccard:0.057258
 |
Correlation:0.13614
  |
Correlation:0.1612
  |
Correlation:0.1612
  |
Correlation:0.24332
  |
Correlation:0.24332
  |
Correlation:0.32937
  |
Correlation:0.45514
  |
Component:145; Jaccard:0.07289
 |
Component:292; Jaccard:0.087363
 |
Component:292; Jaccard:0.10074
 |
Component:145; Jaccard:0.10789
 |
Component:145; Jaccard:0.10328
 |
Component:111; Jaccard:0.082577
 |
Component:278; Jaccard:0.055672
 |
Correlation:0.1612
  |
Correlation:0.24332
  |
Correlation:0.24332
  |
Correlation:0.1612
  |
Correlation:0.1612
  |
Correlation:0.45514
  |
Correlation:0.27911
  |
Component:292; Jaccard:0.072468
 |
Component:111; Jaccard:0.081757
 |
Component:111; Jaccard:0.09693
 |
Component:111; Jaccard:0.10669
 |
Component:111; Jaccard:0.10305
 |
Component:278; Jaccard:0.081133
 |
Component:388; Jaccard:0.049472
 |
Correlation:0.24332
  |
Correlation:0.45514
  |
Correlation:0.45514
  |
Correlation:0.45514
  |
Correlation:0.45514
  |
Correlation:0.27911
  |
Correlation:0.32937
  |
Component:111; Jaccard:0.068468
 |
Component:138; Jaccard:0.07749
 |
Component:138; Jaccard:0.092383
 |
Component:138; Jaccard:0.10203
 |
Component:138; Jaccard:0.097404
 |
Component:145; Jaccard:0.080658
 |
Component:145; Jaccard:0.047423
 |
Correlation:0.45514
  |
Correlation:0.31702
  |
Correlation:0.31702
  |
Correlation:0.31702
  |
Correlation:0.31702
  |
Correlation:0.1612
  |
Correlation:0.1612
  |
Component:144; Jaccard:0.06815
 |
Component:251; Jaccard:0.073526
 |
Component:278; Jaccard:0.08462
 |
Component:278; Jaccard:0.093325
 |
Component:278; Jaccard:0.096043
 |
Component:138; Jaccard:0.077243
 |
Component:138; Jaccard:0.045161
 |
Correlation:0.10263
  |
Correlation:0.43518
  |
Correlation:0.27911
  |
Correlation:0.27911
  |
Correlation:0.27911
  |
Correlation:0.31702
  |
Correlation:0.31702
  |
Component:385; Jaccard:0.067518
 |
Component:278; Jaccard:0.07293
 |
Component:251; Jaccard:0.082362
 |
Component:251; Jaccard:0.088116
 |
Component:251; Jaccard:0.084468
 |
Component:251; Jaccard:0.067179
 |
Component:251; Jaccard:0.041517
 |
Correlation:0.27006
  |
Correlation:0.27911
  |
Correlation:0.43518
  |
Correlation:0.43518
  |
Correlation:0.43518
  |
Correlation:0.43518
  |
Correlation:0.43518
  |
Component:138; Jaccard:0.064946
 |
Component:258; Jaccard:0.072635
 |
Component:317; Jaccard:0.072104
 |
Component:317; Jaccard:0.077034
 |
Component:317; Jaccard:0.073304
 |
Component:317; Jaccard:0.056196
 |
Component:043; Jaccard:0.034917
 |
Correlation:0.31702
  |
Correlation:0.13614
  |
Correlation:0.3111
  |
Correlation:0.3111
  |
Correlation:0.3111
  |
Correlation:0.3111
  |
Correlation:0.26687
  |
Component:343; Jaccard:0.064251
 |
Component:144; Jaccard:0.069885
 |
Component:266; Jaccard:0.071555
 |
Component:043; Jaccard:0.072821
 |
Component:043; Jaccard:0.068233
 |
Component:043; Jaccard:0.054661
 |
Component:317; Jaccard:0.034798
 |
Correlation:0.043939
  |
Correlation:0.10263
  |
Correlation:0.11793
  |
Correlation:0.26687
  |
Correlation:0.26687
  |
Correlation:0.26687
  |
Correlation:0.3111
  |
Component:346; Jaccard:0.063567
 |
Component:266; Jaccard:0.068153
 |
Component:158; Jaccard:0.071483
 |
Component:209; Jaccard:0.069934
 |
Component:209; Jaccard:0.060699
 |
Component:048; Jaccard:0.046598
 |
Component:048; Jaccard:0.028284
 |
Correlation:0.0043739
  |
Correlation:0.11793
  |
Correlation:0.060767
  |
Correlation:0.20675
  |
Correlation:0.20675
  |
Correlation:0.1725
  |
Correlation:0.1725
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































