Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATH BACK
|
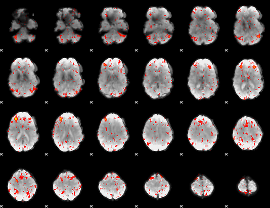
GLM result group-wise
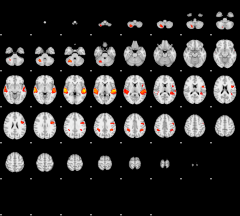 |
GLM binary result thresholded by 1
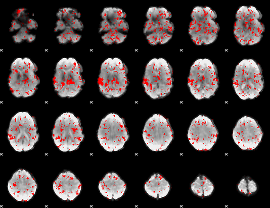 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
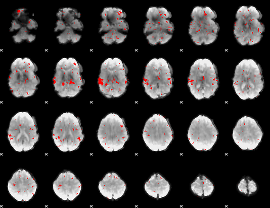 |
GLM binary result thresholded by 2.5
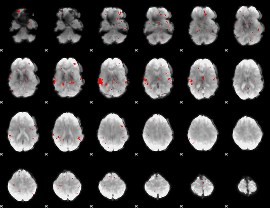 |
GLM binary result thresholded by 3
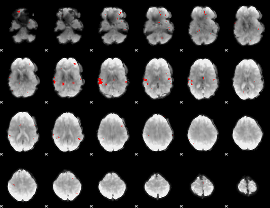 |
GLM binary result thresholded by 3.5
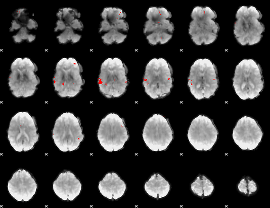 |
GLM binary result thresholded by 4
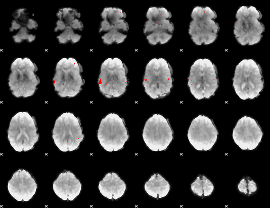 |
GLM Z-score result thresholded by 1
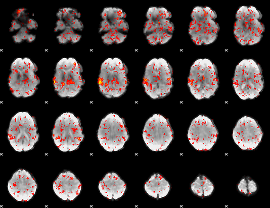 |
GLM Z-score result thresholded by 1.5
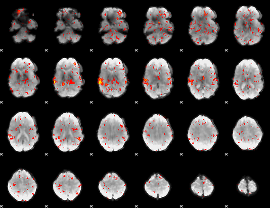 |
GLM Z-score result thresholded by 2
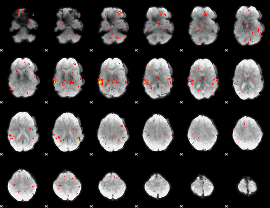 |
GLM Z-score result thresholded by 2.5
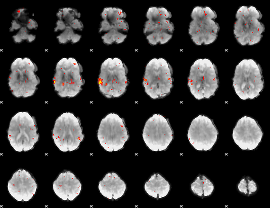 |
GLM Z-score result thresholded by 3
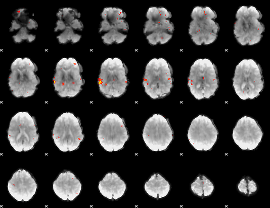 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:353; Jaccard:0.14491
 |
Component:353; Jaccard:0.1537
 |
Component:353; Jaccard:0.13674
 |
Component:353; Jaccard:0.09847
 |
Component:353; Jaccard:0.068539
 |
Component:353; Jaccard:0.043652
 |
Component:353; Jaccard:0.029144
 |
Correlation:-0.4003
  |
Correlation:-0.4003
  |
Correlation:-0.4003
  |
Correlation:-0.4003
  |
Correlation:-0.4003
  |
Correlation:-0.4003
  |
Correlation:-0.4003
  |
Component:278; Jaccard:0.092701
 |
Component:278; Jaccard:0.10124
 |
Component:278; Jaccard:0.088513
 |
Component:278; Jaccard:0.057677
 |
Component:278; Jaccard:0.029548
 |
Component:186; Jaccard:0.012968
 |
Component:171; Jaccard:0.009889
 |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:0.21038
  |
Correlation:-0.21519
  |
Correlation:-0.58391
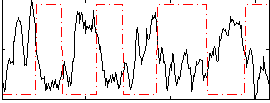 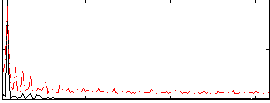 |
Component:341; Jaccard:0.07623
 |
Component:341; Jaccard:0.073678
 |
Component:341; Jaccard:0.059921
 |
Component:341; Jaccard:0.038021
 |
Component:341; Jaccard:0.021543
 |
Component:278; Jaccard:0.012856
 |
Component:186; Jaccard:0.009544
 |
Correlation:0.43857
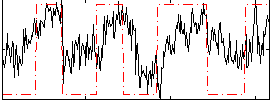 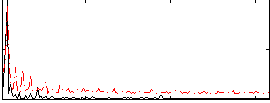 |
Correlation:0.43857
  |
Correlation:0.43857
  |
Correlation:0.43857
  |
Correlation:0.43857
  |
Correlation:0.21038
  |
Correlation:-0.21519
  |
Component:324; Jaccard:0.073728
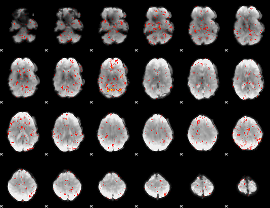 |
Component:145; Jaccard:0.063289
 |
Component:205; Jaccard:0.045864
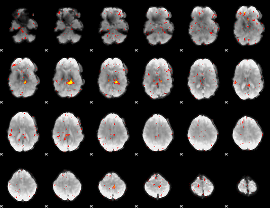 |
Component:205; Jaccard:0.030222
 |
Component:171; Jaccard:0.018435
 |
Component:171; Jaccard:0.012786
 |
Component:051; Jaccard:0.006992
 |
Correlation:0.06052
  |
Correlation:-0.14918
  |
Correlation:0.18225
  |
Correlation:0.18225
  |
Correlation:-0.58391
  |
Correlation:-0.58391
  |
Correlation:-0.32067
  |
Component:093; Jaccard:0.072558
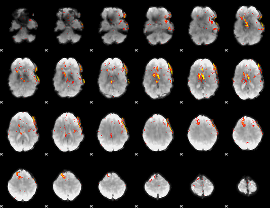 |
Component:324; Jaccard:0.061573
 |
Component:376; Jaccard:0.043871
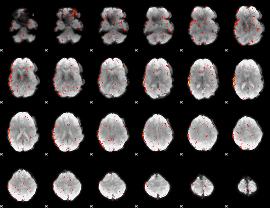 |
Component:376; Jaccard:0.029951
 |
Component:376; Jaccard:0.018051
 |
Component:341; Jaccard:0.01166
 |
Component:159; Jaccard:0.00598
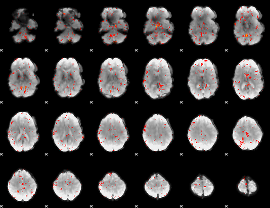 |
Correlation:0.20935
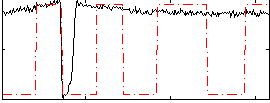  |
Correlation:0.06052
  |
Correlation:0.18826
  |
Correlation:0.18826
  |
Correlation:0.18826
  |
Correlation:0.43857
  |
Correlation:-0.06268
  |
Component:145; Jaccard:0.070722
 |
Component:093; Jaccard:0.060273
 |
Component:145; Jaccard:0.043574
 |
Component:171; Jaccard:0.024902
 |
Component:186; Jaccard:0.017106
 |
Component:376; Jaccard:0.010134
 |
Component:145; Jaccard:0.004803
 |
Correlation:-0.14918
  |
Correlation:0.20935
  |
Correlation:-0.14918
  |
Correlation:-0.58391
  |
Correlation:-0.21519
  |
Correlation:0.18826
  |
Correlation:-0.14918
  |
Component:013; Jaccard:0.069968
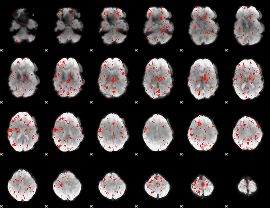 |
Component:013; Jaccard:0.058141
 |
Component:093; Jaccard:0.042042
 |
Component:145; Jaccard:0.024575
 |
Component:145; Jaccard:0.014033
 |
Component:159; Jaccard:0.008659
 |
Component:282; Jaccard:0.004764
 |
Correlation:0.17144
  |
Correlation:0.17144
  |
Correlation:0.20935
  |
Correlation:-0.14918
  |
Correlation:-0.14918
  |
Correlation:-0.06268
  |
Correlation:-0.52192
 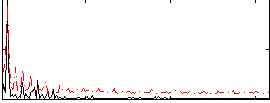 |
Component:316; Jaccard:0.066087
 |
Component:376; Jaccard:0.055487
 |
Component:324; Jaccard:0.041597
 |
Component:186; Jaccard:0.02435
 |
Component:205; Jaccard:0.013772
 |
Component:051; Jaccard:0.007984
 |
Component:341; Jaccard:0.004755
 |
Correlation:0.3781
 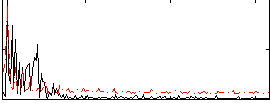 |
Correlation:0.18826
  |
Correlation:0.06052
  |
Correlation:-0.21519
  |
Correlation:0.18225
  |
Correlation:-0.32067
  |
Correlation:0.43857
  |
Component:130; Jaccard:0.065916
 |
Component:316; Jaccard:0.053842
 |
Component:013; Jaccard:0.038126
 |
Component:093; Jaccard:0.021891
 |
Component:188; Jaccard:0.012644
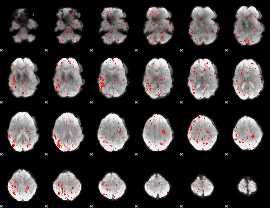 |
Component:282; Jaccard:0.007352
 |
Component:281; Jaccard:0.004672
 |
Correlation:0.24572
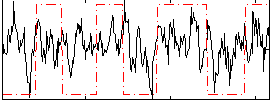 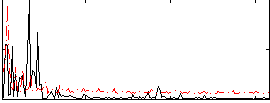 |
Correlation:0.3781
  |
Correlation:0.17144
  |
Correlation:0.20935
  |
Correlation:-0.069936
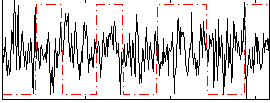  |
Correlation:-0.52192
  |
Correlation:-0.040031
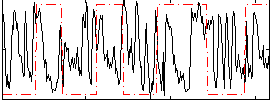 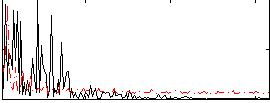 |
Component:338; Jaccard:0.06575
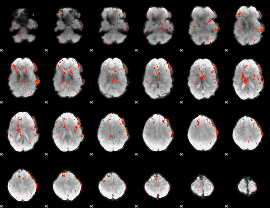 |
Component:329; Jaccard:0.05272
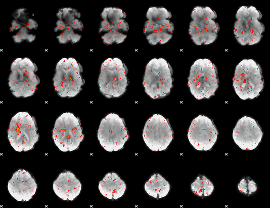 |
Component:316; Jaccard:0.037679
 |
Component:324; Jaccard:0.02161
 |
Component:095; Jaccard:0.011412
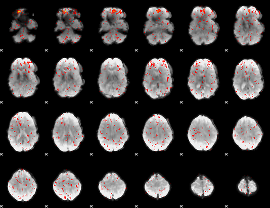 |
Component:145; Jaccard:0.007172
 |
Component:237; Jaccard:0.004575
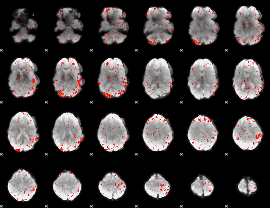 |
Correlation:0.25139
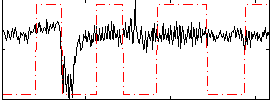 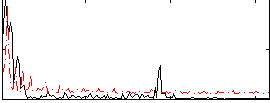 |
Correlation:0.062637
  |
Correlation:0.3781
  |
Correlation:0.06052
  |
Correlation:0.1195
  |
Correlation:-0.14918
  |
Correlation:-0.0011001
 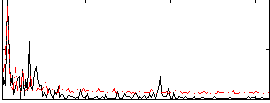 |
Cope: 02:STORY BACK
|
GLM result group-wise
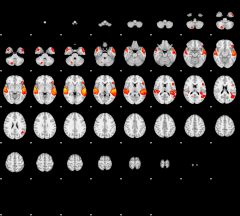 |
GLM binary result thresholded by 1
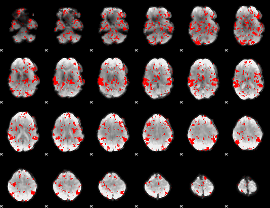 |
GLM binary result thresholded by 1.5
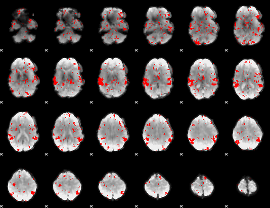 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
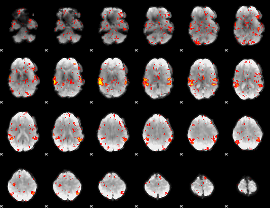 |
GLM Z-score result thresholded by 2
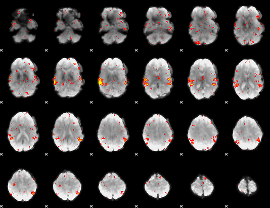 |
GLM Z-score result thresholded by 2.5
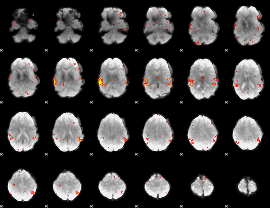 |
GLM Z-score result thresholded by 3
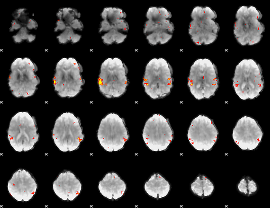 |
GLM Z-score result thresholded by 3.5
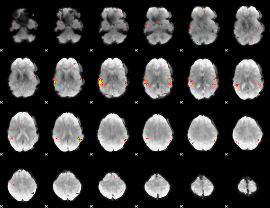 |
GLM Z-score result thresholded by 4
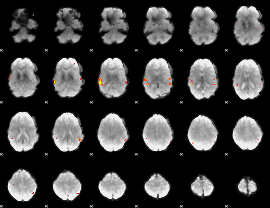 |
Component:353; Jaccard:0.19241
 |
Component:353; Jaccard:0.22556
 |
Component:353; Jaccard:0.2315
 |
Component:353; Jaccard:0.20208
 |
Component:353; Jaccard:0.15537
 |
Component:353; Jaccard:0.11879
 |
Component:353; Jaccard:0.089973
 |
Correlation:0.3525
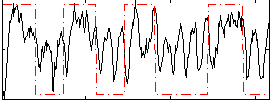 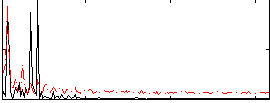 |
Correlation:0.3525
  |
Correlation:0.3525
  |
Correlation:0.3525
  |
Correlation:0.3525
  |
Correlation:0.3525
  |
Correlation:0.3525
  |
Component:171; Jaccard:0.15705
 |
Component:171; Jaccard:0.18408
 |
Component:171; Jaccard:0.18958
 |
Component:171; Jaccard:0.16242
 |
Component:171; Jaccard:0.12073
 |
Component:171; Jaccard:0.082873
 |
Component:171; Jaccard:0.056127
 |
Correlation:0.57195
  |
Correlation:0.57195
  |
Correlation:0.57195
  |
Correlation:0.57195
  |
Correlation:0.57195
  |
Correlation:0.57195
  |
Correlation:0.57195
  |
Component:051; Jaccard:0.11604
 |
Component:051; Jaccard:0.12491
 |
Component:051; Jaccard:0.11951
 |
Component:051; Jaccard:0.10377
 |
Component:051; Jaccard:0.076333
 |
Component:051; Jaccard:0.05177
 |
Component:051; Jaccard:0.036573
 |
Correlation:0.29933
  |
Correlation:0.29933
  |
Correlation:0.29933
  |
Correlation:0.29933
  |
Correlation:0.29933
  |
Correlation:0.29933
  |
Correlation:0.29933
  |
Component:282; Jaccard:0.10065
 |
Component:282; Jaccard:0.10361
 |
Component:282; Jaccard:0.095663
 |
Component:282; Jaccard:0.079827
 |
Component:282; Jaccard:0.061116
 |
Component:282; Jaccard:0.043661
 |
Component:282; Jaccard:0.032129
 |
Correlation:0.52284
  |
Correlation:0.52284
  |
Correlation:0.52284
  |
Correlation:0.52284
  |
Correlation:0.52284
  |
Correlation:0.52284
  |
Correlation:0.52284
  |
Component:336; Jaccard:0.082354
 |
Component:245; Jaccard:0.080782
 |
Component:245; Jaccard:0.073469
 |
Component:245; Jaccard:0.06302
 |
Component:245; Jaccard:0.047105
 |
Component:186; Jaccard:0.036283
 |
Component:186; Jaccard:0.028823
 |
Correlation:0.16362
 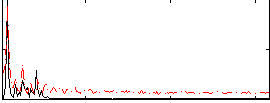 |
Correlation:0.1595
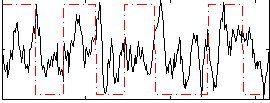  |
Correlation:0.1595
  |
Correlation:0.1595
  |
Correlation:0.1595
  |
Correlation:0.1667
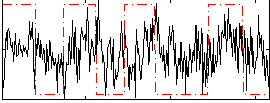 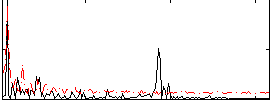 |
Correlation:0.1667
  |
Component:223; Jaccard:0.082154
 |
Component:223; Jaccard:0.077178
 |
Component:186; Jaccard:0.065713
 |
Component:186; Jaccard:0.056368
 |
Component:186; Jaccard:0.045202
 |
Component:245; Jaccard:0.033293
 |
Component:245; Jaccard:0.023476
 |
Correlation:0.18988
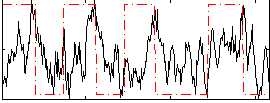  |
Correlation:0.18988
  |
Correlation:0.1667
  |
Correlation:0.1667
  |
Correlation:0.1667
  |
Correlation:0.1595
  |
Correlation:0.1595
  |
Component:245; Jaccard:0.081518
 |
Component:336; Jaccard:0.076841
 |
Component:223; Jaccard:0.065498
 |
Component:336; Jaccard:0.047925
 |
Component:110; Jaccard:0.034236
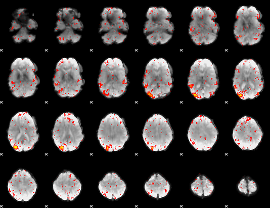 |
Component:110; Jaccard:0.025775
 |
Component:110; Jaccard:0.018809
 |
Correlation:0.1595
  |
Correlation:0.16362
  |
Correlation:0.18988
  |
Correlation:0.16362
  |
Correlation:0.20156
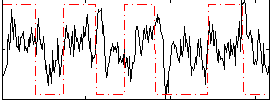  |
Correlation:0.20156
  |
Correlation:0.20156
  |
Component:093; Jaccard:0.07779
 |
Component:145; Jaccard:0.072824
 |
Component:336; Jaccard:0.064203
 |
Component:145; Jaccard:0.047038
 |
Component:313; Jaccard:0.033953
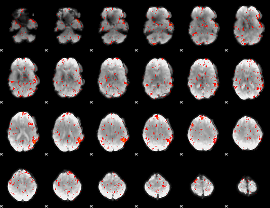 |
Component:313; Jaccard:0.024085
 |
Component:313; Jaccard:0.017319
 |
Correlation:-0.15837
  |
Correlation:0.15585
  |
Correlation:0.16362
  |
Correlation:0.15585
  |
Correlation:0.3529
  |
Correlation:0.3529
  |
Correlation:0.3529
  |
Component:145; Jaccard:0.075954
 |
Component:186; Jaccard:0.071456
 |
Component:145; Jaccard:0.060595
 |
Component:223; Jaccard:0.046602
 |
Component:145; Jaccard:0.032937
 |
Component:376; Jaccard:0.022685
 |
Component:145; Jaccard:0.016002
 |
Correlation:0.15585
  |
Correlation:0.1667
  |
Correlation:0.15585
  |
Correlation:0.18988
  |
Correlation:0.15585
  |
Correlation:-0.24739
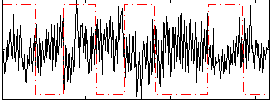 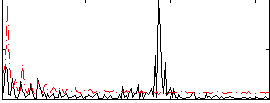 |
Correlation:0.15585
  |
Component:186; Jaccard:0.072149
 |
Component:093; Jaccard:0.070089
 |
Component:376; Jaccard:0.055975
 |
Component:376; Jaccard:0.044617
 |
Component:376; Jaccard:0.032231
 |
Component:145; Jaccard:0.02197
 |
Component:376; Jaccard:0.015996
 |
Correlation:0.1667
  |
Correlation:-0.15837
  |
Correlation:-0.24739
  |
Correlation:-0.24739
  |
Correlation:-0.24739
  |
Correlation:0.15585
  |
Correlation:-0.24739
  |
Cope: 03:MATH-STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
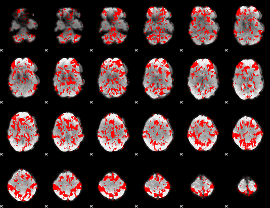 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
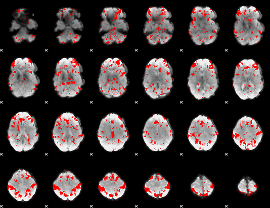 |
GLM binary result thresholded by 2.5
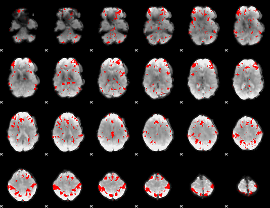 |
GLM binary result thresholded by 3
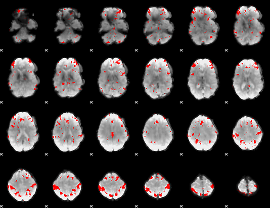 |
GLM binary result thresholded by 3.5
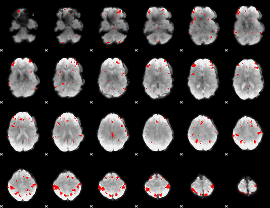 |
GLM binary result thresholded by 4
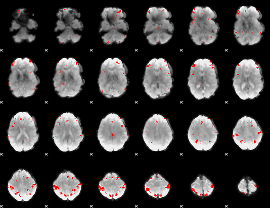 |
GLM Z-score result thresholded by 1
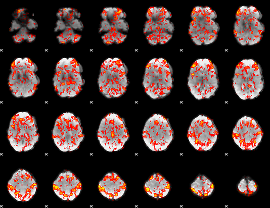 |
GLM Z-score result thresholded by 1.5
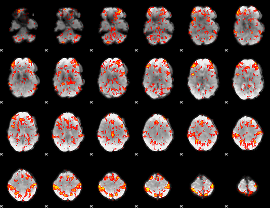 |
GLM Z-score result thresholded by 2
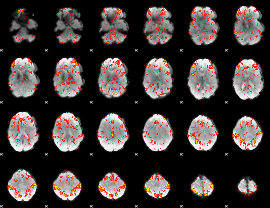 |
GLM Z-score result thresholded by 2.5
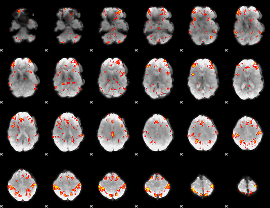 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
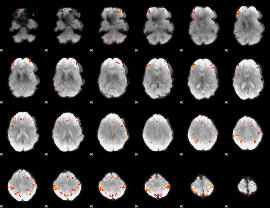 |
Component:321; Jaccard:0.15475
 |
Component:321; Jaccard:0.18237
 |
Component:037; Jaccard:0.21393
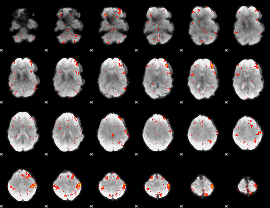 |
Component:037; Jaccard:0.24878
 |
Component:037; Jaccard:0.25656
 |
Component:037; Jaccard:0.24716
 |
Component:037; Jaccard:0.22113
 |
Correlation:0.47436
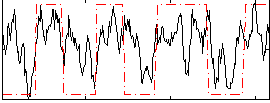 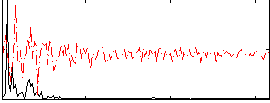 |
Correlation:0.47436
  |
Correlation:0.53142
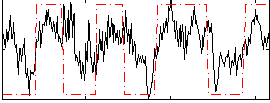 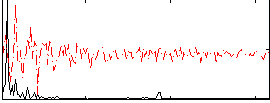 |
Correlation:0.53142
  |
Correlation:0.53142
  |
Correlation:0.53142
  |
Correlation:0.53142
  |
Component:326; Jaccard:0.14566
 |
Component:326; Jaccard:0.17333
 |
Component:321; Jaccard:0.20152
 |
Component:321; Jaccard:0.21383
 |
Component:321; Jaccard:0.20755
 |
Component:326; Jaccard:0.19087
 |
Component:326; Jaccard:0.16447
 |
Correlation:0.45683
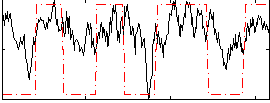 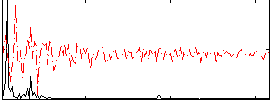 |
Correlation:0.45683
  |
Correlation:0.47436
  |
Correlation:0.47436
  |
Correlation:0.47436
  |
Correlation:0.45683
  |
Correlation:0.45683
  |
Component:037; Jaccard:0.13224
 |
Component:037; Jaccard:0.17153
 |
Component:326; Jaccard:0.19828
 |
Component:326; Jaccard:0.21034
 |
Component:326; Jaccard:0.20695
 |
Component:321; Jaccard:0.18986
 |
Component:321; Jaccard:0.15878
 |
Correlation:0.53142
  |
Correlation:0.53142
  |
Correlation:0.45683
  |
Correlation:0.45683
  |
Correlation:0.45683
  |
Correlation:0.47436
  |
Correlation:0.47436
  |
Component:288; Jaccard:0.11513
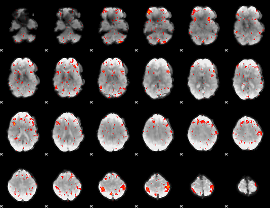 |
Component:288; Jaccard:0.13481
 |
Component:288; Jaccard:0.15113
 |
Component:288; Jaccard:0.15964
 |
Component:288; Jaccard:0.15699
 |
Component:288; Jaccard:0.14343
 |
Component:288; Jaccard:0.1176
 |
Correlation:0.48166
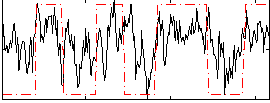 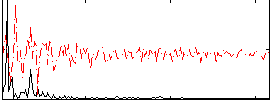 |
Correlation:0.48166
  |
Correlation:0.48166
  |
Correlation:0.48166
  |
Correlation:0.48166
  |
Correlation:0.48166
  |
Correlation:0.48166
  |
Component:116; Jaccard:0.11055
 |
Component:169; Jaccard:0.11984
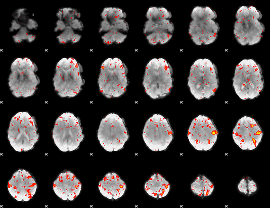 |
Component:169; Jaccard:0.13025
 |
Component:169; Jaccard:0.1349
 |
Component:169; Jaccard:0.12417
 |
Component:169; Jaccard:0.11161
 |
Component:169; Jaccard:0.093866
 |
Correlation:0.24264
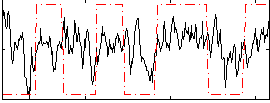 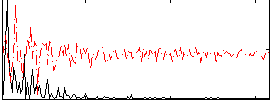 |
Correlation:0.28792
 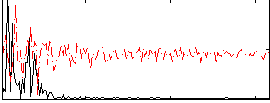 |
Correlation:0.28792
  |
Correlation:0.28792
  |
Correlation:0.28792
  |
Correlation:0.28792
  |
Correlation:0.28792
  |
Component:370; Jaccard:0.10867
 |
Component:116; Jaccard:0.11869
 |
Component:365; Jaccard:0.12322
 |
Component:365; Jaccard:0.12673
 |
Component:365; Jaccard:0.11925
 |
Component:365; Jaccard:0.10627
 |
Component:365; Jaccard:0.085607
 |
Correlation:0.41486
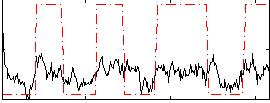  |
Correlation:0.24264
  |
Correlation:0.39209
  |
Correlation:0.39209
  |
Correlation:0.39209
  |
Correlation:0.39209
  |
Correlation:0.39209
  |
Component:169; Jaccard:0.10615
 |
Component:370; Jaccard:0.11575
 |
Component:116; Jaccard:0.12066
 |
Component:341; Jaccard:0.11625
 |
Component:341; Jaccard:0.11093
 |
Component:341; Jaccard:0.098684
 |
Component:341; Jaccard:0.077329
 |
Correlation:0.28792
  |
Correlation:0.41486
  |
Correlation:0.24264
  |
Correlation:0.44569
  |
Correlation:0.44569
  |
Correlation:0.44569
  |
Correlation:0.44569
  |
Component:365; Jaccard:0.10436
 |
Component:365; Jaccard:0.1157
 |
Component:370; Jaccard:0.1196
 |
Component:180; Jaccard:0.11435
 |
Component:388; Jaccard:0.10347
 |
Component:388; Jaccard:0.089026
 |
Component:388; Jaccard:0.072196
 |
Correlation:0.39209
  |
Correlation:0.39209
  |
Correlation:0.41486
  |
Correlation:0.3033
  |
Correlation:0.4014
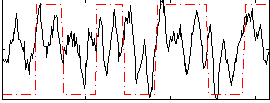 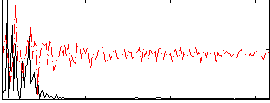 |
Correlation:0.4014
  |
Correlation:0.4014
  |
Component:180; Jaccard:0.10149
 |
Component:388; Jaccard:0.11175
 |
Component:180; Jaccard:0.11618
 |
Component:116; Jaccard:0.11417
 |
Component:180; Jaccard:0.1031
 |
Component:180; Jaccard:0.085934
 |
Component:180; Jaccard:0.068212
 |
Correlation:0.3033
  |
Correlation:0.4014
  |
Correlation:0.3033
  |
Correlation:0.24264
  |
Correlation:0.3033
  |
Correlation:0.3033
  |
Correlation:0.3033
  |
Component:388; Jaccard:0.10094
 |
Component:180; Jaccard:0.11108
 |
Component:388; Jaccard:0.11605
 |
Component:388; Jaccard:0.1139
 |
Component:116; Jaccard:0.099982
 |
Component:370; Jaccard:0.084642
 |
Component:192; Jaccard:0.064655
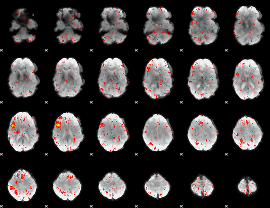 |
Correlation:0.4014
  |
Correlation:0.3033
  |
Correlation:0.4014
  |
Correlation:0.4014
  |
Correlation:0.24264
  |
Correlation:0.41486
  |
Correlation:0.37073
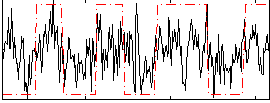 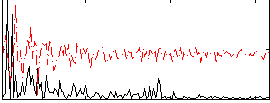 |
Cope: 04:STORY-MATH BACK
|
GLM result group-wise
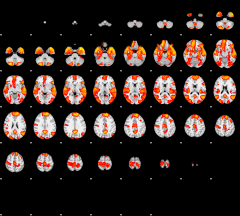 |
GLM binary result thresholded by 1
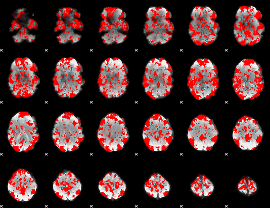 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
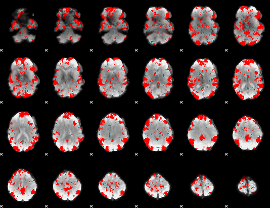 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
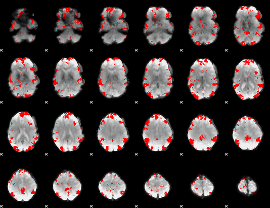 |
GLM binary result thresholded by 3.5
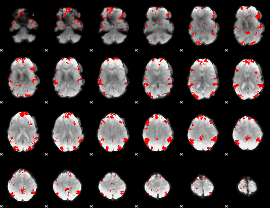 |
GLM binary result thresholded by 4
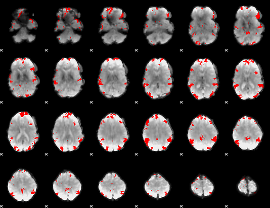 |
GLM Z-score result thresholded by 1
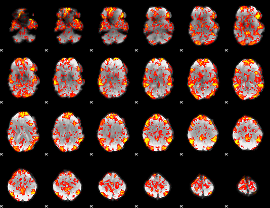 |
GLM Z-score result thresholded by 1.5
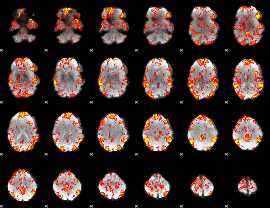 |
GLM Z-score result thresholded by 2
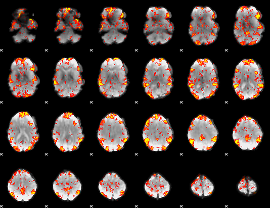 |
GLM Z-score result thresholded by 2.5
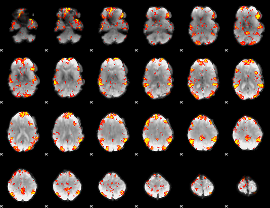 |
GLM Z-score result thresholded by 3
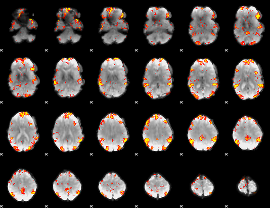 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
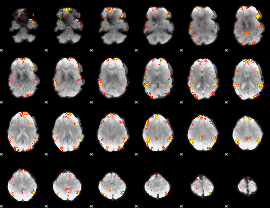 |
Component:336; Jaccard:0.13043
 |
Component:336; Jaccard:0.15207
 |
Component:171; Jaccard:0.18667
 |
Component:171; Jaccard:0.22959
 |
Component:171; Jaccard:0.26843
 |
Component:171; Jaccard:0.29627
 |
Component:171; Jaccard:0.30971
 |
Correlation:0.16324
  |
Correlation:0.16324
  |
Correlation:0.58881
  |
Correlation:0.58881
  |
Correlation:0.58881
  |
Correlation:0.58881
  |
Correlation:0.58881
  |
Component:245; Jaccard:0.11596
 |
Component:171; Jaccard:0.14613
 |
Component:051; Jaccard:0.17255
 |
Component:051; Jaccard:0.20198
 |
Component:051; Jaccard:0.226
 |
Component:051; Jaccard:0.24026
 |
Component:051; Jaccard:0.25054
 |
Correlation:0.17972
  |
Correlation:0.58881
  |
Correlation:0.31583
 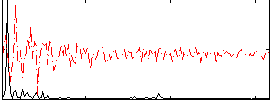 |
Correlation:0.31583
  |
Correlation:0.31583
  |
Correlation:0.31583
  |
Correlation:0.31583
  |
Component:171; Jaccard:0.11302
 |
Component:051; Jaccard:0.14174
 |
Component:336; Jaccard:0.17218
 |
Component:336; Jaccard:0.18814
 |
Component:336; Jaccard:0.19397
 |
Component:336; Jaccard:0.19329
 |
Component:336; Jaccard:0.18327
 |
Correlation:0.58881
  |
Correlation:0.31583
  |
Correlation:0.16324
  |
Correlation:0.16324
  |
Correlation:0.16324
  |
Correlation:0.16324
  |
Correlation:0.16324
  |
Component:051; Jaccard:0.11297
 |
Component:282; Jaccard:0.13202
 |
Component:282; Jaccard:0.14907
 |
Component:282; Jaccard:0.16129
 |
Component:282; Jaccard:0.166
 |
Component:282; Jaccard:0.16537
 |
Component:282; Jaccard:0.15985
 |
Correlation:0.31583
  |
Correlation:0.53221
  |
Correlation:0.53221
  |
Correlation:0.53221
  |
Correlation:0.53221
  |
Correlation:0.53221
  |
Correlation:0.53221
  |
Component:282; Jaccard:0.11203
 |
Component:245; Jaccard:0.13168
 |
Component:245; Jaccard:0.14447
 |
Component:391; Jaccard:0.15725
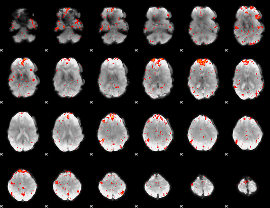 |
Component:391; Jaccard:0.16366
 |
Component:223; Jaccard:0.16484
 |
Component:223; Jaccard:0.15855
 |
Correlation:0.53221
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Correlation:0.3428
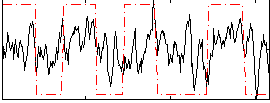  |
Correlation:0.3428
  |
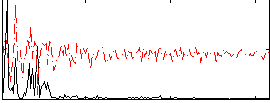
Correlation:0.21452
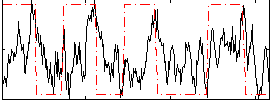  |
Correlation:0.21452
  |
Component:081; Jaccard:0.11159
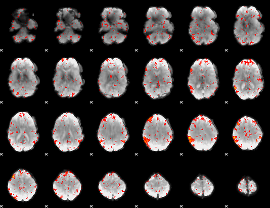 |
Component:081; Jaccard:0.12353
 |
Component:391; Jaccard:0.14166
 |
Component:223; Jaccard:0.15511
 |
Component:223; Jaccard:0.16318
 |
Component:391; Jaccard:0.16121
 |
Component:391; Jaccard:0.15566
 |
Correlation:0.19777
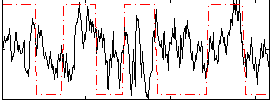  |
Correlation:0.19777
  |
Correlation:0.3428
  |
Correlation:0.21452
  |
Correlation:0.21452
  |
Correlation:0.3428
  |
Correlation:0.3428
  |
Component:313; Jaccard:0.10905
 |
Component:391; Jaccard:0.12345
 |
Component:223; Jaccard:0.14047
 |
Component:245; Jaccard:0.15101
 |
Component:245; Jaccard:0.15083
 |
Component:245; Jaccard:0.14595
 |
Component:110; Jaccard:0.13997
 |
Correlation:0.36869
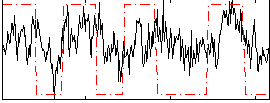 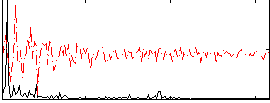 |
Correlation:0.3428
  |
Correlation:0.21452
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Correlation:0.17972
  |
Correlation:0.23397
 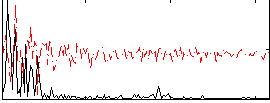 |
Component:178; Jaccard:0.1068
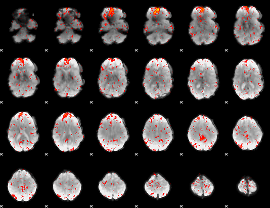 |
Component:223; Jaccard:0.1233
 |
Component:094; Jaccard:0.13259
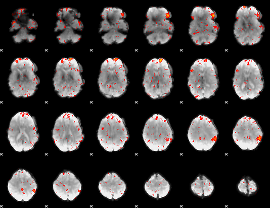 |
Component:094; Jaccard:0.14244
 |
Component:094; Jaccard:0.1478
 |
Component:094; Jaccard:0.14591
 |
Component:094; Jaccard:0.13986
 |
Correlation:0.23166
  |
Correlation:0.21452
  |
Correlation:0.40422
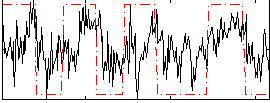 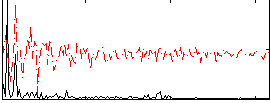 |
Correlation:0.40422
  |
Correlation:0.40422
  |
Correlation:0.40422
  |
Correlation:0.40422
  |
Component:223; Jaccard:0.10543
 |
Component:313; Jaccard:0.11976
 |
Component:081; Jaccard:0.13239
 |
Component:393; Jaccard:0.13951
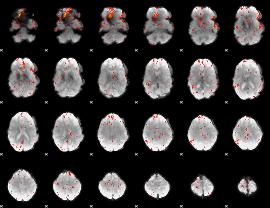 |
Component:110; Jaccard:0.14648
 |
Component:110; Jaccard:0.14579
 |
Component:245; Jaccard:0.13777
 |
Correlation:0.21452
  |
Correlation:0.36869
  |
Correlation:0.19777
  |
Correlation:0.48773
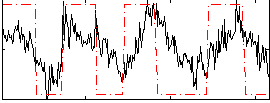  |
Correlation:0.23397
  |
Correlation:0.23397
  |
Correlation:0.17972
  |
Component:391; Jaccard:0.10401
 |
Component:094; Jaccard:0.11678
 |
Component:393; Jaccard:0.13
 |
Component:110; Jaccard:0.13867
 |
Component:393; Jaccard:0.14266
 |
Component:393; Jaccard:0.14112
 |
Component:393; Jaccard:0.13537
 |
Correlation:0.3428
  |
Correlation:0.40422
  |
Correlation:0.48773
  |
Correlation:0.23397
  |
Correlation:0.48773
  |
Correlation:0.48773
  |
Correlation:0.48773
  |
Cope: 05:neg_MATH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
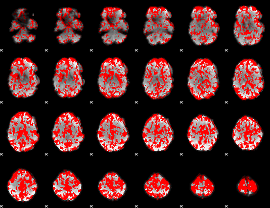 |
GLM binary result thresholded by 1.5
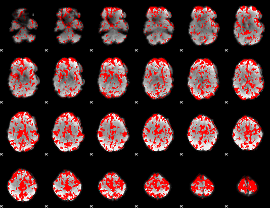 |
GLM binary result thresholded by 2
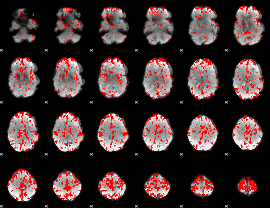 |
GLM binary result thresholded by 2.5
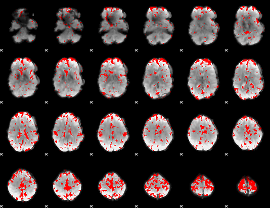 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
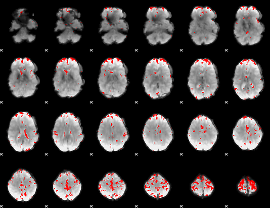 |
GLM binary result thresholded by 4
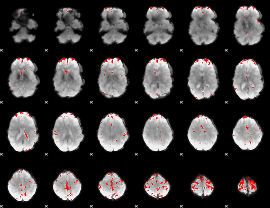 |
GLM Z-score result thresholded by 1
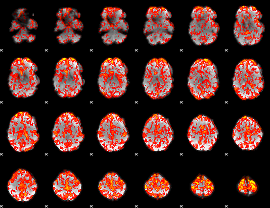 |
GLM Z-score result thresholded by 1.5
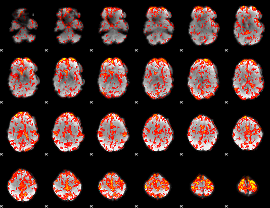 |
GLM Z-score result thresholded by 2
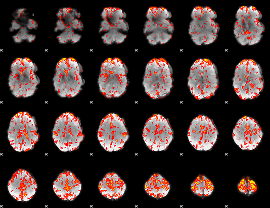 |
GLM Z-score result thresholded by 2.5
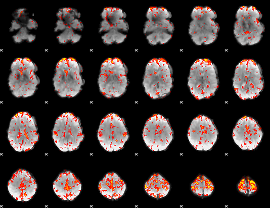 |
GLM Z-score result thresholded by 3
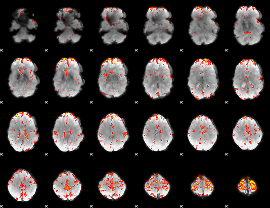 |
GLM Z-score result thresholded by 3.5
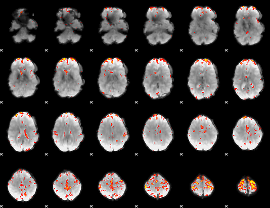 |
GLM Z-score result thresholded by 4
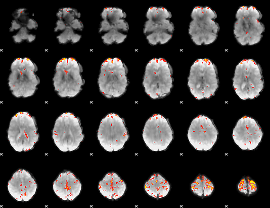 |
Component:072; Jaccard:0.12737
 |
Component:072; Jaccard:0.15721
 |
Component:072; Jaccard:0.19068
 |
Component:072; Jaccard:0.21966
 |
Component:072; Jaccard:0.23247
 |
Component:072; Jaccard:0.22128
 |
Component:072; Jaccard:0.18983
 |
Correlation:0.070809
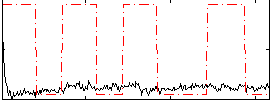 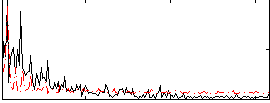 |
Correlation:0.070809
  |
Correlation:0.070809
  |
Correlation:0.070809
  |
Correlation:0.070809
  |
Correlation:0.070809
  |
Correlation:0.070809
  |
Component:385; Jaccard:0.12572
 |
Component:385; Jaccard:0.15306
 |
Component:385; Jaccard:0.17975
 |
Component:385; Jaccard:0.19999
 |
Component:385; Jaccard:0.19734
 |
Component:193; Jaccard:0.18553
 |
Component:193; Jaccard:0.16157
 |
Correlation:0.20544
  |
Correlation:0.20544
  |
Correlation:0.20544
  |
Correlation:0.20544
  |
Correlation:0.20544
  |
Correlation:-0.040286
  |
Correlation:-0.040286
  |
Component:193; Jaccard:0.10626
 |
Component:193; Jaccard:0.13297
 |
Component:193; Jaccard:0.16059
 |
Component:193; Jaccard:0.18315
 |
Component:193; Jaccard:0.19319
 |
Component:385; Jaccard:0.17289
 |
Component:385; Jaccard:0.1388
 |
Correlation:-0.040286
  |
Correlation:-0.040286
  |
Correlation:-0.040286
  |
Correlation:-0.040286
  |
Correlation:-0.040286
  |
Correlation:0.20544
  |
Correlation:0.20544
  |
Component:370; Jaccard:0.10373
 |
Component:370; Jaccard:0.12069
 |
Component:370; Jaccard:0.13647
 |
Component:370; Jaccard:0.14639
 |
Component:287; Jaccard:0.15664
 |
Component:287; Jaccard:0.1516
 |
Component:287; Jaccard:0.13634
 |
Correlation:-0.4019
  |
Correlation:-0.4019
  |
Correlation:-0.4019
  |
Correlation:-0.4019
  |
Correlation:0.18646
  |
Correlation:0.18646
  |
Correlation:0.18646
  |
Component:165; Jaccard:0.094976
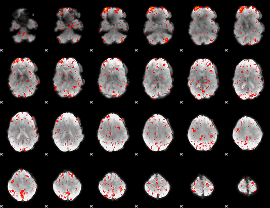 |
Component:287; Jaccard:0.10368
 |
Component:287; Jaccard:0.12369
 |
Component:287; Jaccard:0.14444
 |
Component:370; Jaccard:0.14695
 |
Component:370; Jaccard:0.13498
 |
Component:370; Jaccard:0.1104
 |
Correlation:-0.13583
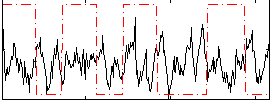  |
Correlation:0.18646
  |
Correlation:0.18646
  |
Correlation:0.18646
  |
Correlation:-0.4019
  |
Correlation:-0.4019
  |
Correlation:-0.4019
  |
Component:359; Jaccard:0.087229
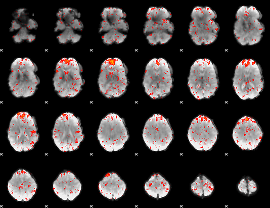 |
Component:165; Jaccard:0.10127
 |
Component:165; Jaccard:0.10456
 |
Component:039; Jaccard:0.10745
 |
Component:039; Jaccard:0.10121
 |
Component:039; Jaccard:0.088274
 |
Component:039; Jaccard:0.070025
 |
Correlation:0.05582
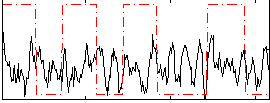 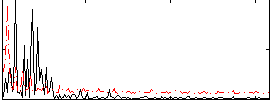 |
Correlation:-0.13583
  |
Correlation:-0.13583
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Component:166; Jaccard:0.087139
 |
Component:039; Jaccard:0.095174
 |
Component:039; Jaccard:0.10425
 |
Component:165; Jaccard:0.10313
 |
Component:301; Jaccard:0.095384
 |
Component:301; Jaccard:0.08113
 |
Component:301; Jaccard:0.061517
 |
Correlation:-0.30352
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:-0.13583
  |
Correlation:-0.052743
  |
Correlation:-0.052743
  |
Correlation:-0.052743
  |
Component:287; Jaccard:0.085344
 |
Component:359; Jaccard:0.093543
 |
Component:301; Jaccard:0.10164
 |
Component:301; Jaccard:0.1022
 |
Component:165; Jaccard:0.094437
 |
Component:165; Jaccard:0.079788
 |
Component:165; Jaccard:0.061219
 |
Correlation:0.18646
  |
Correlation:0.05582
  |
Correlation:-0.052743
  |
Correlation:-0.052743
  |
Correlation:-0.13583
  |
Correlation:-0.13583
  |
Correlation:-0.13583
  |
Component:178; Jaccard:0.085236
 |
Component:301; Jaccard:0.093258
 |
Component:351; Jaccard:0.10062
 |
Component:351; Jaccard:0.10005
 |
Component:113; Jaccard:0.0914
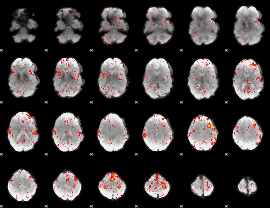 |
Component:113; Jaccard:0.079309
 |
Component:113; Jaccard:0.06063
 |
Correlation:0.24002
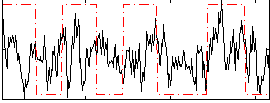 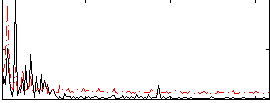 |
Correlation:-0.052743
  |
Correlation:0.11377
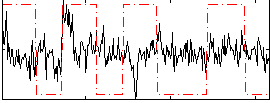  |
Correlation:0.11377
  |
Correlation:0.0047772
  |
Correlation:0.0047772
  |
Correlation:0.0047772
  |
Component:039; Jaccard:0.084932
 |
Component:351; Jaccard:0.092203
 |
Component:359; Jaccard:0.097887
 |
Component:359; Jaccard:0.096773
 |
Component:116; Jaccard:0.091211
 |
Component:116; Jaccard:0.077507
 |
Component:116; Jaccard:0.059815
 |
Correlation:0.075551
  |
Correlation:0.11377
  |
Correlation:0.05582
  |
Correlation:0.05582
  |
Correlation:-0.23173
 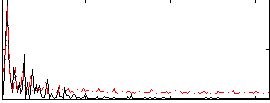 |
Correlation:-0.23173
  |
Correlation:-0.23173
  |
Cope: 06:neg_STORY BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
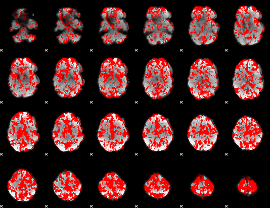 |
GLM binary result thresholded by 1.5
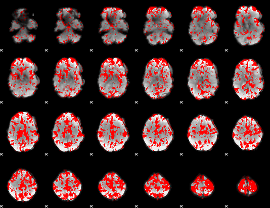 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
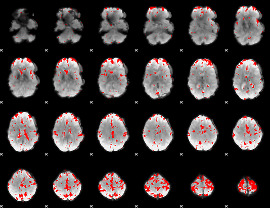 |
GLM binary result thresholded by 3.5
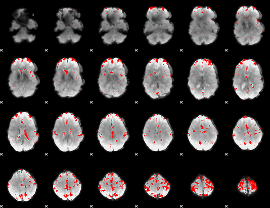 |
GLM binary result thresholded by 4
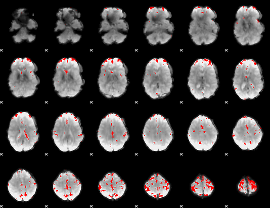 |
GLM Z-score result thresholded by 1
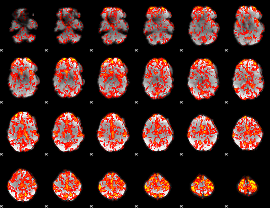 |
GLM Z-score result thresholded by 1.5
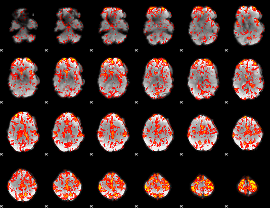 |
GLM Z-score result thresholded by 2
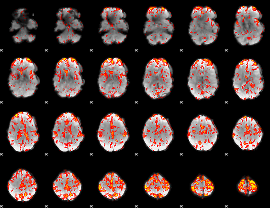 |
GLM Z-score result thresholded by 2.5
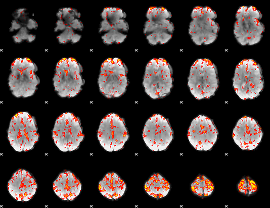 |
GLM Z-score result thresholded by 3
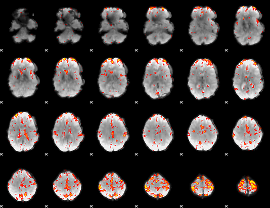 |
GLM Z-score result thresholded by 3.5
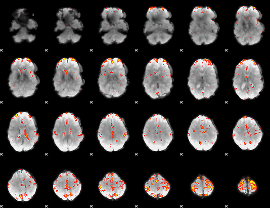 |
GLM Z-score result thresholded by 4
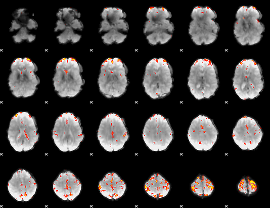 |
Component:072; Jaccard:0.13175
 |
Component:072; Jaccard:0.16194
 |
Component:072; Jaccard:0.19417
 |
Component:072; Jaccard:0.22215
 |
Component:072; Jaccard:0.22665
 |
Component:072; Jaccard:0.21443
 |
Component:072; Jaccard:0.18632
 |
Correlation:-0.072952
  |
Correlation:-0.072952
  |
Correlation:-0.072952
  |
Correlation:-0.072952
  |
Correlation:-0.072952
  |
Correlation:-0.072952
  |
Correlation:-0.072952
  |
Component:370; Jaccard:0.12131
 |
Component:370; Jaccard:0.14738
 |
Component:370; Jaccard:0.17508
 |
Component:370; Jaccard:0.19373
 |
Component:370; Jaccard:0.19964
 |
Component:193; Jaccard:0.19383
 |
Component:193; Jaccard:0.17288
 |
Correlation:0.4125
  |
Correlation:0.4125
  |
Correlation:0.4125
  |
Correlation:0.4125
  |
Correlation:0.4125
  |
Correlation:-0.010066
  |
Correlation:-0.010066
  |
Component:385; Jaccard:0.11937
 |
Component:385; Jaccard:0.14079
 |
Component:385; Jaccard:0.16155
 |
Component:193; Jaccard:0.18262
 |
Component:193; Jaccard:0.19621
 |
Component:370; Jaccard:0.18773
 |
Component:370; Jaccard:0.16332
 |
Correlation:-0.16865
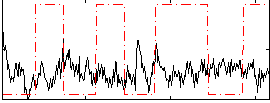 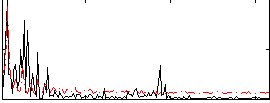 |
Correlation:-0.16865
  |
Correlation:-0.16865
  |
Correlation:-0.010066
  |
Correlation:-0.010066
  |
Correlation:0.4125
  |
Correlation:0.4125
  |
Component:193; Jaccard:0.108
 |
Component:193; Jaccard:0.13282
 |
Component:193; Jaccard:0.15837
 |
Component:385; Jaccard:0.17158
 |
Component:385; Jaccard:0.16238
 |
Component:385; Jaccard:0.14299
 |
Component:385; Jaccard:0.11705
 |
Correlation:-0.010066
  |
Correlation:-0.010066
  |
Correlation:-0.010066
  |
Correlation:-0.16865
  |
Correlation:-0.16865
  |
Correlation:-0.16865
  |
Correlation:-0.16865
  |
Component:365; Jaccard:0.097253
 |
Component:365; Jaccard:0.11293
 |
Component:365; Jaccard:0.12863
 |
Component:365; Jaccard:0.13847
 |
Component:365; Jaccard:0.13867
 |
Component:365; Jaccard:0.12883
 |
Component:365; Jaccard:0.11015
 |
Correlation:0.39066
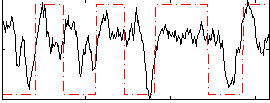 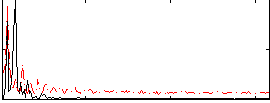 |
Correlation:0.39066
  |
Correlation:0.39066
  |
Correlation:0.39066
  |
Correlation:0.39066
  |
Correlation:0.39066
  |
Correlation:0.39066
  |
Component:116; Jaccard:0.097029
 |
Component:116; Jaccard:0.11235
 |
Component:116; Jaccard:0.12784
 |
Component:116; Jaccard:0.13759
 |
Component:116; Jaccard:0.13721
 |
Component:116; Jaccard:0.12223
 |
Component:116; Jaccard:0.10439
 |
Correlation:0.24459
  |
Correlation:0.24459
  |
Correlation:0.24459
  |
Correlation:0.24459
  |
Correlation:0.24459
  |
Correlation:0.24459
  |
Correlation:0.24459
  |
Component:258; Jaccard:0.090737
 |
Component:301; Jaccard:0.10038
 |
Component:301; Jaccard:0.11028
 |
Component:326; Jaccard:0.11745
 |
Component:326; Jaccard:0.11497
 |
Component:326; Jaccard:0.1046
 |
Component:287; Jaccard:0.088884
 |
Correlation:0.2201
  |
Correlation:0.093419
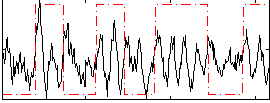 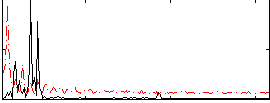 |
Correlation:0.093419
  |
Correlation:0.44953
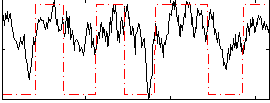 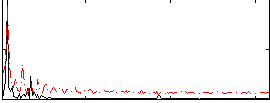 |
Correlation:0.44953
  |
Correlation:0.44953
  |
Correlation:-0.20389
  |
Component:165; Jaccard:0.090645
 |
Component:326; Jaccard:0.10012
 |
Component:326; Jaccard:0.11017
 |
Component:301; Jaccard:0.11565
 |
Component:301; Jaccard:0.11097
 |
Component:287; Jaccard:0.1003
 |
Component:326; Jaccard:0.088623
 |
Correlation:0.10832
  |
Correlation:0.44953
  |
Correlation:0.44953
  |
Correlation:0.093419
  |
Correlation:0.093419
  |
Correlation:-0.20389
  |
Correlation:0.44953
  |
Component:301; Jaccard:0.089317
 |
Component:258; Jaccard:0.099539
 |
Component:039; Jaccard:0.10781
 |
Component:039; Jaccard:0.11056
 |
Component:287; Jaccard:0.10704
 |
Component:301; Jaccard:0.098556
 |
Component:301; Jaccard:0.077675
 |
Correlation:0.093419
  |
Correlation:0.2201
  |
Correlation:-0.036702
  |
Correlation:-0.036702
  |
Correlation:-0.20389
  |
Correlation:0.093419
  |
Correlation:0.093419
  |
Component:326; Jaccard:0.088113
 |
Component:039; Jaccard:0.098357
 |
Component:258; Jaccard:0.106
 |
Component:287; Jaccard:0.10551
 |
Component:039; Jaccard:0.10444
 |
Component:039; Jaccard:0.093151
 |
Component:039; Jaccard:0.074297
 |
Correlation:0.44953
  |
Correlation:-0.036702
  |
Correlation:0.2201
  |
Correlation:-0.20389
  |
Correlation:-0.036702
  |
Correlation:-0.036702
  |
Correlation:-0.036702
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































