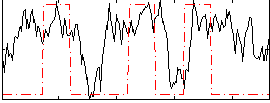
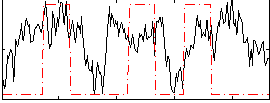
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:341; Jaccard:0.14592
 |
Component:341; Jaccard:0.18413
 |
Component:341; Jaccard:0.22637
 |
Component:341; Jaccard:0.26588
 |
Component:341; Jaccard:0.29427
 |
Component:341; Jaccard:0.3008
 |
Component:341; Jaccard:0.28008
 |
Correlation:0.15621
  |
Correlation:0.15621
  |
Correlation:0.15621
  |
Correlation:0.15621
  |
Correlation:0.15621
  |
Correlation:0.15621
  |
Correlation:0.15621
  |
Component:308; Jaccard:0.13839
 |
Component:308; Jaccard:0.17054
 |
Component:308; Jaccard:0.20025
 |
Component:308; Jaccard:0.22639
 |
Component:308; Jaccard:0.23948
 |
Component:308; Jaccard:0.23342
 |
Component:115; Jaccard:0.20986
 |
Correlation:0.3277
 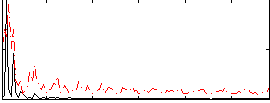 |
Correlation:0.3277
  |
Correlation:0.3277
  |
Correlation:0.3277
  |
Correlation:0.3277
  |
Correlation:0.3277
  |
Correlation:0.15357
  |
Component:115; Jaccard:0.13646
 |
Component:115; Jaccard:0.16734
 |
Component:115; Jaccard:0.19889
 |
Component:115; Jaccard:0.22324
 |
Component:115; Jaccard:0.23599
 |
Component:115; Jaccard:0.23218
 |
Component:308; Jaccard:0.20893
 |
Correlation:0.15357
  |
Correlation:0.15357
  |
Correlation:0.15357
  |
Correlation:0.15357
  |
Correlation:0.15357
  |
Correlation:0.15357
  |
Correlation:0.3277
  |
Component:204; Jaccard:0.11555
 |
Component:204; Jaccard:0.13846
 |
Component:204; Jaccard:0.15821
 |
Component:304; Jaccard:0.17235
 |
Component:304; Jaccard:0.18731
 |
Component:304; Jaccard:0.19137
 |
Component:304; Jaccard:0.18205
 |
Correlation:0.17343
  |
Correlation:0.17343
  |
Correlation:0.17343
  |
Correlation:0.23369
 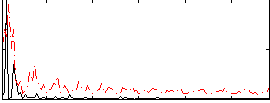 |
Correlation:0.23369
  |
Correlation:0.23369
  |
Correlation:0.23369
  |
Component:192; Jaccard:0.11387
 |
Component:266; Jaccard:0.12958
 |
Component:304; Jaccard:0.1496
 |
Component:204; Jaccard:0.17069
 |
Component:204; Jaccard:0.177
 |
Component:204; Jaccard:0.17119
 |
Component:266; Jaccard:0.15433
 |
Correlation:0.253
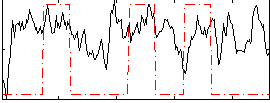  |
Correlation:0.17035
  |
Correlation:0.23369
  |
Correlation:0.17343
  |
Correlation:0.17343
  |
Correlation:0.17343
  |
Correlation:0.17035
  |
Component:384; Jaccard:0.11209
 |
Component:255; Jaccard:0.12661
 |
Component:266; Jaccard:0.14863
 |
Component:266; Jaccard:0.16369
 |
Component:266; Jaccard:0.17287
 |
Component:266; Jaccard:0.1683
 |
Component:204; Jaccard:0.15209
 |
Correlation:0.34448
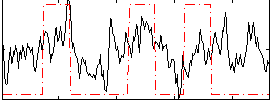  |
Correlation:0.22174
  |
Correlation:0.17035
  |
Correlation:0.17035
  |
Correlation:0.17035
  |
Correlation:0.17035
  |
Correlation:0.17343
  |
Component:255; Jaccard:0.11094
 |
Component:192; Jaccard:0.12522
 |
Component:283; Jaccard:0.14085
 |
Component:283; Jaccard:0.15614
 |
Component:283; Jaccard:0.16447
 |
Component:283; Jaccard:0.15817
 |
Component:283; Jaccard:0.14367
 |
Correlation:0.22174
  |
Correlation:0.253
  |
Correlation:0.24463
  |
Correlation:0.24463
  |
Correlation:0.24463
  |
Correlation:0.24463
  |
Correlation:0.24463
  |
Component:266; Jaccard:0.10902
 |
Component:304; Jaccard:0.12349
 |
Component:255; Jaccard:0.14043
 |
Component:255; Jaccard:0.14821
 |
Component:255; Jaccard:0.14716
 |
Component:255; Jaccard:0.14168
 |
Component:255; Jaccard:0.12346
 |
Correlation:0.17035
  |
Correlation:0.23369
  |
Correlation:0.22174
  |
Correlation:0.22174
  |
Correlation:0.22174
  |
Correlation:0.22174
  |
Correlation:0.22174
  |
Component:283; Jaccard:0.10181
 |
Component:384; Jaccard:0.12277
 |
Component:192; Jaccard:0.13206
 |
Component:192; Jaccard:0.132
 |
Component:192; Jaccard:0.1249
 |
Component:192; Jaccard:0.11368
 |
Component:192; Jaccard:0.095225
 |
Correlation:0.24463
  |
Correlation:0.34448
  |
Correlation:0.253
  |
Correlation:0.253
  |
Correlation:0.253
  |
Correlation:0.253
  |
Correlation:0.253
  |
Component:277; Jaccard:0.10118
 |
Component:283; Jaccard:0.12093
 |
Component:384; Jaccard:0.12973
 |
Component:384; Jaccard:0.12792
 |
Component:384; Jaccard:0.11825
 |
Component:288; Jaccard:0.1056
 |
Component:288; Jaccard:0.08912
 |
Correlation:0.065962
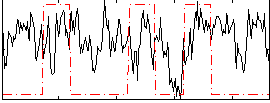  |
Correlation:0.24463
  |
Correlation:0.34448
  |
Correlation:0.34448
  |
Correlation:0.34448
  |
Correlation:-0.087511
  |
Correlation:-0.087511
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
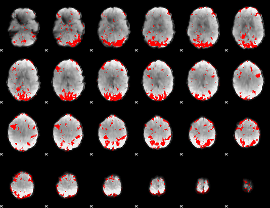 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
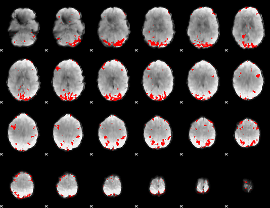 |
GLM binary result thresholded by 3
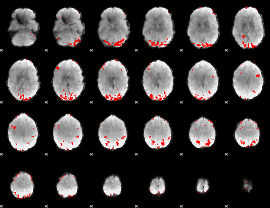 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
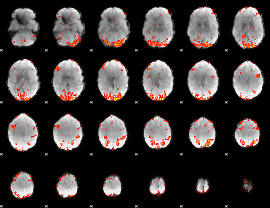 |
GLM Z-score result thresholded by 2.5
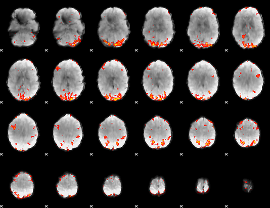 |
GLM Z-score result thresholded by 3
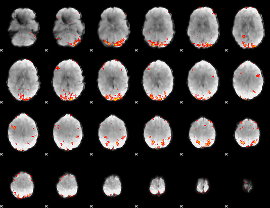 |
GLM Z-score result thresholded by 3.5
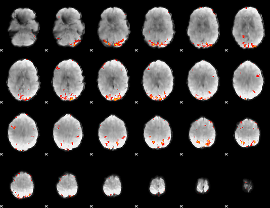 |
GLM Z-score result thresholded by 4
 |
Component:341; Jaccard:0.20389
 |
Component:341; Jaccard:0.24302
 |
Component:341; Jaccard:0.27434
 |
Component:341; Jaccard:0.28636
 |
Component:341; Jaccard:0.27181
 |
Component:341; Jaccard:0.23466
 |
Component:341; Jaccard:0.18355
 |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Correlation:0.27895
  |
Component:115; Jaccard:0.15592
 |
Component:115; Jaccard:0.17346
 |
Component:283; Jaccard:0.1837
 |
Component:283; Jaccard:0.18829
 |
Component:283; Jaccard:0.17741
 |
Component:283; Jaccard:0.15512
 |
Component:283; Jaccard:0.12029
 |
Correlation:0.25261
  |
Correlation:0.25261
  |
Correlation:0.29146
  |
Correlation:0.29146
  |
Correlation:0.29146
  |
Correlation:0.29146
  |
Correlation:0.29146
  |
Component:308; Jaccard:0.15006
 |
Component:283; Jaccard:0.16908
 |
Component:115; Jaccard:0.18102
 |
Component:115; Jaccard:0.17146
 |
Component:304; Jaccard:0.15687
 |
Component:304; Jaccard:0.14015
 |
Component:304; Jaccard:0.11165
 |
Correlation:0.23111
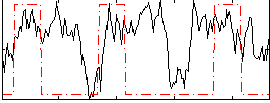 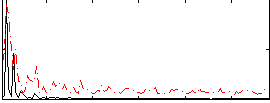 |
Correlation:0.29146
  |
Correlation:0.25261
  |
Correlation:0.25261
  |
Correlation:0.2749
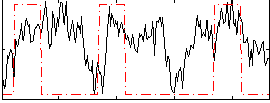 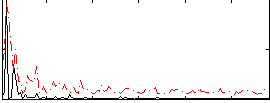 |
Correlation:0.2749
  |
Correlation:0.2749
  |
Component:283; Jaccard:0.1467
 |
Component:308; Jaccard:0.16648
 |
Component:308; Jaccard:0.17407
 |
Component:308; Jaccard:0.1662
 |
Component:115; Jaccard:0.1478
 |
Component:115; Jaccard:0.12003
 |
Component:288; Jaccard:0.086364
 |
Correlation:0.29146
  |
Correlation:0.23111
  |
Correlation:0.23111
  |
Correlation:0.23111
  |
Correlation:0.25261
  |
Correlation:0.25261
  |
Correlation:0.55163
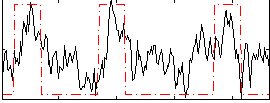  |
Component:266; Jaccard:0.14003
 |
Component:266; Jaccard:0.15523
 |
Component:266; Jaccard:0.16226
 |
Component:304; Jaccard:0.15972
 |
Component:308; Jaccard:0.14579
 |
Component:308; Jaccard:0.11564
 |
Component:115; Jaccard:0.086233
 |
Correlation:0.2445
  |
Correlation:0.2445
  |
Correlation:0.2445
  |
Correlation:0.2749
  |
Correlation:0.23111
  |
Correlation:0.23111
  |
Correlation:0.25261
  |
Component:288; Jaccard:0.13562
 |
Component:288; Jaccard:0.15006
 |
Component:288; Jaccard:0.15614
 |
Component:266; Jaccard:0.15585
 |
Component:266; Jaccard:0.1393
 |
Component:266; Jaccard:0.11302
 |
Component:266; Jaccard:0.084163
 |
Correlation:0.55163
  |
Correlation:0.55163
  |
Correlation:0.55163
  |
Correlation:0.2445
  |
Correlation:0.2445
  |
Correlation:0.2445
  |
Correlation:0.2445
  |
Component:204; Jaccard:0.13226
 |
Component:204; Jaccard:0.14798
 |
Component:204; Jaccard:0.15555
 |
Component:288; Jaccard:0.1508
 |
Component:288; Jaccard:0.1359
 |
Component:288; Jaccard:0.11285
 |
Component:308; Jaccard:0.082146
 |
Correlation:0.22534
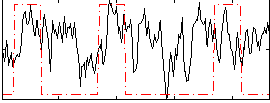  |
Correlation:0.22534
  |
Correlation:0.22534
  |
Correlation:0.55163
  |
Correlation:0.55163
  |
Correlation:0.55163
  |
Correlation:0.23111
  |
Component:304; Jaccard:0.12034
 |
Component:304; Jaccard:0.13955
 |
Component:304; Jaccard:0.15355
 |
Component:204; Jaccard:0.14908
 |
Component:204; Jaccard:0.13331
 |
Component:204; Jaccard:0.10565
 |
Component:204; Jaccard:0.07852
 |
Correlation:0.2749
  |
Correlation:0.2749
  |
Correlation:0.2749
  |
Correlation:0.22534
  |
Correlation:0.22534
  |
Correlation:0.22534
  |
Correlation:0.22534
  |
Component:313; Jaccard:0.10294
 |
Component:313; Jaccard:0.10645
 |
Component:313; Jaccard:0.10413
 |
Component:255; Jaccard:0.096608
 |
Component:218; Jaccard:0.087387
 |
Component:218; Jaccard:0.0787
 |
Component:218; Jaccard:0.065956
 |
Correlation:0.23032
 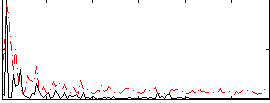 |
Correlation:0.23032
  |
Correlation:0.23032
  |
Correlation:0.040788
  |
Correlation:-0.12444
 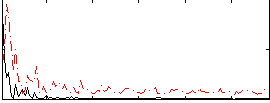 |
Correlation:-0.12444
  |
Correlation:-0.12444
  |
Component:255; Jaccard:0.099364
 |
Component:255; Jaccard:0.10378
 |
Component:255; Jaccard:0.10294
 |
Component:313; Jaccard:0.095046
 |
Component:313; Jaccard:0.084627
 |
Component:313; Jaccard:0.070384
 |
Component:313; Jaccard:0.053661
 |
Correlation:0.040788
  |
Correlation:0.040788
  |
Correlation:0.040788
  |
Correlation:0.23032
  |
Correlation:0.23032
  |
Correlation:0.23032
  |
Correlation:0.23032
  |
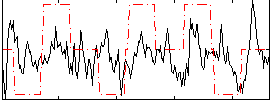
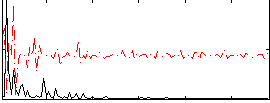
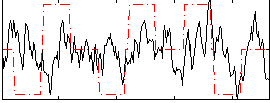
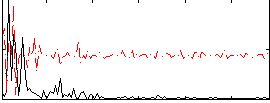
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
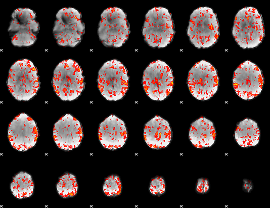 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:317; Jaccard:0.092571
 |
Component:317; Jaccard:0.10965
 |
Component:021; Jaccard:0.13182
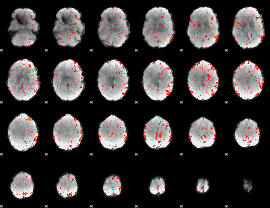 |
Component:021; Jaccard:0.1535
 |
Component:021; Jaccard:0.16687
 |
Component:021; Jaccard:0.16019
 |
Component:021; Jaccard:0.12697
 |
Correlation:0.045647
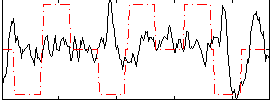 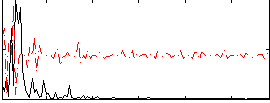 |
Correlation:0.045647
  |
Correlation:0.22131
  |
Correlation:0.22131
  |
Correlation:0.22131
  |
Correlation:0.22131
  |
Correlation:0.22131
  |
Component:093; Jaccard:0.09027
 |
Component:021; Jaccard:0.10824
 |
Component:317; Jaccard:0.12686
 |
Component:317; Jaccard:0.13956
 |
Component:317; Jaccard:0.13754
 |
Component:317; Jaccard:0.11724
 |
Component:160; Jaccard:0.095641
 |
Correlation:-0.010072
  |
Correlation:0.22131
  |
Correlation:0.045647
  |
Correlation:0.045647
  |
Correlation:0.045647
  |
Correlation:0.045647
  |
Correlation:0.29077
  |
Component:356; Jaccard:0.09015
 |
Component:093; Jaccard:0.10338
 |
Component:093; Jaccard:0.11346
 |
Component:329; Jaccard:0.12102
 |
Component:160; Jaccard:0.12439
 |
Component:160; Jaccard:0.11571
 |
Component:302; Jaccard:0.090203
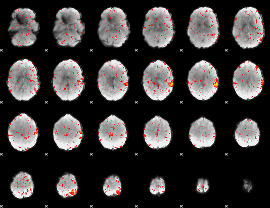 |
Correlation:0.022313
  |
Correlation:-0.010072
  |
Correlation:-0.010072
  |
Correlation:-0.037489
  |
Correlation:0.29077
  |
Correlation:0.29077
  |
Correlation:0.1885
  |
Component:021; Jaccard:0.089321
 |
Component:356; Jaccard:0.10093
 |
Component:329; Jaccard:0.1132
 |
Component:093; Jaccard:0.11832
 |
Component:088; Jaccard:0.11769
 |
Component:302; Jaccard:0.10957
 |
Component:317; Jaccard:0.08358
 |
Correlation:0.22131
  |
Correlation:0.022313
  |
Correlation:-0.037489
  |
Correlation:-0.010072
  |
Correlation:0.18092
  |
Correlation:0.1885
  |
Correlation:0.045647
  |
Component:329; Jaccard:0.087253
 |
Component:329; Jaccard:0.10014
 |
Component:384; Jaccard:0.10829
 |
Component:160; Jaccard:0.11637
 |
Component:329; Jaccard:0.11671
 |
Component:088; Jaccard:0.10528
 |
Component:343; Jaccard:0.079486
 |
Correlation:-0.037489
  |
Correlation:-0.037489
  |
Correlation:0.23591
  |
Correlation:0.29077
  |
Correlation:-0.037489
  |
Correlation:0.18092
  |
Correlation:0.1592
  |
Component:384; Jaccard:0.084542
 |
Component:384; Jaccard:0.097002
 |
Component:356; Jaccard:0.10727
 |
Component:384; Jaccard:0.1155
 |
Component:343; Jaccard:0.11507
 |
Component:343; Jaccard:0.10458
 |
Component:329; Jaccard:0.077854
 |
Correlation:0.23591
  |
Correlation:0.23591
  |
Correlation:0.022313
  |
Correlation:0.23591
  |
Correlation:0.1592
  |
Correlation:0.1592
  |
Correlation:-0.037489
  |
Component:083; Jaccard:0.083532
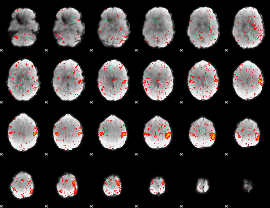 |
Component:083; Jaccard:0.093867
 |
Component:343; Jaccard:0.10526
 |
Component:088; Jaccard:0.11481
 |
Component:384; Jaccard:0.11294
 |
Component:329; Jaccard:0.1032
 |
Component:088; Jaccard:0.077699
 |
Correlation:0.040515
  |
Correlation:0.040515
  |
Correlation:0.1592
  |
Correlation:0.18092
  |
Correlation:0.23591
  |
Correlation:-0.037489
  |
Correlation:0.18092
  |
Component:294; Jaccard:0.083253
 |
Component:192; Jaccard:0.092998
 |
Component:088; Jaccard:0.10503
 |
Component:343; Jaccard:0.11365
 |
Component:302; Jaccard:0.11136
 |
Component:384; Jaccard:0.0993
 |
Component:384; Jaccard:0.071613
 |
Correlation:-0.14441
  |
Correlation:0.095084
  |
Correlation:0.18092
  |
Correlation:0.1592
  |
Correlation:0.1885
  |
Correlation:0.23591
  |
Correlation:0.23591
  |
Component:192; Jaccard:0.082871
 |
Component:343; Jaccard:0.092904
 |
Component:083; Jaccard:0.10441
 |
Component:083; Jaccard:0.111
 |
Component:093; Jaccard:0.11074
 |
Component:083; Jaccard:0.093763
 |
Component:083; Jaccard:0.06847
 |
Correlation:0.095084
  |
Correlation:0.1592
  |
Correlation:0.040515
  |
Correlation:0.040515
  |
Correlation:-0.010072
  |
Correlation:0.040515
  |
Correlation:0.040515
  |
Component:343; Jaccard:0.081166
 |
Component:088; Jaccard:0.092608
 |
Component:192; Jaccard:0.10077
 |
Component:356; Jaccard:0.10656
 |
Component:083; Jaccard:0.10896
 |
Component:093; Jaccard:0.090759
 |
Component:093; Jaccard:0.05984
 |
Correlation:0.1592
  |
Correlation:0.18092
  |
Correlation:0.095084
  |
Correlation:0.022313
  |
Correlation:0.040515
  |
Correlation:-0.010072
  |
Correlation:-0.010072
  |
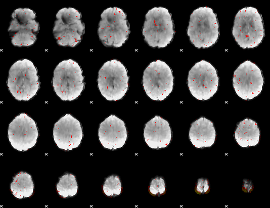
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
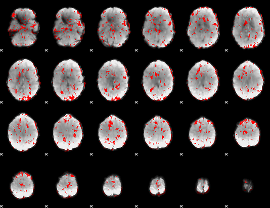 |
GLM binary result thresholded by 1.5
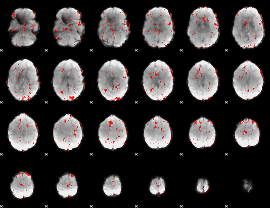 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:033; Jaccard:0.10868
 |
Component:033; Jaccard:0.11506
 |
Component:033; Jaccard:0.09696
 |
Component:033; Jaccard:0.062138
 |
Component:096; Jaccard:0.033964
 |
Component:043; Jaccard:0.014743
 |
Component:058; Jaccard:0.00631
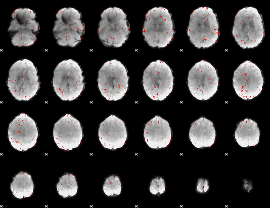 |
Correlation:0.22836
 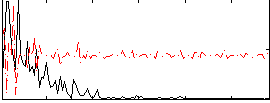 |
Correlation:0.22836
  |
Correlation:0.22836
  |
Correlation:0.22836
  |
Correlation:0.28448
  |
Correlation:0.29329
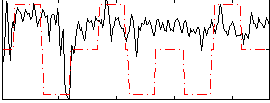  |
Correlation:0.32595
 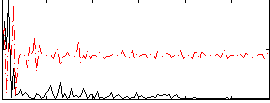 |
Component:096; Jaccard:0.093851
 |
Component:043; Jaccard:0.097777
 |
Component:043; Jaccard:0.090784
 |
Component:096; Jaccard:0.061028
 |
Component:043; Jaccard:0.033134
 |
Component:058; Jaccard:0.013264
 |
Component:316; Jaccard:0.004803
 |
Correlation:0.28448
  |
Correlation:0.29329
  |
Correlation:0.29329
  |
Correlation:0.28448
  |
Correlation:0.29329
  |
Correlation:0.32595
  |
Correlation:-0.1217
  |
Component:043; Jaccard:0.089034
 |
Component:096; Jaccard:0.097242
 |
Component:096; Jaccard:0.084132
 |
Component:043; Jaccard:0.06021
 |
Component:033; Jaccard:0.027839
 |
Component:096; Jaccard:0.012772
 |
Component:288; Jaccard:0.003766
 |
Correlation:0.29329
  |
Correlation:0.28448
  |
Correlation:0.28448
  |
Correlation:0.29329
  |
Correlation:0.22836
  |
Correlation:0.28448
  |
Correlation:0.37882
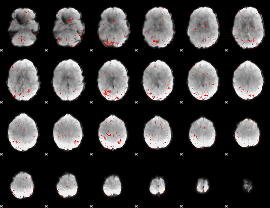
  |
Component:325; Jaccard:0.076528
 |
Component:193; Jaccard:0.071377
 |
Component:193; Jaccard:0.06037
 |
Component:193; Jaccard:0.036737
 |
Component:058; Jaccard:0.022811
 |
Component:033; Jaccard:0.010463
 |
Component:030; Jaccard:0.003623
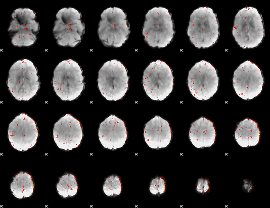
 |
Correlation:0.31899
  |
Correlation:0.13378
  |
Correlation:0.13378
  |
Correlation:0.13378
  |
Correlation:0.32595
  |
Correlation:0.22836
  |
Correlation:0.3644
  |
Component:193; Jaccard:0.072384
 |
Component:325; Jaccard:0.064949
 |
Component:253; Jaccard:0.05373
 |
Component:232; Jaccard:0.036568
 |
Component:316; Jaccard:0.02109
 |
Component:316; Jaccard:0.010382
 |
Component:004; Jaccard:0.003348
 |
Correlation:0.13378
  |
Correlation:0.31899
  |
Correlation:0.089467
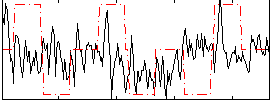 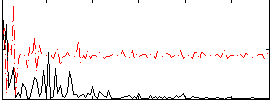 |
Correlation:0.19987
  |
Correlation:-0.1217
  |
Correlation:-0.1217
  |
Correlation:0.08049
  |
Component:256; Jaccard:0.069592
 |
Component:058; Jaccard:0.063416
 |
Component:058; Jaccard:0.050266
 |
Component:058; Jaccard:0.03585
 |
Component:232; Jaccard:0.020811
 |
Component:232; Jaccard:0.00892
 |
Component:043; Jaccard:0.003146
 |
Correlation:0.13117
  |
Correlation:0.32595
  |
Correlation:0.32595
  |
Correlation:0.32595
  |
Correlation:0.19987
  |
Correlation:0.19987
  |
Correlation:0.29329
  |
Component:058; Jaccard:0.068755
 |
Component:253; Jaccard:0.062003
 |
Component:316; Jaccard:0.049489
 |
Component:316; Jaccard:0.035085
 |
Component:193; Jaccard:0.019919
 |
Component:193; Jaccard:0.008554
 |
Component:372; Jaccard:0.002986
 |
Correlation:0.32595
  |
Correlation:0.089467
  |
Correlation:-0.1217
  |
Correlation:-0.1217
  |
Correlation:0.13378
  |
Correlation:0.13378
  |
Correlation:-0.045183
  |
Component:269; Jaccard:0.063536
 |
Component:256; Jaccard:0.061396
 |
Component:232; Jaccard:0.049462
 |
Component:253; Jaccard:0.034483
 |
Component:253; Jaccard:0.016401
 |
Component:288; Jaccard:0.008077
 |
Component:283; Jaccard:0.002853
 |
Correlation:0.32578
 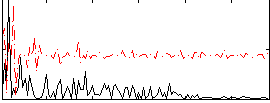 |
Correlation:0.13117
  |
Correlation:0.19987
  |
Correlation:0.089467
  |
Correlation:0.089467
  |
Correlation:0.37882
  |
Correlation:0.027755
 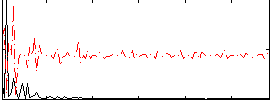 |
Component:323; Jaccard:0.060101
 |
Component:269; Jaccard:0.054862
 |
Component:256; Jaccard:0.046023
 |
Component:256; Jaccard:0.027652
 |
Component:030; Jaccard:0.015575
 |
Component:030; Jaccard:0.006988
 |
Component:232; Jaccard:0.002726
 |
Correlation:0.41734
 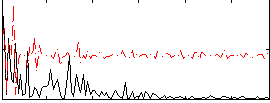 |
Correlation:0.32578
  |
Correlation:0.13117
  |
Correlation:0.13117
  |
Correlation:0.3644
  |
Correlation:0.3644
  |
Correlation:0.19987
  |
Component:288; Jaccard:0.060052
 |
Component:262; Jaccard:0.053076
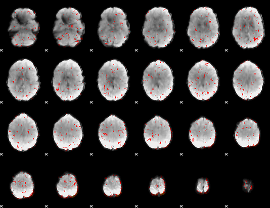 |
Component:325; Jaccard:0.043791
 |
Component:262; Jaccard:0.026232
 |
Component:262; Jaccard:0.014349
 |
Component:262; Jaccard:0.006838
 |
Component:262; Jaccard:0.002725
 |
Correlation:0.37882
  |
Correlation:0.24485
  |
Correlation:0.31899
  |
Correlation:0.24485
  |
Correlation:0.24485
  |
Correlation:0.24485
  |
Correlation:0.24485
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
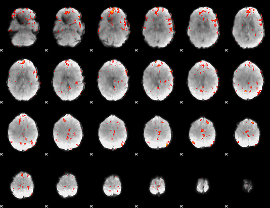
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
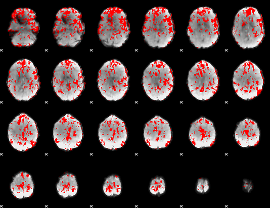 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
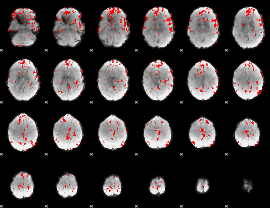 |
GLM binary result thresholded by 3
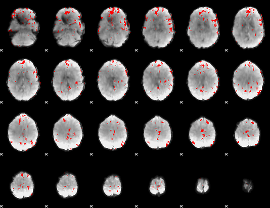 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
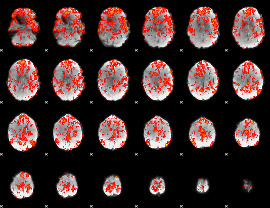 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
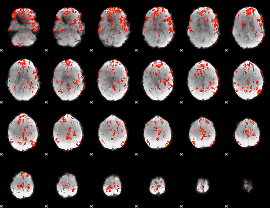 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:342; Jaccard:0.1087
 |
Component:268; Jaccard:0.12736
 |
Component:268; Jaccard:0.15208
 |
Component:268; Jaccard:0.1667
 |
Component:268; Jaccard:0.16399
 |
Component:268; Jaccard:0.13535
 |
Component:268; Jaccard:0.093044
 |
Correlation:0.11738
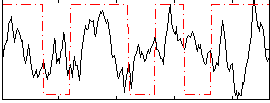  |
Correlation:0.2097
  |
Correlation:0.2097
  |
Correlation:0.2097
  |
Correlation:0.2097
  |
Correlation:0.2097
  |
Correlation:0.2097
  |
Component:268; Jaccard:0.10369
 |
Component:342; Jaccard:0.12683
 |
Component:342; Jaccard:0.14324
 |
Component:342; Jaccard:0.15053
 |
Component:342; Jaccard:0.14098
 |
Component:342; Jaccard:0.11207
 |
Component:342; Jaccard:0.075852
 |
Correlation:0.2097
  |
Correlation:0.11738
  |
Correlation:0.11738
  |
Correlation:0.11738
  |
Correlation:0.11738
  |
Correlation:0.11738
  |
Correlation:0.11738
  |
Component:390; Jaccard:0.096579
 |
Component:104; Jaccard:0.108
 |
Component:104; Jaccard:0.11876
 |
Component:104; Jaccard:0.12123
 |
Component:104; Jaccard:0.10931
 |
Component:104; Jaccard:0.081426
 |
Component:373; Jaccard:0.052766
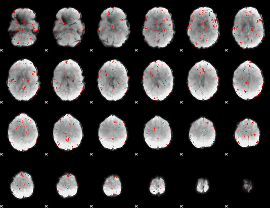 |
Correlation:0.14942
  |
Correlation:0.0027742
  |
Correlation:0.0027742
  |
Correlation:0.0027742
  |
Correlation:0.0027742
  |
Correlation:0.0027742
  |
Correlation:0.15112
  |
Component:177; Jaccard:0.095428
 |
Component:033; Jaccard:0.10559
 |
Component:033; Jaccard:0.11508
 |
Component:033; Jaccard:0.1146
 |
Component:373; Jaccard:0.098237
 |
Component:373; Jaccard:0.07765
 |
Component:039; Jaccard:0.052248
 |
Correlation:0.13178
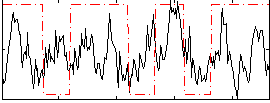  |
Correlation:0.22421
  |
Correlation:0.22421
  |
Correlation:0.22421
  |
Correlation:0.15112
  |
Correlation:0.15112
  |
Correlation:0.22171
  |
Component:104; Jaccard:0.094536
 |
Component:177; Jaccard:0.1011
 |
Component:177; Jaccard:0.10288
 |
Component:373; Jaccard:0.10473
 |
Component:033; Jaccard:0.097247
 |
Component:039; Jaccard:0.075931
 |
Component:058; Jaccard:0.048013
 |
Correlation:0.0027742
  |
Correlation:0.13178
  |
Correlation:0.13178
  |
Correlation:0.15112
  |
Correlation:0.22421
  |
Correlation:0.22171
  |
Correlation:0.36966
  |
Component:033; Jaccard:0.090964
 |
Component:390; Jaccard:0.10081
 |
Component:169; Jaccard:0.10281
 |
Component:039; Jaccard:0.10286
 |
Component:039; Jaccard:0.095015
 |
Component:033; Jaccard:0.06583
 |
Component:104; Jaccard:0.04604
 |
Correlation:0.22421
  |
Correlation:0.14942
  |
Correlation:-0.026525
  |
Correlation:0.22171
  |
Correlation:0.22171
  |
Correlation:0.22421
  |
Correlation:0.0027742
  |
Component:089; Jaccard:0.090542
 |
Component:089; Jaccard:0.097568
 |
Component:373; Jaccard:0.10086
 |
Component:169; Jaccard:0.1019
 |
Component:169; Jaccard:0.08929
 |
Component:305; Jaccard:0.065637
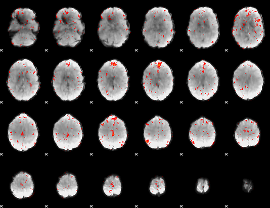 |
Component:256; Jaccard:0.044553
 |
Correlation:0.073019
  |
Correlation:0.073019
  |
Correlation:0.15112
  |
Correlation:-0.026525
  |
Correlation:-0.026525
  |
Correlation:0.22482
  |
Correlation:0.093955
  |
Component:169; Jaccard:0.085848
 |
Component:169; Jaccard:0.095747
 |
Component:039; Jaccard:0.10027
 |
Component:089; Jaccard:0.095834
 |
Component:305; Jaccard:0.083316
 |
Component:324; Jaccard:0.065267
 |
Component:166; Jaccard:0.044512
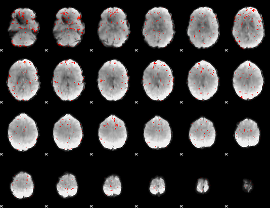 |
Correlation:-0.026525
  |
Correlation:-0.026525
  |
Correlation:0.22171
  |
Correlation:0.073019
  |
Correlation:0.22482
  |
Correlation:0.070896
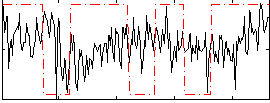  |
Correlation:0.066862
 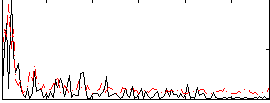 |
Component:223; Jaccard:0.083668
 |
Component:039; Jaccard:0.091139
 |
Component:089; Jaccard:0.10004
 |
Component:177; Jaccard:0.095299
 |
Component:324; Jaccard:0.083006
 |
Component:166; Jaccard:0.065256
 |
Component:148; Jaccard:0.044428
 |
Correlation:0.20274
  |
Correlation:0.22171
  |
Correlation:0.073019
  |
Correlation:0.13178
  |
Correlation:0.070896
  |
Correlation:0.066862
  |
Correlation:-0.020059
  |
Component:230; Jaccard:0.081221
 |
Component:373; Jaccard:0.090937
 |
Component:390; Jaccard:0.099236
 |
Component:390; Jaccard:0.09393
 |
Component:089; Jaccard:0.081355
 |
Component:169; Jaccard:0.064854
 |
Component:305; Jaccard:0.044191
 |
Correlation:-0.098748
  |
Correlation:0.15112
  |
Correlation:0.14942
  |
Correlation:0.14942
  |
Correlation:0.073019
  |
Correlation:-0.026525
  |
Correlation:0.22482
  |
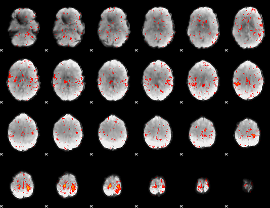
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
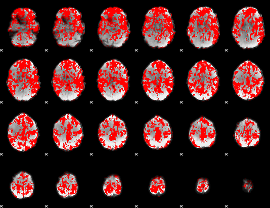 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
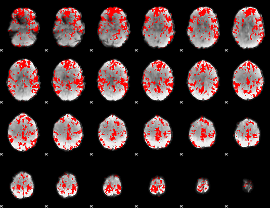 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
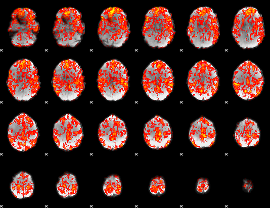 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
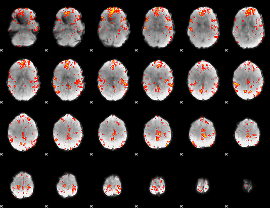 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:317; Jaccard:0.097446
 |
Component:342; Jaccard:0.12112
 |
Component:342; Jaccard:0.15599
 |
Component:342; Jaccard:0.19392
 |
Component:342; Jaccard:0.22745
 |
Component:342; Jaccard:0.24296
 |
Component:342; Jaccard:0.23327
 |
Correlation:0.03379
  |
Correlation:0.15037
  |
Correlation:0.15037
  |
Correlation:0.15037
  |
Correlation:0.15037
  |
Correlation:0.15037
  |
Correlation:0.15037
  |
Component:177; Jaccard:0.096834
 |
Component:317; Jaccard:0.11321
 |
Component:177; Jaccard:0.1292
 |
Component:177; Jaccard:0.141
 |
Component:268; Jaccard:0.15008
 |
Component:268; Jaccard:0.14984
 |
Component:268; Jaccard:0.13194
 |
Correlation:0.067028
  |
Correlation:0.03379
  |
Correlation:0.067028
  |
Correlation:0.067028
  |
Correlation:0.09084
  |
Correlation:0.09084
  |
Correlation:0.09084
  |
Component:342; Jaccard:0.095175
 |
Component:177; Jaccard:0.1127
 |
Component:317; Jaccard:0.12864
 |
Component:268; Jaccard:0.13732
 |
Component:177; Jaccard:0.14477
 |
Component:177; Jaccard:0.13258
 |
Component:177; Jaccard:0.1105
 |
Correlation:0.15037
  |
Correlation:0.067028
  |
Correlation:0.03379
  |
Correlation:0.09084
  |
Correlation:0.067028
  |
Correlation:0.067028
  |
Correlation:0.067028
  |
Component:390; Jaccard:0.091869
 |
Component:089; Jaccard:0.10551
 |
Component:089; Jaccard:0.12045
 |
Component:317; Jaccard:0.13715
 |
Component:317; Jaccard:0.13784
 |
Component:317; Jaccard:0.12793
 |
Component:148; Jaccard:0.10902
 |
Correlation:-0.12732
  |
Correlation:0.078133
  |
Correlation:0.078133
  |
Correlation:0.03379
  |
Correlation:0.03379
  |
Correlation:0.03379
  |
Correlation:0.19688
  |
Component:089; Jaccard:0.090047
 |
Component:390; Jaccard:0.10169
 |
Component:268; Jaccard:0.11904
 |
Component:089; Jaccard:0.1338
 |
Component:089; Jaccard:0.13614
 |
Component:089; Jaccard:0.12641
 |
Component:317; Jaccard:0.10605
 |
Correlation:0.078133
  |
Correlation:-0.12732
  |
Correlation:0.09084
  |
Correlation:0.078133
  |
Correlation:0.078133
  |
Correlation:0.078133
  |
Correlation:0.03379
  |
Component:128; Jaccard:0.086464
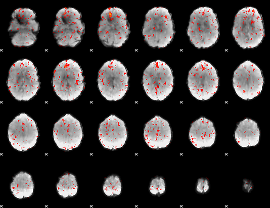 |
Component:128; Jaccard:0.10159
 |
Component:128; Jaccard:0.1149
 |
Component:128; Jaccard:0.12445
 |
Component:128; Jaccard:0.12675
 |
Component:128; Jaccard:0.11943
 |
Component:039; Jaccard:0.1056
 |
Correlation:0.19478
  |
Correlation:0.19478
  |
Correlation:0.19478
  |
Correlation:0.19478
  |
Correlation:0.19478
  |
Correlation:0.19478
  |
Correlation:0.2141
  |
Component:230; Jaccard:0.085346
 |
Component:268; Jaccard:0.097654
 |
Component:390; Jaccard:0.10916
 |
Component:390; Jaccard:0.11122
 |
Component:039; Jaccard:0.11797
 |
Component:148; Jaccard:0.11912
 |
Component:089; Jaccard:0.10495
 |
Correlation:0.12052
  |
Correlation:0.09084
  |
Correlation:-0.12732
  |
Correlation:-0.12732
  |
Correlation:0.2141
  |
Correlation:0.19688
  |
Correlation:0.078133
  |
Component:257; Jaccard:0.083935
 |
Component:257; Jaccard:0.095337
 |
Component:257; Jaccard:0.10523
 |
Component:257; Jaccard:0.11092
 |
Component:148; Jaccard:0.11543
 |
Component:039; Jaccard:0.1175
 |
Component:128; Jaccard:0.10331
 |
Correlation:0.12614
  |
Correlation:0.12614
  |
Correlation:0.12614
  |
Correlation:0.12614
  |
Correlation:0.19688
  |
Correlation:0.2141
  |
Correlation:0.19478
  |
Component:268; Jaccard:0.079675
 |
Component:230; Jaccard:0.094206
 |
Component:230; Jaccard:0.10114
 |
Component:039; Jaccard:0.11039
 |
Component:257; Jaccard:0.10997
 |
Component:169; Jaccard:0.10034
 |
Component:169; Jaccard:0.083155
 |
Correlation:0.09084
  |
Correlation:0.12052
  |
Correlation:0.12052
  |
Correlation:0.2141
  |
Correlation:0.12614
  |
Correlation:0.23008
  |
Correlation:0.23008
  |
Component:116; Jaccard:0.077962
 |
Component:169; Jaccard:0.088742
 |
Component:169; Jaccard:0.10015
 |
Component:169; Jaccard:0.10738
 |
Component:169; Jaccard:0.10927
 |
Component:257; Jaccard:0.09992
 |
Component:257; Jaccard:0.08201
 |
Correlation:0.0016208
  |
Correlation:0.23008
  |
Correlation:0.23008
  |
Correlation:0.23008
  |
Correlation:0.23008
  |
Correlation:0.12614
  |
Correlation:0.12614
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































