Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
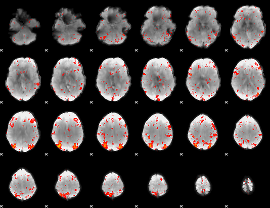
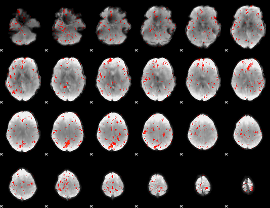
GLM result group-wise
 |
GLM binary result thresholded by 1
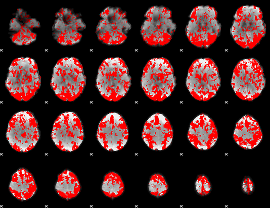 |
GLM binary result thresholded by 1.5
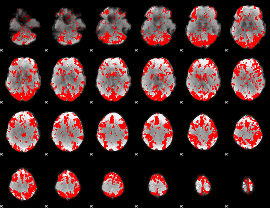 |
GLM binary result thresholded by 2
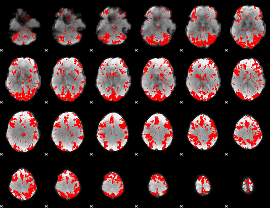 |
GLM binary result thresholded by 2.5
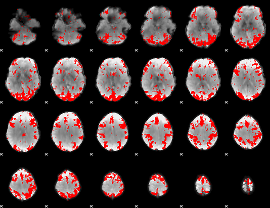 |
GLM binary result thresholded by 3
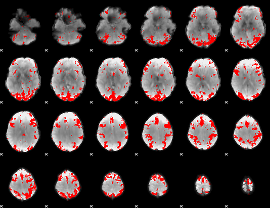 |
GLM binary result thresholded by 3.5
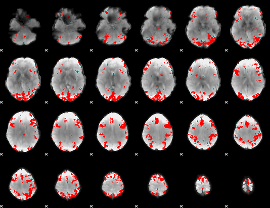 |
GLM binary result thresholded by 4
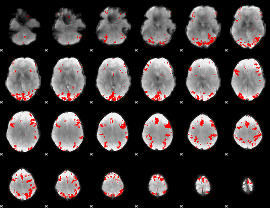 |
GLM Z-score result thresholded by 1
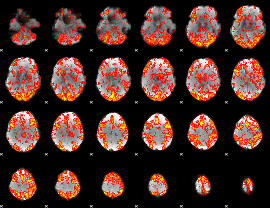 |
GLM Z-score result thresholded by 1.5
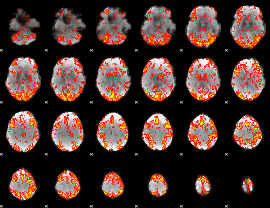 |
GLM Z-score result thresholded by 2
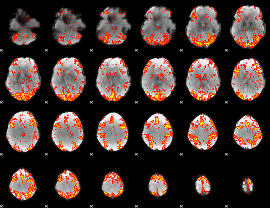 |
GLM Z-score result thresholded by 2.5
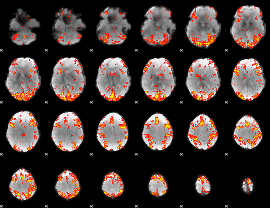 |
GLM Z-score result thresholded by 3
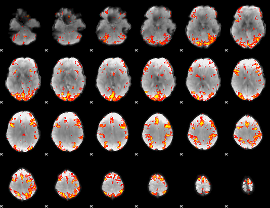 |
GLM Z-score result thresholded by 3.5
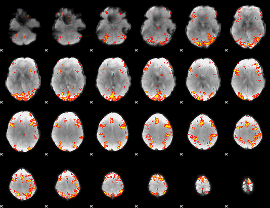 |
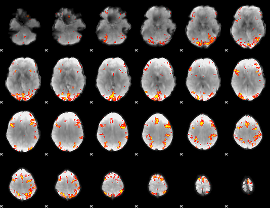
GLM Z-score result thresholded by 4
 |
Component:397; Jaccard:0.15271
 |
Component:397; Jaccard:0.19271
 |
Component:397; Jaccard:0.23853
 |
Component:397; Jaccard:0.28547
 |
Component:397; Jaccard:0.32074
 |
Component:397; Jaccard:0.33559
 |
Component:397; Jaccard:0.33093
 |
Correlation:0.36184
  |
Correlation:0.36184
  |
Correlation:0.36184
  |
Correlation:0.36184
  |
Correlation:0.36184
  |
Correlation:0.36184
  |
Correlation:0.36184
  |
Component:328; Jaccard:0.12575
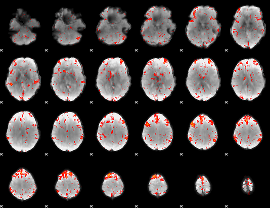 |
Component:353; Jaccard:0.14031
 |
Component:353; Jaccard:0.17293
 |
Component:353; Jaccard:0.20941
 |
Component:379; Jaccard:0.24483
 |
Component:379; Jaccard:0.27666
 |
Component:379; Jaccard:0.29366
 |
Correlation:0.16947
  |
Correlation:0.33719
 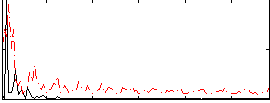 |
Correlation:0.33719
  |
Correlation:0.33719
  |
Correlation:0.22849
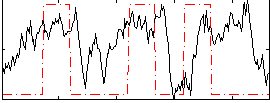  |
Correlation:0.22849
  |
Correlation:0.22849
  |
Component:353; Jaccard:0.11182
 |
Component:328; Jaccard:0.13787
 |
Component:379; Jaccard:0.16615
 |
Component:379; Jaccard:0.20517
 |
Component:353; Jaccard:0.24358
 |
Component:353; Jaccard:0.2654
 |
Component:353; Jaccard:0.26994
 |
Correlation:0.33719
  |
Correlation:0.16947
  |
Correlation:0.22849
  |
Correlation:0.22849
  |
Correlation:0.33719
  |
Correlation:0.33719
  |
Correlation:0.33719
  |
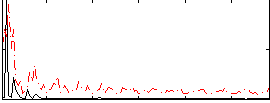
Component:394; Jaccard:0.11167
 |
Component:168; Jaccard:0.13224
 |
Component:186; Jaccard:0.15484
 |
Component:186; Jaccard:0.17673
 |
Component:186; Jaccard:0.19254
 |
Component:186; Jaccard:0.2015
 |
Component:186; Jaccard:0.1985
 |
Correlation:0.31029
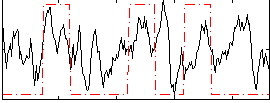 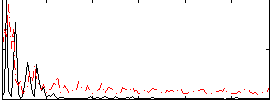 |
Correlation:0.18497
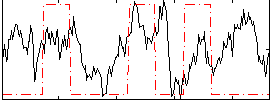 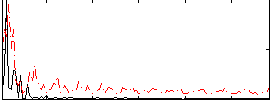 |
Correlation:0.34844
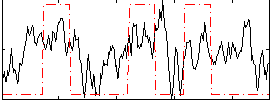 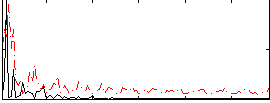 |
Correlation:0.34844
  |
Correlation:0.34844
  |
Correlation:0.34844
  |
Correlation:0.34844
  |
Component:168; Jaccard:0.11093
 |
Component:379; Jaccard:0.13134
 |
Component:168; Jaccard:0.1531
 |
Component:150; Jaccard:0.17242
 |
Component:150; Jaccard:0.18226
 |
Component:150; Jaccard:0.18553
 |
Component:150; Jaccard:0.17566
 |
Correlation:0.18497
  |
Correlation:0.22849
  |
Correlation:0.18497
  |
Correlation:0.24239
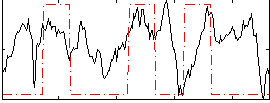 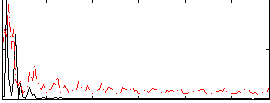 |
Correlation:0.24239
  |
Correlation:0.24239
  |
Correlation:0.24239
  |
Component:150; Jaccard:0.10938
 |
Component:150; Jaccard:0.13102
 |
Component:150; Jaccard:0.15304
 |
Component:168; Jaccard:0.16965
 |
Component:168; Jaccard:0.17901
 |
Component:168; Jaccard:0.18014
 |
Component:168; Jaccard:0.17201
 |
Correlation:0.24239
  |
Correlation:0.24239
  |
Correlation:0.24239
  |
Correlation:0.18497
  |
Correlation:0.18497
  |
Correlation:0.18497
  |
Correlation:0.18497
  |
Component:186; Jaccard:0.10695
 |
Component:186; Jaccard:0.13064
 |
Component:328; Jaccard:0.14493
 |
Component:394; Jaccard:0.15376
 |
Component:394; Jaccard:0.15741
 |
Component:396; Jaccard:0.15918
 |
Component:396; Jaccard:0.15177
 |
Correlation:0.34844
  |
Correlation:0.34844
  |
Correlation:0.16947
  |
Correlation:0.31029
  |
Correlation:0.31029
  |
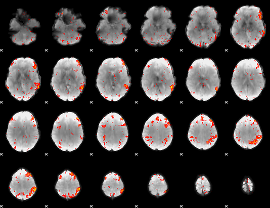
Correlation:0.38896
  |
Correlation:0.38896
  |
Component:215; Jaccard:0.10405
 |
Component:394; Jaccard:0.12831
 |
Component:394; Jaccard:0.14445
 |
Component:215; Jaccard:0.1493
 |
Component:396; Jaccard:0.15507
 |
Component:162; Jaccard:0.15374
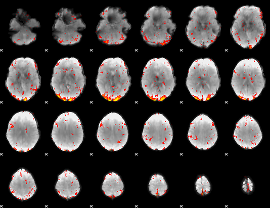 |
Component:277; Jaccard:0.14174
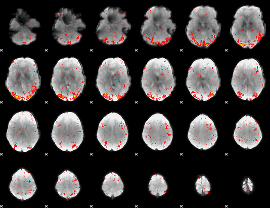 |
Correlation:0.29682
  |
Correlation:0.31029
  |
Correlation:0.31029
  |
Correlation:0.29682
  |
Correlation:0.38896
  |
Correlation:0.16292
  |
Correlation:0.19835
  |
Component:379; Jaccard:0.10264
 |
Component:215; Jaccard:0.12179
 |
Component:215; Jaccard:0.13807
 |
Component:162; Jaccard:0.14815
 |
Component:162; Jaccard:0.1547
 |
Component:394; Jaccard:0.15138
 |
Component:162; Jaccard:0.14135
 |
Correlation:0.22849
  |
Correlation:0.29682
  |
Correlation:0.29682
  |
Correlation:0.16292
  |
Correlation:0.16292
  |
Correlation:0.31029
  |
Correlation:0.16292
  |
Component:301; Jaccard:0.099909
 |
Component:162; Jaccard:0.11624
 |
Component:162; Jaccard:0.13244
 |
Component:396; Jaccard:0.14541
 |
Component:277; Jaccard:0.15199
 |
Component:277; Jaccard:0.15071
 |
Component:215; Jaccard:0.13862
 |
Correlation:0.23389
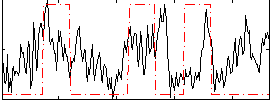 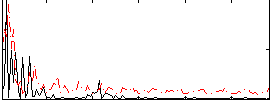 |
Correlation:0.16292
  |
Correlation:0.16292
  |
Correlation:0.38896
  |
Correlation:0.19835
  |
Correlation:0.19835
  |
Correlation:0.29682
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
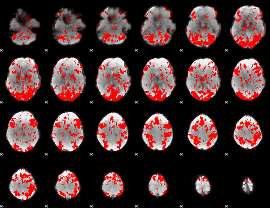 |
GLM binary result thresholded by 1.5
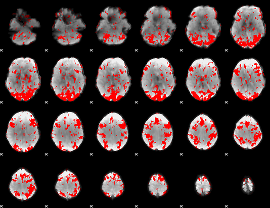 |
GLM binary result thresholded by 2
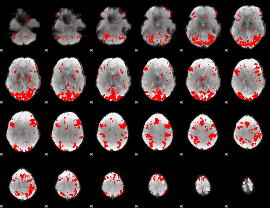 |
GLM binary result thresholded by 2.5
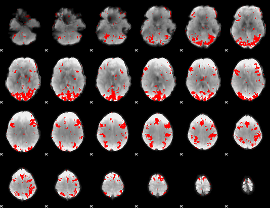 |
GLM binary result thresholded by 3
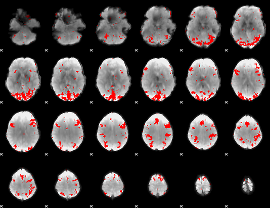 |
GLM binary result thresholded by 3.5
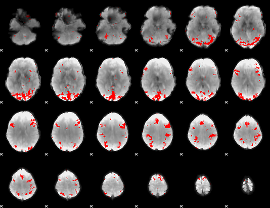 |
GLM binary result thresholded by 4
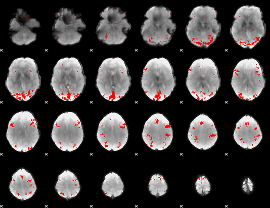 |
GLM Z-score result thresholded by 1
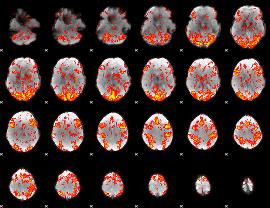 |
GLM Z-score result thresholded by 1.5
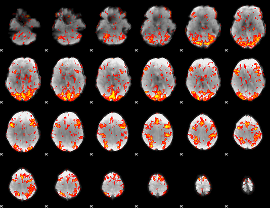 |
GLM Z-score result thresholded by 2
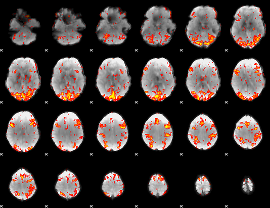 |
GLM Z-score result thresholded by 2.5
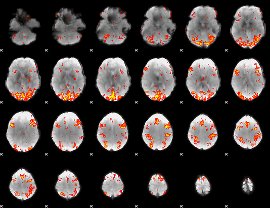 |
GLM Z-score result thresholded by 3
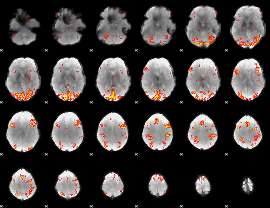 |
GLM Z-score result thresholded by 3.5
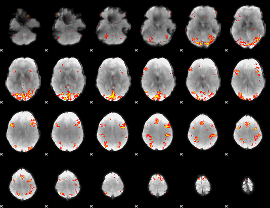 |
GLM Z-score result thresholded by 4
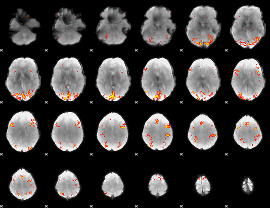 |
Component:397; Jaccard:0.16544
 |
Component:353; Jaccard:0.21222
 |
Component:353; Jaccard:0.26601
 |
Component:353; Jaccard:0.31418
 |
Component:353; Jaccard:0.34732
 |
Component:353; Jaccard:0.35177
 |
Component:353; Jaccard:0.32807
 |
Correlation:-0.23547
  |
Correlation:-0.047484
  |
Correlation:-0.047484
  |
Correlation:-0.047484
  |
Correlation:-0.047484
  |
Correlation:-0.047484
  |
Correlation:-0.047484
  |
Component:353; Jaccard:0.16432
 |
Component:397; Jaccard:0.19178
 |
Component:379; Jaccard:0.2309
 |
Component:379; Jaccard:0.27088
 |
Component:379; Jaccard:0.29928
 |
Component:379; Jaccard:0.29844
 |
Component:379; Jaccard:0.27431
 |
Correlation:-0.047484
  |
Correlation:-0.23547
  |
Correlation:0.059687
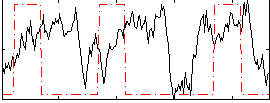 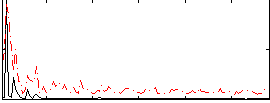 |
Correlation:0.059687
  |
Correlation:0.059687
  |
Correlation:0.059687
  |
Correlation:0.059687
  |
Component:379; Jaccard:0.14613
 |
Component:379; Jaccard:0.18568
 |
Component:397; Jaccard:0.21393
 |
Component:397; Jaccard:0.22438
 |
Component:397; Jaccard:0.21721
 |
Component:397; Jaccard:0.19705
 |
Component:186; Jaccard:0.17168
 |
Correlation:0.059687
  |
Correlation:0.059687
  |
Correlation:-0.23547
  |
Correlation:-0.23547
  |
Correlation:-0.23547
  |
Correlation:-0.23547
  |
Correlation:-0.078896
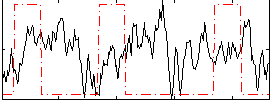 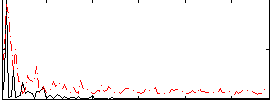 |
Component:186; Jaccard:0.13567
 |
Component:186; Jaccard:0.16026
 |
Component:186; Jaccard:0.18364
 |
Component:186; Jaccard:0.19858
 |
Component:186; Jaccard:0.20153
 |
Component:186; Jaccard:0.19292
 |
Component:397; Jaccard:0.16566
 |
Correlation:-0.078896
  |
Correlation:-0.078896
  |
Correlation:-0.078896
  |
Correlation:-0.078896
  |
Correlation:-0.078896
  |
Correlation:-0.078896
  |
Correlation:-0.23547
  |
Component:168; Jaccard:0.13279
 |
Component:168; Jaccard:0.14929
 |
Component:168; Jaccard:0.16318
 |
Component:277; Jaccard:0.16683
 |
Component:277; Jaccard:0.16584
 |
Component:277; Jaccard:0.15283
 |
Component:168; Jaccard:0.12844
 |
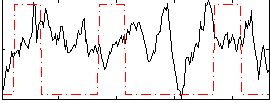
Correlation:-0.13605
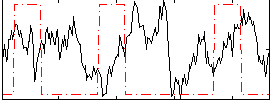  |
Correlation:-0.13605
  |
Correlation:-0.13605
  |
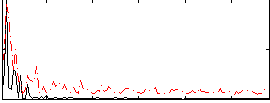
Correlation:8.7494e-05
 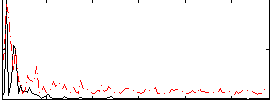 |
Correlation:8.7494e-05
  |
Correlation:8.7494e-05
  |
Correlation:-0.13605
  |
Component:150; Jaccard:0.11943
 |
Component:277; Jaccard:0.13909
 |
Component:277; Jaccard:0.15607
 |
Component:168; Jaccard:0.16675
 |
Component:168; Jaccard:0.16358
 |
Component:168; Jaccard:0.15081
 |
Component:277; Jaccard:0.12834
 |
Correlation:-0.084925
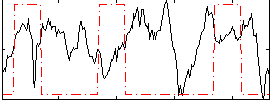  |
Correlation:8.7494e-05
  |
Correlation:8.7494e-05
  |
Correlation:-0.13605
  |
Correlation:-0.13605
  |
Correlation:-0.13605
  |
Correlation:8.7494e-05
  |
Component:277; Jaccard:0.11933
 |
Component:150; Jaccard:0.13633
 |
Component:150; Jaccard:0.14867
 |
Component:150; Jaccard:0.15386
 |
Component:150; Jaccard:0.14958
 |
Component:150; Jaccard:0.1341
 |
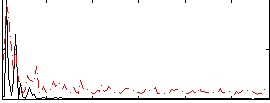
Component:213; Jaccard:0.1172
 |
Correlation:8.7494e-05
  |
Correlation:-0.084925
  |
Correlation:-0.084925
  |
Correlation:-0.084925
  |
Correlation:-0.084925
  |
Correlation:-0.084925
  |
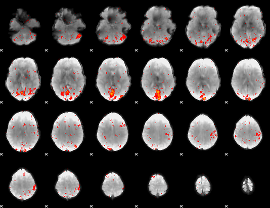
Correlation:-0.11089
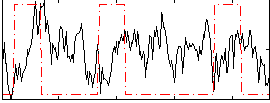  |
Component:394; Jaccard:0.114
 |
Component:396; Jaccard:0.12737
 |
Component:396; Jaccard:0.13934
 |
Component:396; Jaccard:0.14515
 |
Component:396; Jaccard:0.14344
 |
Component:213; Jaccard:0.13177
 |
Component:138; Jaccard:0.11515
 |
Correlation:-0.21191
  |
Correlation:-0.26232
  |
Correlation:-0.26232
  |
Correlation:-0.26232
  |
Correlation:-0.26232
  |
Correlation:-0.11089
  |
Correlation:-0.11492
  |
Component:396; Jaccard:0.11287
 |
Component:394; Jaccard:0.12237
 |
Component:213; Jaccard:0.13173
 |
Component:213; Jaccard:0.14158
 |
Component:213; Jaccard:0.14013
 |
Component:396; Jaccard:0.13164
 |
Component:396; Jaccard:0.11414
 |
Correlation:-0.26232
  |
Correlation:-0.21191
  |
Correlation:-0.11089
  |
Correlation:-0.11089
  |
Correlation:-0.11089
  |
Correlation:-0.26232
  |
Correlation:-0.26232
  |
Component:301; Jaccard:0.1101
 |
Component:213; Jaccard:0.11803
 |
Component:394; Jaccard:0.12746
 |
Component:162; Jaccard:0.12819
 |
Component:138; Jaccard:0.12964
 |
Component:138; Jaccard:0.12754
 |
Component:150; Jaccard:0.11131
 |
Correlation:-0.27828
  |
Correlation:-0.11089
  |
Correlation:-0.21191
  |
Correlation:-0.19817
  |
Correlation:-0.11492
  |
Correlation:-0.11492
  |
Correlation:-0.084925
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
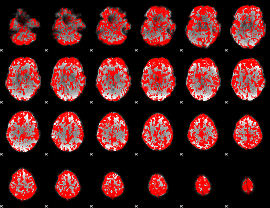 |
GLM binary result thresholded by 1.5
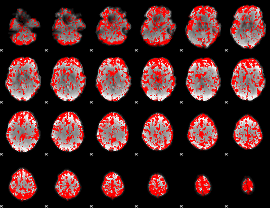 |
GLM binary result thresholded by 2
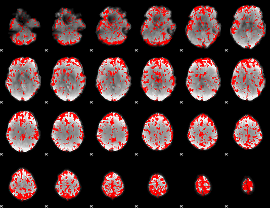 |
GLM binary result thresholded by 2.5
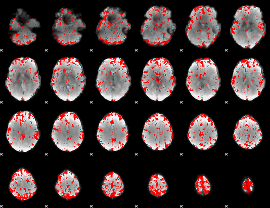 |
GLM binary result thresholded by 3
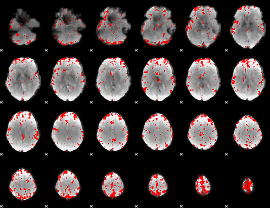 |
GLM binary result thresholded by 3.5
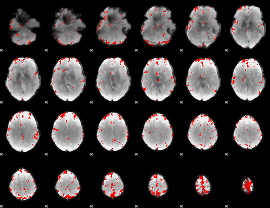 |
GLM binary result thresholded by 4
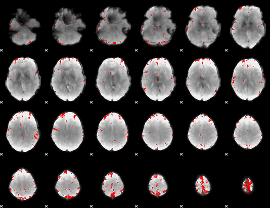 |
GLM Z-score result thresholded by 1
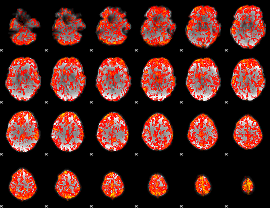 |
GLM Z-score result thresholded by 1.5
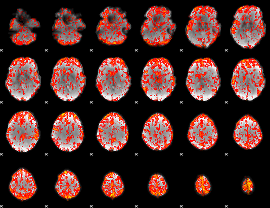 |
GLM Z-score result thresholded by 2
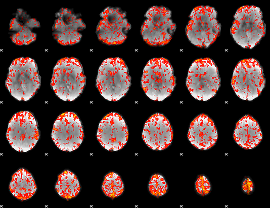 |
GLM Z-score result thresholded by 2.5
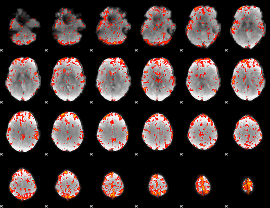 |
GLM Z-score result thresholded by 3
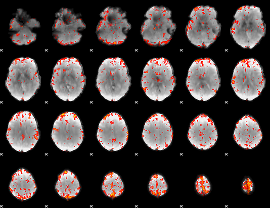 |
GLM Z-score result thresholded by 3.5
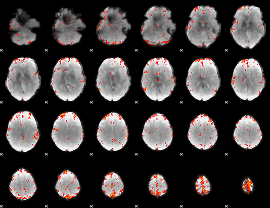 |
GLM Z-score result thresholded by 4
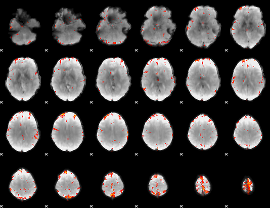 |
Component:201; Jaccard:0.098941
 |
Component:201; Jaccard:0.11934
 |
Component:201; Jaccard:0.14128
 |
Component:064; Jaccard:0.16133
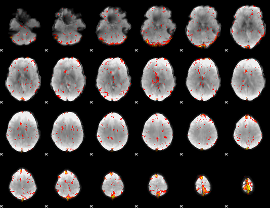 |
Component:064; Jaccard:0.18171
 |
Component:064; Jaccard:0.18204
 |
Component:064; Jaccard:0.15721
 |
Correlation:0.2874
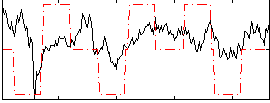  |
Correlation:0.2874
  |
Correlation:0.2874
  |
Correlation:0.34192
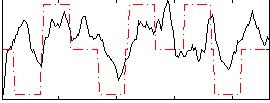 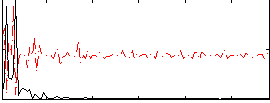 |
Correlation:0.34192
  |
Correlation:0.34192
  |
Correlation:0.34192
  |
Component:328; Jaccard:0.098519
 |
Component:064; Jaccard:0.11371
 |
Component:064; Jaccard:0.13746
 |
Component:201; Jaccard:0.15948
 |
Component:201; Jaccard:0.16225
 |
Component:201; Jaccard:0.14662
 |
Component:010; Jaccard:0.11664
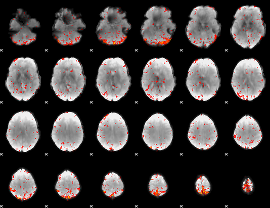 |
Correlation:0.35338
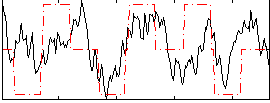 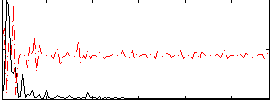 |
Correlation:0.34192
  |
Correlation:0.34192
  |
Correlation:0.2874
  |
Correlation:0.2874
  |
Correlation:0.2874
  |
Correlation:0.53588
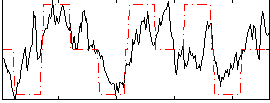 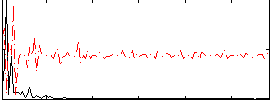 |
Component:064; Jaccard:0.094431
 |
Component:328; Jaccard:0.11151
 |
Component:328; Jaccard:0.12263
 |
Component:332; Jaccard:0.13166
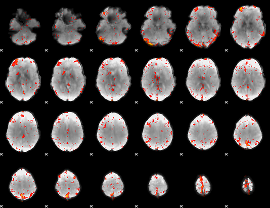 |
Component:010; Jaccard:0.14176
 |
Component:010; Jaccard:0.13787
 |
Component:201; Jaccard:0.11159
 |
Correlation:0.34192
  |
Correlation:0.35338
  |
Correlation:0.35338
  |
Correlation:0.52931
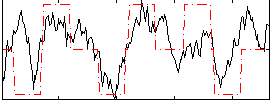 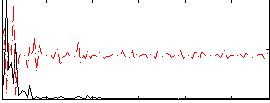 |
Correlation:0.53588
  |
Correlation:0.53588
  |
Correlation:0.2874
  |
Component:397; Jaccard:0.089163
 |
Component:332; Jaccard:0.10251
 |
Component:332; Jaccard:0.11839
 |
Component:010; Jaccard:0.12929
 |
Component:332; Jaccard:0.13716
 |
Component:332; Jaccard:0.1277
 |
Component:332; Jaccard:0.10535
 |
Correlation:0.35403
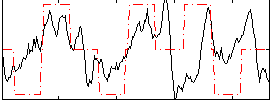 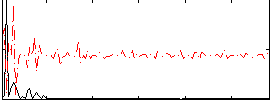 |
Correlation:0.52931
  |
Correlation:0.52931
  |
Correlation:0.53588
  |
Correlation:0.52931
  |
Correlation:0.52931
  |
Correlation:0.52931
  |
Component:332; Jaccard:0.087652
 |
Component:010; Jaccard:0.096472
 |
Component:010; Jaccard:0.11234
 |
Component:328; Jaccard:0.12812
 |
Component:328; Jaccard:0.12459
 |
Component:282; Jaccard:0.11517
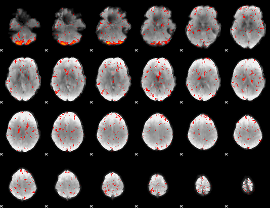 |
Component:122; Jaccard:0.093546
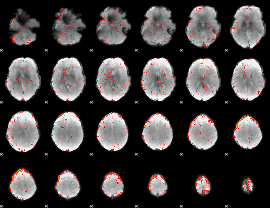 |
Correlation:0.52931
  |
Correlation:0.53588
  |
Correlation:0.53588
  |
Correlation:0.35338
  |
Correlation:0.35338
  |
Correlation:0.35788
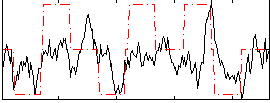 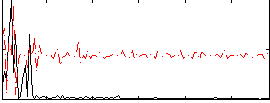 |
Correlation:0.37716
  |
Component:375; Jaccard:0.084925
 |
Component:375; Jaccard:0.096284
 |
Component:282; Jaccard:0.10868
 |
Component:282; Jaccard:0.12045
 |
Component:282; Jaccard:0.12394
 |
Component:122; Jaccard:0.11399
 |
Component:282; Jaccard:0.090189
 |
Correlation:0.088852
  |
Correlation:0.088852
  |
Correlation:0.35788
  |
Correlation:0.35788
  |
Correlation:0.35788
  |
Correlation:0.37716
  |
Correlation:0.35788
  |
Component:234; Jaccard:0.084382
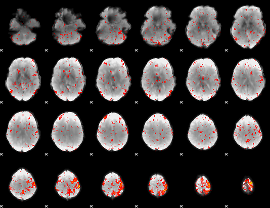 |
Component:397; Jaccard:0.094825
 |
Component:375; Jaccard:0.1062
 |
Component:122; Jaccard:0.11546
 |
Component:122; Jaccard:0.12173
 |
Component:328; Jaccard:0.11034
 |
Component:328; Jaccard:0.085614
 |
Correlation:0.17632
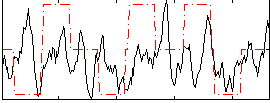 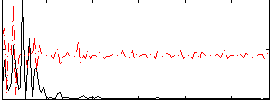 |
Correlation:0.35403
  |
Correlation:0.088852
  |
Correlation:0.37716
  |
Correlation:0.37716
  |
Correlation:0.35338
  |
Correlation:0.35338
  |
Component:345; Jaccard:0.084355
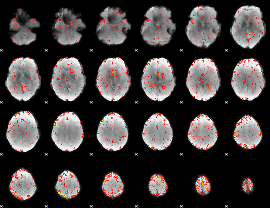 |
Component:282; Jaccard:0.094304
 |
Component:122; Jaccard:0.10222
 |
Component:207; Jaccard:0.10797
 |
Component:207; Jaccard:0.10972
 |
Component:207; Jaccard:0.10366
 |
Component:207; Jaccard:0.082927
 |
Correlation:0.21455
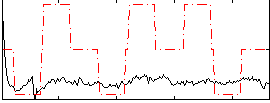 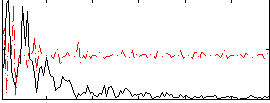 |
Correlation:0.35788
  |
Correlation:0.37716
  |
Correlation:0.34583
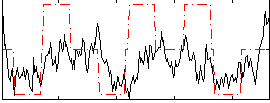 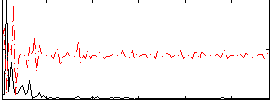 |
Correlation:0.34583
  |
Correlation:0.34583
  |
Correlation:0.34583
  |
Component:010; Jaccard:0.082598
 |
Component:234; Jaccard:0.09236
 |
Component:207; Jaccard:0.10095
 |
Component:375; Jaccard:0.10588
 |
Component:234; Jaccard:0.10321
 |
Component:234; Jaccard:0.095883
 |
Component:234; Jaccard:0.079609
 |
Correlation:0.53588
  |
Correlation:0.17632
  |
Correlation:0.34583
  |
Correlation:0.088852
  |
Correlation:0.17632
  |
Correlation:0.17632
  |
Correlation:0.17632
  |
Component:282; Jaccard:0.081174
 |
Component:207; Jaccard:0.090819
 |
Component:234; Jaccard:0.1
 |
Component:234; Jaccard:0.10359
 |
Component:375; Jaccard:0.096849
 |
Component:390; Jaccard:0.089001
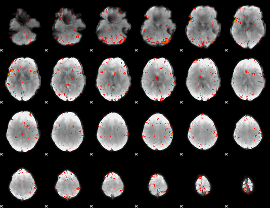 |
Component:390; Jaccard:0.078033
 |
Correlation:0.35788
  |
Correlation:0.34583
  |
Correlation:0.17632
  |
Correlation:0.17632
  |
Correlation:0.088852
  |
Correlation:0.32228
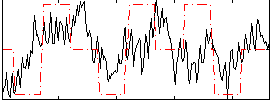 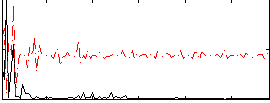 |
Correlation:0.32228
  |
Cope: 04:REL-MATCH BACK
|
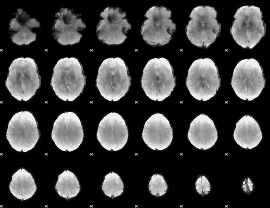
GLM result group-wise
 |
GLM binary result thresholded by 1
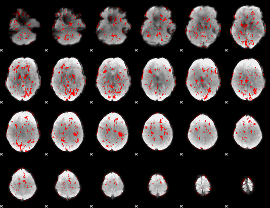 |
GLM binary result thresholded by 1.5
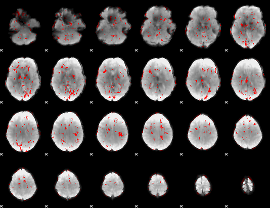 |
GLM binary result thresholded by 2
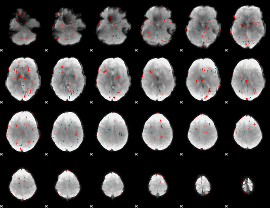 |
GLM binary result thresholded by 2.5
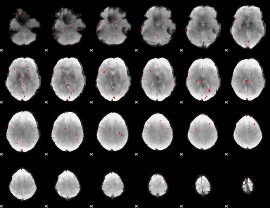 |
GLM binary result thresholded by 3
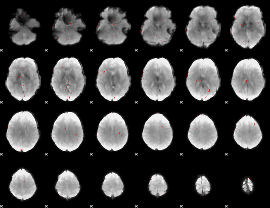 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
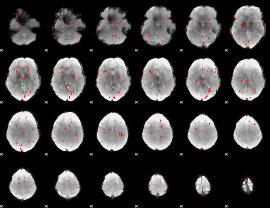 |
GLM Z-score result thresholded by 2.5
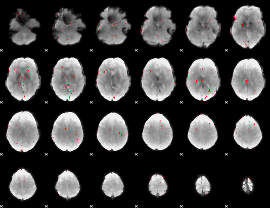 |
GLM Z-score result thresholded by 3
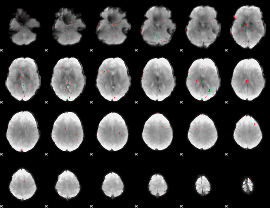 |
GLM Z-score result thresholded by 3.5
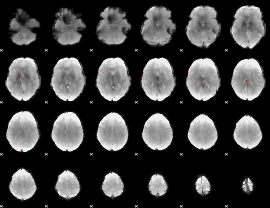 |
GLM Z-score result thresholded by 4
 |
Component:292; Jaccard:0.097925
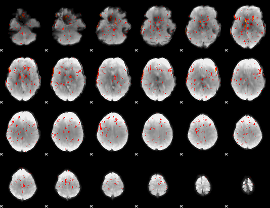 |
Component:292; Jaccard:0.10271
 |
Component:292; Jaccard:0.090072
 |
Component:292; Jaccard:0.069288
 |
Component:292; Jaccard:0.037568
 |
Component:292; Jaccard:0.01848
 |
Component:292; Jaccard:0.007411
 |
Correlation:0.20975
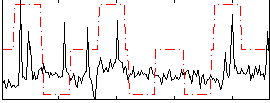 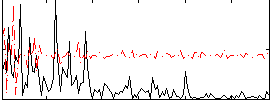 |
Correlation:0.20975
  |
Correlation:0.20975
  |
Correlation:0.20975
  |
Correlation:0.20975
  |
Correlation:0.20975
  |
Correlation:0.20975
  |
Component:342; Jaccard:0.074199
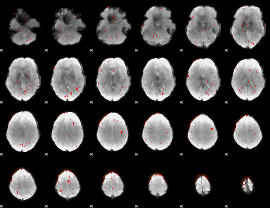 |
Component:342; Jaccard:0.077102
 |
Component:199; Jaccard:0.071792
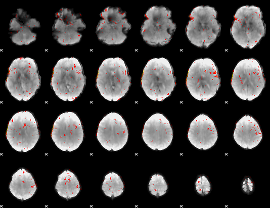 |
Component:199; Jaccard:0.05337
 |
Component:055; Jaccard:0.025253
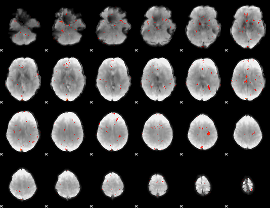 |
Component:055; Jaccard:0.014383
 |
Component:055; Jaccard:0.005742
 |
Correlation:0.054931
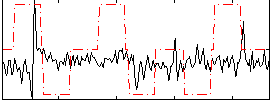  |
Correlation:0.054931
  |
Correlation:0.047285
  |
Correlation:0.047285
  |
Correlation:0.52459
  |
Correlation:0.52459
  |
Correlation:0.52459
  |
Component:298; Jaccard:0.071084
 |
Component:199; Jaccard:0.075546
 |
Component:342; Jaccard:0.068109
 |
Component:342; Jaccard:0.045529
 |
Component:199; Jaccard:0.024711
 |
Component:199; Jaccard:0.010395
 |
Component:199; Jaccard:0.005294
 |
Correlation:0.063328
  |
Correlation:0.047285
  |
Correlation:0.054931
  |
Correlation:0.054931
  |
Correlation:0.047285
  |
Correlation:0.047285
  |
Correlation:0.047285
  |
Component:199; Jaccard:0.070971
 |
Component:315; Jaccard:0.063399
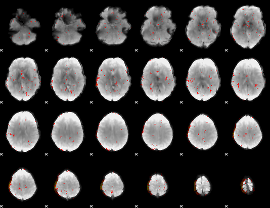 |
Component:315; Jaccard:0.055586
 |
Component:055; Jaccard:0.038583
 |
Component:342; Jaccard:0.022588
 |
Component:342; Jaccard:0.008741
 |
Component:051; Jaccard:0.003341
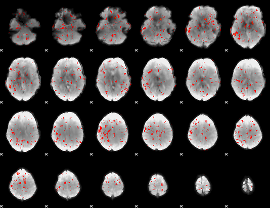 |
Correlation:0.047285
  |
Correlation:0.21268
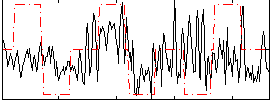 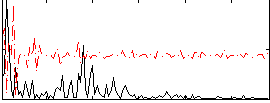 |
Correlation:0.21268
  |
Correlation:0.52459
  |
Correlation:0.054931
  |
Correlation:0.054931
  |
Correlation:0.10978
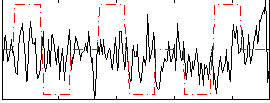  |
Component:051; Jaccard:0.068952
 |
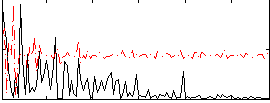
Component:112; Jaccard:0.062645
 |
Component:051; Jaccard:0.048643
 |
Component:315; Jaccard:0.036204
 |
Component:315; Jaccard:0.018737
 |
Component:051; Jaccard:0.00828
 |
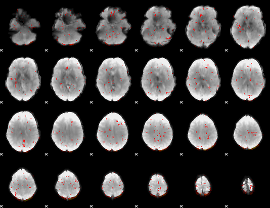
Component:247; Jaccard:0.002945
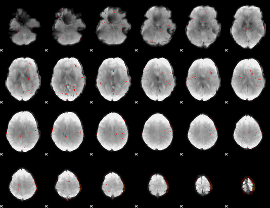 |
Correlation:0.10978
  |
Correlation:0.3921
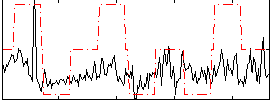 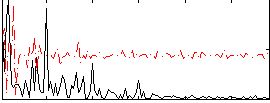 |
Correlation:0.10978
  |
Correlation:0.21268
  |
Correlation:0.21268
  |
Correlation:0.10978
  |
Correlation:0.25135
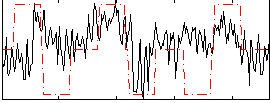 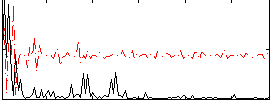 |
Component:112; Jaccard:0.064646
 |
Component:298; Jaccard:0.061222
 |
Component:112; Jaccard:0.048042
 |
Component:300; Jaccard:0.032228
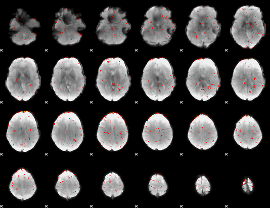 |
Component:051; Jaccard:0.017621
 |
Component:020; Jaccard:0.007779
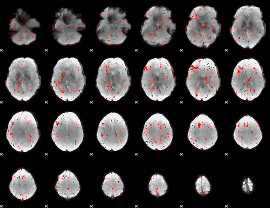 |
Component:381; Jaccard:0.002928
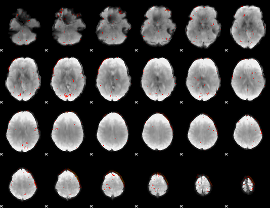 |
Correlation:0.3921
  |
Correlation:0.063328
  |
Correlation:0.3921
  |
Correlation:0.15763
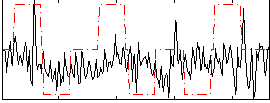 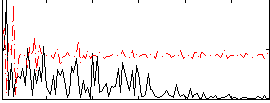 |
Correlation:0.10978
  |
Correlation:0.10249
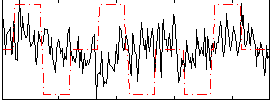 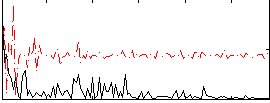 |
Correlation:-0.088707
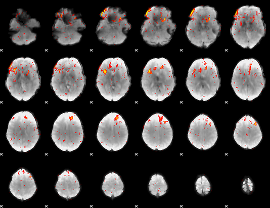
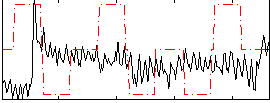 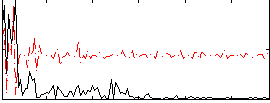 |
Component:121; Jaccard:0.063464
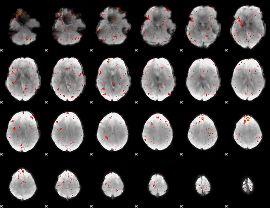
 |
Component:398; Jaccard:0.05939
 |
Component:300; Jaccard:0.046481
 |
Component:051; Jaccard:0.032124
 |
Component:271; Jaccard:0.016712
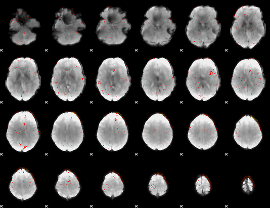 |
Component:300; Jaccard:0.007607
 |
Component:020; Jaccard:0.002781
 |
Correlation:0.3346
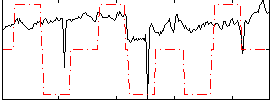 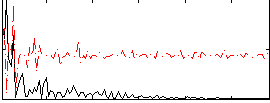 |
Correlation:-0.071442
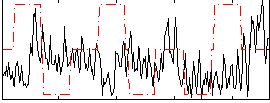 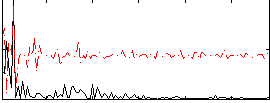 |
Correlation:0.15763
  |
Correlation:0.10978
  |
Correlation:0.068992
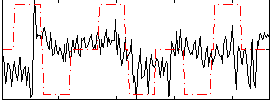 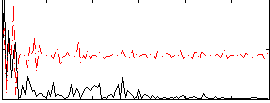 |
Correlation:0.15763
  |
Correlation:0.10249
  |
Component:398; Jaccard:0.063363
 |
Component:051; Jaccard:0.058774
 |
Component:398; Jaccard:0.045367
 |
Component:271; Jaccard:0.031301
 |
Component:300; Jaccard:0.015722
 |
Component:271; Jaccard:0.00721
 |
Component:342; Jaccard:0.002533
 |
Correlation:-0.071442
  |
Correlation:0.10978
  |
Correlation:-0.071442
  |
Correlation:0.068992
  |
Correlation:0.15763
  |
Correlation:0.068992
  |
Correlation:0.054931
  |
Component:315; Jaccard:0.062
 |
Component:258; Jaccard:0.055359
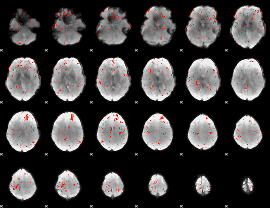 |
Component:121; Jaccard:0.044824
 |
Component:112; Jaccard:0.029253
 |
Component:247; Jaccard:0.014241
 |
Component:247; Jaccard:0.006369
 |
Component:300; Jaccard:0.002506
 |
Correlation:0.21268
  |
Correlation:0.31115
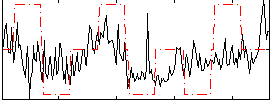  |
Correlation:0.3346
  |
Correlation:0.3921
  |
Correlation:0.25135
  |
Correlation:0.25135
  |
Correlation:0.15763
  |
Component:258; Jaccard:0.05986
 |
Component:300; Jaccard:0.055036
 |
Component:055; Jaccard:0.044502
 |
Component:121; Jaccard:0.029031
 |
Component:381; Jaccard:0.014146
 |
Component:381; Jaccard:0.006209
 |
Component:271; Jaccard:0.002506
 |
Correlation:0.31115
  |
Correlation:0.15763
  |
Correlation:0.52459
  |
Correlation:0.3346
  |
Correlation:-0.088707
  |
Correlation:-0.088707
  |
Correlation:0.068992
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
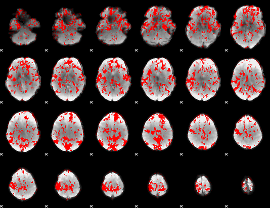 |
GLM binary result thresholded by 1.5
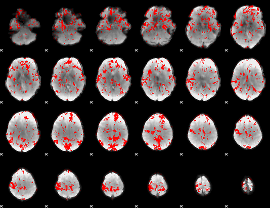 |
GLM binary result thresholded by 2
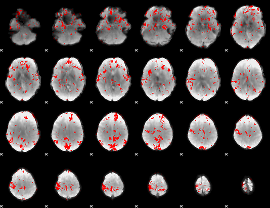 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
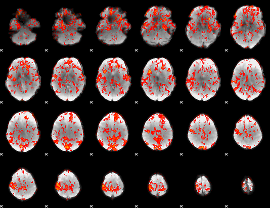 |
GLM Z-score result thresholded by 1.5
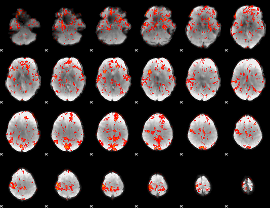 |
GLM Z-score result thresholded by 2
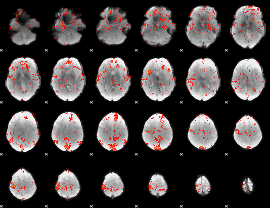 |
GLM Z-score result thresholded by 2.5
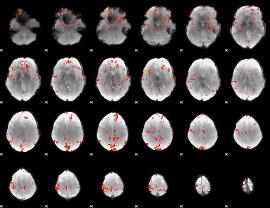 |
GLM Z-score result thresholded by 3
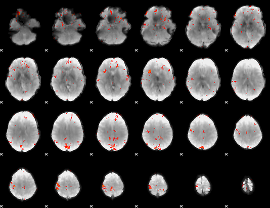 |
GLM Z-score result thresholded by 3.5
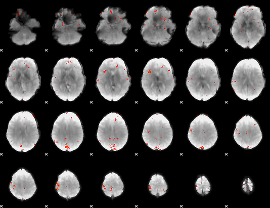 |
GLM Z-score result thresholded by 4
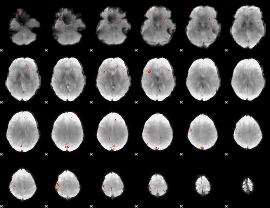 |
Component:212; Jaccard:0.1175
 |
Component:212; Jaccard:0.12876
 |
Component:212; Jaccard:0.12868
 |
Component:212; Jaccard:0.11286
 |
Component:291; Jaccard:0.092226
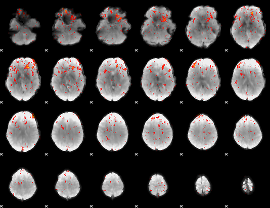 |
Component:291; Jaccard:0.066501
 |
Component:291; Jaccard:0.042708
 |
Correlation:0.037291
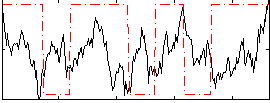 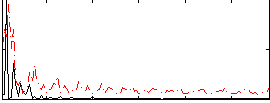 |
Correlation:0.037291
  |
Correlation:0.037291
  |
Correlation:0.037291
  |
Correlation:0.3025
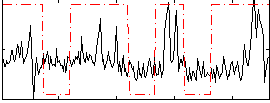 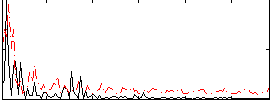 |
Correlation:0.3025
  |
Correlation:0.3025
  |
Component:375; Jaccard:0.1143
 |
Component:375; Jaccard:0.11628
 |
Component:291; Jaccard:0.10823
 |
Component:291; Jaccard:0.10807
 |
Component:212; Jaccard:0.082008
 |
Component:108; Jaccard:0.056561
 |
Component:121; Jaccard:0.035383
 |
Correlation:0.074567
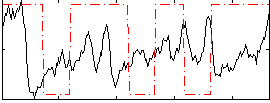 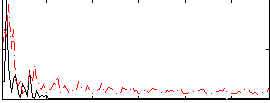 |
Correlation:0.074567
  |
Correlation:0.3025
  |
Correlation:0.3025
  |
Correlation:0.037291
  |
Correlation:-0.07673
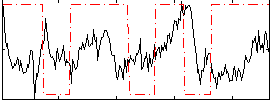 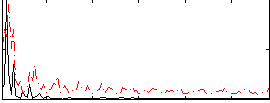 |
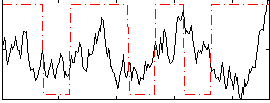
Correlation:0.40697
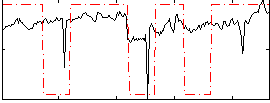  |
Component:365; Jaccard:0.095519
 |
Component:365; Jaccard:0.10274
 |
Component:375; Jaccard:0.10327
 |
Component:108; Jaccard:0.098627
 |
Component:108; Jaccard:0.078529
 |
Component:212; Jaccard:0.055419
 |
Component:108; Jaccard:0.033431
 |
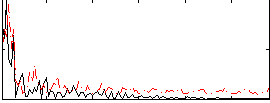
Correlation:0.097399
 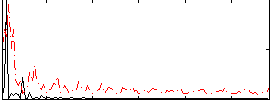 |
Correlation:0.097399
  |
Correlation:0.074567
  |
Correlation:-0.07673
  |
Correlation:-0.07673
  |
Correlation:0.037291
  |
Correlation:-0.07673
  |
Component:387; Jaccard:0.090298
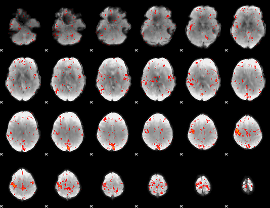 |
Component:291; Jaccard:0.1005
 |
Component:108; Jaccard:0.10299
 |
Component:121; Jaccard:0.088329
 |
Component:121; Jaccard:0.072305
 |
Component:121; Jaccard:0.053617
 |
Component:398; Jaccard:0.031186
 |
Correlation:-0.0095994
  |
Correlation:0.3025
  |
Correlation:-0.07673
  |
Correlation:0.40697
  |
Correlation:0.40697
  |
Correlation:0.40697
  |
Correlation:0.09489
 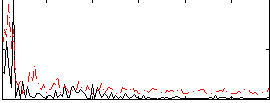 |
Component:106; Jaccard:0.090253
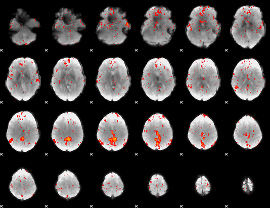 |
Component:108; Jaccard:0.099692
 |
Component:365; Jaccard:0.10174
 |
Component:365; Jaccard:0.085348
 |
Component:365; Jaccard:0.062579
 |
Component:398; Jaccard:0.044617
 |
Component:212; Jaccard:0.030194
 |
Correlation:0.051035
  |
Correlation:-0.07673
  |
Correlation:0.097399
  |
Correlation:0.097399
  |
Correlation:0.097399
  |
Correlation:0.09489
  |
Correlation:0.037291
  |
Component:291; Jaccard:0.087556
 |
Component:106; Jaccard:0.094016
 |
Component:121; Jaccard:0.093582
 |
Component:375; Jaccard:0.078382
 |
Component:387; Jaccard:0.059669
 |
Component:387; Jaccard:0.042479
 |
Component:226; Jaccard:0.027485
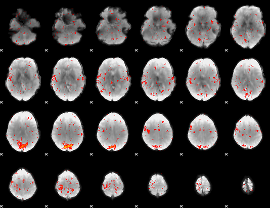 |
Correlation:0.3025
  |
Correlation:0.051035
  |
Correlation:0.40697
  |
Correlation:0.074567
  |
Correlation:-0.0095994
  |
Correlation:-0.0095994
  |
Correlation:-0.041426
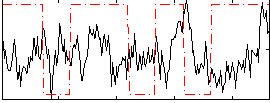 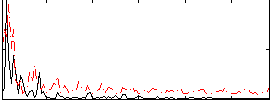 |
Component:108; Jaccard:0.087464
 |
Component:387; Jaccard:0.093217
 |
Component:335; Jaccard:0.089158
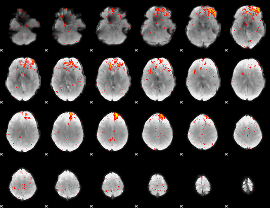 |
Component:387; Jaccard:0.076825
 |
Component:258; Jaccard:0.057297
 |
Component:226; Jaccard:0.041396
 |
Component:270; Jaccard:0.026549
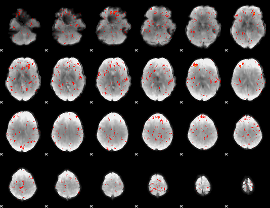 |
Correlation:-0.07673
  |
Correlation:-0.0095994
  |
Correlation:0.26789
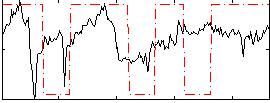 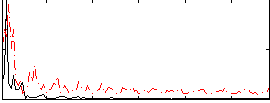 |
Correlation:-0.0095994
  |
Correlation:0.32381
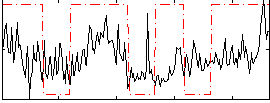  |
Correlation:-0.041426
  |
Correlation:0.21261
  |
Component:335; Jaccard:0.084638
 |
Component:335; Jaccard:0.090693
 |
Component:387; Jaccard:0.088386
 |
Component:335; Jaccard:0.076172
 |
Component:226; Jaccard:0.056809
 |
Component:270; Jaccard:0.04063
 |
Component:387; Jaccard:0.024128
 |
Correlation:0.26789
  |
Correlation:0.26789
  |
Correlation:-0.0095994
  |
Correlation:0.26789
  |
Correlation:-0.041426
  |
Correlation:0.21261
  |
Correlation:-0.0095994
  |
Component:121; Jaccard:0.080206
 |
Component:121; Jaccard:0.089825
 |
Component:106; Jaccard:0.085929
 |
Component:258; Jaccard:0.073087
 |
Component:398; Jaccard:0.052905
 |
Component:365; Jaccard:0.03938
 |
Component:258; Jaccard:0.022717
 |
Correlation:0.40697
  |
Correlation:0.40697
  |
Correlation:0.051035
  |
Correlation:0.32381
  |
Correlation:0.09489
  |
Correlation:0.097399
  |
Correlation:0.32381
  |
Component:231; Jaccard:0.078171
 |
Component:258; Jaccard:0.080841
 |
Component:258; Jaccard:0.080919
 |
Component:226; Jaccard:0.069955
 |
Component:270; Jaccard:0.052842
 |
Component:258; Jaccard:0.03715
 |
Component:365; Jaccard:0.021623
 |
Correlation:0.144
  |
Correlation:0.32381
  |
Correlation:0.32381
  |
Correlation:-0.041426
  |
Correlation:0.21261
  |
Correlation:0.32381
  |
Correlation:0.097399
  |
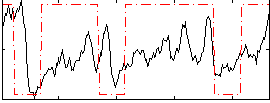
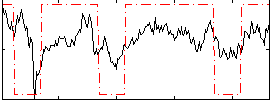
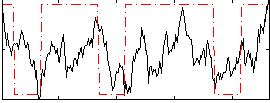
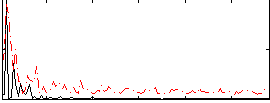
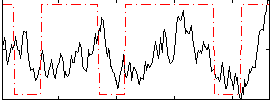
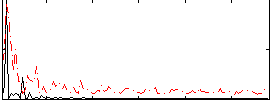
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
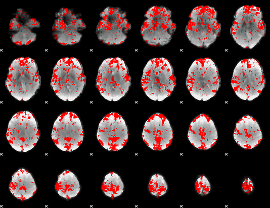 |
GLM binary result thresholded by 2.5
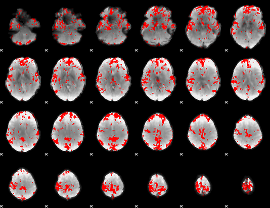 |
GLM binary result thresholded by 3
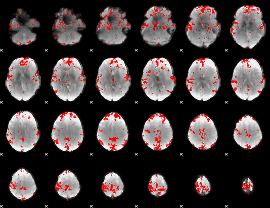 |
GLM binary result thresholded by 3.5
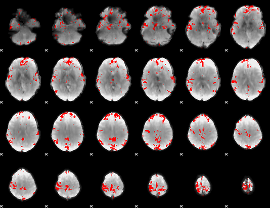 |
GLM binary result thresholded by 4
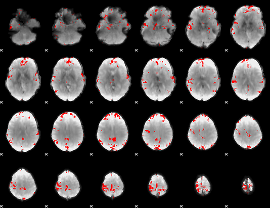 |
GLM Z-score result thresholded by 1
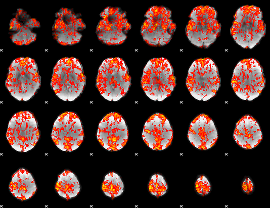 |
GLM Z-score result thresholded by 1.5
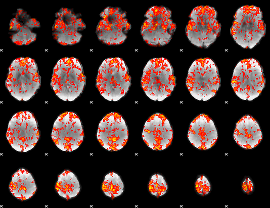 |
GLM Z-score result thresholded by 2
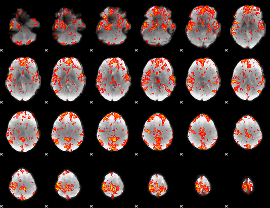 |
GLM Z-score result thresholded by 2.5
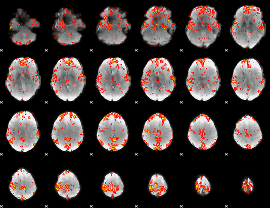 |
GLM Z-score result thresholded by 3
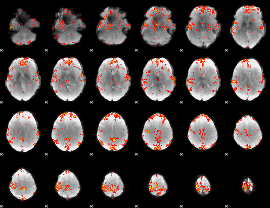 |
GLM Z-score result thresholded by 3.5
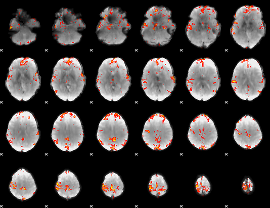 |
GLM Z-score result thresholded by 4
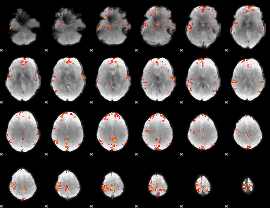 |
Component:375; Jaccard:0.12278
 |
Component:375; Jaccard:0.14685
 |
Component:375; Jaccard:0.17446
 |
Component:375; Jaccard:0.19936
 |
Component:375; Jaccard:0.21463
 |
Component:375; Jaccard:0.21536
 |
Component:375; Jaccard:0.18777
 |
Correlation:0.22448
 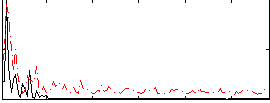 |
Correlation:0.22448
  |
Correlation:0.22448
  |
Correlation:0.22448
  |
Correlation:0.22448
  |
Correlation:0.22448
  |
Correlation:0.22448
  |
Component:201; Jaccard:0.1184
 |
Component:201; Jaccard:0.13549
 |
Component:201; Jaccard:0.15295
 |
Component:201; Jaccard:0.16579
 |
Component:212; Jaccard:0.17269
 |
Component:212; Jaccard:0.17351
 |
Component:212; Jaccard:0.1621
 |
Correlation:0.45667
 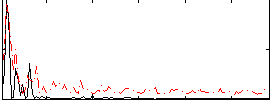 |
Correlation:0.45667
  |
Correlation:0.45667
  |
Correlation:0.45667
  |
Correlation:0.29766
  |
Correlation:0.29766
  |
Correlation:0.29766
  |
Component:212; Jaccard:0.10245
 |
Component:212; Jaccard:0.12075
 |
Component:212; Jaccard:0.14019
 |
Component:212; Jaccard:0.15851
 |
Component:201; Jaccard:0.16861
 |
Component:365; Jaccard:0.16355
 |
Component:365; Jaccard:0.1543
 |
Correlation:0.29766
  |
Correlation:0.29766
  |
Correlation:0.29766
  |
Correlation:0.29766
  |
Correlation:0.45667
  |
Correlation:0.21784
  |
Correlation:0.21784
  |
Component:207; Jaccard:0.096528
 |
Component:365; Jaccard:0.11198
 |
Component:365; Jaccard:0.13169
 |
Component:365; Jaccard:0.15016
 |
Component:365; Jaccard:0.16317
 |
Component:201; Jaccard:0.15955
 |
Component:201; Jaccard:0.13439
 |
Correlation:0.46777
  |
Correlation:0.21784
  |
Correlation:0.21784
  |
Correlation:0.21784
  |
Correlation:0.21784
  |
Correlation:0.45667
  |
Correlation:0.45667
  |
Component:365; Jaccard:0.094446
 |
Component:207; Jaccard:0.1076
 |
Component:207; Jaccard:0.11712
 |
Component:106; Jaccard:0.12576
 |
Component:106; Jaccard:0.1311
 |
Component:106; Jaccard:0.12596
 |
Component:106; Jaccard:0.11167
 |
Correlation:0.21784
  |
Correlation:0.46777
  |
Correlation:0.46777
  |
Correlation:0.18003
  |
Correlation:0.18003
  |
Correlation:0.18003
  |
Correlation:0.18003
  |
Component:276; Jaccard:0.091856
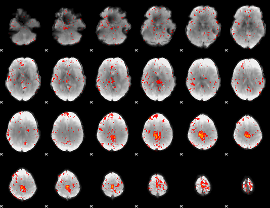 |
Component:276; Jaccard:0.10204
 |
Component:106; Jaccard:0.11499
 |
Component:207; Jaccard:0.12371
 |
Component:207; Jaccard:0.12682
 |
Component:207; Jaccard:0.12452
 |
Component:207; Jaccard:0.10958
 |
Correlation:0.36329
  |
Correlation:0.36329
  |
Correlation:0.18003
  |
Correlation:0.46777
  |
Correlation:0.46777
  |
Correlation:0.46777
  |
Correlation:0.46777
  |
Component:182; Jaccard:0.089694
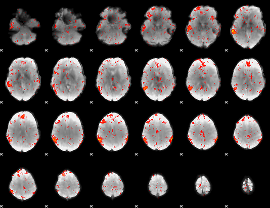 |
Component:106; Jaccard:0.10097
 |
Component:276; Jaccard:0.11124
 |
Component:276; Jaccard:0.11658
 |
Component:108; Jaccard:0.11678
 |
Component:108; Jaccard:0.11618
 |
Component:108; Jaccard:0.10787
 |
Correlation:0.3697
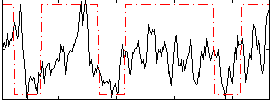 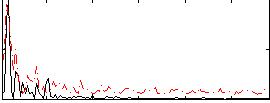 |
Correlation:0.18003
  |
Correlation:0.36329
  |
Correlation:0.36329
  |
Correlation:0.40432
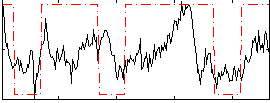 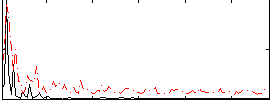 |
Correlation:0.40432
  |
Correlation:0.40432
  |
Component:106; Jaccard:0.087678
 |
Component:099; Jaccard:0.096576
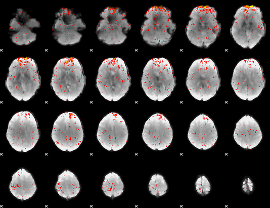 |
Component:099; Jaccard:0.10713
 |
Component:099; Jaccard:0.11439
 |
Component:099; Jaccard:0.11378
 |
Component:099; Jaccard:0.10883
 |
Component:099; Jaccard:0.095646
 |
Correlation:0.18003
  |
Correlation:0.13653
  |
Correlation:0.13653
  |
Correlation:0.13653
  |
Correlation:0.13653
  |
Correlation:0.13653
  |
Correlation:0.13653
  |
Component:115; Jaccard:0.086407
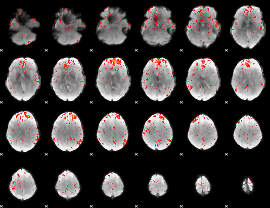 |
Component:182; Jaccard:0.096292
 |
Component:182; Jaccard:0.10047
 |
Component:108; Jaccard:0.11019
 |
Component:276; Jaccard:0.1134
 |
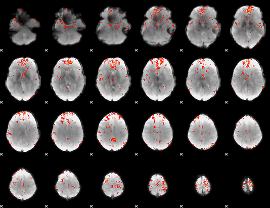
Component:348; Jaccard:0.10558
 |
Component:348; Jaccard:0.091527
 |
Correlation:0.14636
  |
Correlation:0.3697
  |
Correlation:0.3697
  |
Correlation:0.40432
  |
Correlation:0.36329
  |
Correlation:-0.010666
  |
Correlation:-0.010666
  |
Component:099; Jaccard:0.085112
 |
Component:115; Jaccard:0.093634
 |
Component:108; Jaccard:0.10014
 |
Component:115; Jaccard:0.10462
 |
Component:348; Jaccard:0.10726
 |
Component:276; Jaccard:0.10336
 |
Component:276; Jaccard:0.087606
 |
Correlation:0.13653
  |
Correlation:0.14636
  |
Correlation:0.40432
  |
Correlation:0.14636
  |
Correlation:-0.010666
  |
Correlation:0.36329
  |
Correlation:0.36329
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































