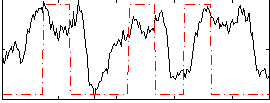
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
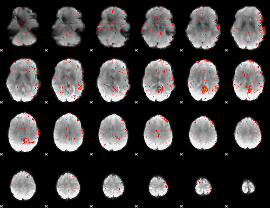
GLM result group-wise
 |
GLM binary result thresholded by 1
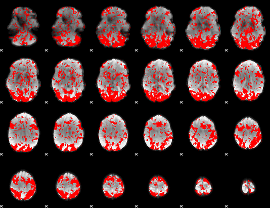 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
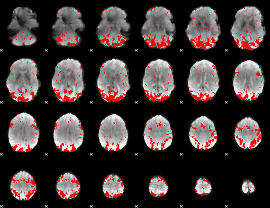 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
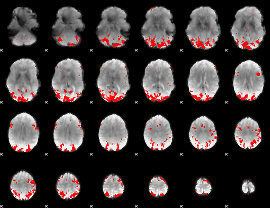 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
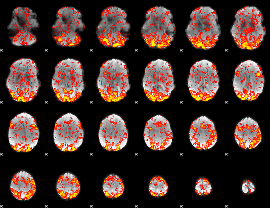 |
GLM Z-score result thresholded by 1.5
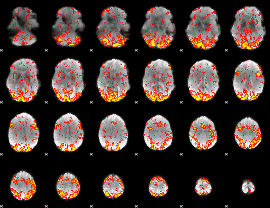 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
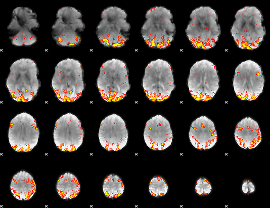 |
GLM Z-score result thresholded by 3.5
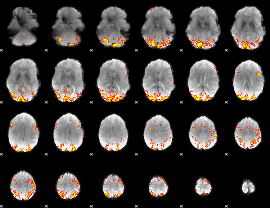 |
GLM Z-score result thresholded by 4
 |
Component:151; Jaccard:0.1561
 |
Component:305; Jaccard:0.1952
 |
Component:305; Jaccard:0.24635
 |
Component:305; Jaccard:0.29557
 |
Component:305; Jaccard:0.33309
 |
Component:305; Jaccard:0.35663
 |
Component:305; Jaccard:0.36322
 |
Correlation:0.21978
  |
Correlation:0.28085
  |
Correlation:0.28085
  |
Correlation:0.28085
  |
Correlation:0.28085
  |
Correlation:0.28085
  |
Correlation:0.28085
  |
Component:275; Jaccard:0.15108
 |
Component:280; Jaccard:0.19019
 |
Component:275; Jaccard:0.22954
 |
Component:275; Jaccard:0.2644
 |
Component:275; Jaccard:0.28675
 |
Component:275; Jaccard:0.29626
 |
Component:275; Jaccard:0.29223
 |
Correlation:0.24167
 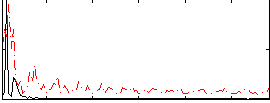 |
Correlation:0.18524
  |
Correlation:0.24167
  |
Correlation:0.24167
  |
Correlation:0.24167
  |
Correlation:0.24167
  |
Correlation:0.24167
  |
Component:280; Jaccard:0.14935
 |
Component:275; Jaccard:0.18963
 |
Component:280; Jaccard:0.22875
 |
Component:280; Jaccard:0.26122
 |
Component:280; Jaccard:0.28283
 |
Component:280; Jaccard:0.28833
 |
Component:264; Jaccard:0.28441
 |
Correlation:0.18524
  |
Correlation:0.24167
  |
Correlation:0.18524
  |
Correlation:0.18524
  |
Correlation:0.18524
  |
Correlation:0.18524
  |
Correlation:0.1702
  |
Component:297; Jaccard:0.14913
 |
Component:297; Jaccard:0.18539
 |
Component:297; Jaccard:0.21922
 |
Component:264; Jaccard:0.248
 |
Component:264; Jaccard:0.27246
 |
Component:264; Jaccard:0.28457
 |
Component:280; Jaccard:0.28341
 |
Correlation:0.14109
  |
Correlation:0.14109
  |
Correlation:0.14109
  |
Correlation:0.1702
  |
Correlation:0.1702
  |
Correlation:0.1702
  |
Correlation:0.18524
  |
Component:305; Jaccard:0.14912
 |
Component:151; Jaccard:0.18226
 |
Component:264; Jaccard:0.21037
 |
Component:297; Jaccard:0.24129
 |
Component:297; Jaccard:0.24782
 |
Component:297; Jaccard:0.23836
 |
Component:297; Jaccard:0.22228
 |
Correlation:0.28085
  |
Correlation:0.21978
  |
Correlation:0.1702
  |
Correlation:0.14109
  |
Correlation:0.14109
  |
Correlation:0.14109
  |
Correlation:0.14109
  |
Component:264; Jaccard:0.12973
 |
Component:264; Jaccard:0.16789
 |
Component:151; Jaccard:0.20466
 |
Component:151; Jaccard:0.21767
 |
Component:151; Jaccard:0.21902
 |
Component:257; Jaccard:0.22075
 |
Component:257; Jaccard:0.21905
 |
Correlation:0.1702
  |
Correlation:0.1702
  |
Correlation:0.21978
  |
Correlation:0.21978
  |
Correlation:0.21978
  |
Correlation:0.26023
  |
Correlation:0.26023
  |
Component:346; Jaccard:0.12531
 |
Component:346; Jaccard:0.1503
 |
Component:257; Jaccard:0.17652
 |
Component:257; Jaccard:0.19914
 |
Component:257; Jaccard:0.21487
 |
Component:151; Jaccard:0.21148
 |
Component:151; Jaccard:0.19568
 |
Correlation:0.3334
  |
Correlation:0.3334
  |
Correlation:0.26023
  |
Correlation:0.26023
  |
Correlation:0.26023
  |
Correlation:0.21978
  |
Correlation:0.21978
  |
Component:257; Jaccard:0.12025
 |
Component:257; Jaccard:0.14878
 |
Component:346; Jaccard:0.17102
 |
Component:346; Jaccard:0.1833
 |
Component:346; Jaccard:0.18658
 |
Component:346; Jaccard:0.17894
 |
Component:346; Jaccard:0.16706
 |
Correlation:0.26023
  |
Correlation:0.26023
  |
Correlation:0.3334
  |
Correlation:0.3334
  |
Correlation:0.3334
  |
Correlation:0.3334
  |
Correlation:0.3334
  |
Component:291; Jaccard:0.1163
 |
Component:291; Jaccard:0.12963
 |
Component:393; Jaccard:0.14034
 |
Component:393; Jaccard:0.14641
 |
Component:241; Jaccard:0.14668
 |
Component:241; Jaccard:0.14357
 |
Component:241; Jaccard:0.13682
 |
Correlation:0.15739
  |
Correlation:0.15739
  |
Correlation:0.30242
 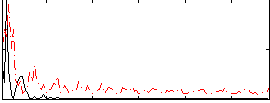 |
Correlation:0.30242
  |
Correlation:0.17435
  |
Correlation:0.17435
  |
Correlation:0.17435
  |
Component:393; Jaccard:0.1129
 |
Component:393; Jaccard:0.12872
 |
Component:291; Jaccard:0.1376
 |
Component:241; Jaccard:0.13949
 |
Component:393; Jaccard:0.14202
 |
Component:393; Jaccard:0.13333
 |
Component:393; Jaccard:0.12313
 |
Correlation:0.30242
  |
Correlation:0.30242
  |
Correlation:0.15739
  |
Correlation:0.17435
  |
Correlation:0.30242
  |
Correlation:0.30242
  |
Correlation:0.30242
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
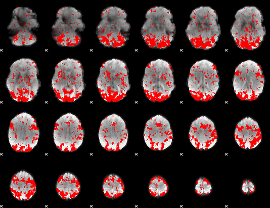 |
GLM binary result thresholded by 2
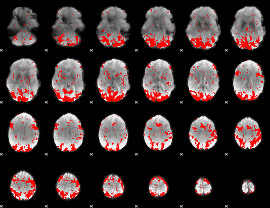 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
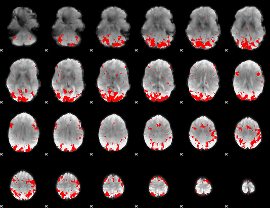 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
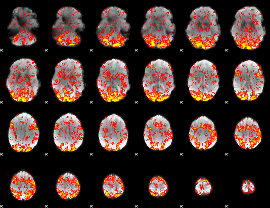 |
GLM Z-score result thresholded by 1.5
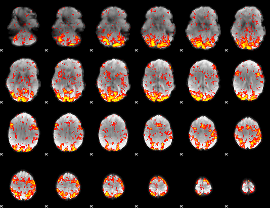 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
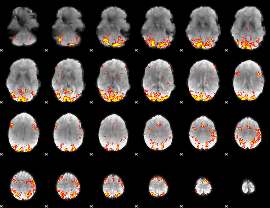 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:305; Jaccard:0.15936
 |
Component:305; Jaccard:0.21257
 |
Component:305; Jaccard:0.2714
 |
Component:305; Jaccard:0.32629
 |
Component:305; Jaccard:0.36793
 |
Component:305; Jaccard:0.39115
 |
Component:305; Jaccard:0.39652
 |
Correlation:0.28678
 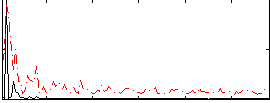 |
Correlation:0.28678
  |
Correlation:0.28678
  |
Correlation:0.28678
  |
Correlation:0.28678
  |
Correlation:0.28678
  |
Correlation:0.28678
  |
Component:275; Jaccard:0.15564
 |
Component:275; Jaccard:0.1944
 |
Component:275; Jaccard:0.23452
 |
Component:275; Jaccard:0.26528
 |
Component:275; Jaccard:0.28438
 |
Component:275; Jaccard:0.29173
 |
Component:264; Jaccard:0.28723
 |
Correlation:0.041772
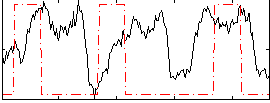 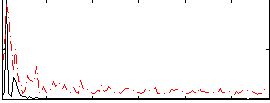 |
Correlation:0.041772
  |
Correlation:0.041772
  |
Correlation:0.041772
  |
Correlation:0.041772
  |
Correlation:0.041772
  |
Correlation:0.31833
  |
Component:297; Jaccard:0.15519
 |
Component:297; Jaccard:0.19228
 |
Component:297; Jaccard:0.22319
 |
Component:264; Jaccard:0.25137
 |
Component:264; Jaccard:0.27675
 |
Component:264; Jaccard:0.2878
 |
Component:275; Jaccard:0.28356
 |
Correlation:0.17181
 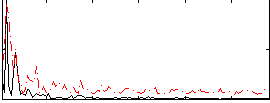 |
Correlation:0.17181
  |
Correlation:0.17181
  |
Correlation:0.31833
  |
Correlation:0.31833
  |
Correlation:0.31833
  |
Correlation:0.041772
  |
Component:151; Jaccard:0.14562
 |
Component:280; Jaccard:0.17769
 |
Component:264; Jaccard:0.21584
 |
Component:297; Jaccard:0.24387
 |
Component:280; Jaccard:0.25364
 |
Component:280; Jaccard:0.25436
 |
Component:280; Jaccard:0.24569
 |
Correlation:-0.046114
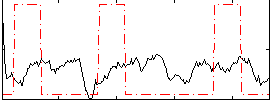  |
Correlation:0.046923
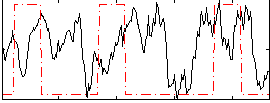  |
Correlation:0.31833
  |
Correlation:0.17181
  |
Correlation:0.046923
  |
Correlation:0.046923
  |
Correlation:0.046923
  |
Component:280; Jaccard:0.14415
 |
Component:264; Jaccard:0.17342
 |
Component:280; Jaccard:0.21204
 |
Component:280; Jaccard:0.23781
 |
Component:297; Jaccard:0.24829
 |
Component:297; Jaccard:0.23993
 |
Component:297; Jaccard:0.22065
 |
Correlation:0.046923
  |
Correlation:0.31833
  |
Correlation:0.046923
  |
Correlation:0.046923
  |
Correlation:0.17181
  |
Correlation:0.17181
  |
Correlation:0.17181
  |
Component:264; Jaccard:0.13386
 |
Component:151; Jaccard:0.16789
 |
Component:151; Jaccard:0.1875
 |
Component:257; Jaccard:0.20461
 |
Component:257; Jaccard:0.21774
 |
Component:257; Jaccard:0.21808
 |
Component:257; Jaccard:0.2085
 |
Correlation:0.31833
  |
Correlation:-0.046114
  |
Correlation:-0.046114
  |
Correlation:0.098612
  |
Correlation:0.098612
  |
Correlation:0.098612
  |
Correlation:0.098612
  |
Component:291; Jaccard:0.12548
 |
Component:257; Jaccard:0.15274
 |
Component:257; Jaccard:0.18277
 |
Component:151; Jaccard:0.19903
 |
Component:151; Jaccard:0.20027
 |
Component:151; Jaccard:0.18836
 |
Component:151; Jaccard:0.1711
 |
Correlation:-0.062846
  |
Correlation:0.098612
  |
Correlation:0.098612
  |
Correlation:-0.046114
  |
Correlation:-0.046114
  |
Correlation:-0.046114
  |
Correlation:-0.046114
  |
Component:257; Jaccard:0.12337
 |
Component:291; Jaccard:0.14028
 |
Component:291; Jaccard:0.15004
 |
Component:346; Jaccard:0.15978
 |
Component:346; Jaccard:0.16121
 |
Component:241; Jaccard:0.15734
 |
Component:241; Jaccard:0.15035
 |
Correlation:0.098612
  |
Correlation:-0.062846
  |
Correlation:-0.062846
  |
Correlation:-0.14614
  |
Correlation:-0.14614
  |
Correlation:0.3207
  |
Correlation:0.3207
  |
Component:346; Jaccard:0.11234
 |
Component:346; Jaccard:0.13181
 |
Component:346; Jaccard:0.14911
 |
Component:241; Jaccard:0.15335
 |
Component:241; Jaccard:0.15905
 |
Component:346; Jaccard:0.15422
 |
Component:346; Jaccard:0.14369
 |
Correlation:-0.14614
  |
Correlation:-0.14614
  |
Correlation:-0.14614
  |
Correlation:0.3207
  |
Correlation:0.3207
  |
Correlation:-0.14614
  |
Correlation:-0.14614
  |
Component:092; Jaccard:0.10904
 |
Component:092; Jaccard:0.12316
 |
Component:241; Jaccard:0.14089
 |
Component:291; Jaccard:0.15033
 |
Component:291; Jaccard:0.14183
 |
Component:291; Jaccard:0.13123
 |
Component:291; Jaccard:0.11513
 |
Correlation:-0.17168
  |
Correlation:-0.17168
  |
Correlation:0.3207
  |
Correlation:-0.062846
  |
Correlation:-0.062846
  |
Correlation:-0.062846
  |
Correlation:-0.062846
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
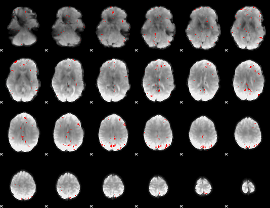 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:072; Jaccard:0.16268
 |
Component:072; Jaccard:0.2052
 |
Component:072; Jaccard:0.21749
 |
Component:072; Jaccard:0.17389
 |
Component:072; Jaccard:0.10079
 |
Component:072; Jaccard:0.047611
 |
Component:072; Jaccard:0.020381
 |
Correlation:0.4124
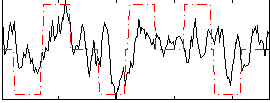  |
Correlation:0.4124
  |
Correlation:0.4124
  |
Correlation:0.4124
  |
Correlation:0.4124
  |
Correlation:0.4124
  |
Correlation:0.4124
  |
Component:059; Jaccard:0.087269
 |
Component:165; Jaccard:0.089071
 |
Component:165; Jaccard:0.081912
 |
Component:165; Jaccard:0.055919
 |
Component:165; Jaccard:0.029755
 |
Component:191; Jaccard:0.014618
 |
Component:191; Jaccard:0.007796
 |
Correlation:0.28544
 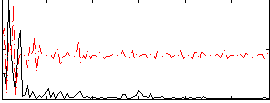 |
Correlation:0.16723
  |
Correlation:0.16723
  |
Correlation:0.16723
  |
Correlation:0.16723
  |
Correlation:-0.022011
  |
Correlation:-0.022011
  |
Component:165; Jaccard:0.084172
 |
Component:059; Jaccard:0.08478
 |
Component:059; Jaccard:0.070776
 |
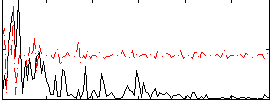
Component:259; Jaccard:0.049829
 |
Component:259; Jaccard:0.029315
 |
Component:259; Jaccard:0.013815
 |
Component:259; Jaccard:0.006613
 |
Correlation:0.16723
  |
Correlation:0.28544
  |
Correlation:0.28544
  |
Correlation:0.13342
  |
Correlation:0.13342
  |
Correlation:0.13342
  |
Correlation:0.13342
  |
Component:144; Jaccard:0.070065
 |
Component:259; Jaccard:0.070598
 |
Component:259; Jaccard:0.067855
 |
Component:379; Jaccard:0.046558
 |
Component:191; Jaccard:0.026433
 |
Component:165; Jaccard:0.012955
 |
Component:099; Jaccard:0.006473
 |
Correlation:-0.06319
  |
Correlation:0.13342
  |
Correlation:0.13342
  |
Correlation:-0.095407
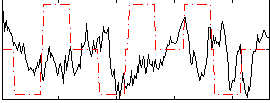  |
Correlation:-0.022011
  |
Correlation:0.16723
  |
Correlation:-0.11121
 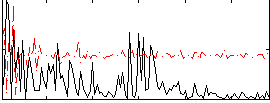 |
Component:151; Jaccard:0.069745
 |
Component:127; Jaccard:0.069134
 |
Component:379; Jaccard:0.062329
 |
Component:059; Jaccard:0.04648
 |
Component:379; Jaccard:0.024795
 |
Component:379; Jaccard:0.011574
 |
Component:165; Jaccard:0.006007
 |
Correlation:0.15759
  |
Correlation:0.18168
 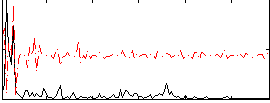 |
Correlation:-0.095407
  |
Correlation:0.28544
  |
Correlation:-0.095407
  |
Correlation:-0.095407
  |
Correlation:0.16723
  |
Component:127; Jaccard:0.068076
 |
Component:002; Jaccard:0.068212
 |
Component:127; Jaccard:0.057893
 |
Component:293; Jaccard:0.040943
 |
Component:059; Jaccard:0.023743
 |
Component:338; Jaccard:0.011468
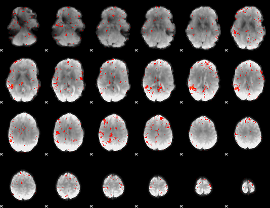 |
Component:338; Jaccard:0.005782
 |
Correlation:0.18168
  |
Correlation:0.12774
  |
Correlation:0.18168
  |
Correlation:-0.026288
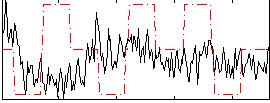 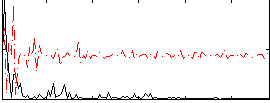 |
Correlation:0.28544
  |
Correlation:0.066191
  |
Correlation:0.066191
  |
Component:002; Jaccard:0.067933
 |
Component:379; Jaccard:0.067149
 |
Component:002; Jaccard:0.057399
 |
Component:211; Jaccard:0.039264
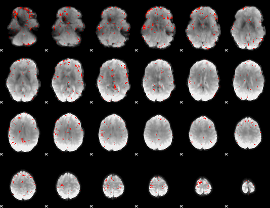 |
Component:293; Jaccard:0.022428
 |
Component:293; Jaccard:0.011317
 |
Component:379; Jaccard:0.005048
 |
Correlation:0.12774
  |
Correlation:-0.095407
  |
Correlation:0.12774
  |
Correlation:-0.041652
  |
Correlation:-0.026288
  |
Correlation:-0.026288
  |
Correlation:-0.095407
  |
Component:271; Jaccard:0.067468
 |
Component:271; Jaccard:0.06536
 |
Component:263; Jaccard:0.055024
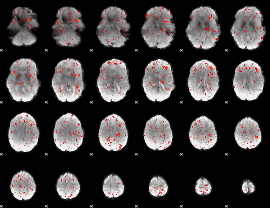 |
Component:002; Jaccard:0.038699
 |
Component:338; Jaccard:0.021849
 |
Component:099; Jaccard:0.010392
 |
Component:344; Jaccard:0.005019
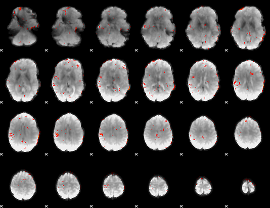 |
Correlation:0.35329
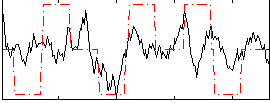  |
Correlation:0.35329
  |
Correlation:0.16032
  |
Correlation:0.12774
  |
Correlation:0.066191
  |
Correlation:-0.11121
  |
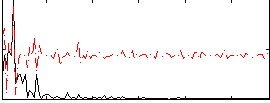
Correlation:-0.11489
  |
Component:263; Jaccard:0.067201
 |
Component:114; Jaccard:0.065333
 |
Component:271; Jaccard:0.054664
 |
Component:127; Jaccard:0.038519
 |
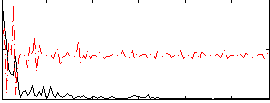
Component:143; Jaccard:0.021181
 |
Component:317; Jaccard:0.009832
 |
Component:293; Jaccard:0.004905
 |
Correlation:0.16032
  |
Correlation:0.00075694
 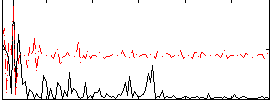 |
Correlation:0.35329
  |
Correlation:0.18168
  |
Correlation:-0.051415
  |
Correlation:0.00014446
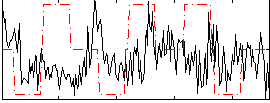  |
Correlation:-0.026288
  |
Component:304; Jaccard:0.066408
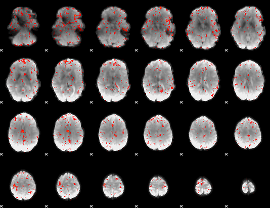 |
Component:144; Jaccard:0.064634
 |
Component:211; Jaccard:0.053276
 |
Component:263; Jaccard:0.03844
 |
Component:317; Jaccard:0.020555
 |
Component:382; Jaccard:0.009579
 |
Component:317; Jaccard:0.004705
 |
Correlation:0.052068
  |
Correlation:-0.06319
  |
Correlation:-0.041652
  |
Correlation:0.16032
  |
Correlation:0.00014446
  |
Correlation:0.01207
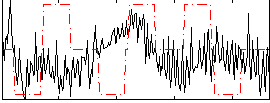  |
Correlation:0.00014446
  |
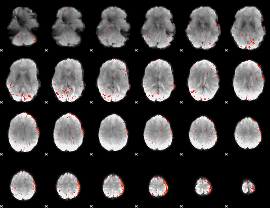
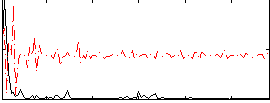
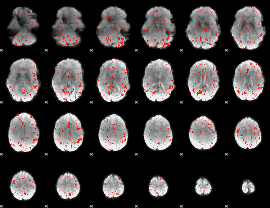
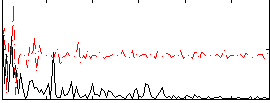
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
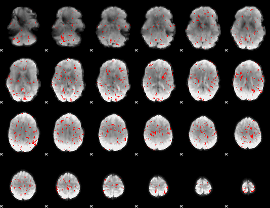 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
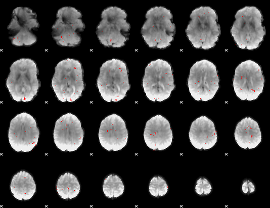 |
GLM binary result thresholded by 3.5
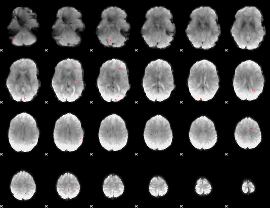 |
GLM binary result thresholded by 4
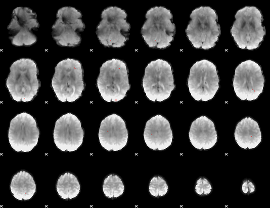 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
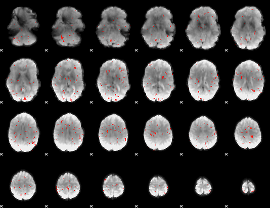 |
GLM Z-score result thresholded by 3
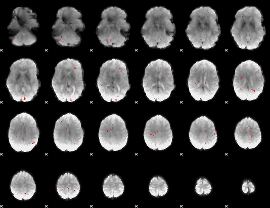 |
GLM Z-score result thresholded by 3.5
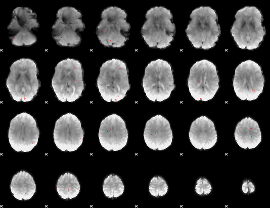 |
GLM Z-score result thresholded by 4
 |
Component:321; Jaccard:0.079388
 |
Component:321; Jaccard:0.075506
 |
Component:321; Jaccard:0.058281
 |
Component:025; Jaccard:0.035348
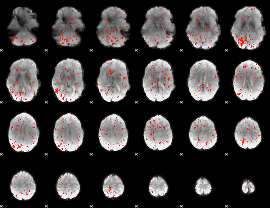 |
Component:321; Jaccard:0.013888
 |
Component:029; Jaccard:0.006697
 |
Component:029; Jaccard:0.002581
 |
Correlation:0.079895
  |
Correlation:0.079895
  |
Correlation:0.079895
  |
Correlation:0.077945
  |
Correlation:0.079895
  |
Correlation:0.21392
  |
Correlation:0.21392
  |
Component:285; Jaccard:0.076616
 |
Component:025; Jaccard:0.071036
 |
Component:025; Jaccard:0.056731
 |
Component:321; Jaccard:0.031677
 |
Component:076; Jaccard:0.013198
 |
Component:076; Jaccard:0.005227
 |
Component:076; Jaccard:0.002125
 |
Correlation:0.15006
  |
Correlation:0.077945
  |
Correlation:0.077945
  |
Correlation:0.079895
  |
Correlation:0.050363
  |
Correlation:0.050363
  |
Correlation:0.050363
  |
Component:286; Jaccard:0.07333
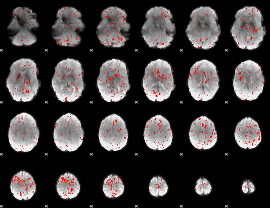 |
Component:285; Jaccard:0.070002
 |
Component:285; Jaccard:0.052791
 |
Component:285; Jaccard:0.029663
 |
Component:025; Jaccard:0.012957
 |
Component:025; Jaccard:0.004882
 |
Component:241; Jaccard:0.001682
 |
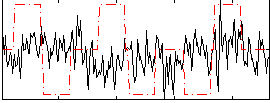
Correlation:-0.00027862
  |
Correlation:0.15006
  |
Correlation:0.15006
  |
Correlation:0.15006
  |
Correlation:0.077945
  |
Correlation:0.077945
  |
Correlation:0.086743
  |
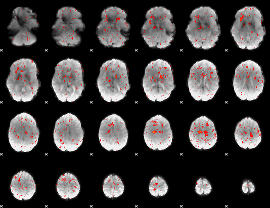
Component:025; Jaccard:0.072067
 |
Component:286; Jaccard:0.066543
 |
Component:286; Jaccard:0.051846
 |
Component:076; Jaccard:0.029132
 |
Component:029; Jaccard:0.012726
 |
Component:285; Jaccard:0.004123
 |
Component:025; Jaccard:0.00161
 |
Correlation:0.077945
  |
Correlation:-0.00027862
  |
Correlation:-0.00027862
  |
Correlation:0.050363
  |
Correlation:0.21392
  |
Correlation:0.15006
  |
Correlation:0.077945
  |
Component:139; Jaccard:0.069589
 |
Component:139; Jaccard:0.063122
 |
Component:076; Jaccard:0.051629
 |
Component:094; Jaccard:0.027502
 |
Component:285; Jaccard:0.01205
 |
Component:241; Jaccard:0.004071
 |
Component:193; Jaccard:0.00157
 |
Correlation:0.15859
  |
Correlation:0.15859
  |
Correlation:0.050363
  |
Correlation:0.0085414
  |
Correlation:0.15006
  |
Correlation:0.086743
  |
Correlation:-0.084791
  |
Component:246; Jaccard:0.068637
 |
Component:094; Jaccard:0.062647
 |
Component:094; Jaccard:0.04796
 |
Component:286; Jaccard:0.027489
 |
Component:326; Jaccard:0.011136
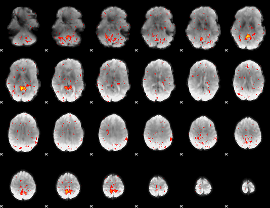 |
Component:139; Jaccard:0.004013
 |
Component:065; Jaccard:0.001525
 |
Correlation:0.02943
  |
Correlation:0.0085414
  |
Correlation:0.0085414
  |
Correlation:-0.00027862
  |
Correlation:0.079527
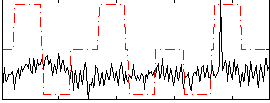  |
Correlation:0.15859
  |
Correlation:0.023952
  |
Component:094; Jaccard:0.064473
 |
Component:076; Jaccard:0.06256
 |
Component:139; Jaccard:0.046435
 |
Component:029; Jaccard:0.025588
 |
Component:094; Jaccard:0.011127
 |
Component:326; Jaccard:0.003992
 |
Component:305; Jaccard:0.001414
 |
Correlation:0.0085414
  |
Correlation:0.050363
  |
Correlation:0.15859
  |
Correlation:0.21392
  |
Correlation:0.0085414
  |
Correlation:0.079527
  |
Correlation:0.0035135
  |
Component:076; Jaccard:0.063235
 |
Component:246; Jaccard:0.058375
 |
Component:065; Jaccard:0.044932
 |
Component:316; Jaccard:0.025507
 |
Component:241; Jaccard:0.011055
 |
Component:090; Jaccard:0.003932
 |
Component:139; Jaccard:0.00132
 |
Correlation:0.050363
  |
Correlation:0.02943
  |
Correlation:0.023952
  |
Correlation:-0.0078153
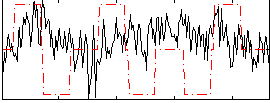  |
Correlation:0.086743
  |
Correlation:0.23237
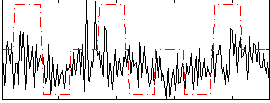  |
Correlation:0.15859
  |
Component:233; Jaccard:0.062712
 |
Component:065; Jaccard:0.056385
 |
Component:316; Jaccard:0.044724
 |
Component:326; Jaccard:0.025479
 |
Component:286; Jaccard:0.010694
 |
Component:321; Jaccard:0.003859
 |
Component:090; Jaccard:0.001289
 |
Correlation:-0.17957
  |
Correlation:0.023952
  |
Correlation:-0.0078153
  |
Correlation:0.079527
  |
Correlation:-0.00027862
  |
Correlation:0.079895
  |
Correlation:0.23237
  |
Component:298; Jaccard:0.061056
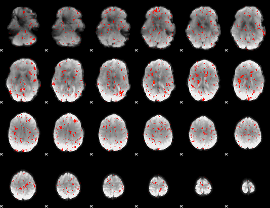 |
Component:233; Jaccard:0.056281
 |
Component:246; Jaccard:0.04308
 |
Component:065; Jaccard:0.024721
 |
Component:193; Jaccard:0.010172
 |
Component:193; Jaccard:0.003844
 |
Component:087; Jaccard:0.001248
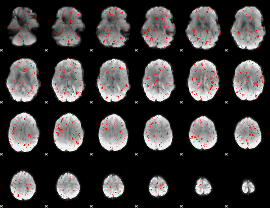 |
Correlation:0.06002
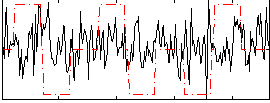  |
Correlation:-0.17957
  |
Correlation:0.02943
  |
Correlation:0.023952
  |
Correlation:-0.084791
  |
Correlation:-0.084791
  |
Correlation:0.086618
  |
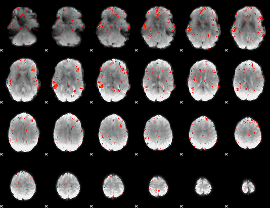
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
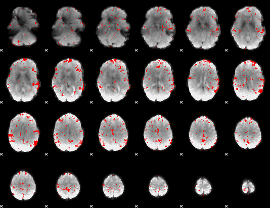 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
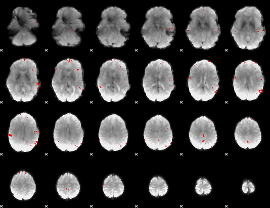 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
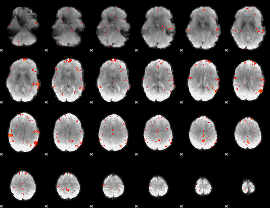 |
GLM Z-score result thresholded by 3.5
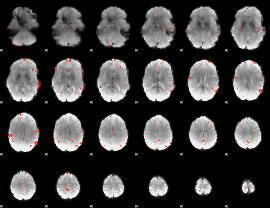 |
GLM Z-score result thresholded by 4
 |
Component:379; Jaccard:0.10474
 |
Component:379; Jaccard:0.13092
 |
Component:379; Jaccard:0.15415
 |
Component:379; Jaccard:0.16326
 |
Component:379; Jaccard:0.14697
 |
Component:379; Jaccard:0.10667
 |
Component:379; Jaccard:0.06966
 |
Correlation:0.21873
  |
Correlation:0.21873
  |
Correlation:0.21873
  |
Correlation:0.21873
  |
Correlation:0.21873
  |
Correlation:0.21873
  |
Correlation:0.21873
  |
Component:128; Jaccard:0.10194
 |
Component:128; Jaccard:0.11959
 |
Component:128; Jaccard:0.12917
 |
Component:128; Jaccard:0.12234
 |
Component:128; Jaccard:0.09813
 |
Component:128; Jaccard:0.06879
 |
Component:128; Jaccard:0.043724
 |
Correlation:0.069865
  |
Correlation:0.069865
  |
Correlation:0.069865
  |
Correlation:0.069865
  |
Correlation:0.069865
  |
Correlation:0.069865
  |
Correlation:0.069865
  |
Component:203; Jaccard:0.10068
 |
Component:203; Jaccard:0.11246
 |
Component:203; Jaccard:0.1171
 |
Component:203; Jaccard:0.10704
 |
Component:203; Jaccard:0.081845
 |
Component:122; Jaccard:0.058057
 |
Component:122; Jaccard:0.03623
 |
Correlation:0.19854
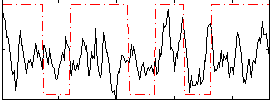  |
Correlation:0.19854
  |
Correlation:0.19854
  |
Correlation:0.19854
  |
Correlation:0.19854
  |
Correlation:0.16206
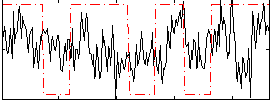 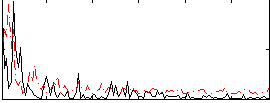 |
Correlation:0.16206
  |
Component:246; Jaccard:0.097466
 |
Component:246; Jaccard:0.10404
 |
Component:246; Jaccard:0.099945
 |
Component:122; Jaccard:0.089372
 |
Component:122; Jaccard:0.072493
 |
Component:203; Jaccard:0.054506
 |
Component:203; Jaccard:0.031235
 |
Correlation:0.12412
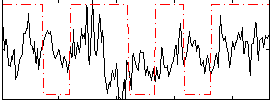 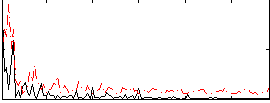 |
Correlation:0.12412
  |
Correlation:0.12412
  |
Correlation:0.16206
  |
Correlation:0.16206
  |
Correlation:0.19854
  |
Correlation:0.19854
  |
Component:202; Jaccard:0.091617
 |
Component:202; Jaccard:0.094893
 |
Component:122; Jaccard:0.092376
 |
Component:246; Jaccard:0.083236
 |
Component:056; Jaccard:0.060059
 |
Component:163; Jaccard:0.037359
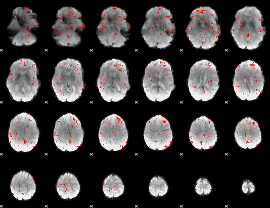 |
Component:163; Jaccard:0.023557
 |
Correlation:-0.013711
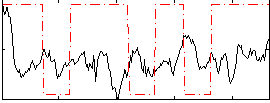  |
Correlation:-0.013711
  |
Correlation:0.16206
  |
Correlation:0.12412
  |
Correlation:0.21584
  |
Correlation:0.12204
  |
Correlation:0.12204
  |
Component:080; Jaccard:0.081143
 |
Component:122; Jaccard:0.086154
 |
Component:202; Jaccard:0.091474
 |
Component:056; Jaccard:0.080267
 |
Component:163; Jaccard:0.057446
 |
Component:056; Jaccard:0.036621
 |
Component:056; Jaccard:0.022083
 |
Correlation:-0.10229
  |
Correlation:0.16206
  |
Correlation:-0.013711
  |
Correlation:0.21584
  |
Correlation:0.12204
  |
Correlation:0.21584
  |
Correlation:0.21584
  |
Component:144; Jaccard:0.081054
 |
Component:056; Jaccard:0.084876
 |
Component:056; Jaccard:0.08935
 |
Component:202; Jaccard:0.073185
 |
Component:246; Jaccard:0.054165
 |
Component:199; Jaccard:0.035938
 |
Component:350; Jaccard:0.021081
 |
Correlation:0.1601
  |
Correlation:0.21584
  |
Correlation:0.21584
  |
Correlation:-0.013711
  |
Correlation:0.12412
  |
Correlation:0.14069
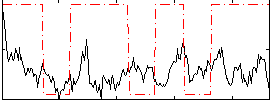  |
Correlation:0.10528
  |
Component:157; Jaccard:0.078404
 |
Component:144; Jaccard:0.081668
 |
Component:295; Jaccard:0.079548
 |
Component:295; Jaccard:0.072143
 |
Component:199; Jaccard:0.054128
 |
Component:350; Jaccard:0.035707
 |
Component:246; Jaccard:0.019411
 |
Correlation:-0.08429
  |
Correlation:0.1601
  |
Correlation:0.16641
  |
Correlation:0.16641
  |
Correlation:0.14069
  |
Correlation:0.10528
  |
Correlation:0.12412
  |
Component:056; Jaccard:0.077964
 |
Component:157; Jaccard:0.081602
 |
Component:157; Jaccard:0.079453
 |
Component:163; Jaccard:0.072058
 |
Component:202; Jaccard:0.053598
 |
Component:277; Jaccard:0.032991
 |
Component:199; Jaccard:0.019296
 |
Correlation:0.21584
  |
Correlation:-0.08429
  |
Correlation:-0.08429
  |
Correlation:0.12204
  |
Correlation:-0.013711
  |
Correlation:0.07542
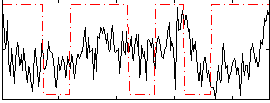  |
Correlation:0.14069
  |
Component:271; Jaccard:0.076378
 |
Component:295; Jaccard:0.07972
 |
Component:199; Jaccard:0.079419
 |
Component:199; Jaccard:0.07094
 |
Component:295; Jaccard:0.053162
 |
Component:246; Jaccard:0.032341
 |
Component:202; Jaccard:0.019099
 |
Correlation:-0.14785
 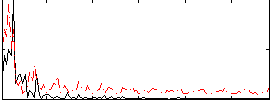 |
Correlation:0.16641
  |
Correlation:0.14069
  |
Correlation:0.14069
  |
Correlation:0.16641
  |
Correlation:0.12412
  |
Correlation:-0.013711
  |
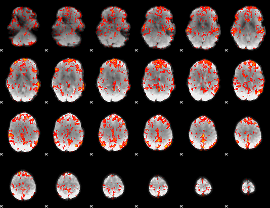
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
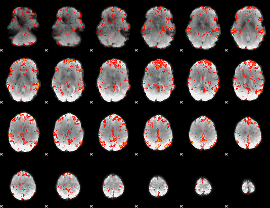 |
GLM Z-score result thresholded by 2.5
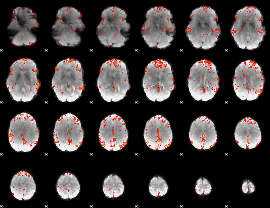 |
GLM Z-score result thresholded by 3
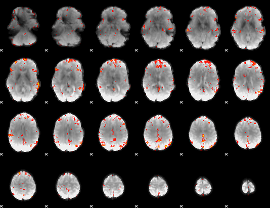 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:072; Jaccard:0.11728
 |
Component:379; Jaccard:0.14552
 |
Component:379; Jaccard:0.18283
 |
Component:379; Jaccard:0.21505
 |
Component:379; Jaccard:0.22353
 |
Component:379; Jaccard:0.19801
 |
Component:379; Jaccard:0.14537
 |
Correlation:0.39177
  |
Correlation:0.057763
 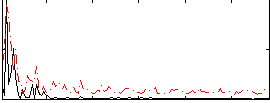 |
Correlation:0.057763
  |
Correlation:0.057763
  |
Correlation:0.057763
  |
Correlation:0.057763
  |
Correlation:0.057763
  |
Component:379; Jaccard:0.11202
 |
Component:128; Jaccard:0.13638
 |
Component:128; Jaccard:0.15785
 |
Component:128; Jaccard:0.16386
 |
Component:128; Jaccard:0.15044
 |
Component:128; Jaccard:0.11839
 |
Component:128; Jaccard:0.082923
 |
Correlation:0.057763
  |
Correlation:0.26901
  |
Correlation:0.26901
  |
Correlation:0.26901
  |
Correlation:0.26901
  |
Correlation:0.26901
  |
Correlation:0.26901
  |
Component:128; Jaccard:0.11194
 |
Component:072; Jaccard:0.13266
 |
Component:072; Jaccard:0.14073
 |
Component:072; Jaccard:0.14046
 |
Component:072; Jaccard:0.12181
 |
Component:072; Jaccard:0.091642
 |
Component:072; Jaccard:0.059896
 |
Correlation:0.26901
  |
Correlation:0.39177
  |
Correlation:0.39177
  |
Correlation:0.39177
  |
Correlation:0.39177
  |
Correlation:0.39177
  |
Correlation:0.39177
  |
Component:202; Jaccard:0.1039
 |
Component:144; Jaccard:0.11151
 |
Component:144; Jaccard:0.11262
 |
Component:203; Jaccard:0.10713
 |
Component:203; Jaccard:0.092776
 |
Component:203; Jaccard:0.072708
 |
Component:203; Jaccard:0.050434
 |
Correlation:0.36146
  |
Correlation:0.053487
  |
Correlation:0.053487
  |
Correlation:0.14658
  |
Correlation:0.14658
  |
Correlation:0.14658
  |
Correlation:0.14658
  |
Component:144; Jaccard:0.10271
 |
Component:202; Jaccard:0.11003
 |
Component:203; Jaccard:0.10975
 |
Component:144; Jaccard:0.10293
 |
Component:127; Jaccard:0.090903
 |
Component:163; Jaccard:0.069118
 |
Component:163; Jaccard:0.046796
 |
Correlation:0.053487
  |
Correlation:0.36146
  |
Correlation:0.14658
  |
Correlation:0.053487
  |
Correlation:0.29948
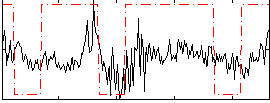 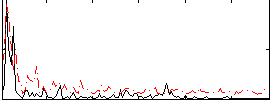 |
Correlation:0.2062
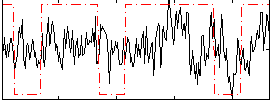  |
Correlation:0.2062
  |
Component:203; Jaccard:0.094477
 |
Component:203; Jaccard:0.10439
 |
Component:202; Jaccard:0.10954
 |
Component:127; Jaccard:0.10245
 |
Component:144; Jaccard:0.083736
 |
Component:127; Jaccard:0.068241
 |
Component:099; Jaccard:0.044804
 |
Correlation:0.14658
  |
Correlation:0.14658
  |
Correlation:0.36146
  |
Correlation:0.29948
  |
Correlation:0.053487
  |
Correlation:0.29948
  |
Correlation:-0.0014817
  |
Component:271; Jaccard:0.0921
 |
Component:271; Jaccard:0.10102
 |
Component:127; Jaccard:0.1042
 |
Component:202; Jaccard:0.099744
 |
Component:163; Jaccard:0.083592
 |
Component:099; Jaccard:0.063934
 |
Component:127; Jaccard:0.04434
 |
Correlation:0.44822
  |
Correlation:0.44822
  |
Correlation:0.29948
  |
Correlation:0.36146
  |
Correlation:0.2062
  |
Correlation:-0.0014817
  |
Correlation:0.29948
  |
Component:127; Jaccard:0.085118
 |
Component:127; Jaccard:0.095933
 |
Component:271; Jaccard:0.10288
 |
Component:271; Jaccard:0.093526
 |
Component:202; Jaccard:0.081116
 |
Component:144; Jaccard:0.063503
 |
Component:259; Jaccard:0.043179
 |
Correlation:0.29948
  |
Correlation:0.29948
  |
Correlation:0.44822
  |
Correlation:0.44822
  |
Correlation:0.36146
  |
Correlation:0.053487
  |
Correlation:0.23204
 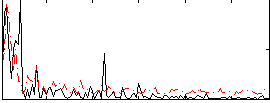 |
Component:309; Jaccard:0.082503
 |
Component:309; Jaccard:0.084961
 |
Component:163; Jaccard:0.091397
 |
Component:163; Jaccard:0.092351
 |
Component:099; Jaccard:0.078565
 |
Component:259; Jaccard:0.06138
 |
Component:144; Jaccard:0.042834
 |
Correlation:0.4662
  |
Correlation:0.4662
  |
Correlation:0.2062
  |
Correlation:0.2062
  |
Correlation:-0.0014817
  |
Correlation:0.23204
  |
Correlation:0.053487
  |
Component:124; Jaccard:0.076908
 |
Component:163; Jaccard:0.083568
 |
Component:099; Jaccard:0.086895
 |
Component:099; Jaccard:0.086112
 |
Component:259; Jaccard:0.076783
 |
Component:202; Jaccard:0.059242
 |
Component:202; Jaccard:0.041352
 |
Correlation:0.24303
  |
Correlation:0.2062
  |
Correlation:-0.0014817
  |
Correlation:-0.0014817
  |
Correlation:0.23204
  |
Correlation:0.36146
  |
Correlation:0.36146
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































