Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
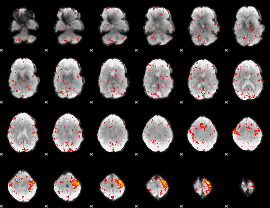
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
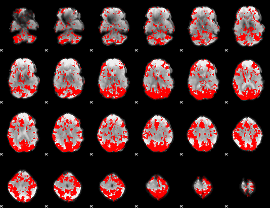 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
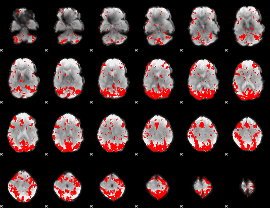 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
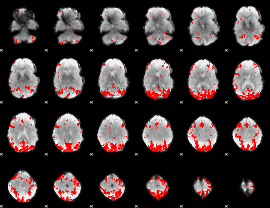 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
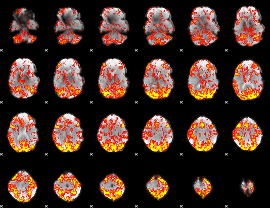 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
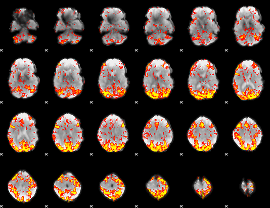 |
GLM Z-score result thresholded by 3
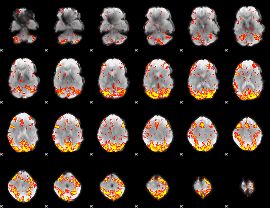 |
GLM Z-score result thresholded by 3.5
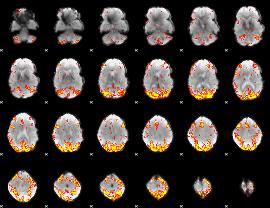 |
GLM Z-score result thresholded by 4
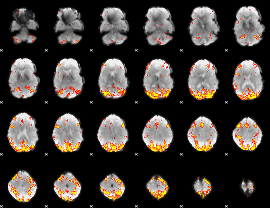 |
Component:212; Jaccard:0.11215
 |
Component:212; Jaccard:0.13415
 |
Component:212; Jaccard:0.15991
 |
Component:331; Jaccard:0.19172
 |
Component:331; Jaccard:0.22413
 |
Component:331; Jaccard:0.2559
 |
Component:331; Jaccard:0.27681
 |
Correlation:0.10519
  |
Correlation:0.10519
  |
Correlation:0.10519
  |
Correlation:0.28865
  |
Correlation:0.28865
  |
Correlation:0.28865
  |
Correlation:0.28865
  |
Component:144; Jaccard:0.10996
 |
Component:331; Jaccard:0.12886
 |
Component:331; Jaccard:0.1583
 |
Component:212; Jaccard:0.18555
 |
Component:212; Jaccard:0.20894
 |
Component:142; Jaccard:0.23088
 |
Component:109; Jaccard:0.26333
 |
Correlation:0.046509
  |
Correlation:0.28865
  |
Correlation:0.28865
  |
Correlation:0.10519
  |
Correlation:0.10519
  |
Correlation:0.19764
  |
Correlation:0.23457
  |
Component:331; Jaccard:0.1049
 |
Component:144; Jaccard:0.12828
 |
Component:144; Jaccard:0.14816
 |
Component:142; Jaccard:0.17252
 |
Component:142; Jaccard:0.20247
 |
Component:109; Jaccard:0.23084
 |
Component:142; Jaccard:0.257
 |
Correlation:0.28865
  |
Correlation:0.046509
  |
Correlation:0.046509
  |
Correlation:0.19764
  |
Correlation:0.19764
  |
Correlation:0.23457
  |
Correlation:0.19764
  |
Component:283; Jaccard:0.10086
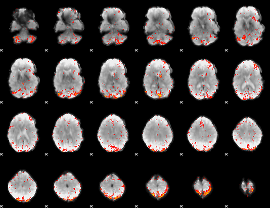 |
Component:142; Jaccard:0.11848
 |
Component:142; Jaccard:0.14354
 |
Component:144; Jaccard:0.16678
 |
Component:109; Jaccard:0.19561
 |
Component:212; Jaccard:0.22649
 |
Component:212; Jaccard:0.2396
 |
Correlation:0.24855
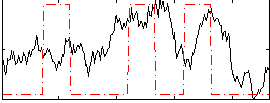  |
Correlation:0.19764
  |
Correlation:0.19764
  |
Correlation:0.046509
  |
Correlation:0.23457
  |
Correlation:0.10519
  |
Correlation:0.10519
  |
Component:089; Jaccard:0.10051
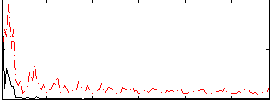
 |
Component:283; Jaccard:0.11558
 |
Component:109; Jaccard:0.13435
 |
Component:109; Jaccard:0.16375
 |
Component:144; Jaccard:0.18102
 |
Component:144; Jaccard:0.19327
 |
Component:144; Jaccard:0.19834
 |
Correlation:0.34811
  |
Correlation:0.24855
  |
Correlation:0.23457
  |
Correlation:0.23457
  |
Correlation:0.046509
  |
Correlation:0.046509
  |
Correlation:0.046509
  |
Component:372; Jaccard:0.099978
 |
Component:224; Jaccard:0.11404
 |
Component:224; Jaccard:0.13083
 |
Component:224; Jaccard:0.14685
 |
Component:330; Jaccard:0.16016
 |
Component:330; Jaccard:0.1716
 |
Component:330; Jaccard:0.17696
 |
Correlation:0.27493
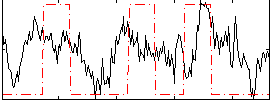  |
Correlation:0.1636
  |
Correlation:0.1636
  |
Correlation:0.1636
  |
Correlation:0.20172
  |
Correlation:0.20172
  |
Correlation:0.20172
  |
Component:224; Jaccard:0.09839
 |
Component:089; Jaccard:0.11338
 |
Component:283; Jaccard:0.13079
 |
Component:330; Jaccard:0.14536
 |
Component:224; Jaccard:0.15943
 |
Component:224; Jaccard:0.16895
 |
Component:224; Jaccard:0.17357
 |
Correlation:0.1636
  |
Correlation:0.34811
  |
Correlation:0.24855
  |
Correlation:0.20172
  |
Correlation:0.1636
  |
Correlation:0.1636
  |
Correlation:0.1636
  |
Component:142; Jaccard:0.097686
 |
Component:372; Jaccard:0.11324
 |
Component:308; Jaccard:0.1292
 |
Component:308; Jaccard:0.1445
 |
Component:308; Jaccard:0.15628
 |
Component:308; Jaccard:0.1625
 |
Component:228; Jaccard:0.16795
 |
Correlation:0.19764
  |
Correlation:0.27493
  |
Correlation:0.2972
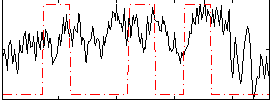  |
Correlation:0.2972
  |
Correlation:0.2972
  |
Correlation:0.2972
  |
Correlation:0.14135
 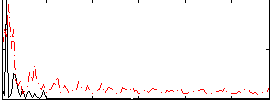 |
Component:308; Jaccard:0.096849
 |
Component:308; Jaccard:0.11217
 |
Component:330; Jaccard:0.12909
 |
Component:283; Jaccard:0.14393
 |
Component:283; Jaccard:0.15377
 |
Component:228; Jaccard:0.15914
 |
Component:308; Jaccard:0.16271
 |
Correlation:0.2972
  |
Correlation:0.2972
  |
Correlation:0.20172
  |
Correlation:0.24855
  |
Correlation:0.24855
  |
Correlation:0.14135
  |
Correlation:0.2972
  |
Component:241; Jaccard:0.09525
 |
Component:330; Jaccard:0.11119
 |
Component:372; Jaccard:0.12776
 |
Component:372; Jaccard:0.14105
 |
Component:372; Jaccard:0.14886
 |
Component:283; Jaccard:0.15732
 |
Component:283; Jaccard:0.15389
 |
Correlation:0.13036
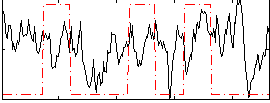  |
Correlation:0.20172
  |
Correlation:0.27493
  |
Correlation:0.27493
  |
Correlation:0.27493
  |
Correlation:0.24855
  |
Correlation:0.24855
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
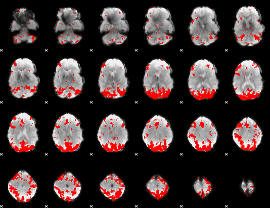 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:144; Jaccard:0.1078
 |
Component:144; Jaccard:0.12673
 |
Component:144; Jaccard:0.14957
 |
Component:331; Jaccard:0.17698
 |
Component:331; Jaccard:0.20956
 |
Component:331; Jaccard:0.23676
 |
Component:109; Jaccard:0.27081
 |
Correlation:0.1832
  |
Correlation:0.1832
  |
Correlation:0.1832
  |
Correlation:0.20172
  |
Correlation:0.20172
  |
Correlation:0.20172
  |
Correlation:0.18823
  |
Component:212; Jaccard:0.10332
 |
Component:212; Jaccard:0.12215
 |
Component:331; Jaccard:0.14651
 |
Component:144; Jaccard:0.17248
 |
Component:142; Jaccard:0.1969
 |
Component:109; Jaccard:0.23495
 |
Component:331; Jaccard:0.25583
 |
Correlation:0.22879
  |
Correlation:0.22879
  |
Correlation:0.20172
  |
Correlation:0.1832
  |
Correlation:0.31601
  |
Correlation:0.18823
  |
Correlation:0.20172
  |
Component:331; Jaccard:0.097475
 |
Component:331; Jaccard:0.11875
 |
Component:212; Jaccard:0.14474
 |
Component:212; Jaccard:0.16886
 |
Component:109; Jaccard:0.1958
 |
Component:142; Jaccard:0.22715
 |
Component:142; Jaccard:0.25244
 |
Correlation:0.20172
  |
Correlation:0.20172
  |
Correlation:0.22879
  |
Correlation:0.22879
  |
Correlation:0.18823
  |
Correlation:0.31601
  |
Correlation:0.31601
  |
Component:372; Jaccard:0.096983
 |
Component:228; Jaccard:0.11292
 |
Component:142; Jaccard:0.13668
 |
Component:142; Jaccard:0.16509
 |
Component:144; Jaccard:0.19275
 |
Component:144; Jaccard:0.20838
 |
Component:144; Jaccard:0.21903
 |
Correlation:0.033696
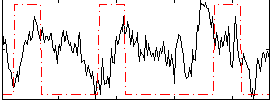  |
Correlation:0.35159
  |
Correlation:0.31601
  |
Correlation:0.31601
  |
Correlation:0.1832
  |
Correlation:0.1832
  |
Correlation:0.1832
  |
Component:224; Jaccard:0.095822
 |
Component:372; Jaccard:0.11218
 |
Component:228; Jaccard:0.1335
 |
Component:109; Jaccard:0.15979
 |
Component:212; Jaccard:0.19007
 |
Component:212; Jaccard:0.20652
 |
Component:212; Jaccard:0.21515
 |
Correlation:0.28128
 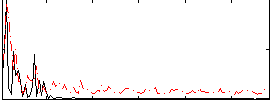 |
Correlation:0.033696
  |
Correlation:0.35159
  |
Correlation:0.18823
  |
Correlation:0.22879
  |
Correlation:0.22879
  |
Correlation:0.22879
  |
Component:241; Jaccard:0.095011
 |
Component:142; Jaccard:0.11204
 |
Component:224; Jaccard:0.12909
 |
Component:228; Jaccard:0.15536
 |
Component:228; Jaccard:0.17544
 |
Component:228; Jaccard:0.191
 |
Component:228; Jaccard:0.20174
 |
Correlation:0.13157
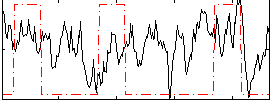 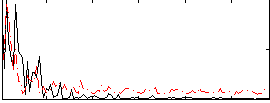 |
Correlation:0.31601
  |
Correlation:0.28128
  |
Correlation:0.35159
  |
Correlation:0.35159
  |
Correlation:0.35159
  |
Correlation:0.35159
  |
Component:228; Jaccard:0.094935
 |
Component:224; Jaccard:0.11197
 |
Component:109; Jaccard:0.12878
 |
Component:224; Jaccard:0.14706
 |
Component:224; Jaccard:0.16377
 |
Component:224; Jaccard:0.17468
 |
Component:224; Jaccard:0.18112
 |
Correlation:0.35159
  |
Correlation:0.28128
  |
Correlation:0.18823
  |
Correlation:0.28128
  |
Correlation:0.28128
  |
Correlation:0.28128
  |
Correlation:0.28128
  |
Component:142; Jaccard:0.092117
 |
Component:241; Jaccard:0.10685
 |
Component:372; Jaccard:0.12785
 |
Component:372; Jaccard:0.14253
 |
Component:372; Jaccard:0.15351
 |
Component:330; Jaccard:0.16303
 |
Component:330; Jaccard:0.17056
 |
Correlation:0.31601
  |
Correlation:0.13157
  |
Correlation:0.033696
  |
Correlation:0.033696
  |
Correlation:0.033696
  |
Correlation:0.2386
  |
Correlation:0.2386
  |
Component:283; Jaccard:0.091584
 |
Component:308; Jaccard:0.10454
 |
Component:308; Jaccard:0.11992
 |
Component:330; Jaccard:0.13688
 |
Component:330; Jaccard:0.15225
 |
Component:372; Jaccard:0.15923
 |
Component:308; Jaccard:0.16148
 |
Correlation:-0.0063738
  |
Correlation:0.055167
  |
Correlation:0.055167
  |
Correlation:0.2386
  |
Correlation:0.2386
  |
Correlation:0.033696
  |
Correlation:0.055167
  |
Component:308; Jaccard:0.090482
 |
Component:283; Jaccard:0.1041
 |
Component:330; Jaccard:0.11976
 |
Component:308; Jaccard:0.13467
 |
Component:308; Jaccard:0.14771
 |
Component:308; Jaccard:0.15647
 |
Component:372; Jaccard:0.15779
 |
Correlation:0.055167
  |
Correlation:-0.0063738
  |
Correlation:0.2386
  |
Correlation:0.055167
  |
Correlation:0.055167
  |
Correlation:0.055167
  |
Correlation:0.033696
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
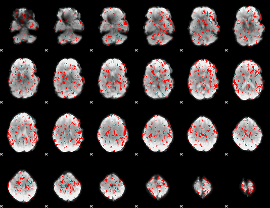 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
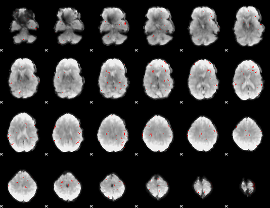 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
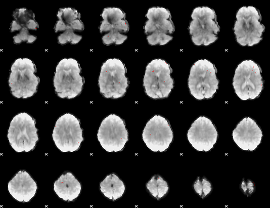 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
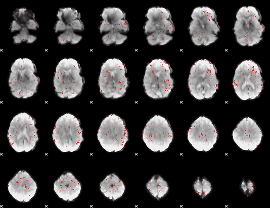 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
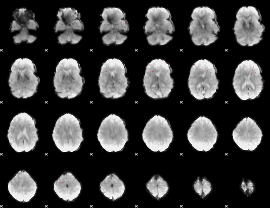 |
Component:234; Jaccard:0.088552
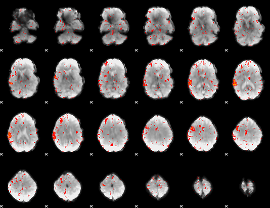 |
Component:097; Jaccard:0.092096
 |
Component:097; Jaccard:0.084226
 |
Component:097; Jaccard:0.057604
 |
Component:097; Jaccard:0.028665
 |
Component:321; Jaccard:0.01258
 |
Component:329; Jaccard:0.004482
 |
Correlation:0.15196
 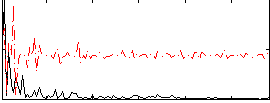 |
Correlation:0.17425
  |
Correlation:0.17425
  |
Correlation:0.17425
  |
Correlation:0.17425
  |
Correlation:0.1171
  |
Correlation:0.14822
  |
Component:099; Jaccard:0.087078
 |
Component:234; Jaccard:0.089207
 |
Component:234; Jaccard:0.078395
 |
Component:234; Jaccard:0.050408
 |
Component:321; Jaccard:0.027602
 |
Component:281; Jaccard:0.011778
 |
Component:321; Jaccard:0.004379
 |
Correlation:0.094178
  |
Correlation:0.15196
  |
Correlation:0.15196
  |
Correlation:0.15196
  |
Correlation:0.1171
  |
Correlation:-0.078619
  |
Correlation:0.1171
  |
Component:097; Jaccard:0.08453
 |
Component:099; Jaccard:0.0835
 |
Component:347; Jaccard:0.067553
 |
Component:321; Jaccard:0.048826
 |
Component:281; Jaccard:0.026119
 |
Component:097; Jaccard:0.011065
 |
Component:281; Jaccard:0.003251
 |
Correlation:0.17425
  |
Correlation:0.094178
  |
Correlation:0.09043
  |
Correlation:0.1171
  |
Correlation:-0.078619
  |
Correlation:0.17425
  |
Correlation:-0.078619
  |
Component:368; Jaccard:0.083799
 |
Component:347; Jaccard:0.083315
 |
Component:281; Jaccard:0.066022
 |
Component:281; Jaccard:0.048067
 |
Component:234; Jaccard:0.024895
 |
Component:234; Jaccard:0.010315
 |
Component:101; Jaccard:0.003006
 |
Correlation:0.27275
  |
Correlation:0.09043
  |
Correlation:-0.078619
  |
Correlation:-0.078619
  |
Correlation:0.15196
  |
Correlation:0.15196
  |
Correlation:0.073208
  |
Component:347; Jaccard:0.083493
 |
Component:075; Jaccard:0.078673
 |
Component:075; Jaccard:0.06517
 |
Component:347; Jaccard:0.047306
 |
Component:347; Jaccard:0.022817
 |
Component:101; Jaccard:0.008752
 |
Component:097; Jaccard:0.002928
 |
Correlation:0.09043
  |
Correlation:0.18148
  |
Correlation:0.18148
  |
Correlation:0.09043
  |
Correlation:0.09043
  |
Correlation:0.073208
  |
Correlation:0.17425
  |
Component:075; Jaccard:0.078339
 |
Component:368; Jaccard:0.078043
 |
Component:099; Jaccard:0.064862
 |
Component:075; Jaccard:0.043223
 |
Component:075; Jaccard:0.020097
 |
Component:329; Jaccard:0.008403
 |
Component:234; Jaccard:0.00289
 |
Correlation:0.18148
  |
Correlation:0.27275
  |
Correlation:0.094178
  |
Correlation:0.18148
  |
Correlation:0.18148
  |
Correlation:0.14822
  |
Correlation:0.15196
  |
Component:299; Jaccard:0.073774
 |
Component:299; Jaccard:0.073648
 |
Component:299; Jaccard:0.064694
 |
Component:299; Jaccard:0.042713
 |
Component:299; Jaccard:0.019903
 |
Component:347; Jaccard:0.008034
 |
Component:232; Jaccard:0.002699
 |
Correlation:0.11785
  |
Correlation:0.11785
  |
Correlation:0.11785
  |
Correlation:0.11785
  |
Correlation:0.11785
  |
Correlation:0.09043
  |
Correlation:0.19259
  |
Component:089; Jaccard:0.072555
 |
Component:281; Jaccard:0.070938
 |
Component:368; Jaccard:0.063575
 |
Component:368; Jaccard:0.040632
 |
Component:101; Jaccard:0.019799
 |
Component:263; Jaccard:0.007271
 |
Component:261; Jaccard:0.002406
 |
Correlation:0.26093
  |
Correlation:-0.078619
  |
Correlation:0.27275
  |
Correlation:0.27275
  |
Correlation:0.073208
  |
Correlation:0.16574
  |
Correlation:0.12456
  |
Component:006; Jaccard:0.07152
 |
Component:089; Jaccard:0.068018
 |
Component:321; Jaccard:0.060338
 |
Component:093; Jaccard:0.040432
 |
Component:367; Jaccard:0.019522
 |
Component:299; Jaccard:0.007189
 |
Component:367; Jaccard:0.002338
 |
Correlation:0.089509
  |
Correlation:0.26093
  |
Correlation:0.1171
  |
Correlation:0.2614
  |
Correlation:0.2175
  |
Correlation:0.11785
  |
Correlation:0.2175
  |
Component:101; Jaccard:0.068413
 |
Component:006; Jaccard:0.067449
 |
Component:093; Jaccard:0.055471
 |
Component:099; Jaccard:0.039664
 |
Component:093; Jaccard:0.018801
 |
Component:314; Jaccard:0.007135
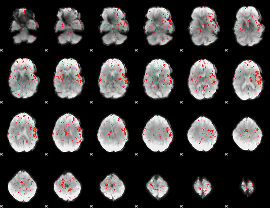 |
Component:006; Jaccard:0.002199
 |
Correlation:0.073208
  |
Correlation:0.089509
  |
Correlation:0.2614
  |
Correlation:0.094178
  |
Correlation:0.2614
  |
Correlation:0.058784
  |
Correlation:0.089509
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
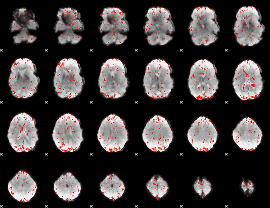 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:113; Jaccard:0.094172
 |
Component:066; Jaccard:0.1109
 |
Component:066; Jaccard:0.11594
 |
Component:066; Jaccard:0.093701
 |
Component:066; Jaccard:0.050662
 |
Component:264; Jaccard:0.027448
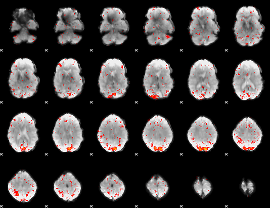 |
Component:264; Jaccard:0.016179
 |
Correlation:0.19163
  |
Correlation:0.39029
 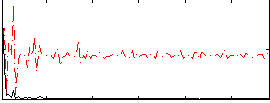 |
Correlation:0.39029
  |
Correlation:0.39029
  |
Correlation:0.39029
  |
Correlation:0.13677
 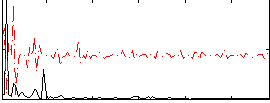 |
Correlation:0.13677
  |
Component:066; Jaccard:0.091667
 |
Component:394; Jaccard:0.10009
 |
Component:187; Jaccard:0.10071
 |
Component:187; Jaccard:0.084079
 |
Component:187; Jaccard:0.048038
 |
Component:253; Jaccard:0.023931
 |
Component:253; Jaccard:0.014366
 |
Correlation:0.39029
  |
Correlation:0.26812
  |
Correlation:0.35337
  |
Correlation:0.35337
  |
Correlation:0.35337
  |
Correlation:-0.076542
  |
Correlation:-0.076542
  |
Component:394; Jaccard:0.088477
 |
Component:113; Jaccard:0.10009
 |
Component:394; Jaccard:0.098293
 |
Component:394; Jaccard:0.079368
 |
Component:264; Jaccard:0.046186
 |
Component:109; Jaccard:0.022744
 |
Component:109; Jaccard:0.012228
 |
Correlation:0.26812
  |
Correlation:0.19163
  |
Correlation:0.26812
  |
Correlation:0.26812
  |
Correlation:0.13677
  |
Correlation:-0.02746
  |
Correlation:-0.02746
  |
Component:228; Jaccard:0.08778
 |
Component:187; Jaccard:0.094475
 |
Component:113; Jaccard:0.09258
 |
Component:228; Jaccard:0.069136
 |
Component:394; Jaccard:0.04603
 |
Component:066; Jaccard:0.020706
 |
Component:142; Jaccard:0.009257
 |
Correlation:0.12461
  |
Correlation:0.35337
  |
Correlation:0.19163
  |
Correlation:0.12461
  |
Correlation:0.26812
  |
Correlation:0.39029
  |
Correlation:0.070157
  |
Component:187; Jaccard:0.079235
 |
Component:228; Jaccard:0.092412
 |
Component:228; Jaccard:0.088357
 |
Component:113; Jaccard:0.067618
 |
Component:109; Jaccard:0.041749
 |
Component:394; Jaccard:0.019177
 |
Component:338; Jaccard:0.008872
 |
Correlation:0.35337
  |
Correlation:0.12461
  |
Correlation:0.12461
  |
Correlation:0.19163
  |
Correlation:-0.02746
  |
Correlation:0.26812
  |
Correlation:-0.062186
  |
Component:276; Jaccard:0.076538
 |
Component:004; Jaccard:0.074075
 |
Component:004; Jaccard:0.074754
 |
Component:109; Jaccard:0.062765
 |
Component:004; Jaccard:0.040816
 |
Component:187; Jaccard:0.018659
 |
Component:228; Jaccard:0.008296
 |
Correlation:0.14455
  |
Correlation:0.069737
  |
Correlation:0.069737
  |
Correlation:-0.02746
  |
Correlation:0.069737
  |
Correlation:0.35337
  |
Correlation:0.12461
  |
Component:264; Jaccard:0.070005
 |
Component:109; Jaccard:0.074043
 |
Component:109; Jaccard:0.073778
 |
Component:004; Jaccard:0.06272
 |
Component:253; Jaccard:0.039029
 |
Component:228; Jaccard:0.016775
 |
Component:205; Jaccard:0.006297
 |
Correlation:0.13677
  |
Correlation:-0.02746
  |
Correlation:-0.02746
  |
Correlation:0.069737
  |
Correlation:-0.076542
  |
Correlation:0.12461
  |
Correlation:-0.0095731
  |
Component:109; Jaccard:0.069801
 |
Component:264; Jaccard:0.073866
 |
Component:264; Jaccard:0.07279
 |
Component:264; Jaccard:0.062407
 |
Component:113; Jaccard:0.038469
 |
Component:142; Jaccard:0.015718
 |
Component:316; Jaccard:0.006181
 |
Correlation:-0.02746
  |
Correlation:0.13677
  |
Correlation:0.13677
  |
Correlation:0.13677
  |
Correlation:0.19163
  |
Correlation:0.070157
  |
Correlation:0.18127
  |
Component:399; Jaccard:0.068924
 |
Component:276; Jaccard:0.073586
 |
Component:225; Jaccard:0.061609
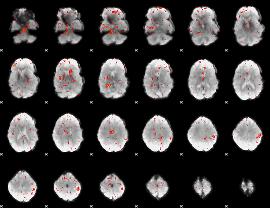 |
Component:253; Jaccard:0.054999
 |
Component:228; Jaccard:0.038398
 |
Component:004; Jaccard:0.01555
 |
Component:372; Jaccard:0.005836
 |
Correlation:0.22123
  |
Correlation:0.14455
  |
Correlation:0.12554
  |
Correlation:-0.076542
  |
Correlation:0.12461
  |
Correlation:0.069737
  |
Correlation:-0.14298
  |
Component:004; Jaccard:0.067423
 |
Component:399; Jaccard:0.06867
 |
Component:253; Jaccard:0.061562
 |
Component:225; Jaccard:0.050709
 |
Component:225; Jaccard:0.032523
 |
Component:338; Jaccard:0.014874
 |
Component:394; Jaccard:0.005399
 |
Correlation:0.069737
  |
Correlation:0.22123
  |
Correlation:-0.076542
  |
Correlation:0.12554
  |
Correlation:0.12554
  |
Correlation:-0.062186
  |
Correlation:0.26812
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
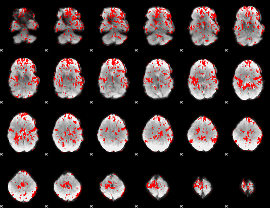 |
GLM binary result thresholded by 1.5
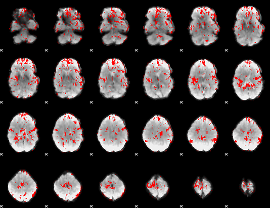 |
GLM binary result thresholded by 2
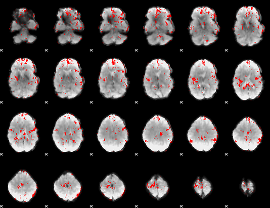 |
GLM binary result thresholded by 2.5
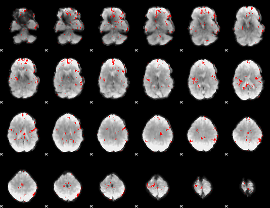 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
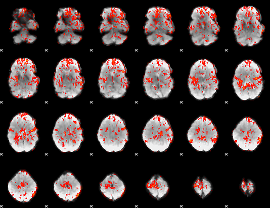 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
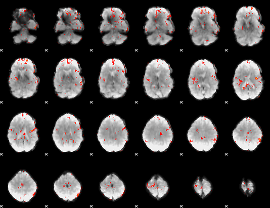 |
GLM Z-score result thresholded by 3
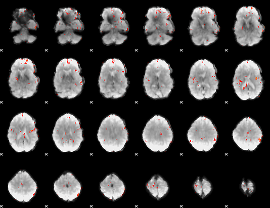 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:047; Jaccard:0.10569
 |
Component:047; Jaccard:0.11525
 |
Component:047; Jaccard:0.11595
 |
Component:047; Jaccard:0.099003
 |
Component:326; Jaccard:0.077042
 |
Component:326; Jaccard:0.058221
 |
Component:326; Jaccard:0.037854
 |
Correlation:0.064359
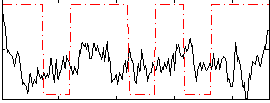  |
Correlation:0.064359
  |
Correlation:0.064359
  |
Correlation:0.064359
  |
Correlation:0.18011
 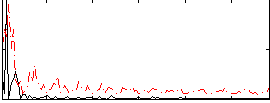 |
Correlation:0.18011
  |
Correlation:0.18011
  |
Component:150; Jaccard:0.090835
 |
Component:150; Jaccard:0.0911
 |
Component:326; Jaccard:0.08769
 |
Component:326; Jaccard:0.089674
 |
Component:092; Jaccard:0.070572
 |
Component:092; Jaccard:0.051217
 |
Component:092; Jaccard:0.030169
 |
Correlation:0.063305
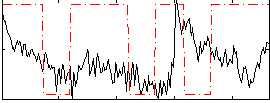  |
Correlation:0.063305
  |
Correlation:0.18011
  |
Correlation:0.18011
  |
Correlation:0.12714
  |
Correlation:0.12714
  |
Correlation:0.12714
  |
Component:187; Jaccard:0.084018
 |
Component:187; Jaccard:0.087611
 |
Component:390; Jaccard:0.085816
 |
Component:092; Jaccard:0.078381
 |
Component:047; Jaccard:0.068163
 |
Component:047; Jaccard:0.044126
 |
Component:047; Jaccard:0.022219
 |
Correlation:0.45558
  |
Correlation:0.45558
  |
Correlation:0.24874
  |
Correlation:0.12714
  |
Correlation:0.064359
  |
Correlation:0.064359
  |
Correlation:0.064359
  |
Component:120; Jaccard:0.081322
 |
Component:390; Jaccard:0.086638
 |
Component:150; Jaccard:0.083622
 |
Component:390; Jaccard:0.075191
 |
Component:390; Jaccard:0.05465
 |
Component:390; Jaccard:0.033057
 |
Component:390; Jaccard:0.017321
 |
Correlation:-0.050725
  |
Correlation:0.24874
  |
Correlation:0.063305
  |
Correlation:0.24874
  |
Correlation:0.24874
  |
Correlation:0.24874
  |
Correlation:0.24874
  |
Component:390; Jaccard:0.080225
 |
Component:120; Jaccard:0.083538
 |
Component:092; Jaccard:0.082554
 |
Component:150; Jaccard:0.068731
 |
Component:150; Jaccard:0.047336
 |
Component:150; Jaccard:0.029918
 |
Component:001; Jaccard:0.016123
 |
Correlation:0.24874
  |
Correlation:-0.050725
  |
Correlation:0.12714
  |
Correlation:0.063305
  |
Correlation:0.063305
  |
Correlation:0.063305
  |
Correlation:0.057533
  |
Component:136; Jaccard:0.074893
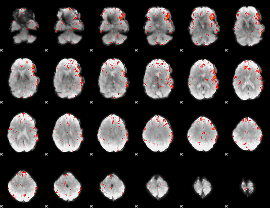 |
Component:326; Jaccard:0.080041
 |
Component:187; Jaccard:0.081342
 |
Component:120; Jaccard:0.063448
 |
Component:004; Jaccard:0.045564
 |
Component:004; Jaccard:0.027931
 |
Component:150; Jaccard:0.014961
 |
Correlation:-0.099597
  |
Correlation:0.18011
  |
Correlation:0.45558
  |
Correlation:-0.050725
  |
Correlation:0.19513
  |
Correlation:0.19513
  |
Correlation:0.063305
  |
Component:004; Jaccard:0.072872
 |
Component:092; Jaccard:0.07596
 |
Component:120; Jaccard:0.077403
 |
Component:187; Jaccard:0.062361
 |
Component:001; Jaccard:0.044682
 |
Component:001; Jaccard:0.027188
 |
Component:305; Jaccard:0.013012
 |
Correlation:0.19513
  |
Correlation:0.12714
  |
Correlation:-0.050725
  |
Correlation:0.45558
  |
Correlation:0.057533
  |
Correlation:0.057533
  |
Correlation:-0.046666
  |
Component:170; Jaccard:0.071983
 |
Component:004; Jaccard:0.075362
 |
Component:004; Jaccard:0.072043
 |
Component:004; Jaccard:0.062157
 |
Component:120; Jaccard:0.040674
 |
Component:120; Jaccard:0.025357
 |
Component:120; Jaccard:0.011999
 |
Correlation:0.042622
  |
Correlation:0.19513
  |
Correlation:0.19513
  |
Correlation:0.19513
  |
Correlation:-0.050725
  |
Correlation:-0.050725
  |
Correlation:-0.050725
  |
Component:300; Jaccard:0.069738
 |
Component:136; Jaccard:0.073712
 |
Component:001; Jaccard:0.070893
 |
Component:001; Jaccard:0.06086
 |
Component:187; Jaccard:0.039717
 |
Component:242; Jaccard:0.023538
 |
Component:004; Jaccard:0.011919
 |
Correlation:0.055586
  |
Correlation:-0.099597
  |
Correlation:0.057533
  |
Correlation:0.057533
  |
Correlation:0.45558
  |
Correlation:0.0010755
  |
Correlation:0.19513
  |
Component:022; Jaccard:0.069715
 |
Component:001; Jaccard:0.069598
 |
Component:136; Jaccard:0.066371
 |
Component:242; Jaccard:0.051503
 |
Component:242; Jaccard:0.037243
 |
Component:187; Jaccard:0.023297
 |
Component:242; Jaccard:0.010815
 |
Correlation:0.22874
  |
Correlation:0.057533
  |
Correlation:-0.099597
  |
Correlation:0.0010755
  |
Correlation:0.0010755
  |
Correlation:0.45558
  |
Correlation:0.0010755
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
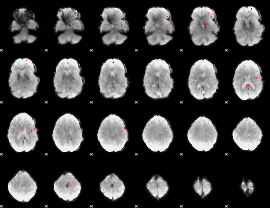 |
GLM Z-score result thresholded by 1
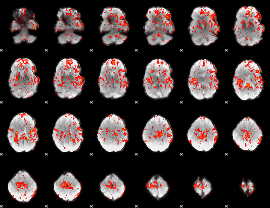 |
GLM Z-score result thresholded by 1.5
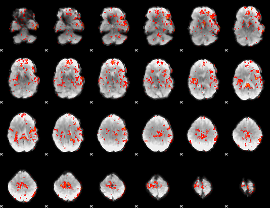 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
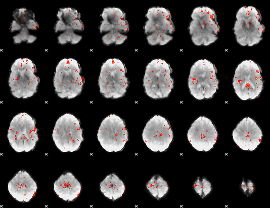 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
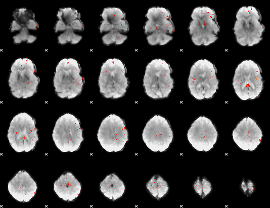 |
GLM Z-score result thresholded by 4
 |
Component:047; Jaccard:0.11813
 |
Component:047; Jaccard:0.12684
 |
Component:047; Jaccard:0.1227
 |
Component:047; Jaccard:0.10608
 |
Component:047; Jaccard:0.071618
 |
Component:326; Jaccard:0.051195
 |
Component:326; Jaccard:0.037488
 |
Correlation:0.24817
  |
Correlation:0.24817
  |
Correlation:0.24817
  |
Correlation:0.24817
  |
Correlation:0.24817
  |
Correlation:0.041414
  |
Correlation:0.041414
  |
Component:136; Jaccard:0.094875
 |
Component:136; Jaccard:0.096655
 |
Component:390; Jaccard:0.090745
 |
Component:326; Jaccard:0.084984
 |
Component:236; Jaccard:0.06994
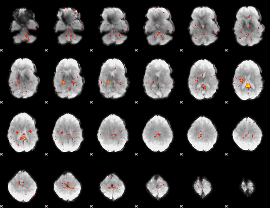 |
Component:236; Jaccard:0.04936
 |
Component:236; Jaccard:0.027555
 |
Correlation:0.17519
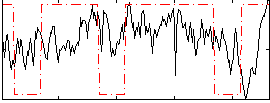 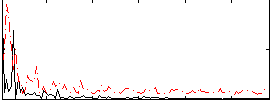 |
Correlation:0.17519
  |
Correlation:0.11197
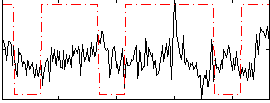 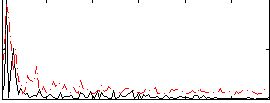 |
Correlation:0.041414
  |
Correlation:0.28699
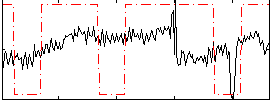 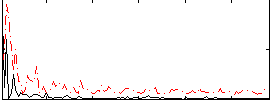 |
Correlation:0.28699
  |
Correlation:0.28699
  |
Component:390; Jaccard:0.084631
 |
Component:390; Jaccard:0.091497
 |
Component:326; Jaccard:0.090078
 |
Component:236; Jaccard:0.082799
 |
Component:326; Jaccard:0.068942
 |
Component:047; Jaccard:0.044101
 |
Component:047; Jaccard:0.025491
 |
Correlation:0.11197
  |
Correlation:0.11197
  |
Correlation:0.041414
  |
Correlation:0.28699
  |
Correlation:0.041414
  |
Correlation:0.24817
  |
Correlation:0.24817
  |
Component:314; Jaccard:0.082602
 |
Component:326; Jaccard:0.085117
 |
Component:136; Jaccard:0.08461
 |
Component:390; Jaccard:0.081616
 |
Component:390; Jaccard:0.05696
 |
Component:083; Jaccard:0.039096
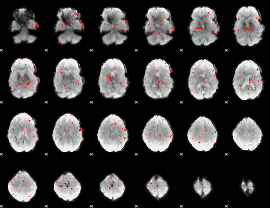 |
Component:083; Jaccard:0.02478
 |
Correlation:0.12076
  |
Correlation:0.041414
  |
Correlation:0.17519
  |
Correlation:0.11197
  |
Correlation:0.11197
  |
Correlation:0.17601
  |
Correlation:0.17601
  |
Component:022; Jaccard:0.078498
 |
Component:314; Jaccard:0.082075
 |
Component:236; Jaccard:0.081215
 |
Component:211; Jaccard:0.066936
 |
Component:083; Jaccard:0.055164
 |
Component:268; Jaccard:0.037682
 |
Component:390; Jaccard:0.023329
 |
Correlation:-0.060724
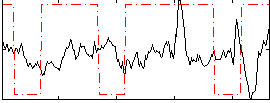 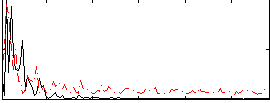 |
Correlation:0.12076
  |
Correlation:0.28699
  |
Correlation:0.41362
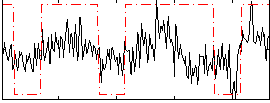  |
Correlation:0.17601
  |
Correlation:0.21993
  |
Correlation:0.11197
  |
Component:157; Jaccard:0.075192
 |
Component:236; Jaccard:0.075506
 |
Component:314; Jaccard:0.074554
 |
Component:136; Jaccard:0.065857
 |
Component:268; Jaccard:0.054054
 |
Component:390; Jaccard:0.037248
 |
Component:092; Jaccard:0.022527
 |
Correlation:0.31038
  |
Correlation:0.28699
  |
Correlation:0.12076
  |
Correlation:0.17519
  |
Correlation:0.21993
  |
Correlation:0.11197
  |
Correlation:0.24434
  |
Component:326; Jaccard:0.074291
 |
Component:157; Jaccard:0.074918
 |
Component:001; Jaccard:0.071745
 |
Component:083; Jaccard:0.065741
 |
Component:329; Jaccard:0.05315
 |
Component:329; Jaccard:0.036015
 |
Component:268; Jaccard:0.021945
 |
Correlation:0.041414
  |
Correlation:0.31038
  |
Correlation:0.28588
  |
Correlation:0.17601
  |
Correlation:0.2351
  |
Correlation:0.2351
  |
Correlation:0.21993
  |
Component:070; Jaccard:0.072496
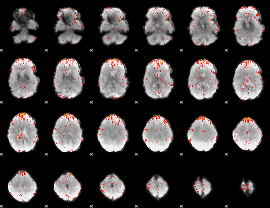 |
Component:001; Jaccard:0.07381
 |
Component:211; Jaccard:0.071143
 |
Component:268; Jaccard:0.063894
 |
Component:211; Jaccard:0.050945
 |
Component:211; Jaccard:0.033762
 |
Component:329; Jaccard:0.021224
 |
Correlation:0.29078
  |
Correlation:0.28588
  |
Correlation:0.41362
  |
Correlation:0.21993
  |
Correlation:0.41362
  |
Correlation:0.41362
  |
Correlation:0.2351
  |
Component:170; Jaccard:0.072457
 |
Component:022; Jaccard:0.072779
 |
Component:083; Jaccard:0.070911
 |
Component:001; Jaccard:0.063053
 |
Component:001; Jaccard:0.050535
 |
Component:092; Jaccard:0.033682
 |
Component:211; Jaccard:0.019206
 |
Correlation:-0.002643
  |
Correlation:-0.060724
  |
Correlation:0.17601
  |
Correlation:0.28588
  |
Correlation:0.28588
  |
Correlation:0.24434
  |
Correlation:0.41362
  |
Component:001; Jaccard:0.071889
 |
Component:211; Jaccard:0.070307
 |
Component:157; Jaccard:0.067567
 |
Component:329; Jaccard:0.062828
 |
Component:092; Jaccard:0.046743
 |
Component:001; Jaccard:0.03129
 |
Component:001; Jaccard:0.015778
 |
Correlation:0.28588
  |
Correlation:0.41362
  |
Correlation:0.31038
  |
Correlation:0.2351
  |
Correlation:0.24434
  |
Correlation:0.28588
  |
Correlation:0.28588
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































