Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
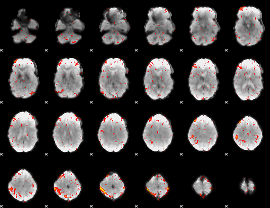
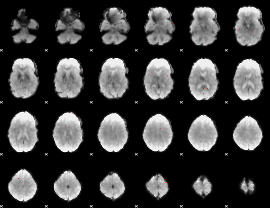
GLM result group-wise
 |
GLM binary result thresholded by 1
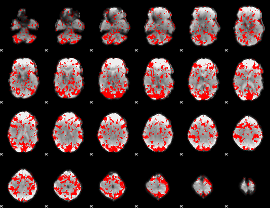 |
GLM binary result thresholded by 1.5
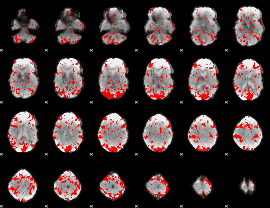 |
GLM binary result thresholded by 2
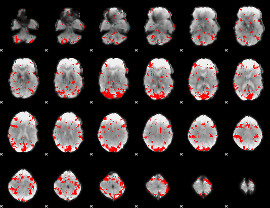 |
GLM binary result thresholded by 2.5
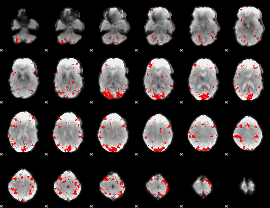 |
GLM binary result thresholded by 3
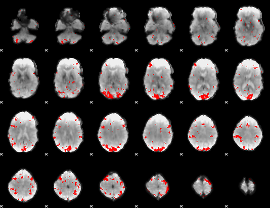 |
GLM binary result thresholded by 3.5
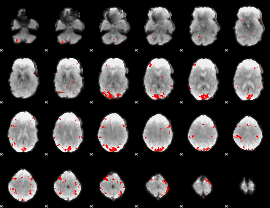 |
GLM binary result thresholded by 4
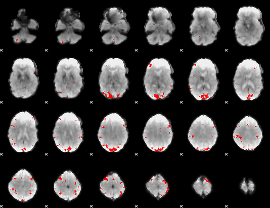 |
GLM Z-score result thresholded by 1
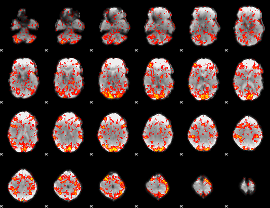 |
GLM Z-score result thresholded by 1.5
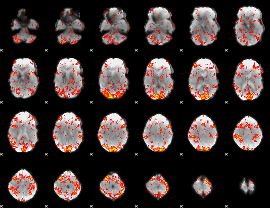 |
GLM Z-score result thresholded by 2
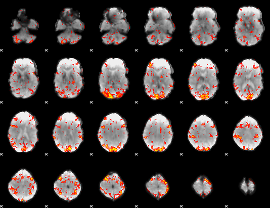 |
GLM Z-score result thresholded by 2.5
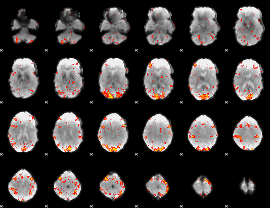 |
GLM Z-score result thresholded by 3
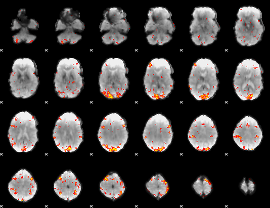 |
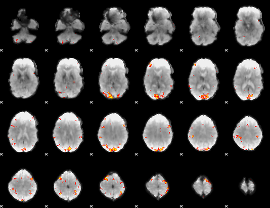
GLM Z-score result thresholded by 3.5
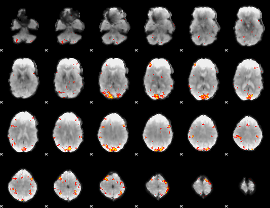 |
GLM Z-score result thresholded by 4
 |
Component:120; Jaccard:0.13946
 |
Component:120; Jaccard:0.1761
 |
Component:120; Jaccard:0.21455
 |
Component:120; Jaccard:0.23898
 |
Component:120; Jaccard:0.24445
 |
Component:204; Jaccard:0.23283
 |
Component:204; Jaccard:0.20397
 |
Correlation:0.22157
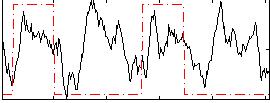 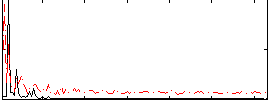 |
Correlation:0.22157
  |
Correlation:0.22157
  |
Correlation:0.22157
  |
Correlation:0.22157
  |
Correlation:0.15821
  |
Correlation:0.15821
  |
Component:260; Jaccard:0.13028
 |
Component:260; Jaccard:0.16155
 |
Component:204; Jaccard:0.19964
 |
Component:204; Jaccard:0.23248
 |
Component:204; Jaccard:0.2444
 |
Component:120; Jaccard:0.23195
 |
Component:120; Jaccard:0.20009
 |
Correlation:0.33498
  |
Correlation:0.33498
  |
Correlation:0.15821
  |
Correlation:0.15821
  |
Correlation:0.15821
  |
Correlation:0.22157
  |
Correlation:0.22157
  |
Component:050; Jaccard:0.1264
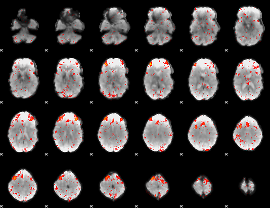 |
Component:204; Jaccard:0.15587
 |
Component:260; Jaccard:0.19158
 |
Component:260; Jaccard:0.20774
 |
Component:260; Jaccard:0.21066
 |
Component:260; Jaccard:0.198
 |
Component:260; Jaccard:0.17454
 |
Correlation:0.11666
  |
Correlation:0.15821
  |
Correlation:0.33498
  |
Correlation:0.33498
  |
Correlation:0.33498
  |
Correlation:0.33498
  |
Correlation:0.33498
  |
Component:204; Jaccard:0.11782
 |
Component:050; Jaccard:0.14289
 |
Component:050; Jaccard:0.15184
 |
Component:091; Jaccard:0.15255
 |
Component:091; Jaccard:0.14185
 |
Component:091; Jaccard:0.12428
 |
Component:091; Jaccard:0.10102
 |
Correlation:0.15821
  |
Correlation:0.11666
  |
Correlation:0.11666
  |
Correlation:0.095114
  |
Correlation:0.095114
  |
Correlation:0.095114
  |
Correlation:0.095114
  |
Component:089; Jaccard:0.11252
 |
Component:091; Jaccard:0.13103
 |
Component:091; Jaccard:0.14711
 |
Component:050; Jaccard:0.14873
 |
Component:089; Jaccard:0.1347
 |
Component:089; Jaccard:0.11429
 |
Component:089; Jaccard:0.092784
 |
Correlation:-0.0030641
  |
Correlation:0.095114
  |
Correlation:0.095114
  |
Correlation:0.11666
  |
Correlation:-0.0030641
  |
Correlation:-0.0030641
  |
Correlation:-0.0030641
  |
Component:144; Jaccard:0.11139
 |
Component:089; Jaccard:0.13009
 |
Component:089; Jaccard:0.14499
 |
Component:089; Jaccard:0.14649
 |
Component:050; Jaccard:0.13435
 |
Component:050; Jaccard:0.1118
 |
Component:050; Jaccard:0.087685
 |
Correlation:0.20137
  |
Correlation:-0.0030641
  |
Correlation:-0.0030641
  |
Correlation:-0.0030641
  |
Correlation:0.11666
  |
Correlation:0.11666
  |
Correlation:0.11666
  |
Component:091; Jaccard:0.10928
 |
Component:144; Jaccard:0.12936
 |
Component:144; Jaccard:0.14002
 |
Component:144; Jaccard:0.14138
 |
Component:144; Jaccard:0.13406
 |
Component:144; Jaccard:0.11089
 |
Component:144; Jaccard:0.086758
 |
Correlation:0.095114
  |
Correlation:0.20137
  |
Correlation:0.20137
  |
Correlation:0.20137
  |
Correlation:0.20137
  |
Correlation:0.20137
  |
Correlation:0.20137
  |
Component:354; Jaccard:0.10192
 |
Component:354; Jaccard:0.11206
 |
Component:393; Jaccard:0.11515
 |
Component:393; Jaccard:0.11203
 |
Component:354; Jaccard:0.10398
 |
Component:354; Jaccard:0.089999
 |
Component:354; Jaccard:0.073061
 |
Correlation:-0.18161
  |
Correlation:-0.18161
  |
Correlation:-0.077797
  |
Correlation:-0.077797
  |
Correlation:-0.18161
  |
Correlation:-0.18161
  |
Correlation:-0.18161
  |
Component:393; Jaccard:0.094475
 |
Component:393; Jaccard:0.10808
 |
Component:354; Jaccard:0.1142
 |
Component:354; Jaccard:0.11181
 |
Component:393; Jaccard:0.099047
 |
Component:393; Jaccard:0.081678
 |
Component:393; Jaccard:0.064763
 |
Correlation:-0.077797
  |
Correlation:-0.077797
  |
Correlation:-0.18161
  |
Correlation:-0.18161
  |
Correlation:-0.077797
  |
Correlation:-0.077797
  |
Correlation:-0.077797
  |
Component:015; Jaccard:0.091431
 |
Component:015; Jaccard:0.10085
 |
Component:015; Jaccard:0.10511
 |
Component:015; Jaccard:0.10216
 |
Component:015; Jaccard:0.093548
 |
Component:054; Jaccard:0.079079
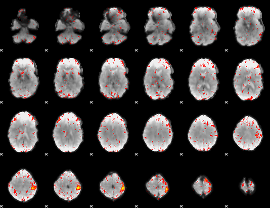 |
Component:054; Jaccard:0.063026
 |
Correlation:0.035425
  |
Correlation:0.035425
  |
Correlation:0.035425
  |
Correlation:0.035425
  |
Correlation:0.035425
  |
Correlation:-0.02901
  |
Correlation:-0.02901
  |
Cope: 02:REWARD BACK
|
GLM result group-wise
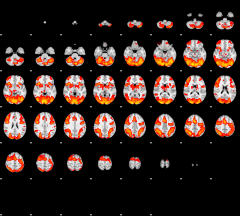 |
GLM binary result thresholded by 1
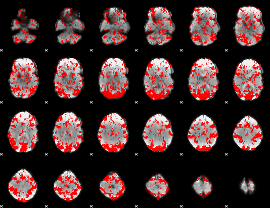 |
GLM binary result thresholded by 1.5
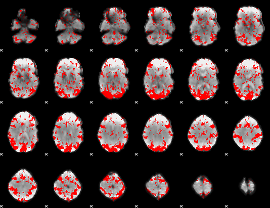 |
GLM binary result thresholded by 2
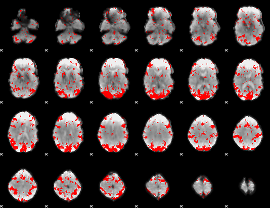 |
GLM binary result thresholded by 2.5
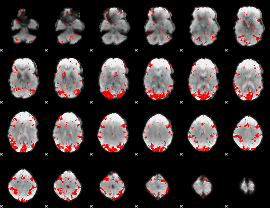 |
GLM binary result thresholded by 3
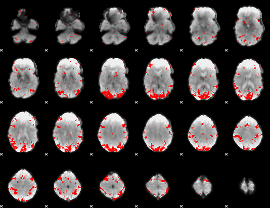 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
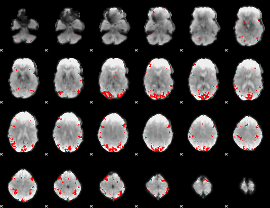 |
GLM Z-score result thresholded by 1
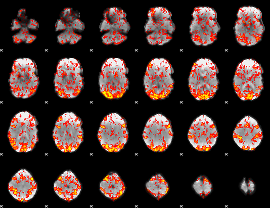 |
GLM Z-score result thresholded by 1.5
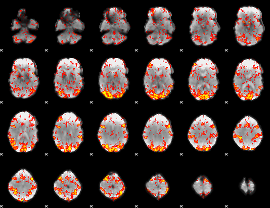 |
GLM Z-score result thresholded by 2
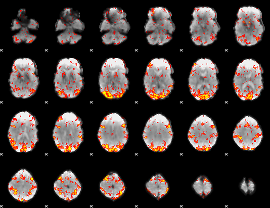 |
GLM Z-score result thresholded by 2.5
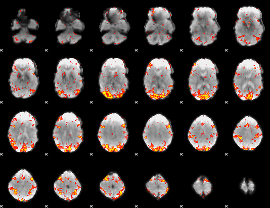 |
GLM Z-score result thresholded by 3
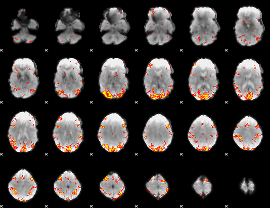 |
GLM Z-score result thresholded by 3.5
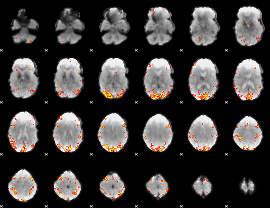 |
GLM Z-score result thresholded by 4
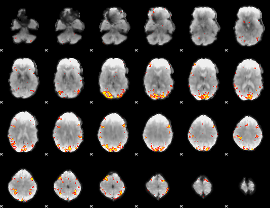 |
Component:120; Jaccard:0.13648
 |
Component:120; Jaccard:0.17521
 |
Component:120; Jaccard:0.21732
 |
Component:120; Jaccard:0.25483
 |
Component:204; Jaccard:0.28319
 |
Component:204; Jaccard:0.30354
 |
Component:204; Jaccard:0.30583
 |
Correlation:0.36291
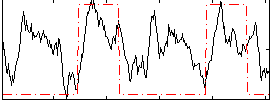 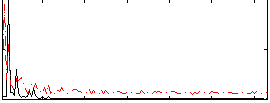 |
Correlation:0.36291
  |
Correlation:0.36291
  |
Correlation:0.36291
  |
Correlation:0.26739
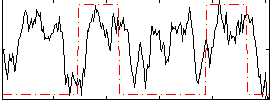 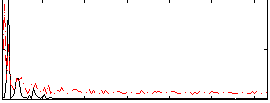 |
Correlation:0.26739
  |
Correlation:0.26739
  |
Component:354; Jaccard:0.13323
 |
Component:089; Jaccard:0.15668
 |
Component:204; Jaccard:0.20001
 |
Component:204; Jaccard:0.24511
 |
Component:120; Jaccard:0.27925
 |
Component:120; Jaccard:0.28823
 |
Component:120; Jaccard:0.27835
 |
Correlation:0.2775
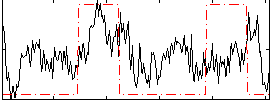  |
Correlation:0.16222
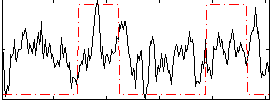 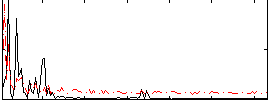 |
Correlation:0.26739
  |
Correlation:0.26739
  |
Correlation:0.36291
  |
Correlation:0.36291
  |
Correlation:0.36291
  |
Component:089; Jaccard:0.13206
 |
Component:354; Jaccard:0.15427
 |
Component:260; Jaccard:0.18672
 |
Component:260; Jaccard:0.21021
 |
Component:260; Jaccard:0.22652
 |
Component:260; Jaccard:0.22481
 |
Component:260; Jaccard:0.21799
 |
Correlation:0.16222
  |
Correlation:0.2775
  |
Correlation:0.20629
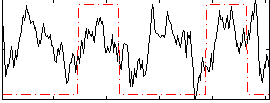 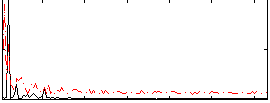 |
Correlation:0.20629
  |
Correlation:0.20629
  |
Correlation:0.20629
  |
Correlation:0.20629
  |
Component:260; Jaccard:0.12359
 |
Component:260; Jaccard:0.15349
 |
Component:089; Jaccard:0.17869
 |
Component:089; Jaccard:0.18933
 |
Component:089; Jaccard:0.19022
 |
Component:089; Jaccard:0.18224
 |
Component:089; Jaccard:0.16726
 |
Correlation:0.20629
  |
Correlation:0.20629
  |
Correlation:0.16222
  |
Correlation:0.16222
  |
Correlation:0.16222
  |
Correlation:0.16222
  |
Correlation:0.16222
  |
Component:204; Jaccard:0.1147
 |
Component:204; Jaccard:0.15287
 |
Component:354; Jaccard:0.16909
 |
Component:354; Jaccard:0.17111
 |
Component:354; Jaccard:0.16533
 |
Component:354; Jaccard:0.15376
 |
Component:354; Jaccard:0.13598
 |
Correlation:0.26739
  |
Correlation:0.26739
  |
Correlation:0.2775
  |
Correlation:0.2775
  |
Correlation:0.2775
  |
Correlation:0.2775
  |
Correlation:0.2775
  |
Component:252; Jaccard:0.1134
 |
Component:252; Jaccard:0.13149
 |
Component:090; Jaccard:0.14592
 |
Component:090; Jaccard:0.15321
 |
Component:252; Jaccard:0.14919
 |
Component:252; Jaccard:0.13926
 |
Component:252; Jaccard:0.12202
 |
Correlation:0.32142
  |
Correlation:0.32142
  |
Correlation:0.27343
  |
Correlation:0.27343
  |
Correlation:0.32142
  |
Correlation:0.32142
  |
Correlation:0.32142
  |
Component:090; Jaccard:0.11282
 |
Component:090; Jaccard:0.13028
 |
Component:252; Jaccard:0.14452
 |
Component:252; Jaccard:0.15133
 |
Component:090; Jaccard:0.14827
 |
Component:090; Jaccard:0.13737
 |
Component:090; Jaccard:0.11965
 |
Correlation:0.27343
  |
Correlation:0.27343
  |
Correlation:0.32142
  |
Correlation:0.32142
  |
Correlation:0.27343
  |
Correlation:0.27343
  |
Correlation:0.27343
  |
Component:393; Jaccard:0.10707
 |
Component:393; Jaccard:0.12525
 |
Component:393; Jaccard:0.13879
 |
Component:393; Jaccard:0.14512
 |
Component:393; Jaccard:0.14382
 |
Component:393; Jaccard:0.13425
 |
Component:393; Jaccard:0.11819
 |
Correlation:0.26877
  |
Correlation:0.26877
  |
Correlation:0.26877
  |
Correlation:0.26877
  |
Correlation:0.26877
  |
Correlation:0.26877
  |
Correlation:0.26877
  |
Component:050; Jaccard:0.10145
 |
Component:091; Jaccard:0.11442
 |
Component:091; Jaccard:0.12618
 |
Component:091; Jaccard:0.13206
 |
Component:091; Jaccard:0.1288
 |
Component:091; Jaccard:0.11986
 |
Component:091; Jaccard:0.10973
 |
Correlation:-0.032856
  |
Correlation:0.06683
  |
Correlation:0.06683
  |
Correlation:0.06683
  |
Correlation:0.06683
  |
Correlation:0.06683
  |
Correlation:0.06683
  |
Component:091; Jaccard:0.10004
 |
Component:050; Jaccard:0.11189
 |
Component:144; Jaccard:0.12182
 |
Component:144; Jaccard:0.12601
 |
Component:144; Jaccard:0.12287
 |
Component:083; Jaccard:0.114
 |
Component:144; Jaccard:0.10126
 |
Correlation:0.06683
  |
Correlation:-0.032856
  |
Correlation:0.081521
  |
Correlation:0.081521
  |
Correlation:0.081521
  |
Correlation:0.079001
 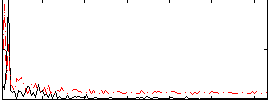 |
Correlation:0.081521
  |
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
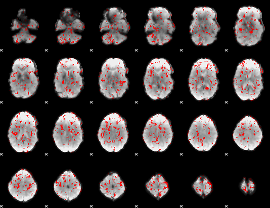 |
GLM binary result thresholded by 2
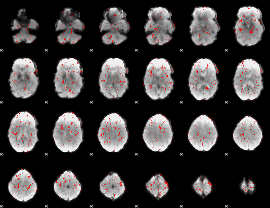 |
GLM binary result thresholded by 2.5
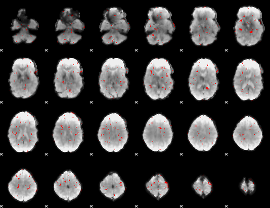 |
GLM binary result thresholded by 3
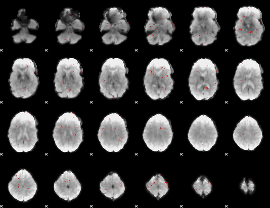 |
GLM binary result thresholded by 3.5
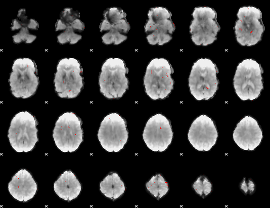 |
GLM binary result thresholded by 4
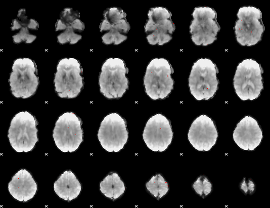 |
GLM Z-score result thresholded by 1
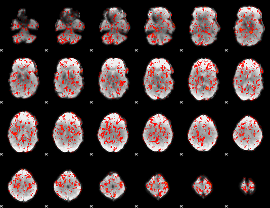 |
GLM Z-score result thresholded by 1.5
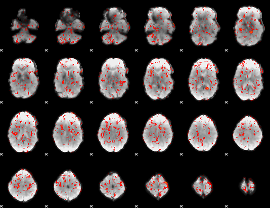 |
GLM Z-score result thresholded by 2
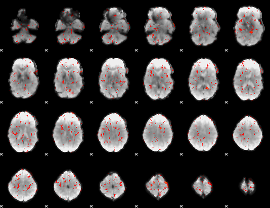 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
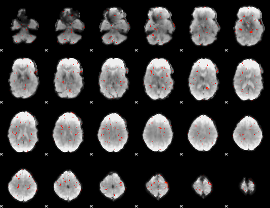
GLM Z-score result thresholded by 3.5
 |
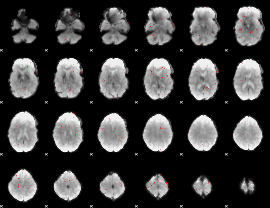
GLM Z-score result thresholded by 4
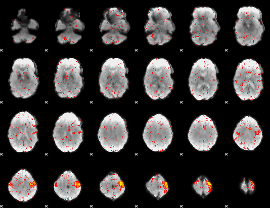
 |
Component:322; Jaccard:0.079901
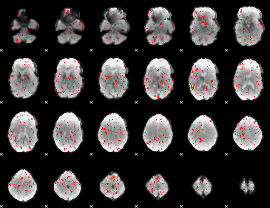
 |
Component:250; Jaccard:0.078767
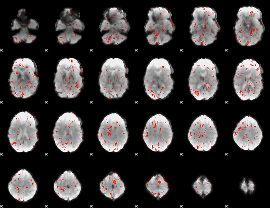
 |
Component:098; Jaccard:0.066965
 |
Component:250; Jaccard:0.043883
 |
Component:175; Jaccard:0.023496
 |
Component:175; Jaccard:0.012014
 |
Component:175; Jaccard:0.005344
 |
Correlation:0.075123
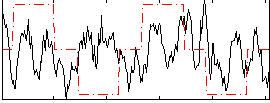 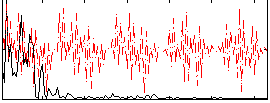 |
Correlation:0.1551
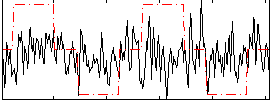 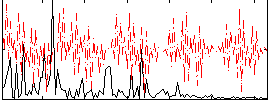 |
Correlation:0.33328
  |
Correlation:0.1551
  |
Correlation:0.16679
  |
Correlation:0.16679
  |
Correlation:0.16679
  |
Component:250; Jaccard:0.079235
 |
Component:322; Jaccard:0.07692
 |
Component:250; Jaccard:0.064961
 |
Component:322; Jaccard:0.042105
 |
Component:011; Jaccard:0.021301
 |
Component:011; Jaccard:0.008686
 |
Component:011; Jaccard:0.00354
 |
Correlation:0.1551
  |
Correlation:0.075123
  |
Correlation:0.1551
  |
Correlation:0.075123
  |
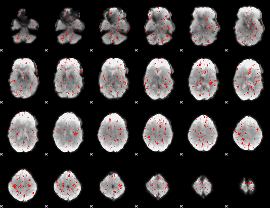
Correlation:-0.07485
  |
Correlation:-0.07485
  |
Correlation:-0.07485
  |
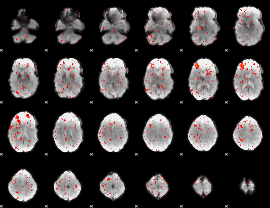
Component:106; Jaccard:0.0726
 |
Component:098; Jaccard:0.07645
 |
Component:322; Jaccard:0.061477
 |
Component:098; Jaccard:0.041906
 |
Component:250; Jaccard:0.021214
 |
Component:250; Jaccard:0.008489
 |
Component:328; Jaccard:0.003492
 |
Correlation:0.2248
  |
Correlation:0.33328
  |
Correlation:0.075123
  |
Correlation:0.33328
  |
Correlation:0.1551
  |
Correlation:0.1551
  |
Correlation:0.17409
  |
Component:109; Jaccard:0.071141
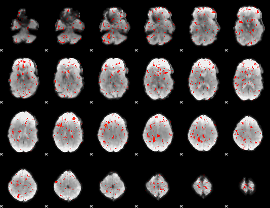 |
Component:106; Jaccard:0.069757
 |
Component:106; Jaccard:0.058924
 |
Component:011; Jaccard:0.040346
 |
Component:322; Jaccard:0.020438
 |
Component:098; Jaccard:0.008399
 |
Component:138; Jaccard:0.003411
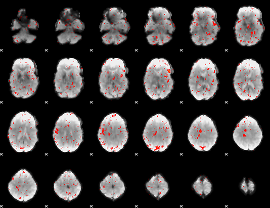 |
Correlation:0.12828
  |
Correlation:0.2248
  |
Correlation:0.2248
  |
Correlation:-0.07485
  |
Correlation:0.075123
  |
Correlation:0.33328
  |
Correlation:0.16348
  |
Component:098; Jaccard:0.070931
 |
Component:377; Jaccard:0.064972
 |
Component:377; Jaccard:0.053269
 |
Component:175; Jaccard:0.038661
 |
Component:098; Jaccard:0.019504
 |
Component:281; Jaccard:0.008291
 |
Component:322; Jaccard:0.003383
 |
Correlation:0.33328
  |
Correlation:0.19443
  |
Correlation:0.19443
  |
Correlation:0.16679
  |
Correlation:0.33328
  |
Correlation:0.27892
  |
Correlation:0.075123
  |
Component:328; Jaccard:0.068647
 |
Component:328; Jaccard:0.064767
 |
Component:281; Jaccard:0.053127
 |
Component:281; Jaccard:0.036717
 |
Component:262; Jaccard:0.017667
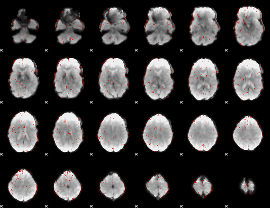 |
Component:322; Jaccard:0.007679
 |
Component:098; Jaccard:0.003167
 |
Correlation:0.17409
  |
Correlation:0.17409
  |
Correlation:0.27892
  |
Correlation:0.27892
  |
Correlation:0.056253
  |
Correlation:0.075123
  |
Correlation:0.33328
  |
Component:060; Jaccard:0.066162
 |
Component:109; Jaccard:0.064365
 |
Component:175; Jaccard:0.05272
 |
Component:106; Jaccard:0.035127
 |
Component:194; Jaccard:0.017381
 |
Component:106; Jaccard:0.007514
 |
Component:106; Jaccard:0.003026
 |
Correlation:0.30081
  |
Correlation:0.12828
  |
Correlation:0.16679
  |
Correlation:0.2248
  |
Correlation:0.10709
  |
Correlation:0.2248
  |
Correlation:0.2248
  |
Component:377; Jaccard:0.065822
 |
Component:175; Jaccard:0.062306
 |
Component:011; Jaccard:0.052107
 |
Component:377; Jaccard:0.03219
 |
Component:106; Jaccard:0.016801
 |
Component:262; Jaccard:0.00744
 |
Component:164; Jaccard:0.002975
 |
Correlation:0.19443
  |
Correlation:0.16679
  |
Correlation:-0.07485
  |
Correlation:0.19443
  |
Correlation:0.2248
  |
Correlation:0.056253
  |
Correlation:0.25674
  |
Component:175; Jaccard:0.063051
 |
Component:060; Jaccard:0.060118
 |
Component:328; Jaccard:0.049196
 |
Component:194; Jaccard:0.031149
 |
Component:281; Jaccard:0.016396
 |
Component:328; Jaccard:0.00706
 |
Component:281; Jaccard:0.002669
 |
Correlation:0.16679
  |
Correlation:0.30081
  |
Correlation:0.17409
  |
Correlation:0.10709
  |
Correlation:0.27892
  |
Correlation:0.17409
  |
Correlation:0.27892
  |
Component:381; Jaccard:0.060051
 |
Component:281; Jaccard:0.057198
 |
Component:060; Jaccard:0.047852
 |
Component:262; Jaccard:0.03114
 |
Component:060; Jaccard:0.015402
 |
Component:138; Jaccard:0.007058
 |
Component:194; Jaccard:0.002601
 |
Correlation:-0.033766
  |
Correlation:0.27892
  |
Correlation:0.30081
  |
Correlation:0.056253
  |
Correlation:0.30081
  |
Correlation:0.16348
  |
Correlation:0.10709
  |
Cope: 04:neg_PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
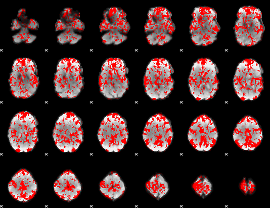 |
GLM binary result thresholded by 1.5
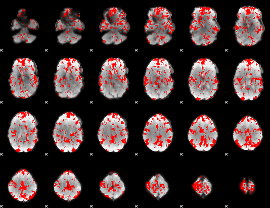 |
GLM binary result thresholded by 2
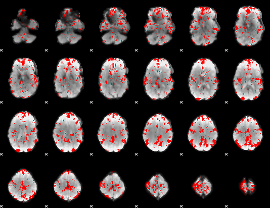 |
GLM binary result thresholded by 2.5
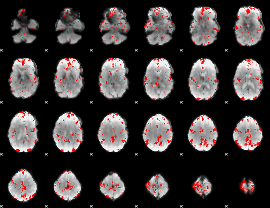 |
GLM binary result thresholded by 3
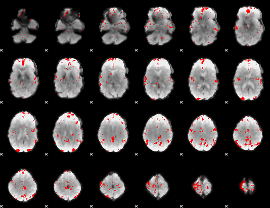 |
GLM binary result thresholded by 3.5
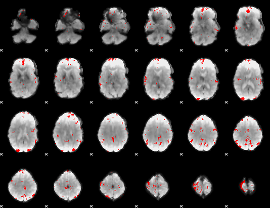 |
GLM binary result thresholded by 4
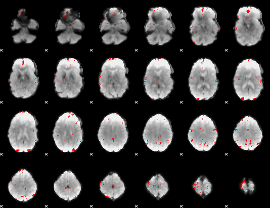 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
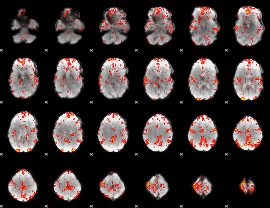 |
GLM Z-score result thresholded by 2.5
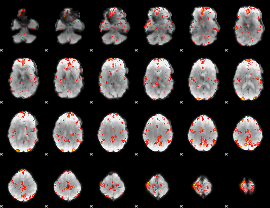 |
GLM Z-score result thresholded by 3
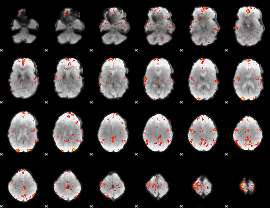 |
GLM Z-score result thresholded by 3.5
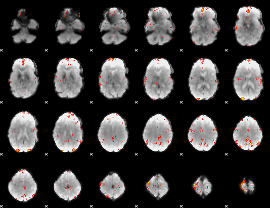 |
GLM Z-score result thresholded by 4
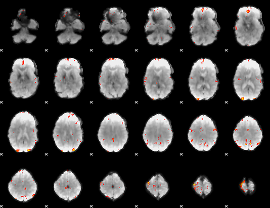 |
Component:218; Jaccard:0.11908
 |
Component:147; Jaccard:0.14546
 |
Component:147; Jaccard:0.17019
 |
Component:147; Jaccard:0.18534
 |
Component:147; Jaccard:0.17684
 |
Component:147; Jaccard:0.14939
 |
Component:147; Jaccard:0.10945
 |
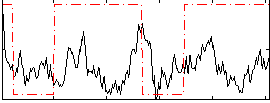
Correlation:0.40265
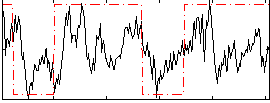 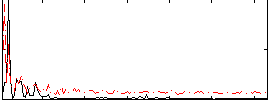 |
Correlation:0.083687
 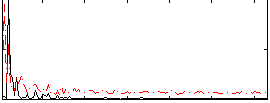 |
Correlation:0.083687
  |
Correlation:0.083687
  |
Correlation:0.083687
  |
Correlation:0.083687
  |
Correlation:0.083687
  |
Component:147; Jaccard:0.1188
 |
Component:218; Jaccard:0.14369
 |
Component:218; Jaccard:0.16418
 |
Component:218; Jaccard:0.1735
 |
Component:218; Jaccard:0.1648
 |
Component:218; Jaccard:0.13546
 |
Component:218; Jaccard:0.09481
 |
Correlation:0.083687
  |
Correlation:0.40265
  |
Correlation:0.40265
  |
Correlation:0.40265
  |
Correlation:0.40265
  |
Correlation:0.40265
  |
Correlation:0.40265
  |
Component:399; Jaccard:0.10946
 |
Component:399; Jaccard:0.13062
 |
Component:399; Jaccard:0.15022
 |
Component:399; Jaccard:0.15566
 |
Component:399; Jaccard:0.14285
 |
Component:399; Jaccard:0.11381
 |
Component:399; Jaccard:0.078483
 |
Correlation:0.092014
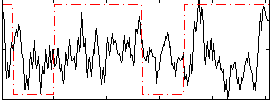  |
Correlation:0.092014
  |
Correlation:0.092014
  |
Correlation:0.092014
  |
Correlation:0.092014
  |
Correlation:0.092014
  |
Correlation:0.092014
  |
Component:012; Jaccard:0.099985
 |
Component:012; Jaccard:0.11863
 |
Component:012; Jaccard:0.13551
 |
Component:012; Jaccard:0.1437
 |
Component:012; Jaccard:0.13622
 |
Component:012; Jaccard:0.11131
 |
Component:012; Jaccard:0.07583
 |
Correlation:0.24313
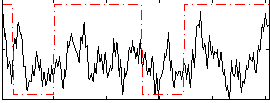  |
Correlation:0.24313
  |
Correlation:0.24313
  |
Correlation:0.24313
  |
Correlation:0.24313
  |
Correlation:0.24313
  |
Correlation:0.24313
  |
Component:247; Jaccard:0.09321
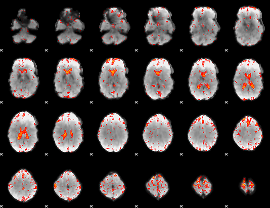 |
Component:105; Jaccard:0.1006
 |
Component:105; Jaccard:0.10578
 |
Component:105; Jaccard:0.098447
 |
Component:343; Jaccard:0.084392
 |
Component:343; Jaccard:0.066828
 |
Component:343; Jaccard:0.045437
 |
Correlation:0.14136
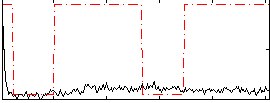 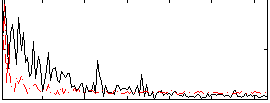 |
Correlation:0.17945
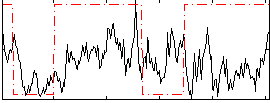  |
Correlation:0.17945
  |
Correlation:0.17945
  |
Correlation:0.18416
  |
Correlation:0.18416
  |
Correlation:0.18416
  |
Component:030; Jaccard:0.092342
 |
Component:030; Jaccard:0.097615
 |
Component:343; Jaccard:0.097786
 |
Component:343; Jaccard:0.095696
 |
Component:356; Jaccard:0.082022
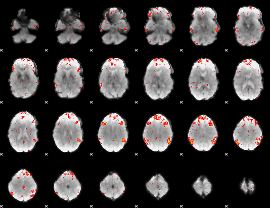 |
Component:233; Jaccard:0.065083
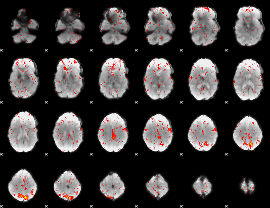 |
Component:233; Jaccard:0.044995
 |
Correlation:0.15503
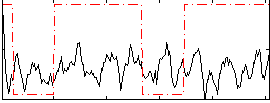 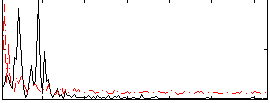 |
Correlation:0.15503
  |
Correlation:0.18416
  |
Correlation:0.18416
  |
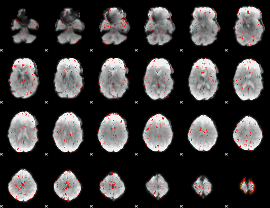
Correlation:0.25102
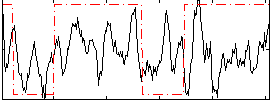  |
Correlation:0.14603
  |
Correlation:0.14603
  |
Component:105; Jaccard:0.087676
 |
Component:343; Jaccard:0.094622
 |
Component:030; Jaccard:0.097707
 |
Component:356; Jaccard:0.092855
 |
Component:233; Jaccard:0.081466
 |
Component:356; Jaccard:0.06261
 |
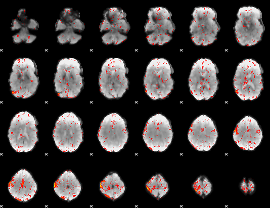
Component:035; Jaccard:0.041538
 |
Correlation:0.17945
  |
Correlation:0.18416
  |
Correlation:0.15503
  |
Correlation:0.25102
  |
Correlation:0.14603
  |
Correlation:0.25102
  |
Correlation:0.057355
  |
Component:343; Jaccard:0.087633
 |
Component:247; Jaccard:0.09386
 |
Component:356; Jaccard:0.095014
 |
Component:233; Jaccard:0.089766
 |
Component:105; Jaccard:0.080284
 |
Component:105; Jaccard:0.060009
 |
Component:356; Jaccard:0.040465
 |
Correlation:0.18416
  |
Correlation:0.14136
  |
Correlation:0.25102
  |
Correlation:0.14603
  |
Correlation:0.17945
  |
Correlation:0.17945
  |
Correlation:0.25102
  |
Component:139; Jaccard:0.084379
 |
Component:139; Jaccard:0.090982
 |
Component:139; Jaccard:0.093301
 |
Component:030; Jaccard:0.087297
 |
Component:202; Jaccard:0.072578
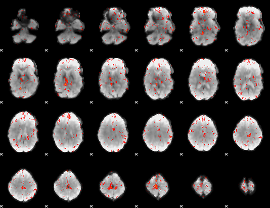 |
Component:035; Jaccard:0.05693
 |
Component:202; Jaccard:0.037564
 |
Correlation:-0.01511
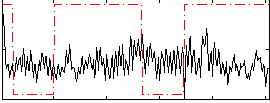 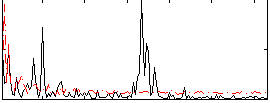 |
Correlation:-0.01511
  |
Correlation:-0.01511
  |
Correlation:0.15503
  |
Correlation:0.016875
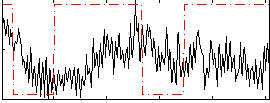  |
Correlation:0.057355
  |
Correlation:0.016875
  |
Component:371; Jaccard:0.083234
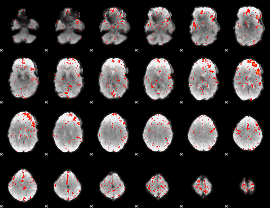 |
Component:356; Jaccard:0.088849
 |
Component:202; Jaccard:0.089573
 |
Component:139; Jaccard:0.085181
 |
Component:035; Jaccard:0.070369
 |
Component:202; Jaccard:0.053679
 |
Component:105; Jaccard:0.037287
 |
Correlation:0.026975
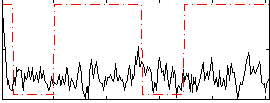 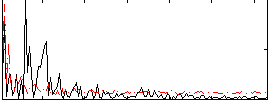 |
Correlation:0.25102
  |
Correlation:0.016875
  |
Correlation:-0.01511
  |
Correlation:0.057355
  |
Correlation:0.016875
  |
Correlation:0.17945
  |
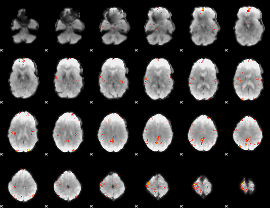
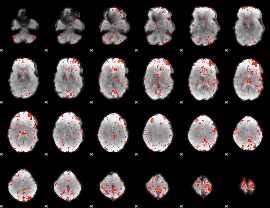
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
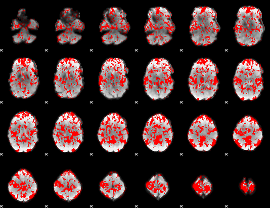 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
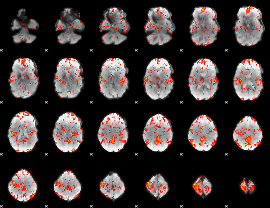 |
GLM Z-score result thresholded by 2.5
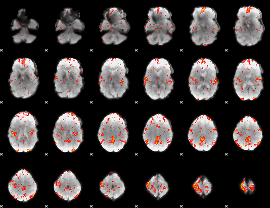 |
GLM Z-score result thresholded by 3
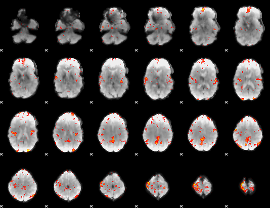 |
GLM Z-score result thresholded by 3.5
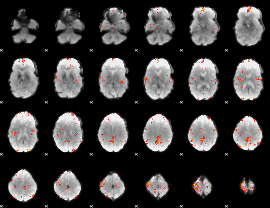 |
GLM Z-score result thresholded by 4
 |
Component:147; Jaccard:0.1345
 |
Component:147; Jaccard:0.17086
 |
Component:147; Jaccard:0.2075
 |
Component:147; Jaccard:0.22986
 |
Component:147; Jaccard:0.22221
 |
Component:147; Jaccard:0.1878
 |
Component:147; Jaccard:0.14394
 |
Correlation:0.17328
 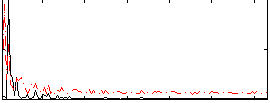 |
Correlation:0.17328
  |
Correlation:0.17328
  |
Correlation:0.17328
  |
Correlation:0.17328
  |
Correlation:0.17328
  |
Correlation:0.17328
  |
Component:399; Jaccard:0.11841
 |
Component:399; Jaccard:0.14262
 |
Component:399; Jaccard:0.16512
 |
Component:399; Jaccard:0.17497
 |
Component:399; Jaccard:0.15934
 |
Component:012; Jaccard:0.12819
 |
Component:012; Jaccard:0.097808
 |
Correlation:0.16151
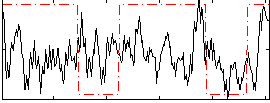 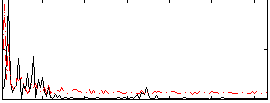 |
Correlation:0.16151
  |
Correlation:0.16151
  |
Correlation:0.16151
  |
Correlation:0.16151
  |
Correlation:0.082342
 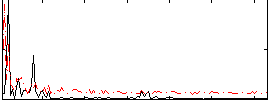 |
Correlation:0.082342
  |
Component:012; Jaccard:0.10731
 |
Component:012; Jaccard:0.12976
 |
Component:012; Jaccard:0.14839
 |
Component:012; Jaccard:0.15839
 |
Component:012; Jaccard:0.15496
 |
Component:399; Jaccard:0.126
 |
Component:399; Jaccard:0.090315
 |
Correlation:0.082342
  |
Correlation:0.082342
  |
Correlation:0.082342
  |
Correlation:0.082342
  |
Correlation:0.082342
  |
Correlation:0.16151
  |
Correlation:0.16151
  |
Component:343; Jaccard:0.095351
 |
Component:343; Jaccard:0.10685
 |
Component:343; Jaccard:0.11671
 |
Component:343; Jaccard:0.11345
 |
Component:343; Jaccard:0.10119
 |
Component:343; Jaccard:0.082647
 |
Component:343; Jaccard:0.060129
 |
Correlation:0.10375
 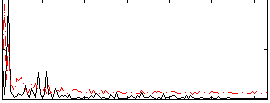 |
Correlation:0.10375
  |
Correlation:0.10375
  |
Correlation:0.10375
  |
Correlation:0.10375
  |
Correlation:0.10375
  |
Correlation:0.10375
  |
Component:218; Jaccard:0.08654
 |
Component:218; Jaccard:0.09667
 |
Component:218; Jaccard:0.10062
 |
Component:218; Jaccard:0.096934
 |
Component:218; Jaccard:0.087076
 |
Component:218; Jaccard:0.069868
 |
Component:218; Jaccard:0.052059
 |
Correlation:0.0021158
  |
Correlation:0.0021158
  |
Correlation:0.0021158
  |
Correlation:0.0021158
  |
Correlation:0.0021158
  |
Correlation:0.0021158
  |
Correlation:0.0021158
  |
Component:124; Jaccard:0.082351
 |
Component:124; Jaccard:0.089934
 |
Component:124; Jaccard:0.095236
 |
Component:124; Jaccard:0.089835
 |
Component:124; Jaccard:0.07973
 |
Component:171; Jaccard:0.064314
 |
Component:171; Jaccard:0.050874
 |
Correlation:0.23747
 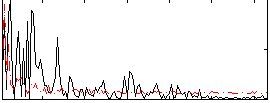 |
Correlation:0.23747
  |
Correlation:0.23747
  |
Correlation:0.23747
  |
Correlation:0.23747
  |
Correlation:0.15807
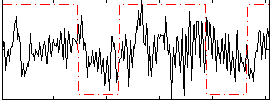  |
Correlation:0.15807
  |
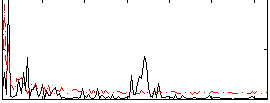
Component:107; Jaccard:0.079979
 |
Component:107; Jaccard:0.080879
 |
Component:171; Jaccard:0.079938
 |
Component:171; Jaccard:0.082328
 |
Component:171; Jaccard:0.076993
 |
Component:124; Jaccard:0.061258
 |
Component:124; Jaccard:0.042945
 |
Correlation:0.14503
  |
Correlation:0.14503
  |
Correlation:0.15807
  |
Correlation:0.15807
  |
Correlation:0.15807
  |
Correlation:0.23747
  |
Correlation:0.23747
  |
Component:371; Jaccard:0.076786
 |
Component:371; Jaccard:0.078387
 |
Component:107; Jaccard:0.078075
 |
Component:098; Jaccard:0.076569
 |
Component:098; Jaccard:0.06481
 |
Component:098; Jaccard:0.048678
 |
Component:139; Jaccard:0.034576
 |
Correlation:0.095821
  |
Correlation:0.095821
  |
Correlation:0.14503
  |
Correlation:0.3887
  |
Correlation:0.3887
  |
Correlation:0.3887
  |
Correlation:0.10737
  |
Component:030; Jaccard:0.075489
 |
Component:177; Jaccard:0.075786
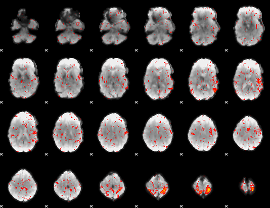 |
Component:098; Jaccard:0.077643
 |
Component:371; Jaccard:0.07065
 |
Component:139; Jaccard:0.060093
 |
Component:139; Jaccard:0.046947
 |
Component:035; Jaccard:0.033785
 |
Correlation:0.044131
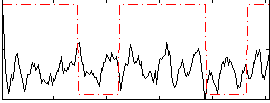 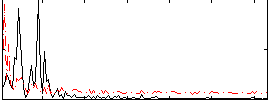 |
Correlation:0.045883
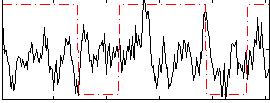 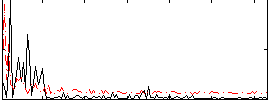 |
Correlation:0.3887
  |
Correlation:0.095821
  |
Correlation:0.10737
  |
Correlation:0.10737
  |
Correlation:0.14558
  |
Component:139; Jaccard:0.073248
 |
Component:356; Jaccard:0.075654
 |
Component:371; Jaccard:0.076613
 |
Component:139; Jaccard:0.070394
 |
Component:035; Jaccard:0.059281
 |
Component:035; Jaccard:0.046396
 |
Component:360; Jaccard:0.03293
 |
Correlation:0.10737
  |
Correlation:0.026867
  |
Correlation:0.095821
  |
Correlation:0.10737
  |
Correlation:0.14558
  |
Correlation:0.14558
  |
Correlation:0.18233
  |
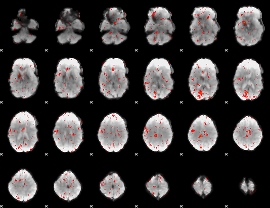
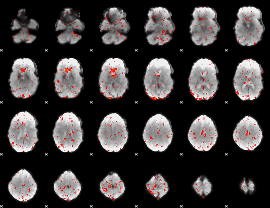
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
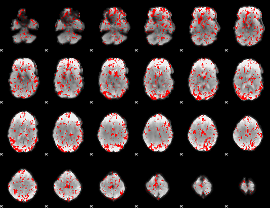 |
GLM binary result thresholded by 2
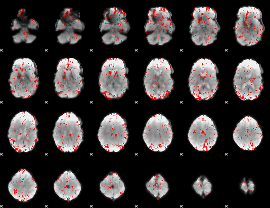 |
GLM binary result thresholded by 2.5
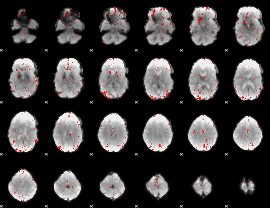 |
GLM binary result thresholded by 3
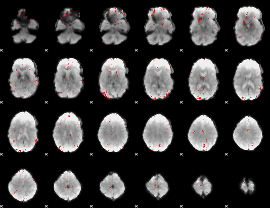 |
GLM binary result thresholded by 3.5
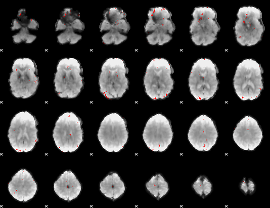 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
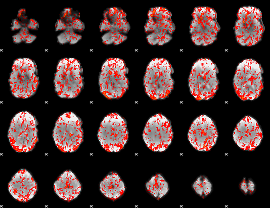 |
GLM Z-score result thresholded by 1.5
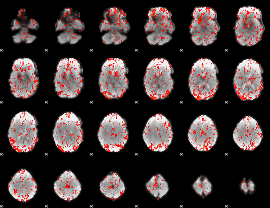 |
GLM Z-score result thresholded by 2
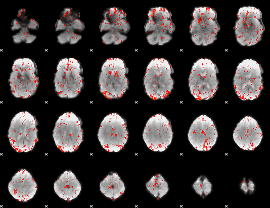 |
GLM Z-score result thresholded by 2.5
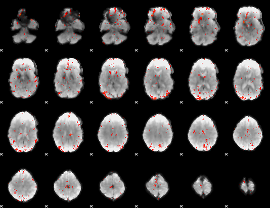 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
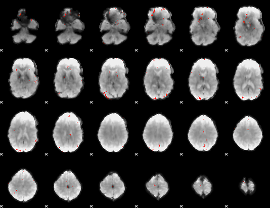 |
GLM Z-score result thresholded by 4
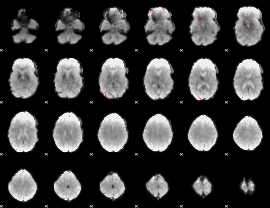 |
Component:252; Jaccard:0.095988
 |
Component:252; Jaccard:0.10613
 |
Component:252; Jaccard:0.1025
 |
Component:252; Jaccard:0.081109
 |
Component:023; Jaccard:0.050148
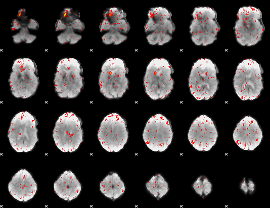 |
Component:023; Jaccard:0.028753
 |
Component:023; Jaccard:0.014378
 |
Correlation:0.16662
  |
Correlation:0.16662
  |
Correlation:0.16662
  |
Correlation:0.16662
  |
Correlation:0.47314
  |
Correlation:0.47314
  |
Correlation:0.47314
  |
Component:090; Jaccard:0.090114
 |
Component:090; Jaccard:0.097386
 |
Component:090; Jaccard:0.088932
 |
Component:023; Jaccard:0.073537
 |
Component:252; Jaccard:0.048836
 |
Component:252; Jaccard:0.024469
 |
Component:252; Jaccard:0.010431
 |
Correlation:0.13362
  |
Correlation:0.13362
  |
Correlation:0.13362
  |
Correlation:0.47314
  |
Correlation:0.16662
  |
Correlation:0.16662
  |
Correlation:0.16662
  |
Component:354; Jaccard:0.088662
 |
Component:354; Jaccard:0.091044
 |
Component:023; Jaccard:0.087239
 |
Component:090; Jaccard:0.067156
 |
Component:025; Jaccard:0.039038
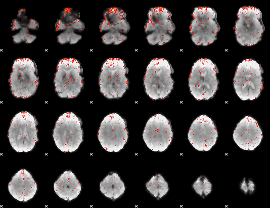 |
Component:218; Jaccard:0.020483
 |
Component:335; Jaccard:0.010204
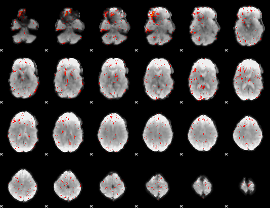 |
Correlation:0.27
  |
Correlation:0.27
  |
Correlation:0.47314
  |
Correlation:0.13362
  |
Correlation:0.25337
  |
Correlation:0.23555
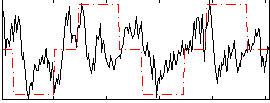 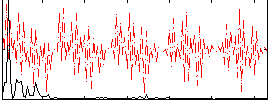 |
Correlation:0.47307
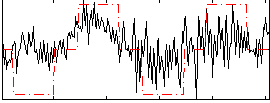 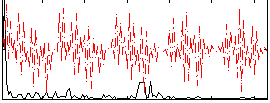 |
Component:023; Jaccard:0.078211
 |
Component:023; Jaccard:0.085657
 |
Component:354; Jaccard:0.08339
 |
Component:354; Jaccard:0.063634
 |
Component:335; Jaccard:0.03701
 |
Component:335; Jaccard:0.020222
 |
Component:324; Jaccard:0.008917
 |
Correlation:0.47314
  |
Correlation:0.47314
  |
Correlation:0.27
  |
Correlation:0.27
  |
Correlation:0.47307
  |
Correlation:0.47307
  |
Correlation:0.24305
  |
Component:082; Jaccard:0.077922
 |
Component:025; Jaccard:0.080312
 |
Component:025; Jaccard:0.076966
 |
Component:025; Jaccard:0.061373
 |
Component:354; Jaccard:0.036461
 |
Component:025; Jaccard:0.018935
 |
Component:006; Jaccard:0.008404
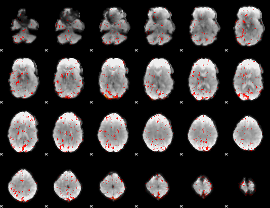 |
Correlation:0.24615
  |
Correlation:0.25337
  |
Correlation:0.25337
  |
Correlation:0.25337
  |
Correlation:0.27
  |
Correlation:0.25337
  |
Correlation:0.32406
  |
Component:131; Jaccard:0.077776
 |
Component:082; Jaccard:0.080269
 |
Component:082; Jaccard:0.073664
 |
Component:218; Jaccard:0.057042
 |
Component:090; Jaccard:0.036285
 |
Component:018; Jaccard:0.01859
 |
Component:018; Jaccard:0.008131
 |
Correlation:0.10221
  |
Correlation:0.24615
  |
Correlation:0.24615
  |
Correlation:0.23555
  |
Correlation:0.13362
  |
Correlation:0.27583
  |
Correlation:0.27583
  |
Component:006; Jaccard:0.076571
 |
Component:131; Jaccard:0.07859
 |
Component:218; Jaccard:0.072715
 |
Component:082; Jaccard:0.05625
 |
Component:218; Jaccard:0.035917
 |
Component:354; Jaccard:0.017979
 |
Component:218; Jaccard:0.008057
 |
Correlation:0.32406
  |
Correlation:0.10221
  |
Correlation:0.23555
  |
Correlation:0.24615
  |
Correlation:0.23555
  |
Correlation:0.27
  |
Correlation:0.23555
  |
Component:247; Jaccard:0.075989
 |
Component:006; Jaccard:0.077137
 |
Component:006; Jaccard:0.070568
 |
Component:335; Jaccard:0.05606
 |
Component:018; Jaccard:0.034847
 |
Component:006; Jaccard:0.01771
 |
Component:082; Jaccard:0.007956
 |
Correlation:0.1131
  |
Correlation:0.32406
  |
Correlation:0.32406
  |
Correlation:0.47307
  |
Correlation:0.27583
  |
Correlation:0.32406
  |
Correlation:0.24615
  |
Component:016; Jaccard:0.075813
 |
Component:018; Jaccard:0.076259
 |
Component:018; Jaccard:0.069671
 |
Component:006; Jaccard:0.053786
 |
Component:082; Jaccard:0.034578
 |
Component:324; Jaccard:0.017234
 |
Component:346; Jaccard:0.007241
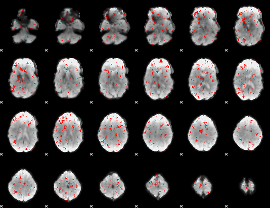 |
Correlation:0.041227
  |
Correlation:0.27583
  |
Correlation:0.27583
  |
Correlation:0.32406
  |
Correlation:0.24615
  |
Correlation:0.24305
  |
Correlation:0.25831
  |
Component:018; Jaccard:0.075324
 |
Component:089; Jaccard:0.076206
 |
Component:131; Jaccard:0.068271
 |
Component:018; Jaccard:0.052408
 |
Component:006; Jaccard:0.034007
 |
Component:090; Jaccard:0.017188
 |
Component:090; Jaccard:0.006929
 |
Correlation:0.27583
  |
Correlation:0.097202
  |
Correlation:0.10221
  |
Correlation:0.27583
  |
Correlation:0.32406
  |
Correlation:0.13362
  |
Correlation:0.13362
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































