Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
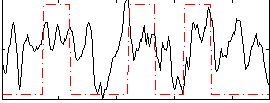
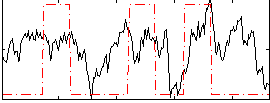
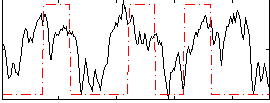
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
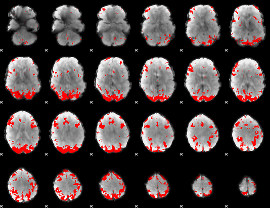 |
GLM Z-score result thresholded by 1
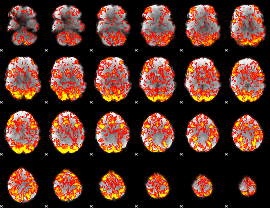 |
GLM Z-score result thresholded by 1.5
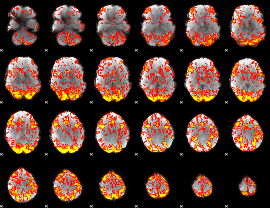 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
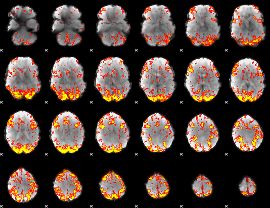 |
GLM Z-score result thresholded by 3
 |
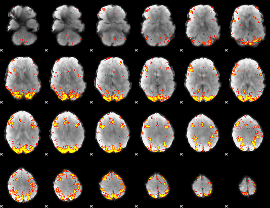
GLM Z-score result thresholded by 3.5
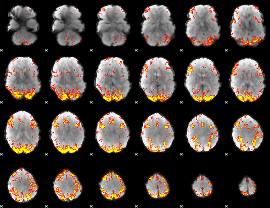 |
GLM Z-score result thresholded by 4
 |
Component:295; Jaccard:0.11396
 |
Component:295; Jaccard:0.13519
 |
Component:383; Jaccard:0.16494
 |
Component:383; Jaccard:0.19856
 |
Component:012; Jaccard:0.23745
 |
Component:012; Jaccard:0.27851
 |
Component:012; Jaccard:0.31531
 |
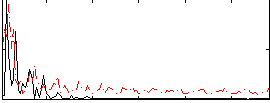
Correlation:0.23005
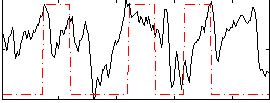 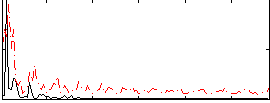 |
Correlation:0.23005
  |
Correlation:0.23215
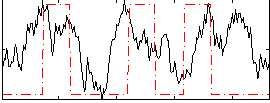  |
Correlation:0.23215
  |
Correlation:0.31935
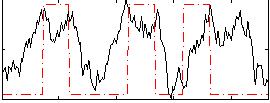  |
Correlation:0.31935
  |
Correlation:0.31935
  |
Component:018; Jaccard:0.1096
 |
Component:383; Jaccard:0.13341
 |
Component:012; Jaccard:0.1612
 |
Component:012; Jaccard:0.19775
 |
Component:383; Jaccard:0.23326
 |
Component:383; Jaccard:0.26174
 |
Component:383; Jaccard:0.28497
 |
Correlation:0.34229
  |
Correlation:0.23215
  |
Correlation:0.31935
  |
Correlation:0.31935
  |
Correlation:0.23215
  |
Correlation:0.23215
  |
Correlation:0.23215
  |
Component:383; Jaccard:0.108
 |
Component:381; Jaccard:0.13061
 |
Component:381; Jaccard:0.15954
 |
Component:381; Jaccard:0.18912
 |
Component:381; Jaccard:0.21624
 |
Component:381; Jaccard:0.23844
 |
Component:274; Jaccard:0.25883
 |
Correlation:0.23215
  |
Correlation:0.28689
  |
Correlation:0.28689
  |
Correlation:0.28689
  |
Correlation:0.28689
  |
Correlation:0.28689
  |
Correlation:0.3113
  |
Component:381; Jaccard:0.1056
 |
Component:018; Jaccard:0.12876
 |
Component:295; Jaccard:0.15837
 |
Component:295; Jaccard:0.17869
 |
Component:274; Jaccard:0.20546
 |
Component:274; Jaccard:0.23345
 |
Component:381; Jaccard:0.24962
 |
Correlation:0.28689
  |
Correlation:0.34229
  |
Correlation:0.23005
  |
Correlation:0.23005
  |
Correlation:0.3113
  |
Correlation:0.3113
  |
Correlation:0.28689
  |
Component:012; Jaccard:0.10278
 |
Component:012; Jaccard:0.12846
 |
Component:018; Jaccard:0.14758
 |
Component:274; Jaccard:0.17521
 |
Component:295; Jaccard:0.19109
 |
Component:295; Jaccard:0.1974
 |
Component:295; Jaccard:0.19369
 |
Correlation:0.31935
  |
Correlation:0.31935
  |
Correlation:0.34229
  |
Correlation:0.3113
  |
Correlation:0.23005
  |
Correlation:0.23005
  |
Correlation:0.23005
  |
Component:397; Jaccard:0.09947
 |
Component:274; Jaccard:0.1197
 |
Component:274; Jaccard:0.14629
 |
Component:018; Jaccard:0.16349
 |
Component:397; Jaccard:0.17683
 |
Component:397; Jaccard:0.18567
 |
Component:068; Jaccard:0.19021
 |
Correlation:0.18378
  |
Correlation:0.3113
  |
Correlation:0.3113
  |
Correlation:0.34229
  |
Correlation:0.18378
  |
Correlation:0.18378
  |
Correlation:0.24303
  |
Component:274; Jaccard:0.097687
 |
Component:397; Jaccard:0.11899
 |
Component:397; Jaccard:0.14011
 |
Component:397; Jaccard:0.1597
 |
Component:018; Jaccard:0.17354
 |
Component:068; Jaccard:0.18313
 |
Component:397; Jaccard:0.19006
 |
Correlation:0.3113
  |
Correlation:0.18378
  |
Correlation:0.18378
  |
Correlation:0.18378
  |
Correlation:0.34229
  |
Correlation:0.24303
  |
Correlation:0.18378
  |
Component:357; Jaccard:0.097257
 |
Component:068; Jaccard:0.11543
 |
Component:068; Jaccard:0.13555
 |
Component:068; Jaccard:0.15459
 |
Component:068; Jaccard:0.17097
 |
Component:018; Jaccard:0.17661
 |
Component:018; Jaccard:0.17234
 |
Correlation:0.33019
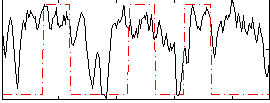 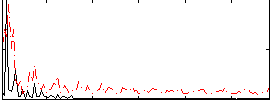 |
Correlation:0.24303
  |
Correlation:0.24303
  |
Correlation:0.24303
  |
Correlation:0.24303
  |
Correlation:0.34229
  |
Correlation:0.34229
  |
Component:068; Jaccard:0.097166
 |
Component:357; Jaccard:0.11451
 |
Component:357; Jaccard:0.13355
 |
Component:357; Jaccard:0.14983
 |
Component:357; Jaccard:0.16223
 |
Component:357; Jaccard:0.16832
 |
Component:357; Jaccard:0.16786
 |
Correlation:0.24303
  |
Correlation:0.33019
  |
Correlation:0.33019
  |
Correlation:0.33019
  |
Correlation:0.33019
  |
Correlation:0.33019
  |
Correlation:0.33019
  |
Component:145; Jaccard:0.093499
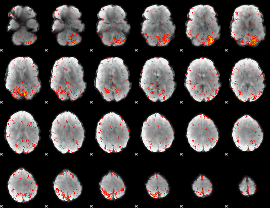 |
Component:145; Jaccard:0.10619
 |
Component:165; Jaccard:0.11962
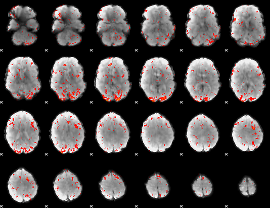 |
Component:165; Jaccard:0.13516
 |
Component:165; Jaccard:0.14806
 |
Component:165; Jaccard:0.159
 |
Component:165; Jaccard:0.16508
 |
Correlation:0.043852
  |
Correlation:0.043852
  |
Correlation:0.29504
  |
Correlation:0.29504
  |
Correlation:0.29504
  |
Correlation:0.29504
  |
Correlation:0.29504
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
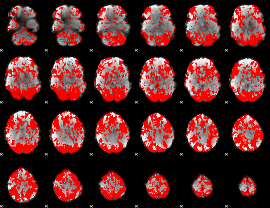 |
GLM binary result thresholded by 1.5
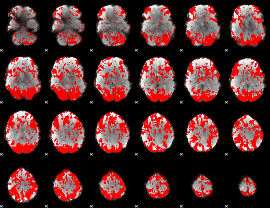 |
GLM binary result thresholded by 2
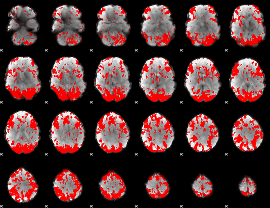 |
GLM binary result thresholded by 2.5
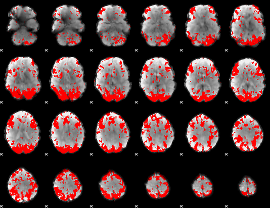 |
GLM binary result thresholded by 3
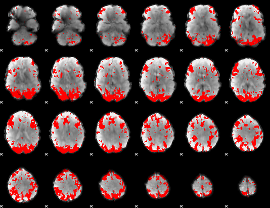 |
GLM binary result thresholded by 3.5
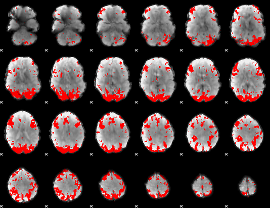 |
GLM binary result thresholded by 4
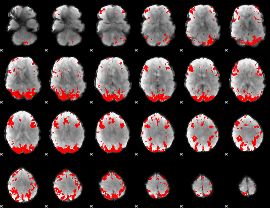 |
GLM Z-score result thresholded by 1
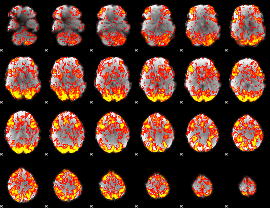 |
GLM Z-score result thresholded by 1.5
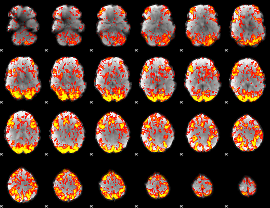 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:295; Jaccard:0.12026
 |
Component:295; Jaccard:0.14307
 |
Component:383; Jaccard:0.17638
 |
Component:383; Jaccard:0.21323
 |
Component:383; Jaccard:0.25054
 |
Component:383; Jaccard:0.28469
 |
Component:383; Jaccard:0.30961
 |
Correlation:-0.086792
  |
Correlation:-0.086792
  |
Correlation:0.10713
  |
Correlation:0.10713
  |
Correlation:0.10713
  |
Correlation:0.10713
  |
Correlation:0.10713
  |
Component:383; Jaccard:0.11471
 |
Component:383; Jaccard:0.14274
 |
Component:295; Jaccard:0.16718
 |
Component:012; Jaccard:0.19794
 |
Component:012; Jaccard:0.23375
 |
Component:012; Jaccard:0.2685
 |
Component:012; Jaccard:0.29978
 |
Correlation:0.10713
  |
Correlation:0.10713
  |
Correlation:-0.086792
  |
Correlation:0.043708
  |
Correlation:0.043708
  |
Correlation:0.043708
  |
Correlation:0.043708
  |
Component:018; Jaccard:0.10958
 |
Component:012; Jaccard:0.13243
 |
Component:012; Jaccard:0.16286
 |
Component:295; Jaccard:0.18889
 |
Component:274; Jaccard:0.22031
 |
Component:274; Jaccard:0.24884
 |
Component:274; Jaccard:0.27457
 |
Correlation:-0.15528
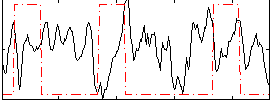 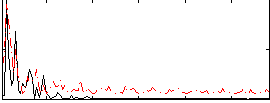 |
Correlation:0.043708
  |
Correlation:0.043708
  |
Correlation:-0.086792
  |
Correlation:0.22388
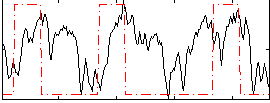 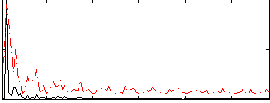 |
Correlation:0.22388
  |
Correlation:0.22388
  |
Component:381; Jaccard:0.10843
 |
Component:381; Jaccard:0.13173
 |
Component:381; Jaccard:0.15765
 |
Component:274; Jaccard:0.18884
 |
Component:381; Jaccard:0.20652
 |
Component:381; Jaccard:0.22313
 |
Component:381; Jaccard:0.23433
 |
Correlation:0.13416
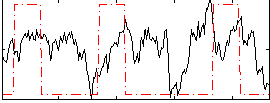 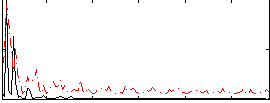 |
Correlation:0.13416
  |
Correlation:0.13416
  |
Correlation:0.22388
  |
Correlation:0.13416
  |
Correlation:0.13416
  |
Correlation:0.13416
  |
Component:012; Jaccard:0.10676
 |
Component:274; Jaccard:0.12866
 |
Component:274; Jaccard:0.15716
 |
Component:381; Jaccard:0.18377
 |
Component:295; Jaccard:0.20336
 |
Component:295; Jaccard:0.20929
 |
Component:068; Jaccard:0.21689
 |
Correlation:0.043708
  |
Correlation:0.22388
  |
Correlation:0.22388
  |
Correlation:0.13416
  |
Correlation:-0.086792
  |
Correlation:-0.086792
  |
Correlation:0.053545
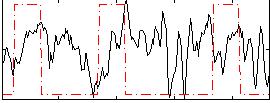 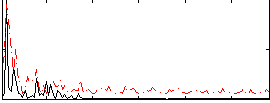 |
Component:068; Jaccard:0.10528
 |
Component:018; Jaccard:0.12665
 |
Component:068; Jaccard:0.14824
 |
Component:068; Jaccard:0.17026
 |
Component:068; Jaccard:0.19035
 |
Component:068; Jaccard:0.207
 |
Component:295; Jaccard:0.2073
 |
Correlation:0.053545
  |
Correlation:-0.15528
  |
Correlation:0.053545
  |
Correlation:0.053545
  |
Correlation:0.053545
  |
Correlation:0.053545
  |
Correlation:-0.086792
  |
Component:397; Jaccard:0.10454
 |
Component:068; Jaccard:0.12548
 |
Component:397; Jaccard:0.148
 |
Component:397; Jaccard:0.16886
 |
Component:397; Jaccard:0.18566
 |
Component:397; Jaccard:0.19568
 |
Component:397; Jaccard:0.19997
 |
Correlation:0.10524
  |
Correlation:0.053545
  |
Correlation:0.10524
  |
Correlation:0.10524
  |
Correlation:0.10524
  |
Correlation:0.10524
  |
Correlation:0.10524
  |
Component:274; Jaccard:0.10421
 |
Component:397; Jaccard:0.12546
 |
Component:018; Jaccard:0.14256
 |
Component:357; Jaccard:0.15787
 |
Component:357; Jaccard:0.17145
 |
Component:357; Jaccard:0.17885
 |
Component:357; Jaccard:0.1809
 |
Correlation:0.22388
  |
Correlation:0.10524
  |
Correlation:-0.15528
  |
Correlation:-0.0030561
  |
Correlation:-0.0030561
  |
Correlation:-0.0030561
  |
Correlation:-0.0030561
  |
Component:145; Jaccard:0.10382
 |
Component:357; Jaccard:0.12145
 |
Component:357; Jaccard:0.14082
 |
Component:018; Jaccard:0.15436
 |
Component:018; Jaccard:0.16096
 |
Component:167; Jaccard:0.1661
 |
Component:167; Jaccard:0.17283
 |
Correlation:-0.014401
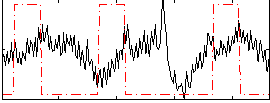  |
Correlation:-0.0030561
  |
Correlation:-0.0030561
  |
Correlation:-0.15528
  |
Correlation:-0.15528
  |
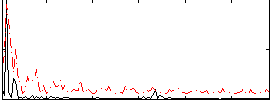
Correlation:0.31757
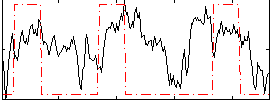  |
Correlation:0.31757
  |
Component:357; Jaccard:0.10265
 |
Component:145; Jaccard:0.11831
 |
Component:145; Jaccard:0.13252
 |
Component:167; Jaccard:0.14352
 |
Component:167; Jaccard:0.15628
 |
Component:165; Jaccard:0.16165
 |
Component:165; Jaccard:0.16711
 |
Correlation:-0.0030561
  |
Correlation:-0.014401
  |
Correlation:-0.014401
  |
Correlation:0.31757
  |
Correlation:0.31757
  |
Correlation:0.16587
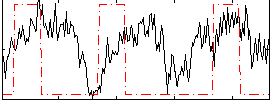  |
Correlation:0.16587
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
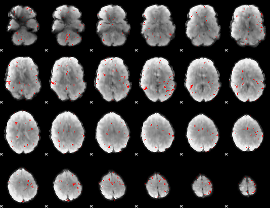 |
GLM binary result thresholded by 3
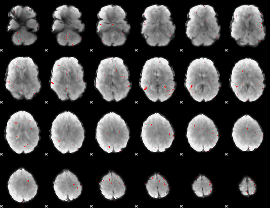 |
GLM binary result thresholded by 3.5
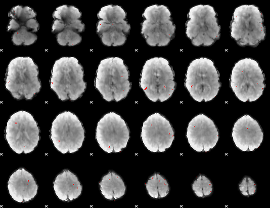 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
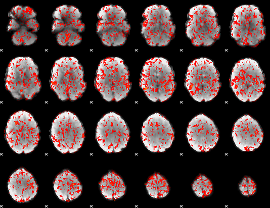 |
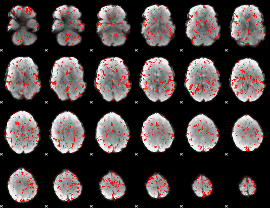
GLM Z-score result thresholded by 1.5
 |
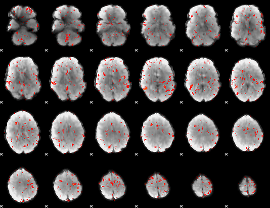
GLM Z-score result thresholded by 2
 |
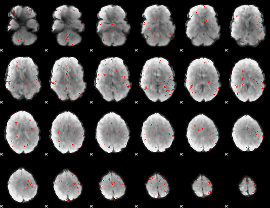
GLM Z-score result thresholded by 2.5
 |
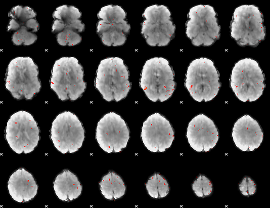
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
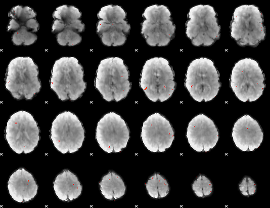 |
GLM Z-score result thresholded by 4
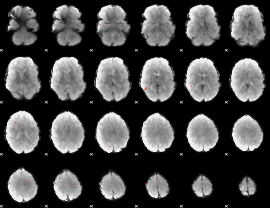 |
Component:195; Jaccard:0.092757
 |
Component:195; Jaccard:0.098979
 |
Component:195; Jaccard:0.084598
 |
Component:195; Jaccard:0.057535
 |
Component:195; Jaccard:0.028337
 |
Component:195; Jaccard:0.01112
 |
Component:195; Jaccard:0.004453
 |
Correlation:0.274
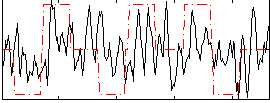 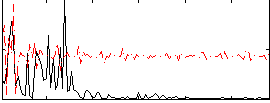 |
Correlation:0.274
  |
Correlation:0.274
  |
Correlation:0.274
  |
Correlation:0.274
  |
Correlation:0.274
  |
Correlation:0.274
  |
Component:197; Jaccard:0.082457
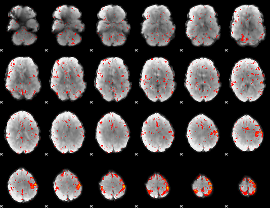 |
Component:197; Jaccard:0.083394
 |
Component:197; Jaccard:0.069431
 |
Component:028; Jaccard:0.043988
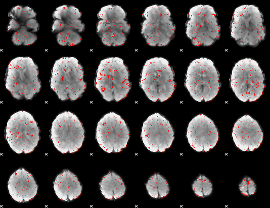 |
Component:028; Jaccard:0.022684
 |
Component:282; Jaccard:0.009708
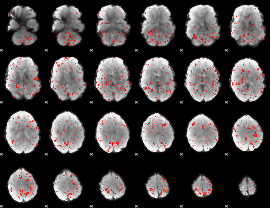 |
Component:028; Jaccard:0.003936
 |
Correlation:0.14135
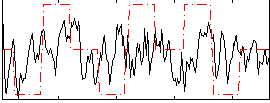 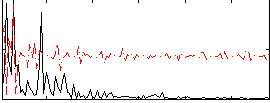 |
Correlation:0.14135
  |
Correlation:0.14135
  |
Correlation:0.29309
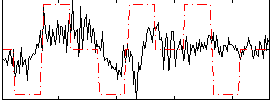  |
Correlation:0.29309
  |
Correlation:0.19002
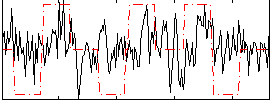  |
Correlation:0.29309
  |
Component:217; Jaccard:0.078651
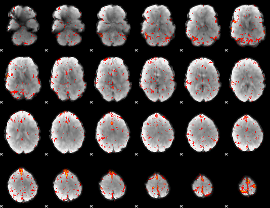 |
Component:028; Jaccard:0.074324
 |
Component:028; Jaccard:0.064956
 |
Component:197; Jaccard:0.042654
 |
Component:282; Jaccard:0.021226
 |
Component:028; Jaccard:0.009195
 |
Component:282; Jaccard:0.003933
 |
Correlation:0.15561
  |
Correlation:0.29309
  |
Correlation:0.29309
  |
Correlation:0.14135
  |
Correlation:0.19002
  |
Correlation:0.29309
  |
Correlation:0.19002
  |
Component:282; Jaccard:0.074081
 |
Component:282; Jaccard:0.073806
 |
Component:282; Jaccard:0.062176
 |
Component:282; Jaccard:0.040634
 |
Component:197; Jaccard:0.017389
 |
Component:197; Jaccard:0.006111
 |
Component:009; Jaccard:0.002215
 |
Correlation:0.19002
  |
Correlation:0.19002
  |
Correlation:0.19002
  |
Correlation:0.19002
  |
Correlation:0.14135
  |
Correlation:0.14135
  |
Correlation:-0.045615
  |
Component:028; Jaccard:0.072024
 |
Component:217; Jaccard:0.070482
 |
Component:370; Jaccard:0.060382
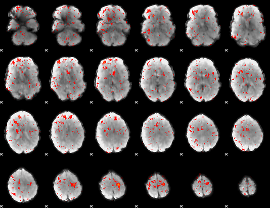 |
Component:370; Jaccard:0.039164
 |
Component:370; Jaccard:0.016756
 |
Component:370; Jaccard:0.005781
 |
Component:370; Jaccard:0.002129
 |
Correlation:0.29309
  |
Correlation:0.15561
  |
Correlation:0.10042
  |
Correlation:0.10042
  |
Correlation:0.10042
  |
Correlation:0.10042
  |
Correlation:0.10042
  |
Component:306; Jaccard:0.070728
 |
Component:370; Jaccard:0.07042
 |
Component:083; Jaccard:0.050027
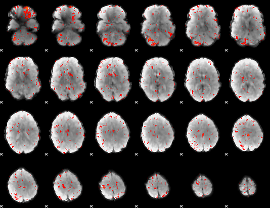 |
Component:009; Jaccard:0.030213
 |
Component:009; Jaccard:0.016552
 |
Component:343; Jaccard:0.005591
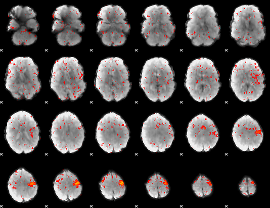 |
Component:366; Jaccard:0.002056
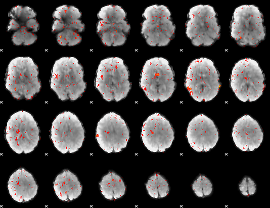 |
Correlation:0.15945
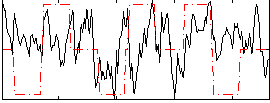  |
Correlation:0.10042
  |
Correlation:0.37404
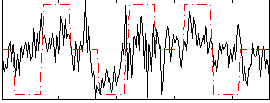 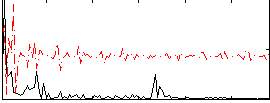 |
Correlation:-0.045615
  |
Correlation:-0.045615
  |
Correlation:0.15048
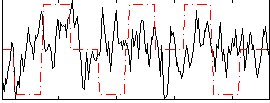 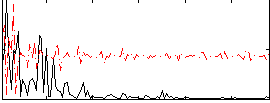 |
Correlation:-0.011637
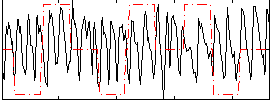 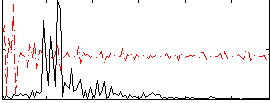 |
Component:370; Jaccard:0.068574
 |
Component:306; Jaccard:0.063404
 |
Component:217; Jaccard:0.049439
 |
Component:202; Jaccard:0.028803
 |
Component:111; Jaccard:0.012197
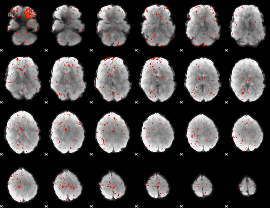 |
Component:201; Jaccard:0.00541
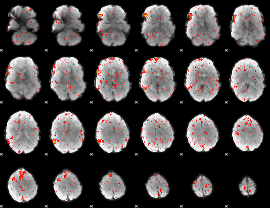 |
Component:201; Jaccard:0.002049
 |
Correlation:0.10042
  |
Correlation:0.15945
  |
Correlation:0.15561
  |
Correlation:0.010761
 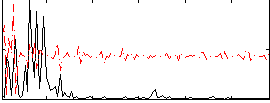 |
Correlation:0.1682
  |
Correlation:0.25506
  |
Correlation:0.25506
  |
Component:092; Jaccard:0.067978
 |
Component:083; Jaccard:0.062014
 |
Component:384; Jaccard:0.046337
 |
Component:083; Jaccard:0.028745
 |
Component:343; Jaccard:0.012148
 |
Component:009; Jaccard:0.005365
 |
Component:087; Jaccard:0.00195
 |
Correlation:0.12248
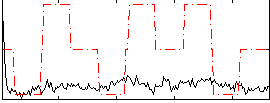  |
Correlation:0.37404
  |
Correlation:-0.029226
  |
Correlation:0.37404
  |
Correlation:0.15048
  |
Correlation:-0.045615
  |
Correlation:-0.056705
  |
Component:363; Jaccard:0.063357
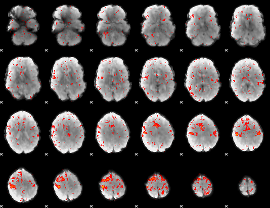 |
Component:092; Jaccard:0.06196
 |
Component:363; Jaccard:0.046296
 |
Component:111; Jaccard:0.027416
 |
Component:171; Jaccard:0.011807
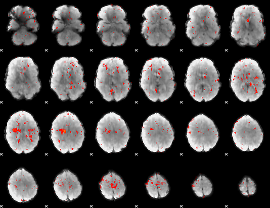 |
Component:363; Jaccard:0.004613
 |
Component:171; Jaccard:0.001864
 |
Correlation:0.07631
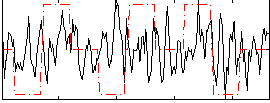 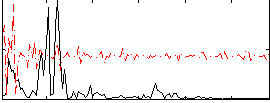 |
Correlation:0.12248
  |
Correlation:0.07631
  |
Correlation:0.1682
  |
Correlation:0.20662
  |
Correlation:0.07631
  |
Correlation:0.20662
  |
Component:384; Jaccard:0.062814
 |
Component:384; Jaccard:0.060656
 |
Component:146; Jaccard:0.045614
 |
Component:369; Jaccard:0.027414
 |
Component:202; Jaccard:0.011713
 |
Component:149; Jaccard:0.004515
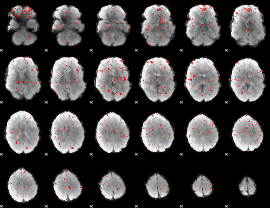 |
Component:148; Jaccard:0.001858
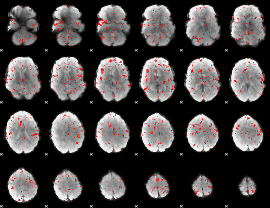 |
Correlation:-0.029226
  |
Correlation:-0.029226
  |
Correlation:0.20081
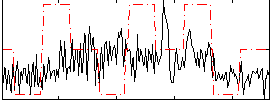 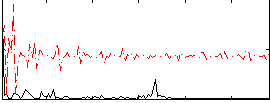 |
Correlation:0.16387
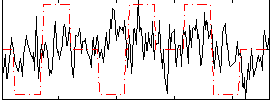 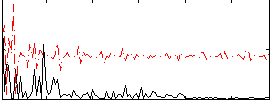 |
Correlation:0.010761
  |
Correlation:0.34629
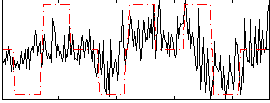 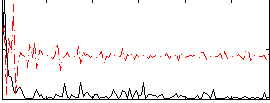 |
Correlation:0.013318
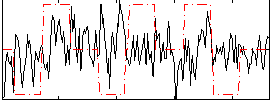 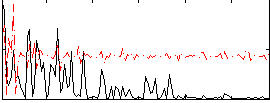 |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
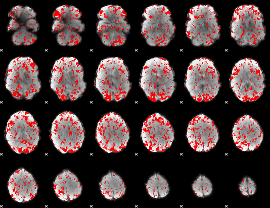 |
GLM binary result thresholded by 1.5
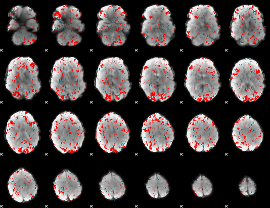 |
GLM binary result thresholded by 2
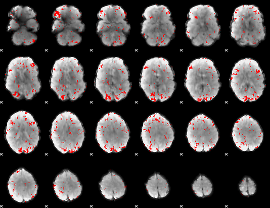 |
GLM binary result thresholded by 2.5
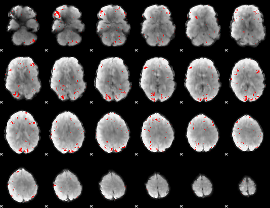 |
GLM binary result thresholded by 3
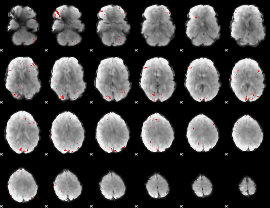 |
GLM binary result thresholded by 3.5
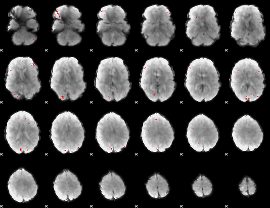 |
GLM binary result thresholded by 4
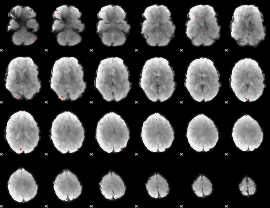 |
GLM Z-score result thresholded by 1
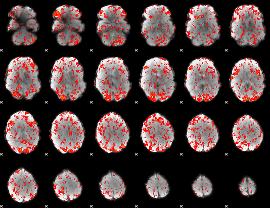 |
GLM Z-score result thresholded by 1.5
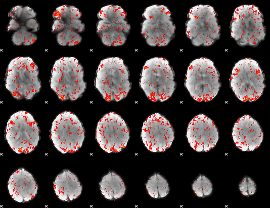 |
GLM Z-score result thresholded by 2
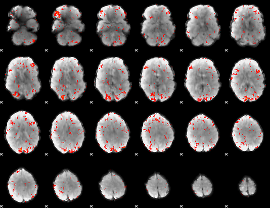 |
GLM Z-score result thresholded by 2.5
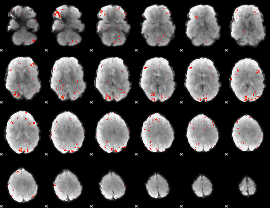 |
GLM Z-score result thresholded by 3
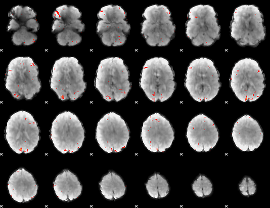 |
GLM Z-score result thresholded by 3.5
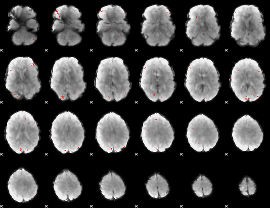 |
GLM Z-score result thresholded by 4
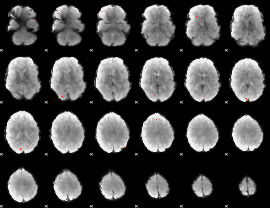 |
Component:388; Jaccard:0.11498
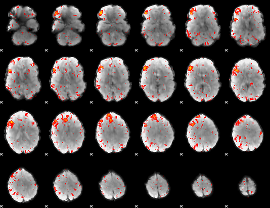 |
Component:388; Jaccard:0.11996
 |
Component:274; Jaccard:0.1042
 |
Component:274; Jaccard:0.074031
 |
Component:264; Jaccard:0.041795
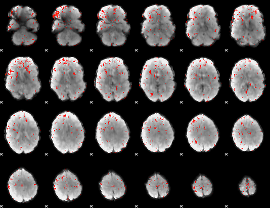 |
Component:264; Jaccard:0.021739
 |
Component:264; Jaccard:0.010778
 |
Correlation:0.02201
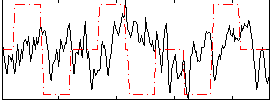 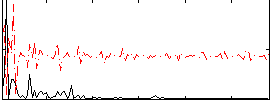 |
Correlation:0.02201
  |
Correlation:-0.051813
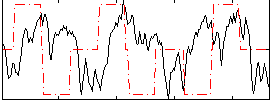 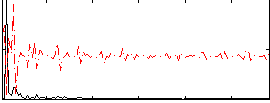 |
Correlation:-0.051813
  |
Correlation:0.17131
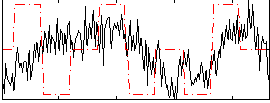 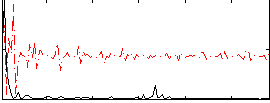 |
Correlation:0.17131
  |
Correlation:0.17131
  |
Component:274; Jaccard:0.099377
 |
Component:274; Jaccard:0.11095
 |
Component:388; Jaccard:0.10412
 |
Component:388; Jaccard:0.069972
 |
Component:274; Jaccard:0.039543
 |
Component:388; Jaccard:0.018906
 |
Component:274; Jaccard:0.00731
 |
Correlation:-0.051813
  |
Correlation:-0.051813
  |
Correlation:0.02201
  |
Correlation:0.02201
  |
Correlation:-0.051813
  |
Correlation:0.02201
  |
Correlation:-0.051813
  |
Component:383; Jaccard:0.099164
 |
Component:383; Jaccard:0.10113
 |
Component:264; Jaccard:0.087623
 |
Component:264; Jaccard:0.066339
 |
Component:388; Jaccard:0.037927
 |
Component:274; Jaccard:0.016608
 |
Component:388; Jaccard:0.00701
 |
Correlation:-0.0741
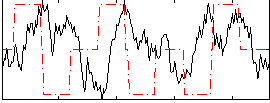 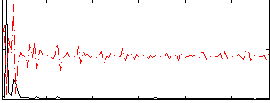 |
Correlation:-0.0741
  |
Correlation:0.17131
  |
Correlation:0.17131
  |
Correlation:0.02201
  |
Correlation:-0.051813
  |
Correlation:0.02201
  |
Component:068; Jaccard:0.087673
 |
Component:264; Jaccard:0.09366
 |
Component:383; Jaccard:0.087509
 |
Component:383; Jaccard:0.056647
 |
Component:383; Jaccard:0.030009
 |
Component:321; Jaccard:0.013701
 |
Component:383; Jaccard:0.005677
 |
Correlation:-0.11231
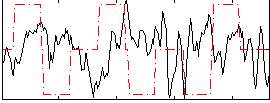 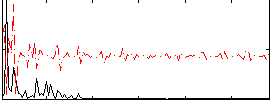 |
Correlation:0.17131
  |
Correlation:-0.0741
  |
Correlation:-0.0741
  |
Correlation:-0.0741
  |
Correlation:0.0027416
  |
Correlation:-0.0741
  |
Component:199; Jaccard:0.084085
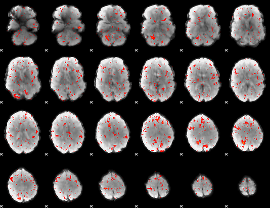 |
Component:068; Jaccard:0.086974
 |
Component:068; Jaccard:0.069459
 |
Component:321; Jaccard:0.044628
 |
Component:321; Jaccard:0.025677
 |
Component:383; Jaccard:0.013155
 |
Component:321; Jaccard:0.005448
 |
Correlation:0.25224
  |
Correlation:-0.11231
  |
Correlation:-0.11231
  |
Correlation:0.0027416
  |
Correlation:0.0027416
  |
Correlation:-0.0741
  |
Correlation:0.0027416
  |
Component:264; Jaccard:0.081107
 |
Component:199; Jaccard:0.082471
 |
Component:199; Jaccard:0.064381
 |
Component:167; Jaccard:0.040593
 |
Component:199; Jaccard:0.021458
 |
Component:199; Jaccard:0.009837
 |
Component:199; Jaccard:0.004557
 |
Correlation:0.17131
  |
Correlation:0.25224
  |
Correlation:0.25224
  |
Correlation:0.016593
  |
Correlation:0.25224
  |
Correlation:0.25224
  |
Correlation:0.25224
  |
Component:131; Jaccard:0.078784
 |
Component:131; Jaccard:0.076646
 |
Component:167; Jaccard:0.061628
 |
Component:068; Jaccard:0.040423
 |
Component:131; Jaccard:0.019478
 |
Component:131; Jaccard:0.00872
 |
Component:262; Jaccard:0.004072
 |
Correlation:-0.1449
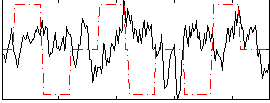 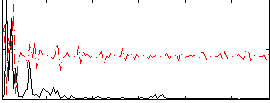 |
Correlation:-0.1449
  |
Correlation:0.016593
  |
Correlation:-0.11231
  |
Correlation:-0.1449
  |
Correlation:-0.1449
  |
Correlation:-0.030244
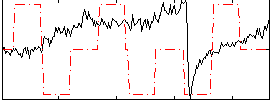 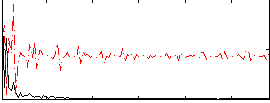 |
Component:147; Jaccard:0.074385
 |
Component:147; Jaccard:0.072191
 |
Component:131; Jaccard:0.060509
 |
Component:199; Jaccard:0.039684
 |
Component:167; Jaccard:0.019381
 |
Component:224; Jaccard:0.008065
 |
Component:387; Jaccard:0.004021
 |
Correlation:0.036827
  |
Correlation:0.036827
  |
Correlation:-0.1449
  |
Correlation:0.25224
  |
Correlation:0.016593
  |
Correlation:-0.19596
  |
Correlation:-0.037642
  |
Component:145; Jaccard:0.070736
 |
Component:167; Jaccard:0.071251
 |
Component:321; Jaccard:0.059584
 |
Component:131; Jaccard:0.037057
 |
Component:068; Jaccard:0.018379
 |
Component:167; Jaccard:0.007285
 |
Component:154; Jaccard:0.003538
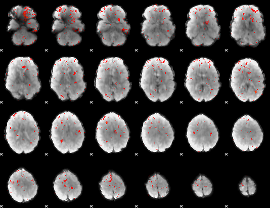 |
Correlation:-0.034526
  |
Correlation:0.016593
  |
Correlation:0.0027416
  |
Correlation:-0.1449
  |
Correlation:-0.11231
  |
Correlation:0.016593
  |
Correlation:0.042418
  |
Component:167; Jaccard:0.070229
 |
Component:012; Jaccard:0.068466
 |
Component:147; Jaccard:0.057806
 |
Component:012; Jaccard:0.034017
 |
Component:012; Jaccard:0.017126
 |
Component:051; Jaccard:0.007143
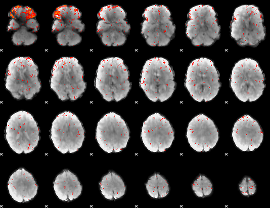 |
Component:367; Jaccard:0.003355
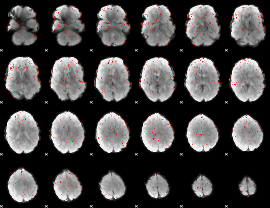 |
Correlation:0.016593
  |
Correlation:-0.16337
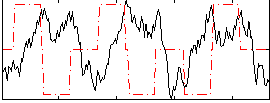 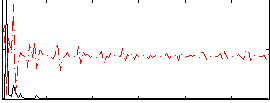 |
Correlation:0.036827
  |
Correlation:-0.16337
  |
Correlation:-0.16337
  |
Correlation:-0.15001
  |
Correlation:0.15873
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
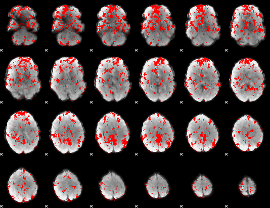 |
GLM binary result thresholded by 1.5
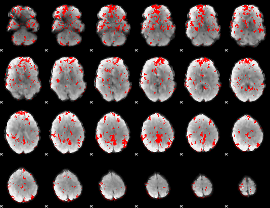 |
GLM binary result thresholded by 2
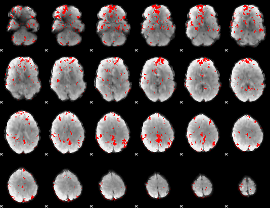 |
GLM binary result thresholded by 2.5
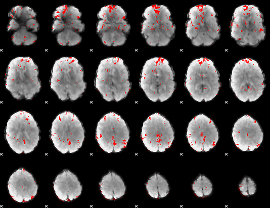 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
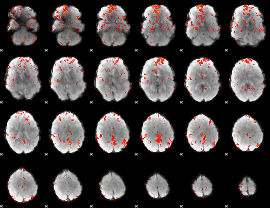 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
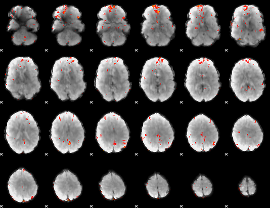 |
GLM Z-score result thresholded by 3.5
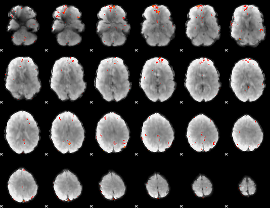 |
GLM Z-score result thresholded by 4
 |
Component:002; Jaccard:0.12101
 |
Component:002; Jaccard:0.13347
 |
Component:002; Jaccard:0.13492
 |
Component:109; Jaccard:0.12971
 |
Component:109; Jaccard:0.11652
 |
Component:109; Jaccard:0.091519
 |
Component:109; Jaccard:0.058342
 |
Correlation:0.042521
 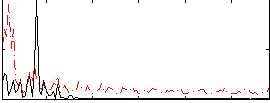 |
Correlation:0.042521
  |
Correlation:0.042521
  |
Correlation:0.21526
 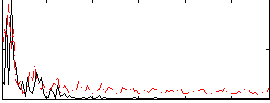 |
Correlation:0.21526
  |
Correlation:0.21526
  |
Correlation:0.21526
  |
Component:093; Jaccard:0.10487
 |
Component:093; Jaccard:0.11545
 |
Component:109; Jaccard:0.12769
 |
Component:002; Jaccard:0.1274
 |
Component:002; Jaccard:0.098594
 |
Component:352; Jaccard:0.072889
 |
Component:352; Jaccard:0.049539
 |
Correlation:0.21259
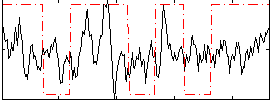 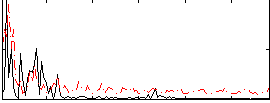 |
Correlation:0.21259
  |
Correlation:0.21526
  |
Correlation:0.042521
  |
Correlation:0.042521
  |
Correlation:0.096386
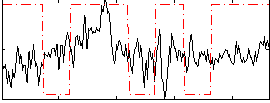 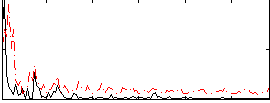 |
Correlation:0.096386
  |
Component:058; Jaccard:0.097858
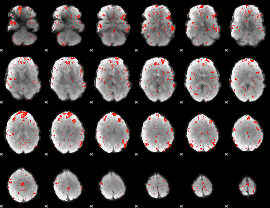 |
Component:109; Jaccard:0.11292
 |
Component:093; Jaccard:0.1159
 |
Component:352; Jaccard:0.10775
 |
Component:352; Jaccard:0.097468
 |
Component:002; Jaccard:0.067018
 |
Component:002; Jaccard:0.042405
 |
Correlation:0.090218
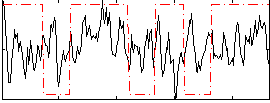  |
Correlation:0.21526
  |
Correlation:0.21259
  |
Correlation:0.096386
  |
Correlation:0.096386
  |
Correlation:0.042521
  |
Correlation:0.042521
  |
Component:109; Jaccard:0.093927
 |
Component:249; Jaccard:0.10524
 |
Component:352; Jaccard:0.10853
 |
Component:249; Jaccard:0.10315
 |
Component:249; Jaccard:0.0847
 |
Component:249; Jaccard:0.060419
 |
Component:206; Jaccard:0.037333
 |
Correlation:0.21526
  |
Correlation:0.30464
  |
Correlation:0.096386
  |
Correlation:0.30464
  |
Correlation:0.30464
  |
Correlation:0.30464
  |
Correlation:0.42477
  |
Component:249; Jaccard:0.093845
 |
Component:352; Jaccard:0.099434
 |
Component:249; Jaccard:0.10817
 |
Component:093; Jaccard:0.10075
 |
Component:093; Jaccard:0.073378
 |
Component:206; Jaccard:0.053739
 |
Component:249; Jaccard:0.035646
 |
Correlation:0.30464
  |
Correlation:0.096386
  |
Correlation:0.30464
  |
Correlation:0.21259
  |
Correlation:0.21259
  |
Correlation:0.42477
  |
Correlation:0.30464
  |
Component:352; Jaccard:0.088678
 |
Component:058; Jaccard:0.096529
 |
Component:058; Jaccard:0.084933
 |
Component:279; Jaccard:0.080177
 |
Component:206; Jaccard:0.069616
 |
Component:279; Jaccard:0.049751
 |
Component:279; Jaccard:0.033478
 |
Correlation:0.096386
  |
Correlation:0.090218
  |
Correlation:0.090218
  |
Correlation:-0.012641
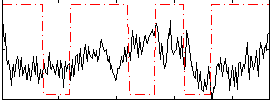 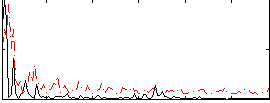 |
Correlation:0.42477
  |
Correlation:-0.012641
  |
Correlation:-0.012641
  |
Component:081; Jaccard:0.079714
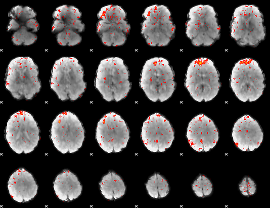 |
Component:081; Jaccard:0.084056
 |
Component:126; Jaccard:0.084201
 |
Component:126; Jaccard:0.078998
 |
Component:279; Jaccard:0.068857
 |
Component:186; Jaccard:0.046853
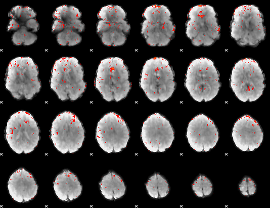 |
Component:186; Jaccard:0.033314
 |
Correlation:0.22994
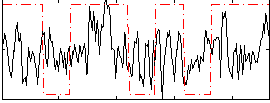 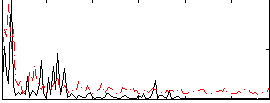 |
Correlation:0.22994
  |
Correlation:0.078393
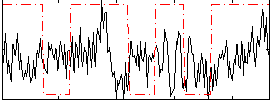 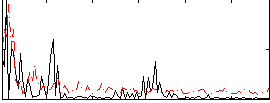 |
Correlation:0.078393
  |
Correlation:-0.012641
  |
Correlation:-0.052545
  |
Correlation:-0.052545
  |
Component:353; Jaccard:0.077998
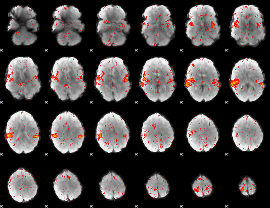 |
Component:126; Jaccard:0.083221
 |
Component:279; Jaccard:0.083569
 |
Component:313; Jaccard:0.074647
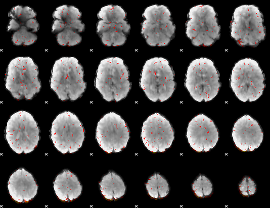 |
Component:126; Jaccard:0.065663
 |
Component:126; Jaccard:0.046344
 |
Component:126; Jaccard:0.02879
 |
Correlation:0.11179
  |
Correlation:0.078393
  |
Correlation:-0.012641
  |
Correlation:0.36406
  |
Correlation:0.078393
  |
Correlation:0.078393
  |
Correlation:0.078393
  |
Component:126; Jaccard:0.076422
 |
Component:279; Jaccard:0.078068
 |
Component:081; Jaccard:0.08234
 |
Component:081; Jaccard:0.074457
 |
Component:313; Jaccard:0.061976
 |
Component:093; Jaccard:0.044905
 |
Component:225; Jaccard:0.025588
 |
Correlation:0.078393
  |
Correlation:-0.012641
  |
Correlation:0.22994
  |
Correlation:0.22994
  |
Correlation:0.36406
  |
Correlation:0.21259
  |
Correlation:0.15781
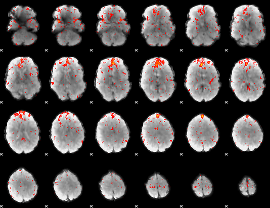
  |
Component:304; Jaccard:0.072989
 |
Component:313; Jaccard:0.075561
 |
Component:313; Jaccard:0.079087
 |
Component:206; Jaccard:0.072121
 |
Component:186; Jaccard:0.058987
 |
Component:225; Jaccard:0.041068
 |
Component:093; Jaccard:0.025203
 |
Correlation:0.097445
  |
Correlation:0.36406
  |
Correlation:0.36406
  |
Correlation:0.42477
  |
Correlation:-0.052545
  |
Correlation:0.15781
  |
Correlation:0.21259
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
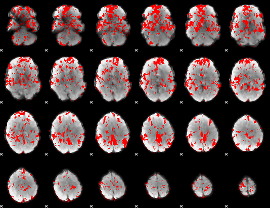 |
GLM binary result thresholded by 1.5
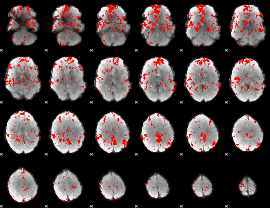 |
GLM binary result thresholded by 2
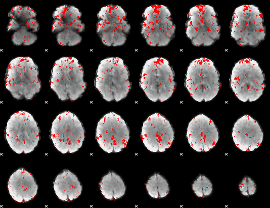 |
GLM binary result thresholded by 2.5
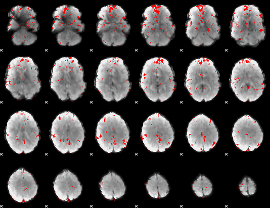 |
GLM binary result thresholded by 3
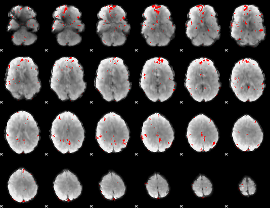 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
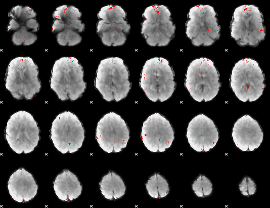 |
GLM Z-score result thresholded by 1
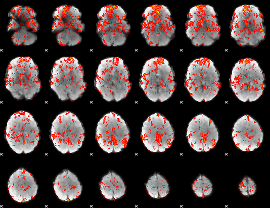 |
GLM Z-score result thresholded by 1.5
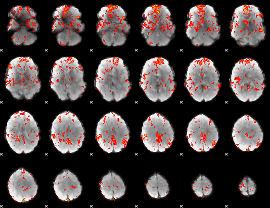 |
GLM Z-score result thresholded by 2
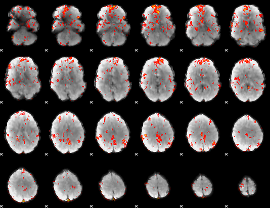 |
GLM Z-score result thresholded by 2.5
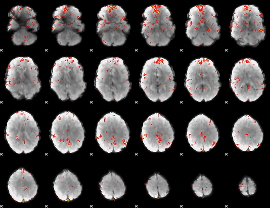 |
GLM Z-score result thresholded by 3
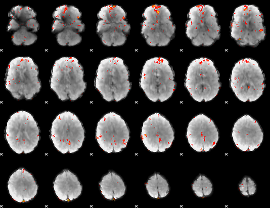 |
GLM Z-score result thresholded by 3.5
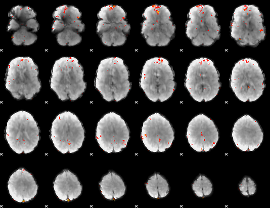 |
GLM Z-score result thresholded by 4
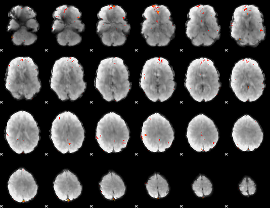 |
Component:002; Jaccard:0.098346
 |
Component:002; Jaccard:0.10606
 |
Component:109; Jaccard:0.119
 |
Component:109; Jaccard:0.12859
 |
Component:109; Jaccard:0.12048
 |
Component:109; Jaccard:0.09666
 |
Component:109; Jaccard:0.067111
 |
Correlation:-0.033099
  |
Correlation:-0.033099
  |
Correlation:-0.11844
  |
Correlation:-0.11844
  |
Correlation:-0.11844
  |
Correlation:-0.11844
  |
Correlation:-0.11844
  |
Component:353; Jaccard:0.092675
 |
Component:109; Jaccard:0.10304
 |
Component:002; Jaccard:0.1071
 |
Component:002; Jaccard:0.097947
 |
Component:352; Jaccard:0.088788
 |
Component:352; Jaccard:0.073775
 |
Component:352; Jaccard:0.05068
 |
Correlation:0.34215
  |
Correlation:-0.11844
  |
Correlation:-0.033099
  |
Correlation:-0.033099
  |
Correlation:-0.1034
  |
Correlation:-0.1034
  |
Correlation:-0.1034
  |
Component:093; Jaccard:0.092559
 |
Component:093; Jaccard:0.10153
 |
Component:093; Jaccard:0.10027
 |
Component:352; Jaccard:0.09687
 |
Component:279; Jaccard:0.081365
 |
Component:279; Jaccard:0.064436
 |
Component:279; Jaccard:0.046076
 |
Correlation:-0.080812
  |
Correlation:-0.080812
  |
Correlation:-0.080812
  |
Correlation:-0.1034
  |
Correlation:0.28964
  |
Correlation:0.28964
  |
Correlation:0.28964
  |
Component:058; Jaccard:0.09037
 |
Component:352; Jaccard:0.090593
 |
Component:352; Jaccard:0.097676
 |
Component:279; Jaccard:0.09086
 |
Component:002; Jaccard:0.078456
 |
Component:249; Jaccard:0.058216
 |
Component:206; Jaccard:0.038248
 |
Correlation:-0.058573
  |
Correlation:-0.1034
  |
Correlation:-0.1034
  |
Correlation:0.28964
  |
Correlation:-0.033099
  |
Correlation:-0.19341
  |
Correlation:-0.16036
  |
Component:209; Jaccard:0.086187
 |
Component:249; Jaccard:0.089644
 |
Component:249; Jaccard:0.091209
 |
Component:093; Jaccard:0.087861
 |
Component:249; Jaccard:0.075617
 |
Component:002; Jaccard:0.054054
 |
Component:202; Jaccard:0.035878
 |
Correlation:0.17003
  |
Correlation:-0.19341
  |
Correlation:-0.19341
  |
Correlation:-0.080812
  |
Correlation:-0.19341
  |
Correlation:-0.033099
  |
Correlation:0.20022
  |
Component:109; Jaccard:0.085371
 |
Component:209; Jaccard:0.089196
 |
Component:279; Jaccard:0.089335
 |
Component:249; Jaccard:0.08748
 |
Component:093; Jaccard:0.06875
 |
Component:206; Jaccard:0.049166
 |
Component:002; Jaccard:0.035234
 |
Correlation:-0.11844
  |
Correlation:0.17003
  |
Correlation:0.28964
  |
Correlation:-0.19341
  |
Correlation:-0.080812
  |
Correlation:-0.16036
  |
Correlation:-0.033099
  |
Component:352; Jaccard:0.081061
 |
Component:058; Jaccard:0.089065
 |
Component:081; Jaccard:0.087872
 |
Component:081; Jaccard:0.076938
 |
Component:126; Jaccard:0.065228
 |
Component:126; Jaccard:0.046561
 |
Component:249; Jaccard:0.034534
 |
Correlation:-0.1034
  |
Correlation:-0.058573
  |
Correlation:-0.127
  |
Correlation:-0.127
  |
Correlation:0.099133
  |
Correlation:0.099133
  |
Correlation:-0.19341
  |
Component:249; Jaccard:0.07999
 |
Component:353; Jaccard:0.088207
 |
Component:209; Jaccard:0.08409
 |
Component:126; Jaccard:0.074539
 |
Component:081; Jaccard:0.061545
 |
Component:225; Jaccard:0.046291
 |
Component:126; Jaccard:0.032467
 |
Correlation:-0.19341
  |
Correlation:0.34215
  |
Correlation:0.17003
  |
Correlation:0.099133
  |
Correlation:-0.127
  |
Correlation:-0.16399
  |
Correlation:0.099133
  |
Component:081; Jaccard:0.077839
 |
Component:081; Jaccard:0.085417
 |
Component:058; Jaccard:0.081543
 |
Component:209; Jaccard:0.071581
 |
Component:225; Jaccard:0.060478
 |
Component:093; Jaccard:0.042138
 |
Component:225; Jaccard:0.032148
 |
Correlation:-0.127
  |
Correlation:-0.127
  |
Correlation:-0.058573
  |
Correlation:0.17003
  |
Correlation:-0.16399
  |
Correlation:-0.080812
  |
Correlation:-0.16399
  |
Component:304; Jaccard:0.076976
 |
Component:279; Jaccard:0.080968
 |
Component:126; Jaccard:0.078555
 |
Component:225; Jaccard:0.070665
 |
Component:206; Jaccard:0.055182
 |
Component:081; Jaccard:0.040945
 |
Component:226; Jaccard:0.031617
 |
Correlation:-0.18722
  |
Correlation:0.28964
  |
Correlation:0.099133
  |
Correlation:-0.16399
  |
Correlation:-0.16036
  |
Correlation:-0.127
  |
Correlation:0.069584
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































