Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
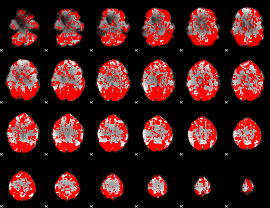 |
GLM binary result thresholded by 1.5
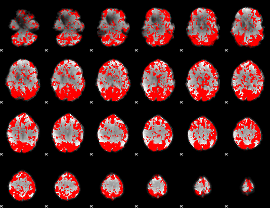 |
GLM binary result thresholded by 2
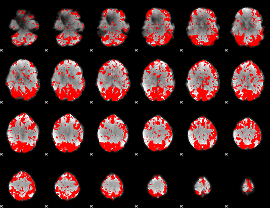 |
GLM binary result thresholded by 2.5
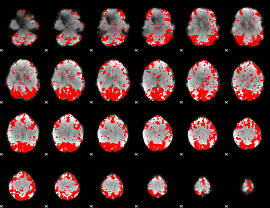 |
GLM binary result thresholded by 3
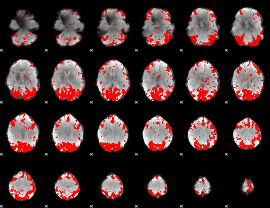 |
GLM binary result thresholded by 3.5
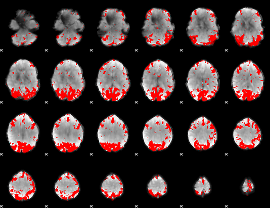 |
GLM binary result thresholded by 4
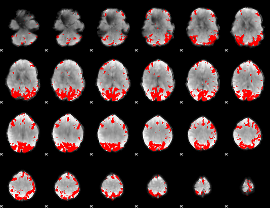 |
GLM Z-score result thresholded by 1
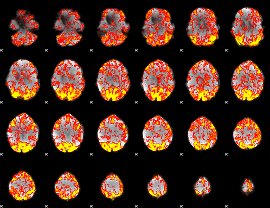 |
GLM Z-score result thresholded by 1.5
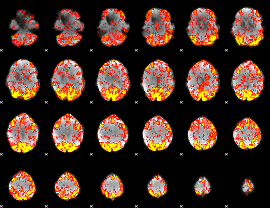 |
GLM Z-score result thresholded by 2
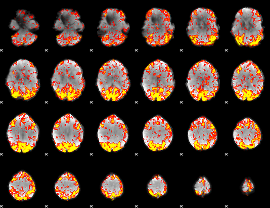 |
GLM Z-score result thresholded by 2.5
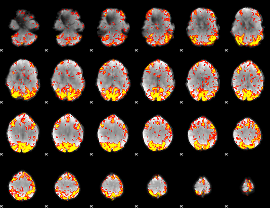 |
GLM Z-score result thresholded by 3
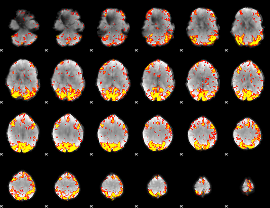 |
GLM Z-score result thresholded by 3.5
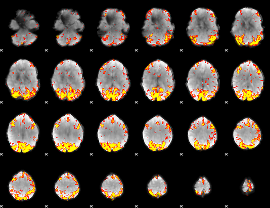 |
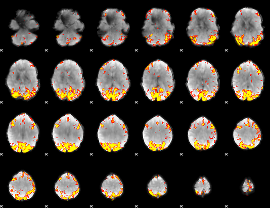
GLM Z-score result thresholded by 4
 |
Component:345; Jaccard:0.129
 |
Component:345; Jaccard:0.15846
 |
Component:345; Jaccard:0.19548
 |
Component:345; Jaccard:0.23787
 |
Component:345; Jaccard:0.28671
 |
Component:345; Jaccard:0.33852
 |
Component:345; Jaccard:0.39013
 |
Correlation:0.27556
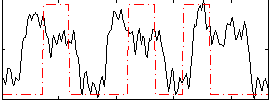 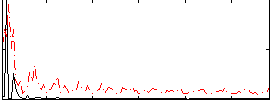 |
Correlation:0.27556
  |
Correlation:0.27556
  |
Correlation:0.27556
  |
Correlation:0.27556
  |
Correlation:0.27556
  |
Correlation:0.27556
  |
Component:107; Jaccard:0.12523
 |
Component:344; Jaccard:0.1487
 |
Component:344; Jaccard:0.17697
 |
Component:344; Jaccard:0.207
 |
Component:344; Jaccard:0.23726
 |
Component:344; Jaccard:0.26571
 |
Component:344; Jaccard:0.28967
 |
Correlation:0.1322
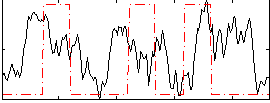 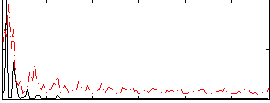 |
Correlation:0.1671
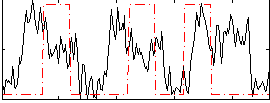 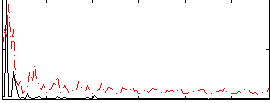 |
Correlation:0.1671
  |
Correlation:0.1671
  |
Correlation:0.1671
  |
Correlation:0.1671
  |
Correlation:0.1671
  |
Component:344; Jaccard:0.1242
 |
Component:107; Jaccard:0.14789
 |
Component:107; Jaccard:0.17285
 |
Component:107; Jaccard:0.19835
 |
Component:107; Jaccard:0.22231
 |
Component:107; Jaccard:0.24345
 |
Component:170; Jaccard:0.26204
 |
Correlation:0.1671
  |
Correlation:0.1322
  |
Correlation:0.1322
  |
Correlation:0.1322
  |
Correlation:0.1322
  |
Correlation:0.1322
  |
Correlation:0.13661
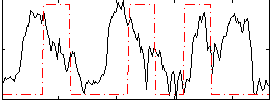 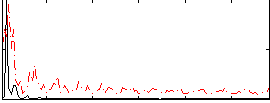 |
Component:153; Jaccard:0.12192
 |
Component:347; Jaccard:0.1431
 |
Component:153; Jaccard:0.16711
 |
Component:153; Jaccard:0.19067
 |
Component:153; Jaccard:0.21253
 |
Component:170; Jaccard:0.23852
 |
Component:107; Jaccard:0.25979
 |
Correlation:0.25026
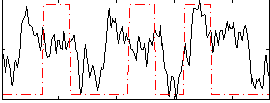 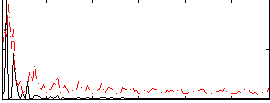 |
Correlation:0.19124
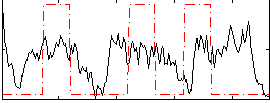 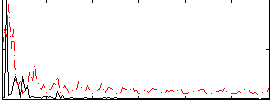 |
Correlation:0.25026
  |
Correlation:0.25026
  |
Correlation:0.25026
  |
Correlation:0.13661
  |
Correlation:0.1322
  |
Component:347; Jaccard:0.12152
 |
Component:153; Jaccard:0.14308
 |
Component:347; Jaccard:0.16581
 |
Component:347; Jaccard:0.18615
 |
Component:170; Jaccard:0.21187
 |
Component:153; Jaccard:0.23193
 |
Component:153; Jaccard:0.24426
 |
Correlation:0.19124
  |
Correlation:0.25026
  |
Correlation:0.19124
  |
Correlation:0.19124
  |
Correlation:0.13661
  |
Correlation:0.25026
  |
Correlation:0.25026
  |
Component:246; Jaccard:0.11052
 |
Component:170; Jaccard:0.13022
 |
Component:170; Jaccard:0.15571
 |
Component:170; Jaccard:0.18318
 |
Component:347; Jaccard:0.20252
 |
Component:347; Jaccard:0.21328
 |
Component:347; Jaccard:0.21676
 |
Correlation:0.32055
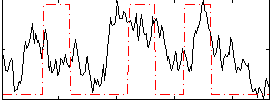 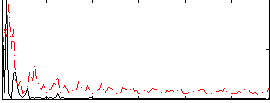 |
Correlation:0.13661
  |
Correlation:0.13661
  |
Correlation:0.13661
  |
Correlation:0.19124
  |
Correlation:0.19124
  |
Correlation:0.19124
  |
Component:170; Jaccard:0.10881
 |
Component:246; Jaccard:0.129
 |
Component:246; Jaccard:0.14955
 |
Component:246; Jaccard:0.17001
 |
Component:246; Jaccard:0.18734
 |
Component:246; Jaccard:0.20146
 |
Component:246; Jaccard:0.21042
 |
Correlation:0.13661
  |
Correlation:0.32055
  |
Correlation:0.32055
  |
Correlation:0.32055
  |
Correlation:0.32055
  |
Correlation:0.32055
  |
Correlation:0.32055
  |
Component:377; Jaccard:0.10491
 |
Component:377; Jaccard:0.12101
 |
Component:265; Jaccard:0.13789
 |
Component:265; Jaccard:0.15613
 |
Component:265; Jaccard:0.17195
 |
Component:265; Jaccard:0.18297
 |
Component:265; Jaccard:0.18854
 |
Correlation:0.1724
  |
Correlation:0.1724
  |
Correlation:0.095769
  |
Correlation:0.095769
  |
Correlation:0.095769
  |
Correlation:0.095769
  |
Correlation:0.095769
  |
Component:265; Jaccard:0.10004
 |
Component:265; Jaccard:0.11807
 |
Component:377; Jaccard:0.13777
 |
Component:377; Jaccard:0.15297
 |
Component:377; Jaccard:0.16325
 |
Component:377; Jaccard:0.16866
 |
Component:329; Jaccard:0.17299
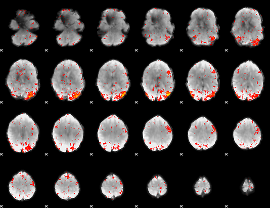 |
Correlation:0.095769
  |
Correlation:0.095769
  |
Correlation:0.1724
  |
Correlation:0.1724
  |
Correlation:0.1724
  |
Correlation:0.1724
  |
Correlation:0.22488
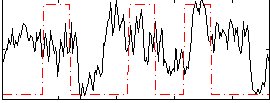 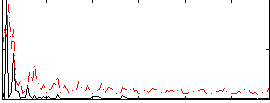 |
Component:113; Jaccard:0.1
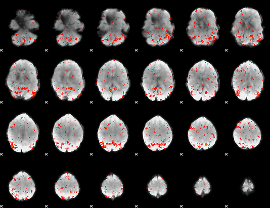 |
Component:113; Jaccard:0.11331
 |
Component:329; Jaccard:0.12903
 |
Component:329; Jaccard:0.14424
 |
Component:329; Jaccard:0.15875
 |
Component:329; Jaccard:0.16821
 |
Component:377; Jaccard:0.17027
 |
Correlation:0.31825
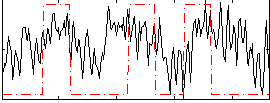 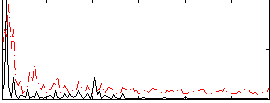 |
Correlation:0.31825
  |
Correlation:0.22488
  |
Correlation:0.22488
  |
Correlation:0.22488
  |
Correlation:0.22488
  |
Correlation:0.1724
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
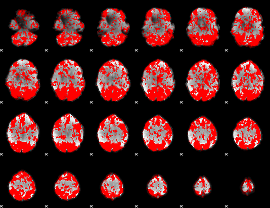 |
GLM binary result thresholded by 1.5
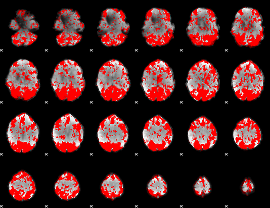 |
GLM binary result thresholded by 2
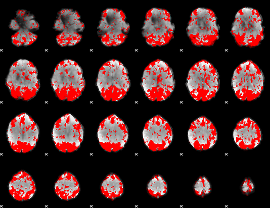 |
GLM binary result thresholded by 2.5
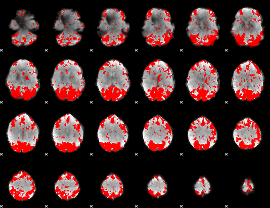 |
GLM binary result thresholded by 3
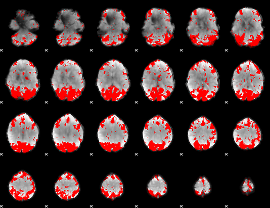 |
GLM binary result thresholded by 3.5
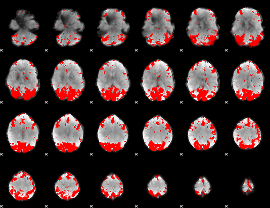 |
GLM binary result thresholded by 4
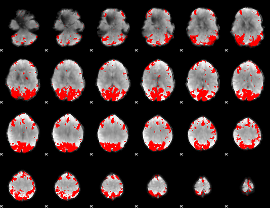 |
GLM Z-score result thresholded by 1
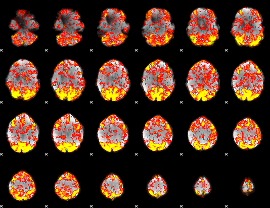 |
GLM Z-score result thresholded by 1.5
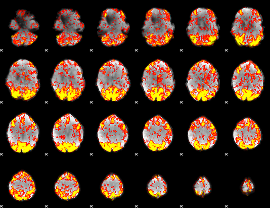 |
GLM Z-score result thresholded by 2
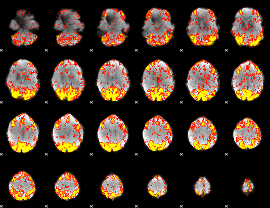 |
GLM Z-score result thresholded by 2.5
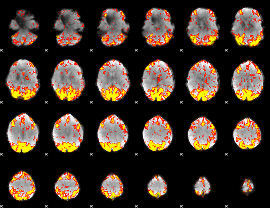 |
GLM Z-score result thresholded by 3
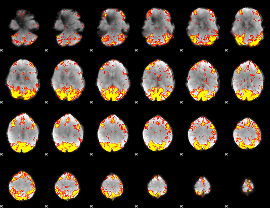 |
GLM Z-score result thresholded by 3.5
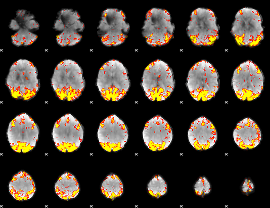 |
GLM Z-score result thresholded by 4
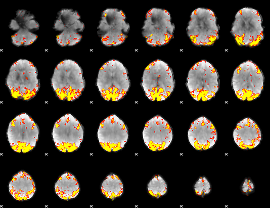 |
Component:107; Jaccard:0.12737
 |
Component:107; Jaccard:0.15086
 |
Component:345; Jaccard:0.18239
 |
Component:345; Jaccard:0.2206
 |
Component:345; Jaccard:0.26321
 |
Component:345; Jaccard:0.30785
 |
Component:345; Jaccard:0.35362
 |
Correlation:0.22887
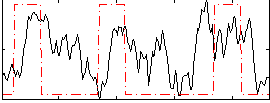 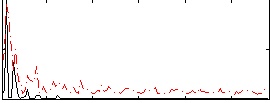 |
Correlation:0.22887
  |
Correlation:0.30028
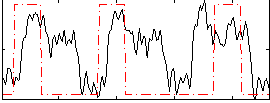 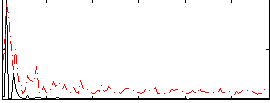 |
Correlation:0.30028
  |
Correlation:0.30028
  |
Correlation:0.30028
  |
Correlation:0.30028
  |
Component:345; Jaccard:0.12417
 |
Component:345; Jaccard:0.15015
 |
Component:107; Jaccard:0.17724
 |
Component:107; Jaccard:0.20543
 |
Component:344; Jaccard:0.23567
 |
Component:344; Jaccard:0.26437
 |
Component:344; Jaccard:0.28953
 |
Correlation:0.30028
  |
Correlation:0.30028
  |
Correlation:0.22887
  |
Correlation:0.22887
  |
Correlation:0.3847
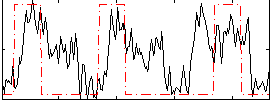 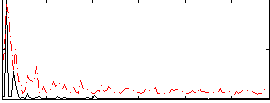 |
Correlation:0.3847
  |
Correlation:0.3847
  |
Component:344; Jaccard:0.12322
 |
Component:344; Jaccard:0.14677
 |
Component:344; Jaccard:0.17454
 |
Component:344; Jaccard:0.20516
 |
Component:107; Jaccard:0.2328
 |
Component:107; Jaccard:0.25667
 |
Component:170; Jaccard:0.28479
 |
Correlation:0.3847
  |
Correlation:0.3847
  |
Correlation:0.3847
  |
Correlation:0.3847
  |
Correlation:0.22887
  |
Correlation:0.22887
  |
Correlation:0.44207
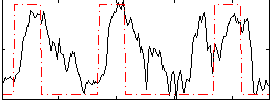 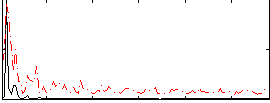 |
Component:153; Jaccard:0.11949
 |
Component:153; Jaccard:0.13949
 |
Component:153; Jaccard:0.16206
 |
Component:170; Jaccard:0.19042
 |
Component:170; Jaccard:0.22291
 |
Component:170; Jaccard:0.25383
 |
Component:107; Jaccard:0.27625
 |
Correlation:0.36298
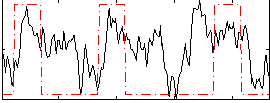 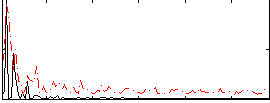 |
Correlation:0.36298
  |
Correlation:0.36298
  |
Correlation:0.44207
  |
Correlation:0.44207
  |
Correlation:0.44207
  |
Correlation:0.22887
  |
Component:347; Jaccard:0.11569
 |
Component:347; Jaccard:0.13348
 |
Component:170; Jaccard:0.15921
 |
Component:153; Jaccard:0.18526
 |
Component:153; Jaccard:0.20702
 |
Component:153; Jaccard:0.22693
 |
Component:153; Jaccard:0.24107
 |
Correlation:0.041913
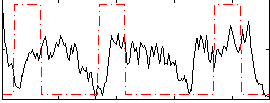 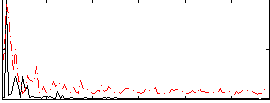 |
Correlation:0.041913
  |
Correlation:0.44207
  |
Correlation:0.36298
  |
Correlation:0.36298
  |
Correlation:0.36298
  |
Correlation:0.36298
  |
Component:170; Jaccard:0.10928
 |
Component:170; Jaccard:0.13183
 |
Component:347; Jaccard:0.15245
 |
Component:347; Jaccard:0.16981
 |
Component:347; Jaccard:0.18402
 |
Component:347; Jaccard:0.1936
 |
Component:347; Jaccard:0.19899
 |
Correlation:0.44207
  |
Correlation:0.44207
  |
Correlation:0.041913
  |
Correlation:0.041913
  |
Correlation:0.041913
  |
Correlation:0.041913
  |
Correlation:0.041913
  |
Component:221; Jaccard:0.1061
 |
Component:246; Jaccard:0.12151
 |
Component:246; Jaccard:0.13853
 |
Component:246; Jaccard:0.15443
 |
Component:246; Jaccard:0.16873
 |
Component:246; Jaccard:0.18031
 |
Component:246; Jaccard:0.19032
 |
Correlation:0.44791
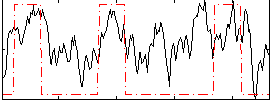 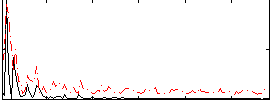 |
Correlation:0.082256
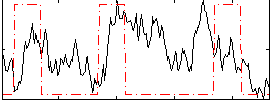 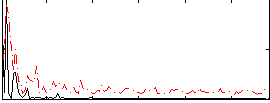 |
Correlation:0.082256
  |
Correlation:0.082256
  |
Correlation:0.082256
  |
Correlation:0.082256
  |
Correlation:0.082256
  |
Component:246; Jaccard:0.10577
 |
Component:221; Jaccard:0.11964
 |
Component:377; Jaccard:0.13471
 |
Component:377; Jaccard:0.14981
 |
Component:377; Jaccard:0.16181
 |
Component:377; Jaccard:0.17044
 |
Component:377; Jaccard:0.17498
 |
Correlation:0.082256
  |
Correlation:0.44791
  |
Correlation:0.15715
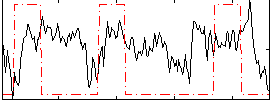 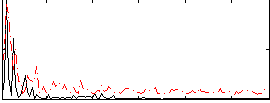 |
Correlation:0.15715
  |
Correlation:0.15715
  |
Correlation:0.15715
  |
Correlation:0.15715
  |
Component:377; Jaccard:0.10297
 |
Component:377; Jaccard:0.11839
 |
Component:221; Jaccard:0.13273
 |
Component:221; Jaccard:0.14431
 |
Component:221; Jaccard:0.15407
 |
Component:265; Jaccard:0.16127
 |
Component:329; Jaccard:0.16862
 |
Correlation:0.15715
  |
Correlation:0.15715
  |
Correlation:0.44791
  |
Correlation:0.44791
  |
Correlation:0.44791
  |
Correlation:0.17688
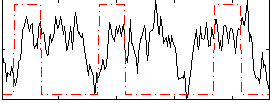 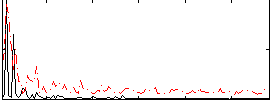 |
Correlation:0.35913
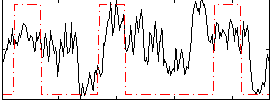 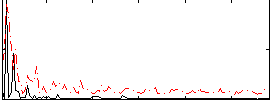 |
Component:181; Jaccard:0.098208
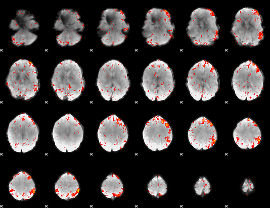 |
Component:181; Jaccard:0.11002
 |
Component:265; Jaccard:0.12466
 |
Component:265; Jaccard:0.13975
 |
Component:265; Jaccard:0.15201
 |
Component:221; Jaccard:0.1612
 |
Component:221; Jaccard:0.16566
 |
Correlation:-0.0035611
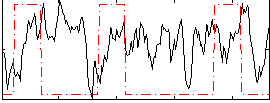  |
Correlation:-0.0035611
  |
Correlation:0.17688
  |
Correlation:0.17688
  |
Correlation:0.17688
  |
Correlation:0.44791
  |
Correlation:0.44791
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
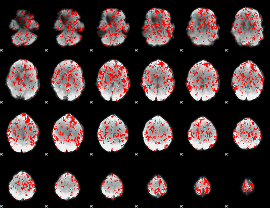 |
GLM binary result thresholded by 1.5
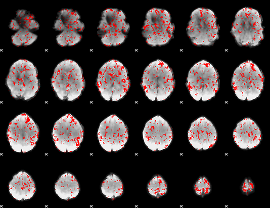 |
GLM binary result thresholded by 2
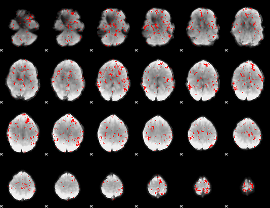 |
GLM binary result thresholded by 2.5
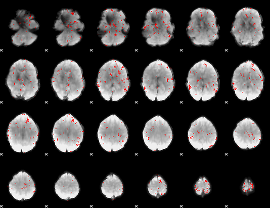 |
GLM binary result thresholded by 3
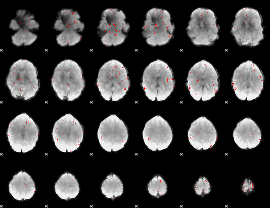 |
GLM binary result thresholded by 3.5
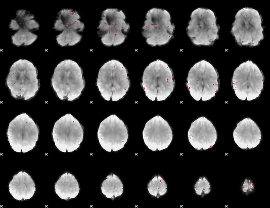 |
GLM binary result thresholded by 4
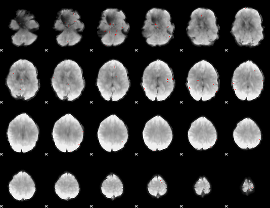 |
GLM Z-score result thresholded by 1
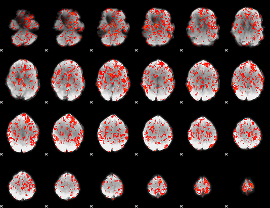 |
GLM Z-score result thresholded by 1.5
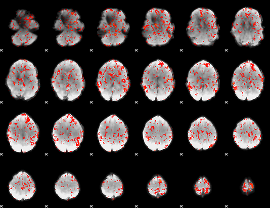 |
GLM Z-score result thresholded by 2
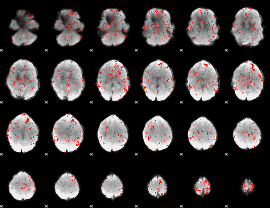 |
GLM Z-score result thresholded by 2.5
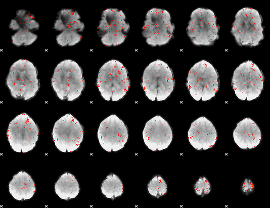 |
GLM Z-score result thresholded by 3
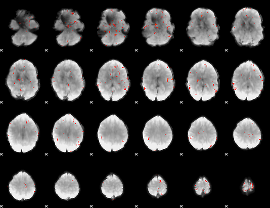 |
GLM Z-score result thresholded by 3.5
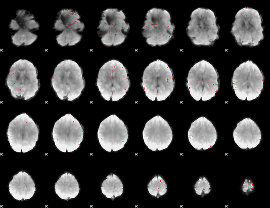 |
GLM Z-score result thresholded by 4
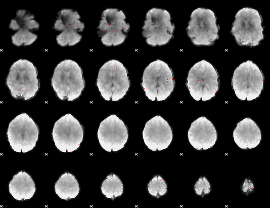 |
Component:355; Jaccard:0.091747
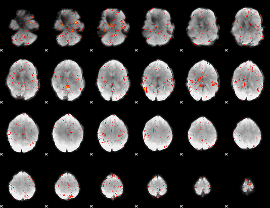 |
Component:355; Jaccard:0.10142
 |
Component:355; Jaccard:0.1019
 |
Component:355; Jaccard:0.085879
 |
Component:355; Jaccard:0.062507
 |
Component:355; Jaccard:0.036775
 |
Component:355; Jaccard:0.021835
 |
Correlation:0.015684
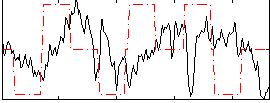  |
Correlation:0.015684
  |
Correlation:0.015684
  |
Correlation:0.015684
  |
Correlation:0.015684
  |
Correlation:0.015684
  |
Correlation:0.015684
  |
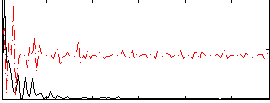
Component:171; Jaccard:0.087085
 |
Component:252; Jaccard:0.092163
 |
Component:252; Jaccard:0.09269
 |
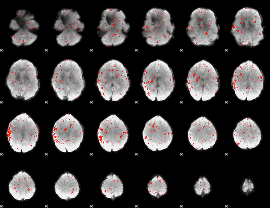
Component:360; Jaccard:0.079137
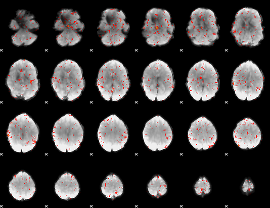 |
Component:360; Jaccard:0.055887
 |
Component:360; Jaccard:0.032964
 |
Component:360; Jaccard:0.016841
 |
Correlation:0.18632
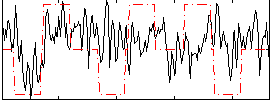 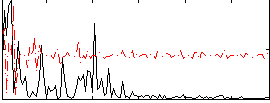 |
Correlation:0.24311
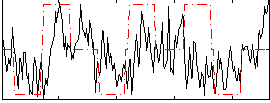 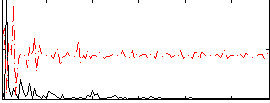 |
Correlation:0.24311
  |
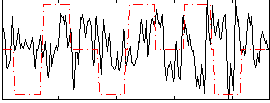
Correlation:0.15224
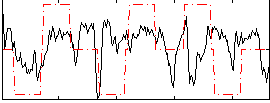  |
Correlation:0.15224
  |
Correlation:0.15224
  |
Correlation:0.15224
  |
Component:360; Jaccard:0.082476
 |
Component:171; Jaccard:0.092043
 |
Component:360; Jaccard:0.092322
 |
Component:252; Jaccard:0.07517
 |
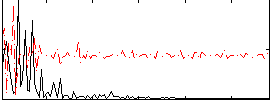
Component:296; Jaccard:0.053428
 |
Component:296; Jaccard:0.030908
 |
Component:296; Jaccard:0.015695
 |
Correlation:0.15224
  |
Correlation:0.18632
  |
Correlation:0.15224
  |
Correlation:0.24311
  |
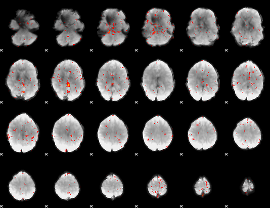
Correlation:0.0040085
 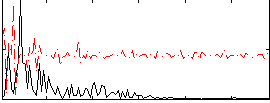 |
Correlation:0.0040085
  |
Correlation:0.0040085
  |
Component:310; Jaccard:0.081958
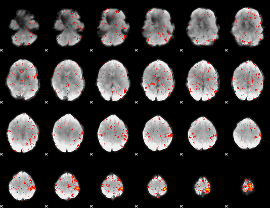 |
Component:360; Jaccard:0.091389
 |
Component:171; Jaccard:0.086353
 |
Component:296; Jaccard:0.073549
 |
Component:171; Jaccard:0.043782
 |
Component:171; Jaccard:0.024118
 |
Component:122; Jaccard:0.012751
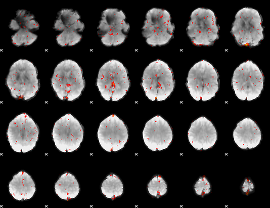 |
Correlation:0.089473
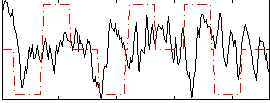 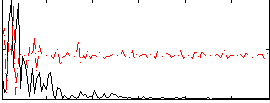 |
Correlation:0.15224
  |
Correlation:0.18632
  |
Correlation:0.0040085
  |
Correlation:0.18632
  |
Correlation:0.18632
  |
Correlation:0.099127
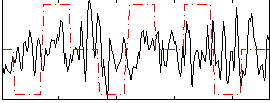 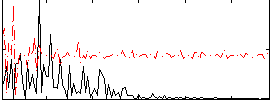 |
Component:008; Jaccard:0.079352
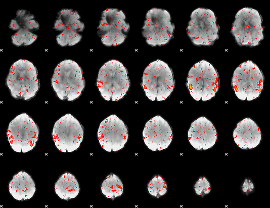 |
Component:018; Jaccard:0.079433
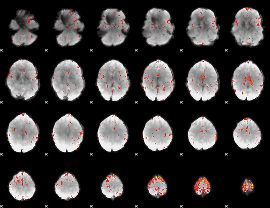 |
Component:296; Jaccard:0.078597
 |
Component:171; Jaccard:0.06816
 |
Component:252; Jaccard:0.041783
 |
Component:252; Jaccard:0.020592
 |
Component:173; Jaccard:0.010921
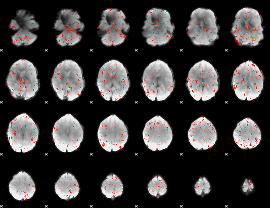 |
Correlation:0.045135
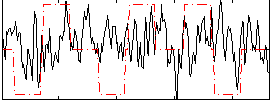 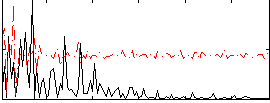 |
Correlation:0.15902
  |
Correlation:0.0040085
  |
Correlation:0.18632
  |
Correlation:0.24311
  |
Correlation:0.24311
  |
Correlation:-0.09434
  |
Component:252; Jaccard:0.079136
 |
Component:008; Jaccard:0.075807
 |
Component:018; Jaccard:0.074406
 |
Component:391; Jaccard:0.058962
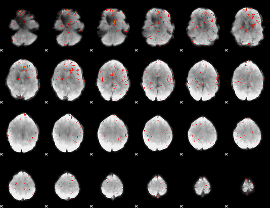 |
Component:391; Jaccard:0.038629
 |
Component:122; Jaccard:0.020502
 |
Component:171; Jaccard:0.010776
 |
Correlation:0.24311
  |
Correlation:0.045135
  |
Correlation:0.15902
  |
Correlation:0.23058
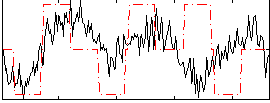 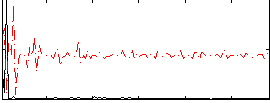 |
Correlation:0.23058
  |
Correlation:0.099127
  |
Correlation:0.18632
  |
Component:161; Jaccard:0.077344
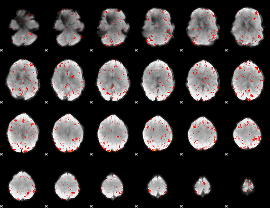 |
Component:333; Jaccard:0.075431
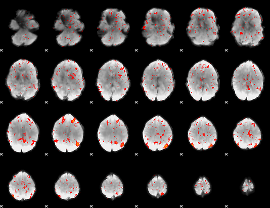 |
Component:173; Jaccard:0.069966
 |
Component:173; Jaccard:0.056625
 |
Component:173; Jaccard:0.037899
 |
Component:173; Jaccard:0.020364
 |
Component:391; Jaccard:0.009515
 |
Correlation:0.25133
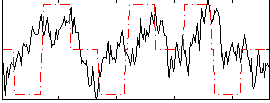 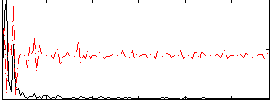 |
Correlation:0.21412
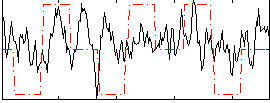 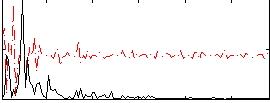 |
Correlation:-0.09434
  |
Correlation:-0.09434
  |
Correlation:-0.09434
  |
Correlation:-0.09434
  |
Correlation:0.23058
  |
Component:333; Jaccard:0.075968
 |
Component:310; Jaccard:0.075313
 |
Component:333; Jaccard:0.066882
 |
Component:018; Jaccard:0.055588
 |
Component:122; Jaccard:0.033579
 |
Component:391; Jaccard:0.020275
 |
Component:198; Jaccard:0.009434
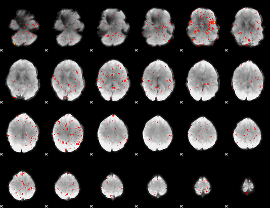 |
Correlation:0.21412
  |
Correlation:0.089473
  |
Correlation:0.21412
  |
Correlation:0.15902
  |
Correlation:0.099127
  |
Correlation:0.23058
  |
Correlation:0.18332
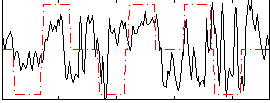 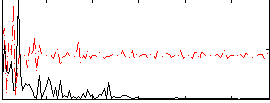 |
Component:018; Jaccard:0.075572
 |
Component:161; Jaccard:0.074675
 |
Component:008; Jaccard:0.065003
 |
Component:302; Jaccard:0.050277
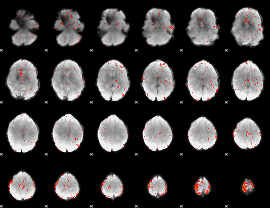 |
Component:302; Jaccard:0.030444
 |
Component:152; Jaccard:0.016591
 |
Component:152; Jaccard:0.009096
 |
Correlation:0.15902
  |
Correlation:0.25133
  |
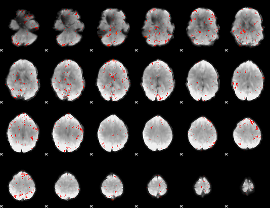
Correlation:0.045135
  |
Correlation:0.25329
  |
Correlation:0.25329
  |
Correlation:0.11829
  |
Correlation:0.11829
  |
Component:145; Jaccard:0.074283
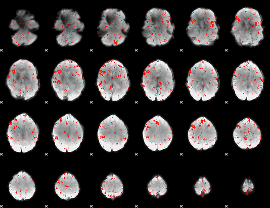 |
Component:173; Jaccard:0.074662
 |
Component:391; Jaccard:0.064231
 |
Component:333; Jaccard:0.047495
 |
Component:018; Jaccard:0.030153
 |
Component:302; Jaccard:0.015221
 |
Component:252; Jaccard:0.007626
 |
Correlation:0.19435
  |
Correlation:-0.09434
  |
Correlation:0.23058
  |
Correlation:0.21412
  |
Correlation:0.15902
  |
Correlation:0.25329
  |
Correlation:0.24311
  |
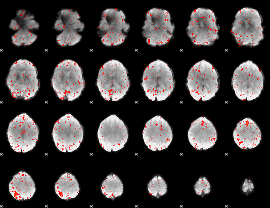
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
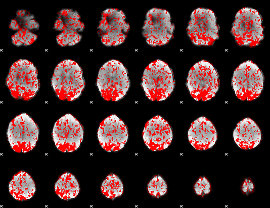 |
GLM binary result thresholded by 1.5
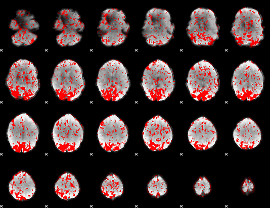 |
GLM binary result thresholded by 2
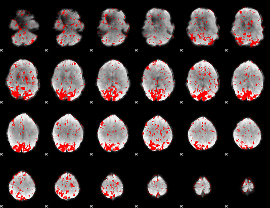 |
GLM binary result thresholded by 2.5
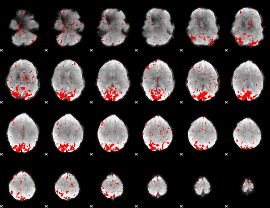 |
GLM binary result thresholded by 3
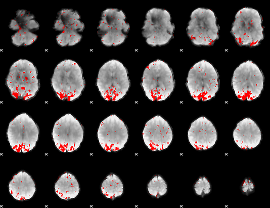 |
GLM binary result thresholded by 3.5
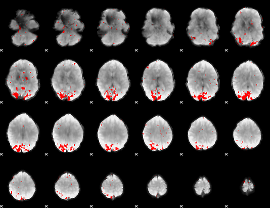 |
GLM binary result thresholded by 4
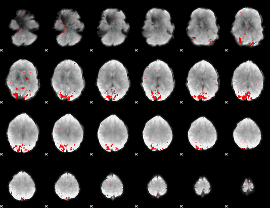 |
GLM Z-score result thresholded by 1
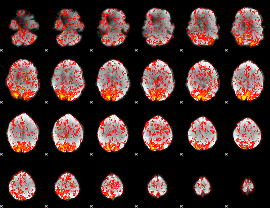 |
GLM Z-score result thresholded by 1.5
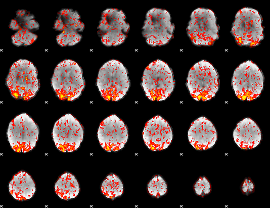 |
GLM Z-score result thresholded by 2
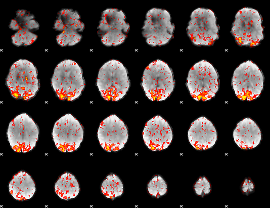 |
GLM Z-score result thresholded by 2.5
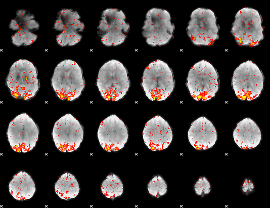 |
GLM Z-score result thresholded by 3
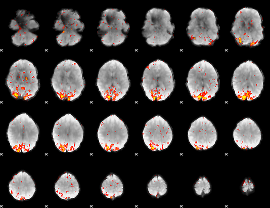 |
GLM Z-score result thresholded by 3.5
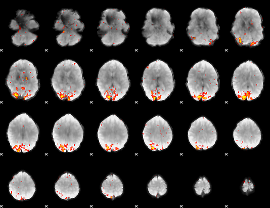 |
GLM Z-score result thresholded by 4
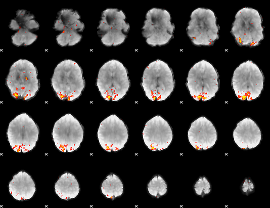 |
Component:170; Jaccard:0.1534
 |
Component:170; Jaccard:0.20559
 |
Component:170; Jaccard:0.26649
 |
Component:170; Jaccard:0.32179
 |
Component:170; Jaccard:0.34027
 |
Component:170; Jaccard:0.31709
 |
Component:170; Jaccard:0.26478
 |
Correlation:0.18104
 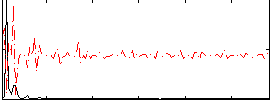 |
Correlation:0.18104
  |
Correlation:0.18104
  |
Correlation:0.18104
  |
Correlation:0.18104
  |
Correlation:0.18104
  |
Correlation:0.18104
  |
Component:345; Jaccard:0.13666
 |
Component:345; Jaccard:0.16737
 |
Component:345; Jaccard:0.19729
 |
Component:345; Jaccard:0.21997
 |
Component:345; Jaccard:0.21499
 |
Component:345; Jaccard:0.19028
 |
Component:345; Jaccard:0.15606
 |
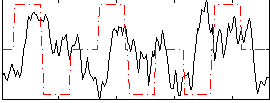
Correlation:0.014651
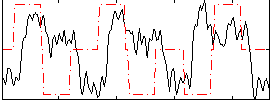  |
Correlation:0.014651
  |
Correlation:0.014651
  |
Correlation:0.014651
  |
Correlation:0.014651
  |
Correlation:0.014651
  |
Correlation:0.014651
  |
Component:107; Jaccard:0.13227
 |
Component:344; Jaccard:0.15607
 |
Component:344; Jaccard:0.18285
 |
Component:344; Jaccard:0.20006
 |
Component:344; Jaccard:0.19498
 |
Component:344; Jaccard:0.17488
 |
Component:344; Jaccard:0.14576
 |
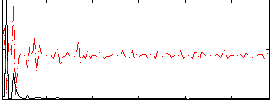
Correlation:0.057294
 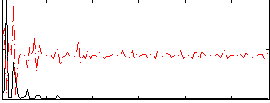 |
Correlation:0.12897
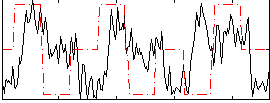  |
Correlation:0.12897
  |
Correlation:0.12897
  |
Correlation:0.12897
  |
Correlation:0.12897
  |
Correlation:0.12897
  |
Component:344; Jaccard:0.13077
 |
Component:107; Jaccard:0.15539
 |
Component:107; Jaccard:0.17305
 |
Component:107; Jaccard:0.18126
 |
Component:107; Jaccard:0.16696
 |
Component:107; Jaccard:0.14355
 |
Component:107; Jaccard:0.115
 |
Correlation:0.12897
  |
Correlation:0.057294
  |
Correlation:0.057294
  |
Correlation:0.057294
  |
Correlation:0.057294
  |
Correlation:0.057294
  |
Correlation:0.057294
  |
Component:221; Jaccard:0.12312
 |
Component:221; Jaccard:0.13973
 |
Component:221; Jaccard:0.15307
 |
Component:221; Jaccard:0.15306
 |
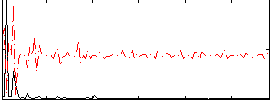
Component:172; Jaccard:0.14907
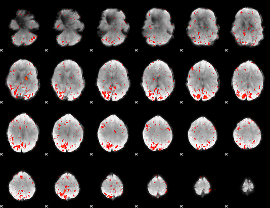 |
Component:172; Jaccard:0.13105
 |
Component:172; Jaccard:0.11063
 |
Correlation:0.26998
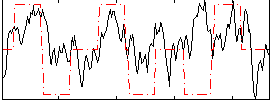 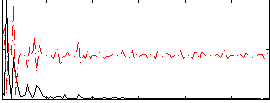 |
Correlation:0.26998
  |
Correlation:0.26998
  |
Correlation:0.26998
  |
Correlation:0.17749
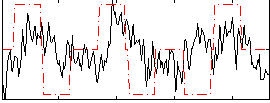 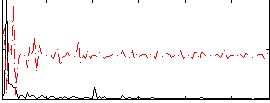 |
Correlation:0.17749
  |
Correlation:0.17749
  |
Component:172; Jaccard:0.11369
 |
Component:172; Jaccard:0.13222
 |
Component:172; Jaccard:0.14652
 |
Component:172; Jaccard:0.15287
 |
Component:221; Jaccard:0.13595
 |
Component:221; Jaccard:0.11042
 |
Component:221; Jaccard:0.085591
 |
Correlation:0.17749
  |
Correlation:0.17749
  |
Correlation:0.17749
  |
Correlation:0.17749
  |
Correlation:0.26998
  |
Correlation:0.26998
  |
Correlation:0.26998
  |
Component:098; Jaccard:0.10777
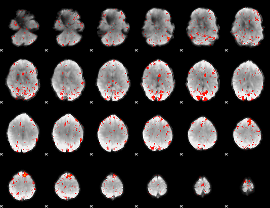 |
Component:098; Jaccard:0.12301
 |
Component:098; Jaccard:0.13226
 |
Component:098; Jaccard:0.13507
 |
Component:098; Jaccard:0.12221
 |
Component:098; Jaccard:0.10239
 |
Component:098; Jaccard:0.08467
 |
Correlation:0.15378
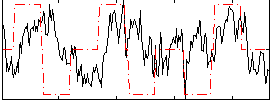 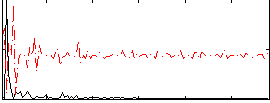 |
Correlation:0.15378
  |
Correlation:0.15378
  |
Correlation:0.15378
  |
Correlation:0.15378
  |
Correlation:0.15378
  |
Correlation:0.15378
  |
Component:153; Jaccard:0.098965
 |
Component:153; Jaccard:0.10831
 |
Component:153; Jaccard:0.11459
 |
Component:153; Jaccard:0.11545
 |
Component:153; Jaccard:0.10865
 |
Component:153; Jaccard:0.094019
 |
Component:153; Jaccard:0.076321
 |
Correlation:0.06681
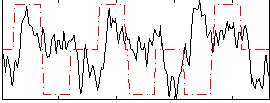 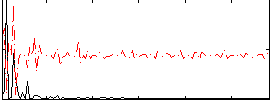 |
Correlation:0.06681
  |
Correlation:0.06681
  |
Correlation:0.06681
  |
Correlation:0.06681
  |
Correlation:0.06681
  |
Correlation:0.06681
  |
Component:316; Jaccard:0.087503
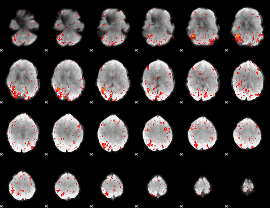 |
Component:316; Jaccard:0.093092
 |
Component:241; Jaccard:0.097348
 |
Component:241; Jaccard:0.1014
 |
Component:241; Jaccard:0.091407
 |
Component:241; Jaccard:0.069494
 |
Component:241; Jaccard:0.049976
 |
Correlation:0.16498
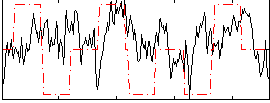 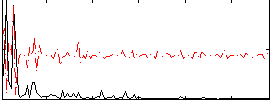 |
Correlation:0.16498
  |
Correlation:0.043878
 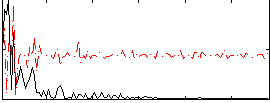 |
Correlation:0.043878
  |
Correlation:0.043878
  |
Correlation:0.043878
  |
Correlation:0.043878
  |
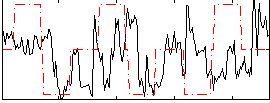
Component:271; Jaccard:0.084493
 |
Component:271; Jaccard:0.088476
 |
Component:316; Jaccard:0.095634
 |
Component:316; Jaccard:0.093132
 |
Component:316; Jaccard:0.077314
 |
Component:246; Jaccard:0.061439
 |
Component:246; Jaccard:0.049131
 |
Correlation:0.17107
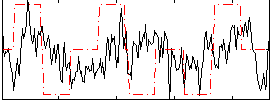  |
Correlation:0.17107
  |
Correlation:0.16498
  |
Correlation:0.16498
  |
Correlation:0.16498
  |
Correlation:-0.14124
  |
Correlation:-0.14124
  |
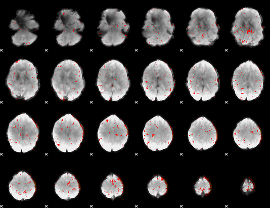
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
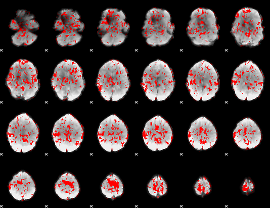 |
GLM binary result thresholded by 1.5
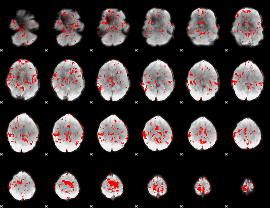 |
GLM binary result thresholded by 2
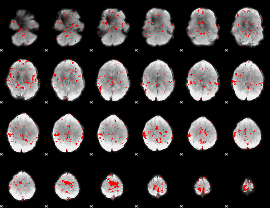 |
GLM binary result thresholded by 2.5
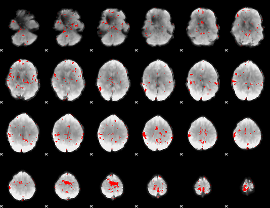 |
GLM binary result thresholded by 3
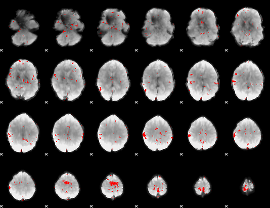 |
GLM binary result thresholded by 3.5
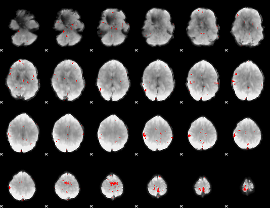 |
GLM binary result thresholded by 4
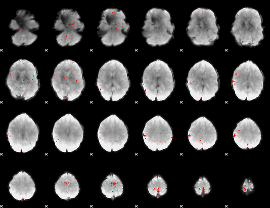 |
GLM Z-score result thresholded by 1
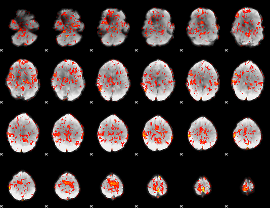 |
GLM Z-score result thresholded by 1.5
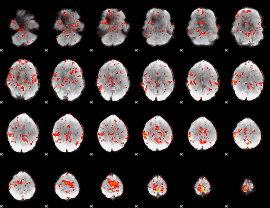 |
GLM Z-score result thresholded by 2
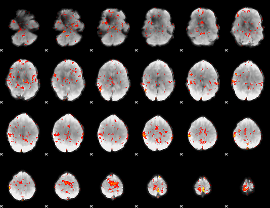 |
GLM Z-score result thresholded by 2.5
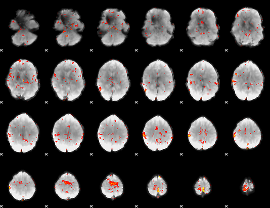 |
GLM Z-score result thresholded by 3
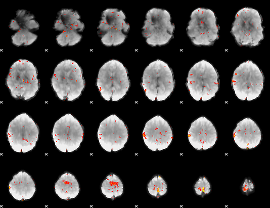 |
GLM Z-score result thresholded by 3.5
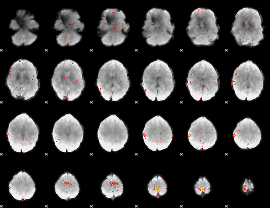 |
GLM Z-score result thresholded by 4
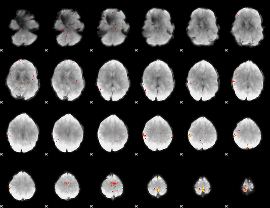 |
Component:242; Jaccard:0.1013
 |
Component:242; Jaccard:0.11606
 |
Component:242; Jaccard:0.12341
 |
Component:130; Jaccard:0.12448
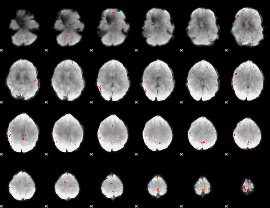 |
Component:130; Jaccard:0.16231
 |
Component:130; Jaccard:0.18297
 |
Component:130; Jaccard:0.18486
 |
Correlation:0.27147
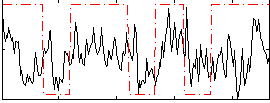 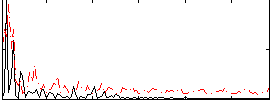 |
Correlation:0.27147
  |
Correlation:0.27147
  |
Correlation:0.26147
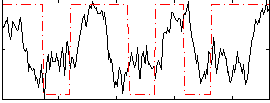 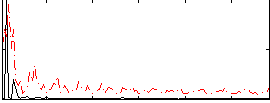 |
Correlation:0.26147
  |
Correlation:0.26147
  |
Correlation:0.26147
  |
Component:215; Jaccard:0.099921
 |
Component:011; Jaccard:0.10456
 |
Component:241; Jaccard:0.10562
 |
Component:242; Jaccard:0.11841
 |
Component:242; Jaccard:0.10178
 |
Component:242; Jaccard:0.082994
 |
Component:242; Jaccard:0.061163
 |
Correlation:0.043063
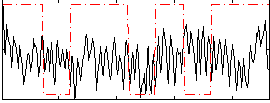 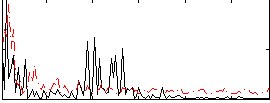 |
Correlation:0.38989
  |
Correlation:0.083556
  |
Correlation:0.27147
  |
Correlation:0.27147
  |
Correlation:0.27147
  |
Correlation:0.27147
  |
Component:011; Jaccard:0.098003
 |
Component:215; Jaccard:0.10335
 |
Component:011; Jaccard:0.10533
 |
Component:241; Jaccard:0.10635
 |
Component:219; Jaccard:0.096685
 |
Component:219; Jaccard:0.077509
 |
Component:219; Jaccard:0.0536
 |
Correlation:0.38989
  |
Correlation:0.043063
  |
Correlation:0.38989
  |
Correlation:0.083556
  |
Correlation:0.21861
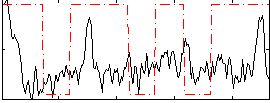  |
Correlation:0.21861
  |
Correlation:0.21861
  |
Component:241; Jaccard:0.08394
 |
Component:241; Jaccard:0.096962
 |
Component:219; Jaccard:0.10352
 |
Component:219; Jaccard:0.1053
 |
Component:241; Jaccard:0.090637
 |
Component:241; Jaccard:0.069017
 |
Component:241; Jaccard:0.05079
 |
Correlation:0.083556
  |
Correlation:0.083556
  |
Correlation:0.21861
  |
Correlation:0.21861
  |
Correlation:0.083556
  |
Correlation:0.083556
  |
Correlation:0.083556
  |
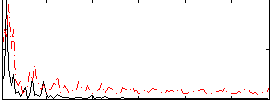
Component:002; Jaccard:0.083798
 |
Component:219; Jaccard:0.0959
 |
Component:215; Jaccard:0.097786
 |
Component:011; Jaccard:0.096548
 |
Component:011; Jaccard:0.074252
 |
Component:011; Jaccard:0.050809
 |
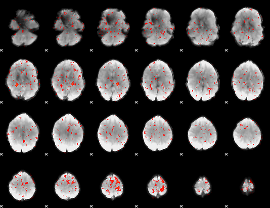
Component:162; Jaccard:0.042334
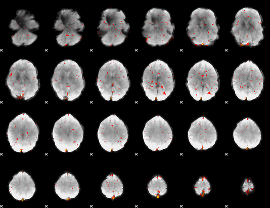 |
Correlation:0.24483
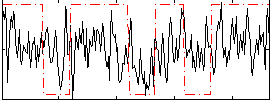 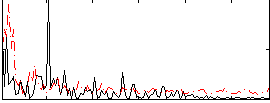 |
Correlation:0.21861
  |
Correlation:0.043063
  |
Correlation:0.38989
  |
Correlation:0.38989
  |
Correlation:0.38989
  |
Correlation:0.10899
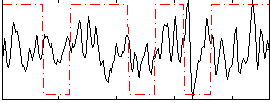 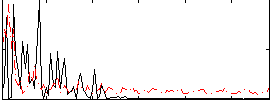 |
Component:219; Jaccard:0.082798
 |
Component:155; Jaccard:0.084084
 |
Component:130; Jaccard:0.090802
 |
Component:215; Jaccard:0.081045
 |
Component:215; Jaccard:0.062045
 |
Component:162; Jaccard:0.050255
 |
Component:215; Jaccard:0.032451
 |
Correlation:0.21861
  |
Correlation:0.34638
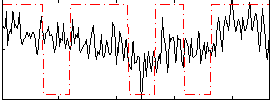 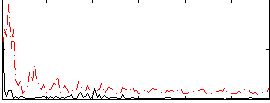 |
Correlation:0.26147
  |
Correlation:0.043063
  |
Correlation:0.043063
  |
Correlation:0.10899
  |
Correlation:0.043063
  |
Component:362; Jaccard:0.081542
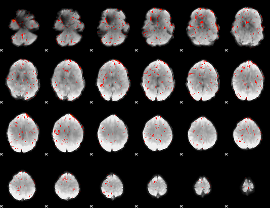 |
Component:002; Jaccard:0.083717
 |
Component:002; Jaccard:0.077004
 |
Component:030; Jaccard:0.063711
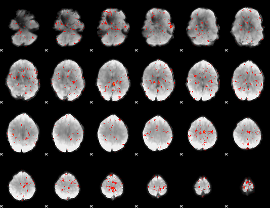 |
Component:162; Jaccard:0.053479
 |
Component:215; Jaccard:0.043182
 |
Component:011; Jaccard:0.032416
 |
Correlation:0.078121
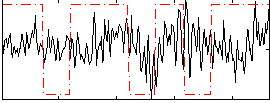 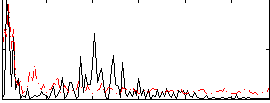 |
Correlation:0.24483
  |
Correlation:0.24483
  |
Correlation:0.22028
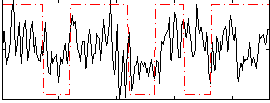  |
Correlation:0.10899
  |
Correlation:0.043063
  |
Correlation:0.38989
  |
Component:155; Jaccard:0.081423
 |
Component:032; Jaccard:0.077965
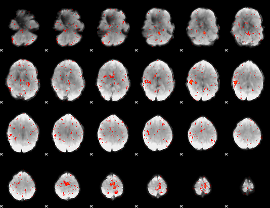 |
Component:155; Jaccard:0.075932
 |
Component:032; Jaccard:0.062798
 |
Component:309; Jaccard:0.051608
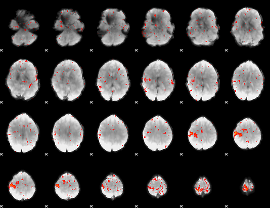 |
Component:309; Jaccard:0.037047
 |
Component:309; Jaccard:0.025964
 |
Correlation:0.34638
  |
Correlation:0.2182
 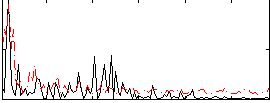 |
Correlation:0.34638
  |
Correlation:0.2182
  |
Correlation:0.25468
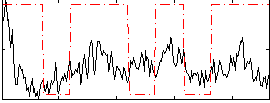 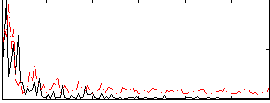 |
Correlation:0.25468
  |
Correlation:0.25468
  |
Component:032; Jaccard:0.07728
 |
Component:362; Jaccard:0.077526
 |
Component:032; Jaccard:0.07288
 |
Component:346; Jaccard:0.061275
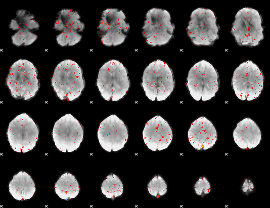 |
Component:030; Jaccard:0.049078
 |
Component:009; Jaccard:0.034706
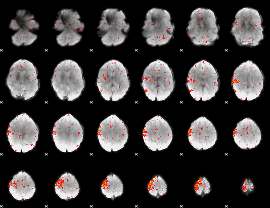 |
Component:030; Jaccard:0.025186
 |
Correlation:0.2182
  |
Correlation:0.078121
  |
Correlation:0.2182
  |
Correlation:0.19575
  |
Correlation:0.22028
  |
Correlation:0.12783
 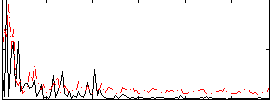 |
Correlation:0.22028
  |
Component:030; Jaccard:0.068639
 |
Component:030; Jaccard:0.072941
 |
Component:030; Jaccard:0.071154
 |
Component:207; Jaccard:0.060745
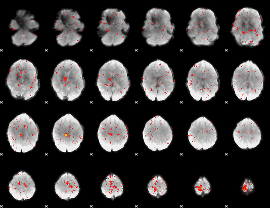 |
Component:032; Jaccard:0.047995
 |
Component:030; Jaccard:0.034538
 |
Component:009; Jaccard:0.024832
 |
Correlation:0.22028
  |
Correlation:0.22028
  |
Correlation:0.22028
  |
Correlation:0.16988
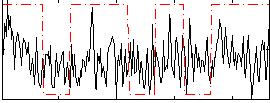 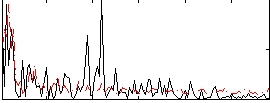 |
Correlation:0.2182
  |
Correlation:0.22028
  |
Correlation:0.12783
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
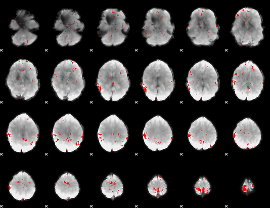 |
GLM binary result thresholded by 3.5
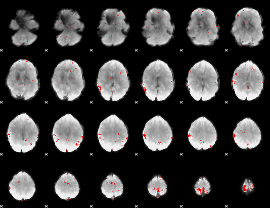 |
GLM binary result thresholded by 4
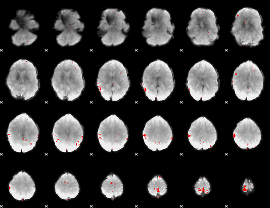 |
GLM Z-score result thresholded by 1
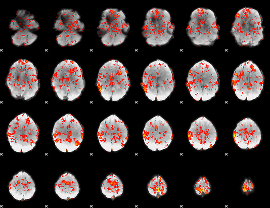 |
GLM Z-score result thresholded by 1.5
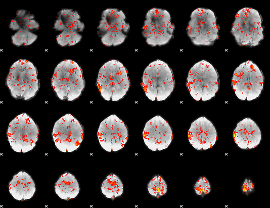 |
GLM Z-score result thresholded by 2
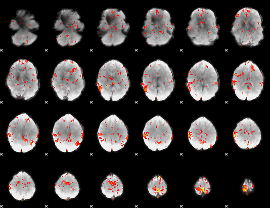 |
GLM Z-score result thresholded by 2.5
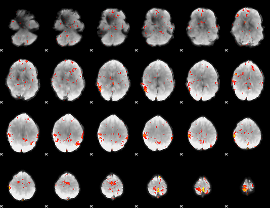 |
GLM Z-score result thresholded by 3
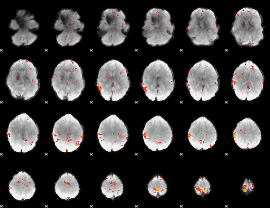 |
GLM Z-score result thresholded by 3.5
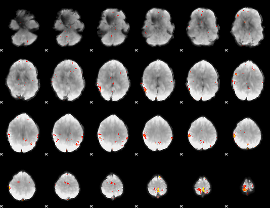 |
GLM Z-score result thresholded by 4
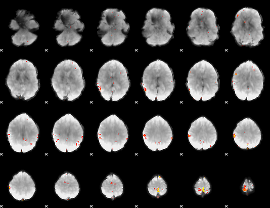 |
Component:219; Jaccard:0.11342
 |
Component:219; Jaccard:0.13775
 |
Component:219; Jaccard:0.15999
 |
Component:219; Jaccard:0.16683
 |
Component:130; Jaccard:0.20185
 |
Component:130; Jaccard:0.2318
 |
Component:130; Jaccard:0.22828
 |
Correlation:0.25592
  |
Correlation:0.25592
  |
Correlation:0.25592
  |
Correlation:0.25592
  |
Correlation:0.22468
  |
Correlation:0.22468
  |
Correlation:0.22468
  |
Component:321; Jaccard:0.10061
 |
Component:321; Jaccard:0.10185
 |
Component:130; Jaccard:0.10587
 |
Component:130; Jaccard:0.15064
 |
Component:219; Jaccard:0.15474
 |
Component:219; Jaccard:0.13441
 |
Component:219; Jaccard:0.097376
 |
Correlation:0.19968
  |
Correlation:0.19968
  |
Correlation:0.22468
  |
Correlation:0.22468
  |
Correlation:0.25592
  |
Correlation:0.25592
  |
Correlation:0.25592
  |
Component:215; Jaccard:0.09163
 |
Component:309; Jaccard:0.094968
 |
Component:309; Jaccard:0.09883
 |
Component:309; Jaccard:0.090392
 |
Component:309; Jaccard:0.075855
 |
Component:309; Jaccard:0.061489
 |
Component:309; Jaccard:0.0443
 |
Correlation:0.10679
  |
Correlation:0.13084
  |
Correlation:0.13084
  |
Correlation:0.13084
  |
Correlation:0.13084
  |
Correlation:0.13084
  |
Correlation:0.13084
  |
Component:309; Jaccard:0.086092
 |
Component:215; Jaccard:0.090705
 |
Component:321; Jaccard:0.095782
 |
Component:321; Jaccard:0.081342
 |
Component:242; Jaccard:0.067281
 |
Component:242; Jaccard:0.05455
 |
Component:162; Jaccard:0.043593
 |
Correlation:0.13084
  |
Correlation:0.10679
  |
Correlation:0.19968
  |
Correlation:0.19968
  |
Correlation:0.19412
  |
Correlation:0.19412
  |
Correlation:0.2643
 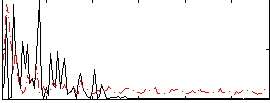 |
Component:089; Jaccard:0.083656
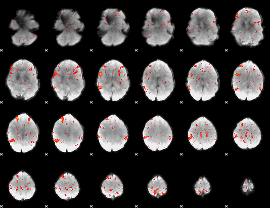 |
Component:089; Jaccard:0.087552
 |
Component:242; Jaccard:0.085816
 |
Component:242; Jaccard:0.079734
 |
Component:011; Jaccard:0.064196
 |
Component:011; Jaccard:0.049245
 |
Component:242; Jaccard:0.039893
 |
Correlation:0.14901
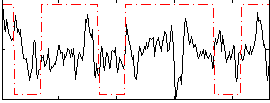 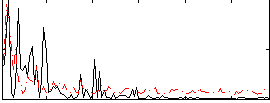 |
Correlation:0.14901
  |
Correlation:0.19412
  |
Correlation:0.19412
  |
Correlation:-0.031995
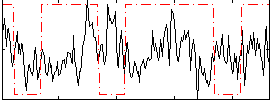 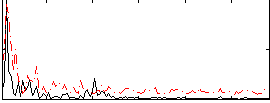 |
Correlation:-0.031995
  |
Correlation:0.19412
  |
Component:242; Jaccard:0.082038
 |
Component:242; Jaccard:0.0865
 |
Component:089; Jaccard:0.085037
 |
Component:011; Jaccard:0.075306
 |
Component:321; Jaccard:0.063538
 |
Component:162; Jaccard:0.048141
 |
Component:011; Jaccard:0.03455
 |
Correlation:0.19412
  |
Correlation:0.19412
  |
Correlation:0.14901
  |
Correlation:-0.031995
  |
Correlation:0.19968
  |
Correlation:0.2643
  |
Correlation:-0.031995
  |
Component:011; Jaccard:0.081923
 |
Component:011; Jaccard:0.08519
 |
Component:215; Jaccard:0.0847
 |
Component:215; Jaccard:0.074068
 |
Component:215; Jaccard:0.058818
 |
Component:321; Jaccard:0.045925
 |
Component:215; Jaccard:0.032322
 |
Correlation:-0.031995
  |
Correlation:-0.031995
  |
Correlation:0.10679
  |
Correlation:0.10679
  |
Correlation:0.10679
  |
Correlation:0.19968
  |
Correlation:0.10679
  |
Component:252; Jaccard:0.080977
 |
Component:252; Jaccard:0.083456
 |
Component:011; Jaccard:0.083242
 |
Component:089; Jaccard:0.072586
 |
Component:089; Jaccard:0.055928
 |
Component:215; Jaccard:0.043759
 |
Component:321; Jaccard:0.031942
 |
Correlation:0.39472
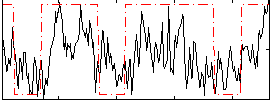 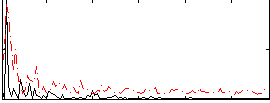 |
Correlation:0.39472
  |
Correlation:-0.031995
  |
Correlation:0.14901
  |
Correlation:0.14901
  |
Correlation:0.10679
  |
Correlation:0.19968
  |
Component:333; Jaccard:0.077805
 |
Component:333; Jaccard:0.077808
 |
Component:252; Jaccard:0.07912
 |
Component:252; Jaccard:0.066042
 |
Component:018; Jaccard:0.052827
 |
Component:009; Jaccard:0.039939
 |
Component:009; Jaccard:0.030038
 |
Correlation:0.22141
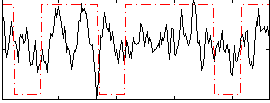 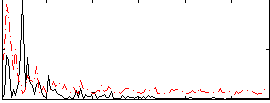 |
Correlation:0.22141
  |
Correlation:0.39472
  |
Correlation:0.39472
  |
Correlation:0.23945
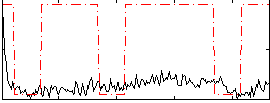 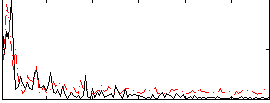 |
Correlation:0.19463
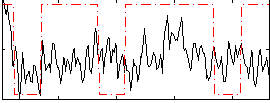 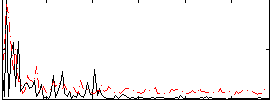 |
Correlation:0.19463
  |
Component:235; Jaccard:0.077224
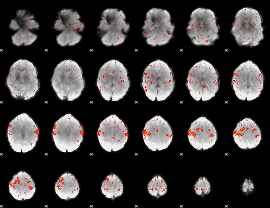 |
Component:235; Jaccard:0.077307
 |
Component:235; Jaccard:0.074118
 |
Component:018; Jaccard:0.064023
 |
Component:162; Jaccard:0.050863
 |
Component:018; Jaccard:0.039848
 |
Component:018; Jaccard:0.02806
 |
Correlation:0.17941
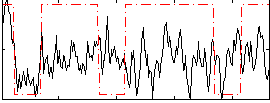  |
Correlation:0.17941
  |
Correlation:0.17941
  |
Correlation:0.23945
  |
Correlation:0.2643
  |
Correlation:0.23945
  |
Correlation:0.23945
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































