Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
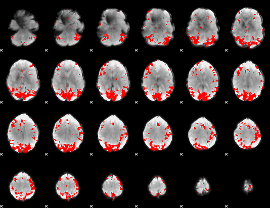 |
GLM binary result thresholded by 3.5
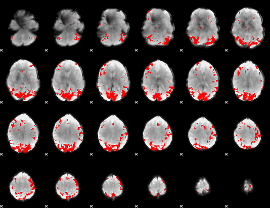 |
GLM binary result thresholded by 4
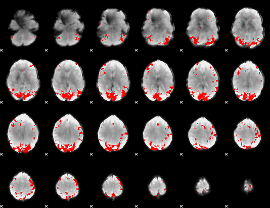 |
GLM Z-score result thresholded by 1
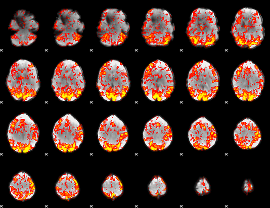 |
GLM Z-score result thresholded by 1.5
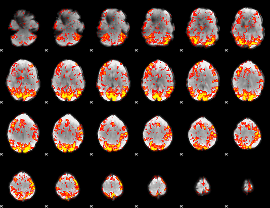 |
GLM Z-score result thresholded by 2
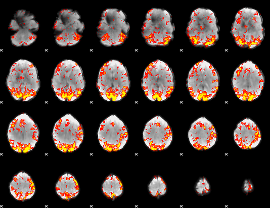 |
GLM Z-score result thresholded by 2.5
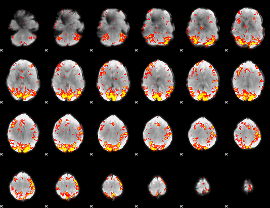 |
GLM Z-score result thresholded by 3
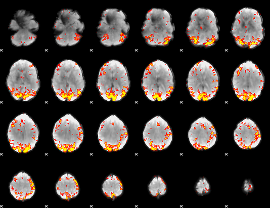 |
GLM Z-score result thresholded by 3.5
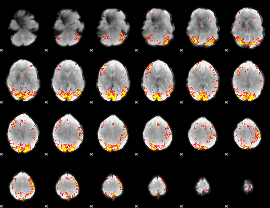 |
GLM Z-score result thresholded by 4
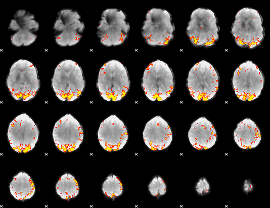 |
Component:208; Jaccard:0.12978
 |
Component:208; Jaccard:0.16276
 |
Component:208; Jaccard:0.20091
 |
Component:205; Jaccard:0.24296
 |
Component:205; Jaccard:0.29792
 |
Component:205; Jaccard:0.34963
 |
Component:205; Jaccard:0.38774
 |
Correlation:0.33701
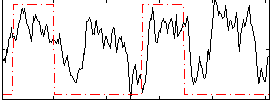 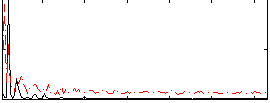 |
Correlation:0.33701
  |
Correlation:0.33701
  |
Correlation:0.29208
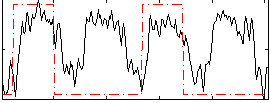  |
Correlation:0.29208
  |
Correlation:0.29208
  |
Correlation:0.29208
  |
Component:012; Jaccard:0.11917
 |
Component:205; Jaccard:0.14943
 |
Component:205; Jaccard:0.19167
 |
Component:208; Jaccard:0.24231
 |
Component:208; Jaccard:0.28156
 |
Component:208; Jaccard:0.30608
 |
Component:208; Jaccard:0.31187
 |
Correlation:0.26177
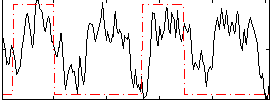  |
Correlation:0.29208
  |
Correlation:0.29208
  |
Correlation:0.33701
  |
Correlation:0.33701
  |
Correlation:0.33701
  |
Correlation:0.33701
  |
Component:381; Jaccard:0.11823
 |
Component:012; Jaccard:0.14827
 |
Component:012; Jaccard:0.18195
 |
Component:012; Jaccard:0.21872
 |
Component:012; Jaccard:0.24979
 |
Component:012; Jaccard:0.2733
 |
Component:012; Jaccard:0.28449
 |
Correlation:0.26518
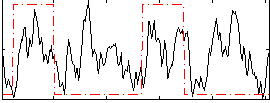 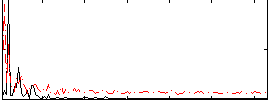 |
Correlation:0.26177
  |
Correlation:0.26177
  |
Correlation:0.26177
  |
Correlation:0.26177
  |
Correlation:0.26177
  |
Correlation:0.26177
  |
Component:205; Jaccard:0.11625
 |
Component:381; Jaccard:0.13316
 |
Component:236; Jaccard:0.14837
 |
Component:236; Jaccard:0.1643
 |
Component:010; Jaccard:0.17735
 |
Component:010; Jaccard:0.18889
 |
Component:010; Jaccard:0.19196
 |
Correlation:0.29208
  |
Correlation:0.26518
  |
Correlation:0.1667
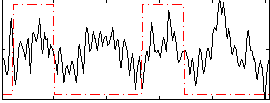 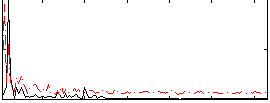 |
Correlation:0.1667
  |
Correlation:0.13311
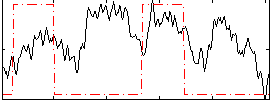 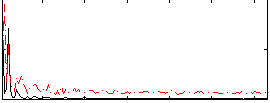 |
Correlation:0.13311
  |
Correlation:0.13311
  |
Component:056; Jaccard:0.1142
 |
Component:056; Jaccard:0.13037
 |
Component:381; Jaccard:0.14727
 |
Component:378; Jaccard:0.16255
 |
Component:378; Jaccard:0.17646
 |
Component:378; Jaccard:0.18188
 |
Component:378; Jaccard:0.17959
 |
Correlation:0.19712
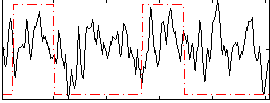 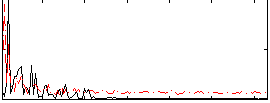 |
Correlation:0.19712
  |
Correlation:0.26518
  |
Correlation:0.25911
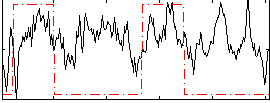 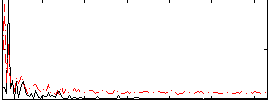 |
Correlation:0.25911
  |
Correlation:0.25911
  |
Correlation:0.25911
  |
Component:236; Jaccard:0.11213
 |
Component:236; Jaccard:0.12977
 |
Component:056; Jaccard:0.14633
 |
Component:010; Jaccard:0.15985
 |
Component:236; Jaccard:0.1735
 |
Component:236; Jaccard:0.17442
 |
Component:236; Jaccard:0.16773
 |
Correlation:0.1667
  |
Correlation:0.1667
  |
Correlation:0.19712
  |
Correlation:0.13311
  |
Correlation:0.1667
  |
Correlation:0.1667
  |
Correlation:0.1667
  |
Component:378; Jaccard:0.10621
 |
Component:378; Jaccard:0.12537
 |
Component:378; Jaccard:0.14446
 |
Component:381; Jaccard:0.15839
 |
Component:056; Jaccard:0.16412
 |
Component:056; Jaccard:0.16145
 |
Component:056; Jaccard:0.15196
 |
Correlation:0.25911
  |
Correlation:0.25911
  |
Correlation:0.25911
  |
Correlation:0.26518
  |
Correlation:0.19712
  |
Correlation:0.19712
  |
Correlation:0.19712
  |
Component:282; Jaccard:0.10582
 |
Component:010; Jaccard:0.12116
 |
Component:010; Jaccard:0.14106
 |
Component:056; Jaccard:0.158
 |
Component:381; Jaccard:0.16223
 |
Component:381; Jaccard:0.15967
 |
Component:381; Jaccard:0.15135
 |
Correlation:0.053821
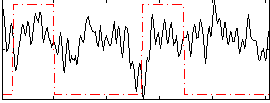 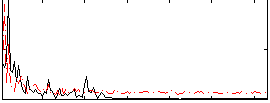 |
Correlation:0.13311
  |
Correlation:0.13311
  |
Correlation:0.19712
  |
Correlation:0.26518
  |
Correlation:0.26518
  |
Correlation:0.26518
  |
Component:193; Jaccard:0.1022
 |
Component:282; Jaccard:0.1178
 |
Component:193; Jaccard:0.1312
 |
Component:193; Jaccard:0.14104
 |
Component:193; Jaccard:0.14366
 |
Component:193; Jaccard:0.13972
 |
Component:391; Jaccard:0.13163
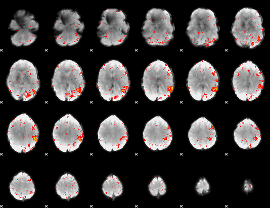 |
Correlation:0.061908
  |
Correlation:0.053821
  |
Correlation:0.061908
  |
Correlation:0.061908
  |
Correlation:0.061908
  |
Correlation:0.061908
  |
Correlation:0.13046
 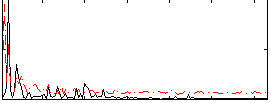 |
Component:010; Jaccard:0.10211
 |
Component:193; Jaccard:0.1175
 |
Component:282; Jaccard:0.12955
 |
Component:282; Jaccard:0.137
 |
Component:391; Jaccard:0.13849
 |
Component:391; Jaccard:0.13723
 |
Component:164; Jaccard:0.12981
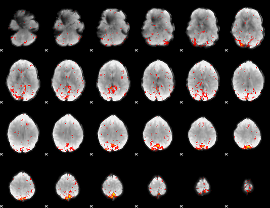 |
Correlation:0.13311
  |
Correlation:0.061908
  |
Correlation:0.053821
  |
Correlation:0.053821
  |
Correlation:0.13046
  |
Correlation:0.13046
  |
Correlation:0.19616
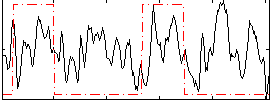 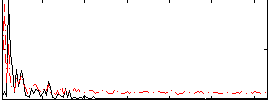 |
Cope: 02:REWARD BACK
|
GLM result group-wise
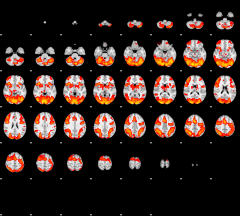 |
GLM binary result thresholded by 1
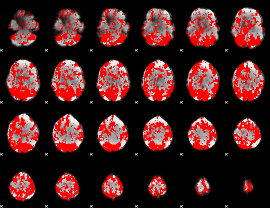 |
GLM binary result thresholded by 1.5
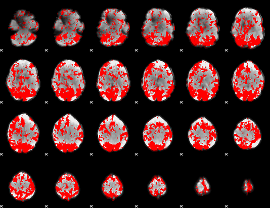 |
GLM binary result thresholded by 2
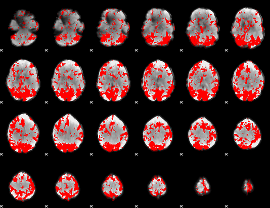 |
GLM binary result thresholded by 2.5
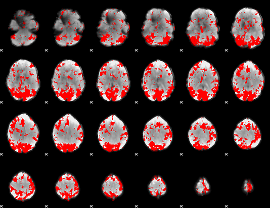 |
GLM binary result thresholded by 3
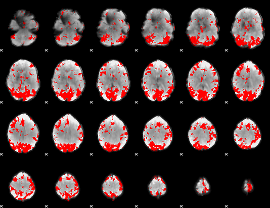 |
GLM binary result thresholded by 3.5
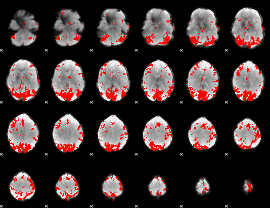 |
GLM binary result thresholded by 4
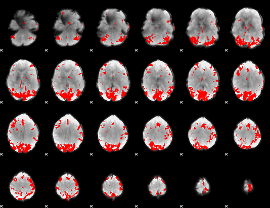 |
GLM Z-score result thresholded by 1
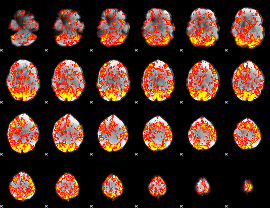 |
GLM Z-score result thresholded by 1.5
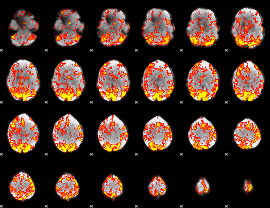 |
GLM Z-score result thresholded by 2
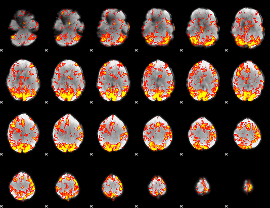 |
GLM Z-score result thresholded by 2.5
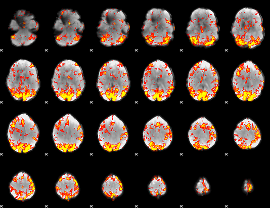 |
GLM Z-score result thresholded by 3
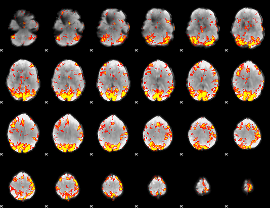 |
GLM Z-score result thresholded by 3.5
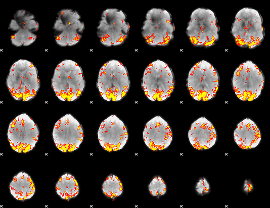 |
GLM Z-score result thresholded by 4
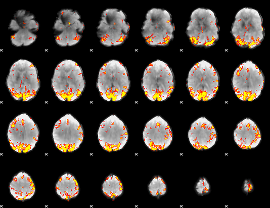 |
Component:282; Jaccard:0.1106
 |
Component:236; Jaccard:0.12865
 |
Component:236; Jaccard:0.15111
 |
Component:208; Jaccard:0.17806
 |
Component:208; Jaccard:0.20593
 |
Component:205; Jaccard:0.24096
 |
Component:205; Jaccard:0.27372
 |
Correlation:0.2508
 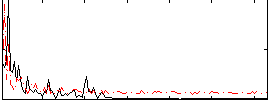 |
Correlation:0.38469
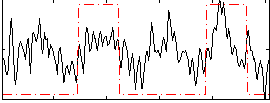 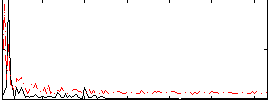 |
Correlation:0.38469
  |
Correlation:0.28509
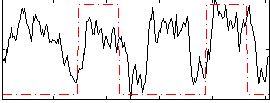 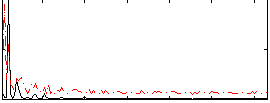 |
Correlation:0.28509
  |
Correlation:0.37658
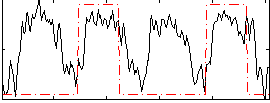  |
Correlation:0.37658
  |
Component:236; Jaccard:0.10893
 |
Component:282; Jaccard:0.12752
 |
Component:208; Jaccard:0.15007
 |
Component:236; Jaccard:0.17425
 |
Component:205; Jaccard:0.20405
 |
Component:208; Jaccard:0.23346
 |
Component:208; Jaccard:0.25545
 |
Correlation:0.38469
  |
Correlation:0.2508
  |
Correlation:0.28509
  |
Correlation:0.38469
  |
Correlation:0.37658
  |
Correlation:0.28509
  |
Correlation:0.28509
  |
Component:056; Jaccard:0.1037
 |
Component:208; Jaccard:0.12472
 |
Component:282; Jaccard:0.14516
 |
Component:205; Jaccard:0.1696
 |
Component:012; Jaccard:0.1974
 |
Component:012; Jaccard:0.22386
 |
Component:012; Jaccard:0.2475
 |
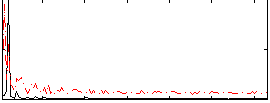
Correlation:0.15927
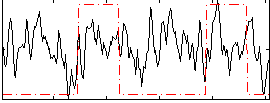  |
Correlation:0.28509
  |
Correlation:0.2508
  |
Correlation:0.37658
  |
Correlation:0.21178
  |
Correlation:0.21178
  |
Correlation:0.21178
  |
Component:208; Jaccard:0.10354
 |
Component:056; Jaccard:0.11981
 |
Component:012; Jaccard:0.14059
 |
Component:012; Jaccard:0.16889
 |
Component:236; Jaccard:0.19601
 |
Component:236; Jaccard:0.21407
 |
Component:236; Jaccard:0.22327
 |
Correlation:0.28509
  |
Correlation:0.15927
  |
Correlation:0.21178
  |
Correlation:0.21178
  |
Correlation:0.38469
  |
Correlation:0.38469
  |
Correlation:0.38469
  |
Component:381; Jaccard:0.10021
 |
Component:012; Jaccard:0.1159
 |
Component:205; Jaccard:0.13828
 |
Component:282; Jaccard:0.15921
 |
Component:010; Jaccard:0.18188
 |
Component:010; Jaccard:0.2024
 |
Component:010; Jaccard:0.21928
 |
Correlation:0.3566
 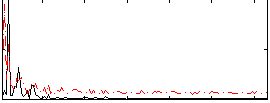 |
Correlation:0.21178
  |
Correlation:0.37658
  |
Correlation:0.2508
  |
Correlation:0.36719
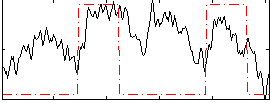 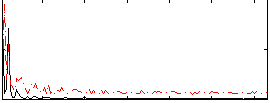 |
Correlation:0.36719
  |
Correlation:0.36719
  |
Component:012; Jaccard:0.095061
 |
Component:381; Jaccard:0.11298
 |
Component:056; Jaccard:0.13647
 |
Component:010; Jaccard:0.15812
 |
Component:282; Jaccard:0.16888
 |
Component:282; Jaccard:0.17395
 |
Component:056; Jaccard:0.17416
 |
Correlation:0.21178
  |
Correlation:0.3566
  |
Correlation:0.15927
  |
Correlation:0.36719
  |
Correlation:0.2508
  |
Correlation:0.2508
  |
Correlation:0.15927
  |
Component:010; Jaccard:0.09398
 |
Component:010; Jaccard:0.11285
 |
Component:010; Jaccard:0.13462
 |
Component:056; Jaccard:0.15155
 |
Component:056; Jaccard:0.16263
 |
Component:056; Jaccard:0.17067
 |
Component:378; Jaccard:0.17284
 |
Correlation:0.36719
  |
Correlation:0.36719
  |
Correlation:0.36719
  |
Correlation:0.15927
  |
Correlation:0.15927
  |
Correlation:0.15927
  |
Correlation:0.25058
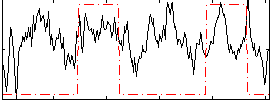 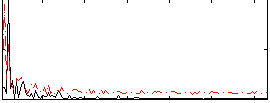 |
Component:193; Jaccard:0.091152
 |
Component:205; Jaccard:0.11187
 |
Component:381; Jaccard:0.12621
 |
Component:381; Jaccard:0.13753
 |
Component:378; Jaccard:0.1525
 |
Component:378; Jaccard:0.16441
 |
Component:282; Jaccard:0.17198
 |
Correlation:0.2949
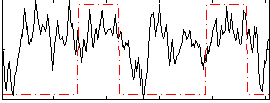 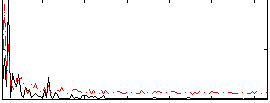 |
Correlation:0.37658
  |
Correlation:0.3566
  |
Correlation:0.3566
  |
Correlation:0.25058
  |
Correlation:0.25058
  |
Correlation:0.2508
  |
Component:205; Jaccard:0.091033
 |
Component:193; Jaccard:0.10595
 |
Component:193; Jaccard:0.12144
 |
Component:378; Jaccard:0.13745
 |
Component:381; Jaccard:0.14605
 |
Component:381; Jaccard:0.15037
 |
Component:223; Jaccard:0.15643
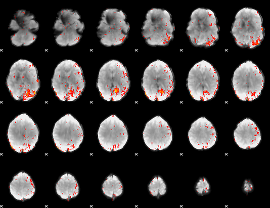 |
Correlation:0.37658
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.25058
  |
Correlation:0.3566
  |
Correlation:0.3566
  |
Correlation:0.39357
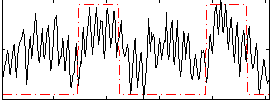 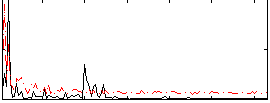 |
Component:378; Jaccard:0.089998
 |
Component:378; Jaccard:0.10458
 |
Component:378; Jaccard:0.1207
 |
Component:193; Jaccard:0.13519
 |
Component:193; Jaccard:0.14388
 |
Component:193; Jaccard:0.15006
 |
Component:193; Jaccard:0.15146
 |
Correlation:0.25058
  |
Correlation:0.25058
  |
Correlation:0.25058
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
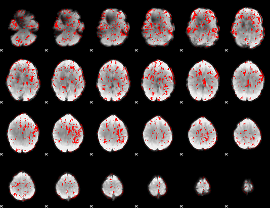 |
GLM binary result thresholded by 1.5
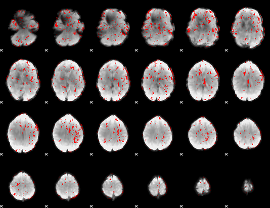 |
GLM binary result thresholded by 2
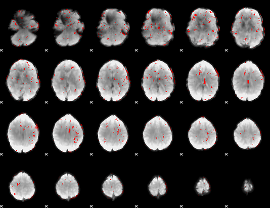 |
GLM binary result thresholded by 2.5
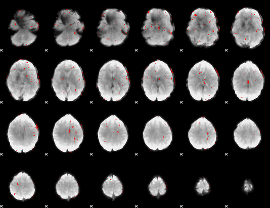 |
GLM binary result thresholded by 3
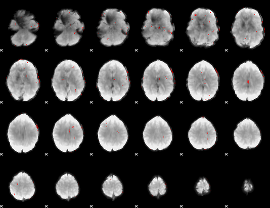 |
GLM binary result thresholded by 3.5
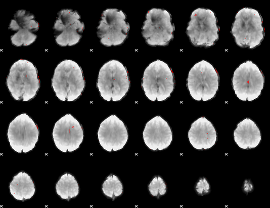 |
GLM binary result thresholded by 4
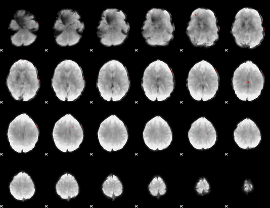 |
GLM Z-score result thresholded by 1
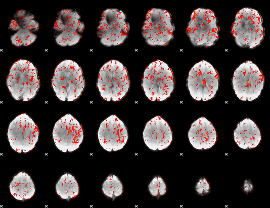 |
GLM Z-score result thresholded by 1.5
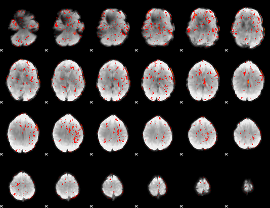 |
GLM Z-score result thresholded by 2
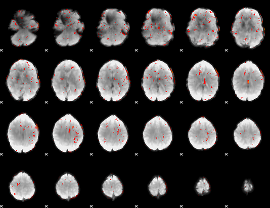 |
GLM Z-score result thresholded by 2.5
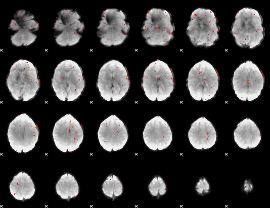 |
GLM Z-score result thresholded by 3
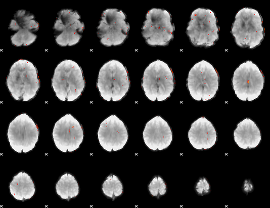 |
GLM Z-score result thresholded by 3.5
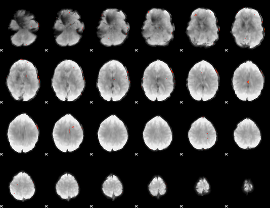 |
GLM Z-score result thresholded by 4
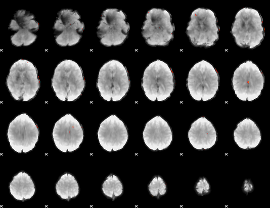 |
Component:104; Jaccard:0.10567
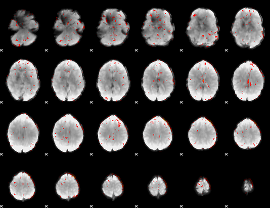 |
Component:395; Jaccard:0.12309
 |
Component:395; Jaccard:0.13552
 |
Component:395; Jaccard:0.12855
 |
Component:395; Jaccard:0.10326
 |
Component:395; Jaccard:0.071635
 |
Component:395; Jaccard:0.047391
 |
Correlation:0.22756
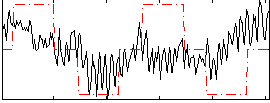 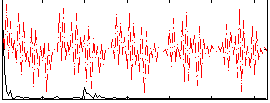 |
Correlation:0.41381
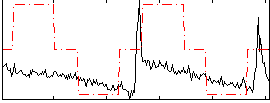  |
Correlation:0.41381
  |
Correlation:0.41381
  |
Correlation:0.41381
  |
Correlation:0.41381
  |
Correlation:0.41381
  |
Component:395; Jaccard:0.10297
 |
Component:104; Jaccard:0.1125
 |
Component:104; Jaccard:0.10392
 |
Component:104; Jaccard:0.075898
 |
Component:104; Jaccard:0.047275
 |
Component:325; Jaccard:0.035965
 |
Component:325; Jaccard:0.026362
 |
Correlation:0.41381
  |
Correlation:0.22756
  |
Correlation:0.22756
  |
Correlation:0.22756
  |
Correlation:0.22756
  |
Correlation:0.17057
  |
Correlation:0.17057
  |
Component:392; Jaccard:0.093647
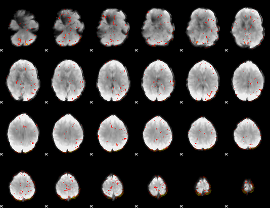 |
Component:392; Jaccard:0.09511
 |
Component:392; Jaccard:0.080434
 |
Component:392; Jaccard:0.054381
 |
Component:325; Jaccard:0.045434
 |
Component:379; Jaccard:0.03212
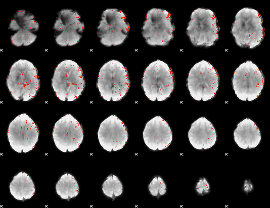 |
Component:379; Jaccard:0.024238
 |
Correlation:0.19486
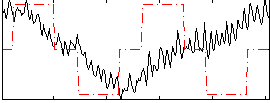 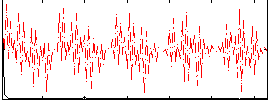 |
Correlation:0.19486
  |
Correlation:0.19486
  |
Correlation:0.19486
  |
Correlation:0.17057
  |
Correlation:0.12382
  |
Correlation:0.12382
  |
Component:080; Jaccard:0.082856
 |
Component:080; Jaccard:0.082259
 |
Component:064; Jaccard:0.065855
 |
Component:325; Jaccard:0.051586
 |
Component:379; Jaccard:0.03751
 |
Component:104; Jaccard:0.02157
 |
Component:104; Jaccard:0.010663
 |
Correlation:0.23719
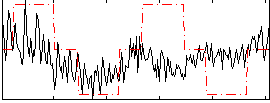  |
Correlation:0.23719
  |
Correlation:0.16522
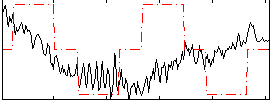  |
Correlation:0.17057
  |
Correlation:0.12382
  |
Correlation:0.22756
  |
Correlation:0.22756
  |
Component:270; Jaccard:0.079515
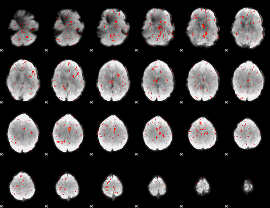 |
Component:270; Jaccard:0.078031
 |
Component:080; Jaccard:0.065638
 |
Component:270; Jaccard:0.046139
 |
Component:392; Jaccard:0.029837
 |
Component:134; Jaccard:0.013827
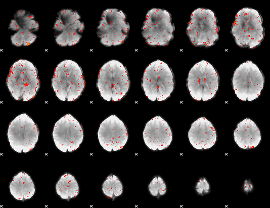 |
Component:134; Jaccard:0.009172
 |
Correlation:0.12578
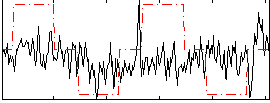 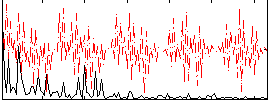 |
Correlation:0.12578
  |
Correlation:0.23719
  |
Correlation:0.12578
  |
Correlation:0.19486
  |
Correlation:0.47587
  |
Correlation:0.47587
  |
Component:220; Jaccard:0.078224
 |
Component:064; Jaccard:0.077672
 |
Component:270; Jaccard:0.065008
 |
Component:379; Jaccard:0.044513
 |
Component:270; Jaccard:0.026238
 |
Component:270; Jaccard:0.01293
 |
Component:118; Jaccard:0.00798
 |
Correlation:0.046364
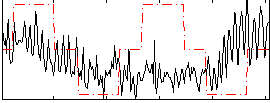 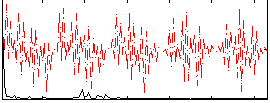 |
Correlation:0.16522
  |
Correlation:0.12578
  |
Correlation:0.12382
  |
Correlation:0.12578
  |
Correlation:0.12578
  |
Correlation:0.34878
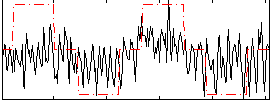 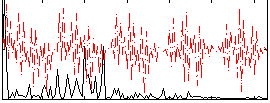 |
Component:064; Jaccard:0.077817
 |
Component:220; Jaccard:0.076014
 |
Component:220; Jaccard:0.062414
 |
Component:064; Jaccard:0.043729
 |
Component:064; Jaccard:0.023745
 |
Component:118; Jaccard:0.012035
 |
Component:094; Jaccard:0.007702
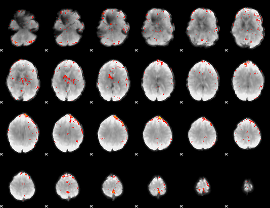 |
Correlation:0.16522
  |
Correlation:0.046364
  |
Correlation:0.046364
  |
Correlation:0.16522
  |
Correlation:0.16522
  |
Correlation:0.34878
  |
Correlation:0.23244
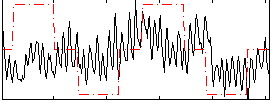 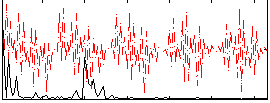 |
Component:087; Jaccard:0.075429
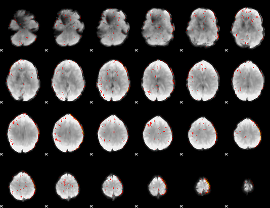 |
Component:087; Jaccard:0.074181
 |
Component:087; Jaccard:0.060992
 |
Component:080; Jaccard:0.042567
 |
Component:080; Jaccard:0.023214
 |
Component:392; Jaccard:0.011985
 |
Component:085; Jaccard:0.007512
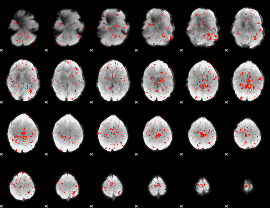 |
Correlation:-0.011797
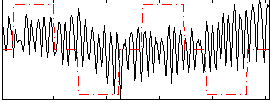 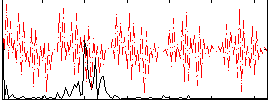 |
Correlation:-0.011797
  |
Correlation:-0.011797
  |
Correlation:0.23719
  |
Correlation:0.23719
  |
Correlation:0.19486
  |
Correlation:0.22416
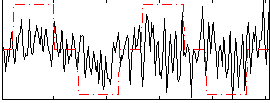  |
Component:095; Jaccard:0.071018
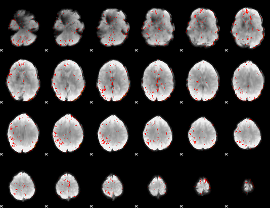 |
Component:112; Jaccard:0.066056
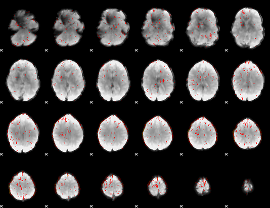 |
Component:112; Jaccard:0.054414
 |
Component:220; Jaccard:0.041992
 |
Component:220; Jaccard:0.022752
 |
Component:085; Jaccard:0.011043
 |
Component:182; Jaccard:0.00633
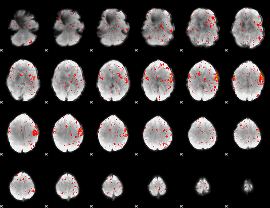 |
Correlation:0.28112
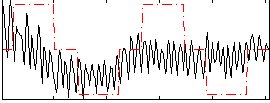 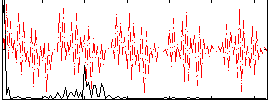 |
Correlation:0.058796
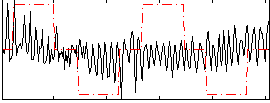 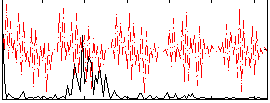 |
Correlation:0.058796
  |
Correlation:0.046364
  |
Correlation:0.046364
  |
Correlation:0.22416
  |
Correlation:0.16271
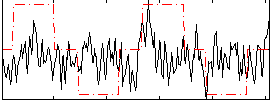 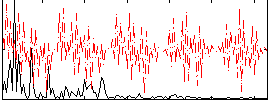 |
Component:112; Jaccard:0.070279
 |
Component:095; Jaccard:0.064459
 |
Component:095; Jaccard:0.052244
 |
Component:087; Jaccard:0.039913
 |
Component:087; Jaccard:0.021993
 |
Component:261; Jaccard:0.01081
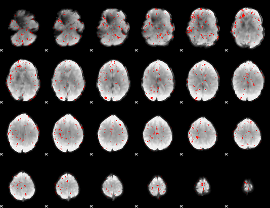 |
Component:261; Jaccard:0.006167
 |
Correlation:0.058796
  |
Correlation:0.28112
  |
Correlation:0.28112
  |
Correlation:-0.011797
  |
Correlation:-0.011797
  |
Correlation:0.30648
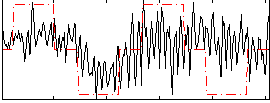  |
Correlation:0.30648
  |
Cope: 04:neg_PUNISH BACK
|
GLM result group-wise
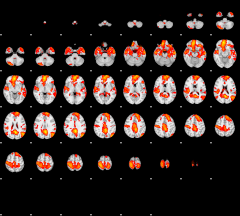 |
GLM binary result thresholded by 1
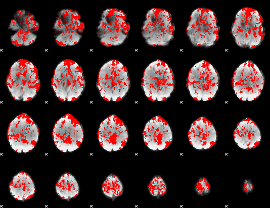 |
GLM binary result thresholded by 1.5
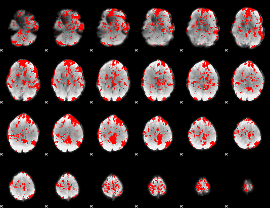 |
GLM binary result thresholded by 2
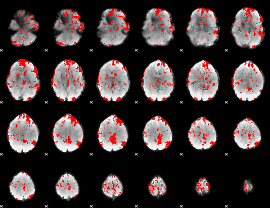 |
GLM binary result thresholded by 2.5
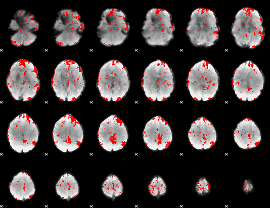 |
GLM binary result thresholded by 3
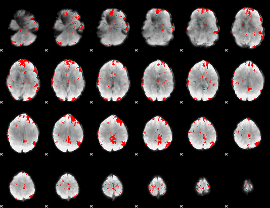 |
GLM binary result thresholded by 3.5
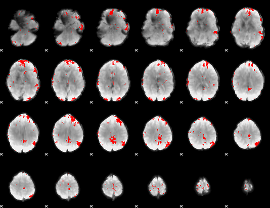 |
GLM binary result thresholded by 4
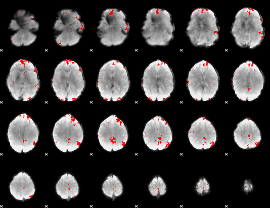 |
GLM Z-score result thresholded by 1
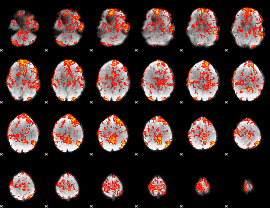 |
GLM Z-score result thresholded by 1.5
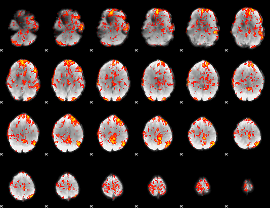 |
GLM Z-score result thresholded by 2
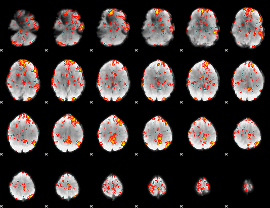 |
GLM Z-score result thresholded by 2.5
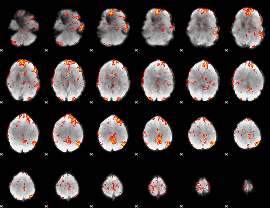 |
GLM Z-score result thresholded by 3
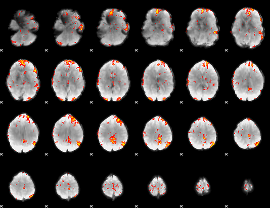 |
GLM Z-score result thresholded by 3.5
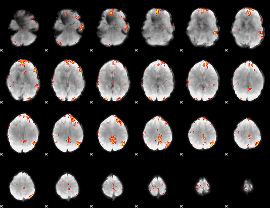 |
GLM Z-score result thresholded by 4
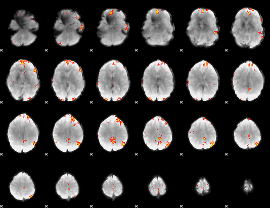 |
Component:234; Jaccard:0.11636
 |
Component:234; Jaccard:0.14136
 |
Component:334; Jaccard:0.16767
 |
Component:334; Jaccard:0.20116
 |
Component:334; Jaccard:0.22845
 |
Component:334; Jaccard:0.23704
 |
Component:334; Jaccard:0.22375
 |
Correlation:0.047182
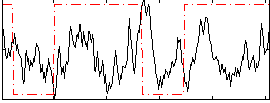 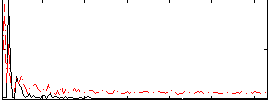 |
Correlation:0.047182
  |
Correlation:0.11681
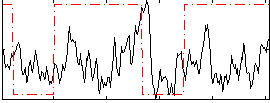 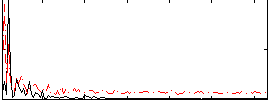 |
Correlation:0.11681
  |
Correlation:0.11681
  |
Correlation:0.11681
  |
Correlation:0.11681
  |
Component:096; Jaccard:0.1081
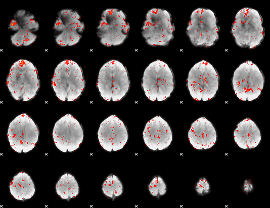 |
Component:334; Jaccard:0.13199
 |
Component:234; Jaccard:0.16745
 |
Component:198; Jaccard:0.19001
 |
Component:198; Jaccard:0.21337
 |
Component:198; Jaccard:0.22701
 |
Component:198; Jaccard:0.21772
 |
Correlation:0.076071
  |
Correlation:0.11681
  |
Correlation:0.047182
  |
Correlation:0.18282
  |
Correlation:0.18282
  |
Correlation:0.18282
  |
Correlation:0.18282
  |
Component:334; Jaccard:0.1029
 |
Component:198; Jaccard:0.12989
 |
Component:198; Jaccard:0.16047
 |
Component:234; Jaccard:0.18965
 |
Component:234; Jaccard:0.19809
 |
Component:234; Jaccard:0.19689
 |
Component:234; Jaccard:0.17973
 |
Correlation:0.11681
  |
Correlation:0.18282
  |
Correlation:0.18282
  |
Correlation:0.047182
  |
Correlation:0.047182
  |
Correlation:0.047182
  |
Correlation:0.047182
  |
Component:198; Jaccard:0.10285
 |
Component:096; Jaccard:0.11567
 |
Component:070; Jaccard:0.12277
 |
Component:070; Jaccard:0.13211
 |
Component:070; Jaccard:0.13675
 |
Component:070; Jaccard:0.13319
 |
Component:070; Jaccard:0.11757
 |
Correlation:0.18282
  |
Correlation:0.076071
  |
Correlation:0.064889
  |
Correlation:0.064889
  |
Correlation:0.064889
  |
Correlation:0.064889
  |
Correlation:0.064889
  |
Component:022; Jaccard:0.095724
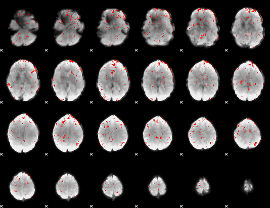 |
Component:070; Jaccard:0.10903
 |
Component:331; Jaccard:0.12087
 |
Component:331; Jaccard:0.12945
 |
Component:331; Jaccard:0.13266
 |
Component:331; Jaccard:0.12949
 |
Component:331; Jaccard:0.11294
 |
Correlation:0.37205
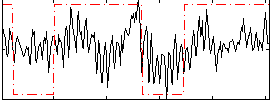 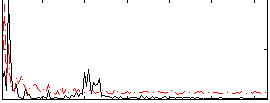 |
Correlation:0.064889
  |
Correlation:0.21284
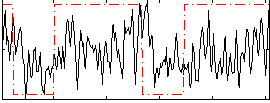 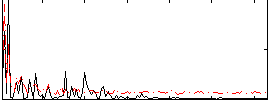 |
Correlation:0.21284
  |
Correlation:0.21284
  |
Correlation:0.21284
  |
Correlation:0.21284
  |
Component:121; Jaccard:0.094081
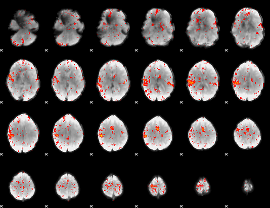 |
Component:331; Jaccard:0.10749
 |
Component:389; Jaccard:0.11658
 |
Component:389; Jaccard:0.12528
 |
Component:389; Jaccard:0.12779
 |
Component:389; Jaccard:0.11517
 |
Component:389; Jaccard:0.10119
 |
Correlation:0.066284
  |
Correlation:0.21284
  |
Correlation:0.075616
 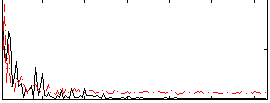 |
Correlation:0.075616
  |
Correlation:0.075616
  |
Correlation:0.075616
  |
Correlation:0.075616
  |
Component:070; Jaccard:0.093865
 |
Component:022; Jaccard:0.10712
 |
Component:096; Jaccard:0.11626
 |
Component:022; Jaccard:0.11951
 |
Component:022; Jaccard:0.11173
 |
Component:022; Jaccard:0.10049
 |
Component:022; Jaccard:0.082767
 |
Correlation:0.064889
  |
Correlation:0.37205
  |
Correlation:0.076071
  |
Correlation:0.37205
  |
Correlation:0.37205
  |
Correlation:0.37205
  |
Correlation:0.37205
  |
Component:331; Jaccard:0.093263
 |
Component:389; Jaccard:0.10254
 |
Component:022; Jaccard:0.11614
 |
Component:096; Jaccard:0.1137
 |
Component:290; Jaccard:0.1064
 |
Component:290; Jaccard:0.095616
 |
Component:290; Jaccard:0.080204
 |
Correlation:0.21284
  |
Correlation:0.075616
  |
Correlation:0.37205
  |
Correlation:0.076071
  |
Correlation:0.11511
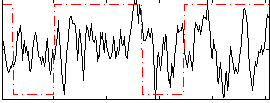 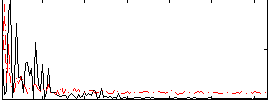 |
Correlation:0.11511
  |
Correlation:0.11511
  |
Component:290; Jaccard:0.089275
 |
Component:327; Jaccard:0.097549
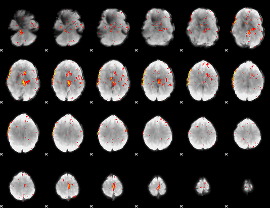 |
Component:327; Jaccard:0.10799
 |
Component:327; Jaccard:0.10896
 |
Component:096; Jaccard:0.10347
 |
Component:096; Jaccard:0.090691
 |
Component:096; Jaccard:0.075797
 |
Correlation:0.11511
  |
Correlation:0.11995
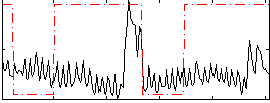 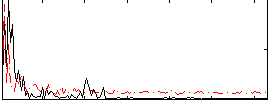 |
Correlation:0.11995
  |
Correlation:0.11995
  |
Correlation:0.076071
  |
Correlation:0.076071
  |
Correlation:0.076071
  |
Component:389; Jaccard:0.089155
 |
Component:290; Jaccard:0.097334
 |
Component:290; Jaccard:0.10487
 |
Component:290; Jaccard:0.10814
 |
Component:149; Jaccard:0.096459
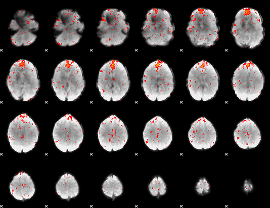 |
Component:141; Jaccard:0.085421
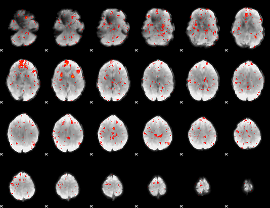 |
Component:149; Jaccard:0.072469
 |
Correlation:0.075616
  |
Correlation:0.11511
  |
Correlation:0.11511
  |
Correlation:0.11511
  |
Correlation:0.044604
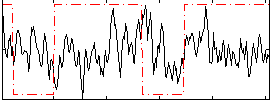 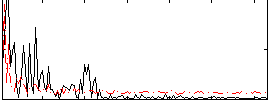 |
Correlation:0.058871
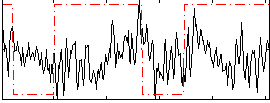 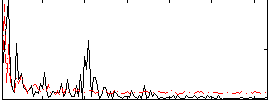 |
Correlation:0.044604
  |
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
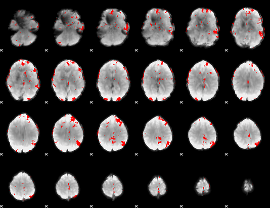
GLM binary result thresholded by 3
 |
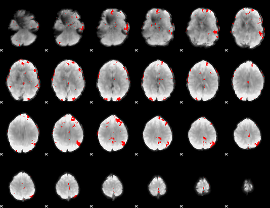
GLM binary result thresholded by 3.5
 |
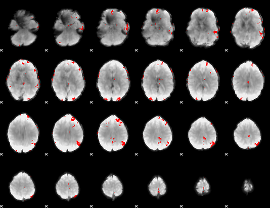
GLM binary result thresholded by 4
 |
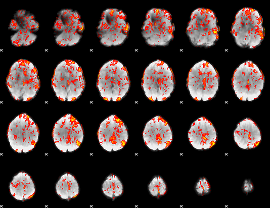
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
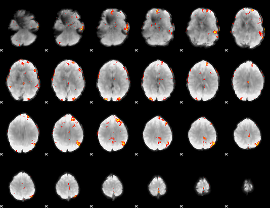 |
GLM Z-score result thresholded by 4
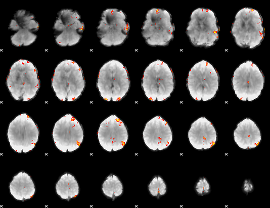 |
Component:198; Jaccard:0.12244
 |
Component:198; Jaccard:0.14951
 |
Component:334; Jaccard:0.17745
 |
Component:334; Jaccard:0.19761
 |
Component:334; Jaccard:0.20664
 |
Component:334; Jaccard:0.1982
 |
Component:334; Jaccard:0.17662
 |
Correlation:0.19947
  |
Correlation:0.19947
  |
Correlation:0.21425
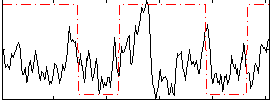 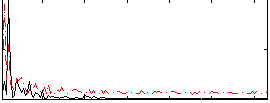 |
Correlation:0.21425
  |
Correlation:0.21425
  |
Correlation:0.21425
  |
Correlation:0.21425
  |
Component:234; Jaccard:0.1211
 |
Component:334; Jaccard:0.14801
 |
Component:198; Jaccard:0.17434
 |
Component:198; Jaccard:0.18839
 |
Component:198; Jaccard:0.19105
 |
Component:198; Jaccard:0.17474
 |
Component:198; Jaccard:0.15129
 |
Correlation:0.10979
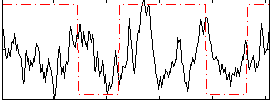 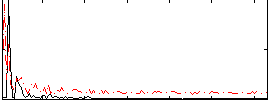 |
Correlation:0.21425
  |
Correlation:0.19947
  |
Correlation:0.19947
  |
Correlation:0.19947
  |
Correlation:0.19947
  |
Correlation:0.19947
  |
Component:334; Jaccard:0.12019
 |
Component:234; Jaccard:0.13825
 |
Component:234; Jaccard:0.15253
 |
Component:234; Jaccard:0.15993
 |
Component:234; Jaccard:0.15543
 |
Component:234; Jaccard:0.14283
 |
Component:234; Jaccard:0.12169
 |
Correlation:0.21425
  |
Correlation:0.10979
  |
Correlation:0.10979
  |
Correlation:0.10979
  |
Correlation:0.10979
  |
Correlation:0.10979
  |
Correlation:0.10979
  |
Component:290; Jaccard:0.098062
 |
Component:395; Jaccard:0.11173
 |
Component:395; Jaccard:0.12437
 |
Component:395; Jaccard:0.13272
 |
Component:395; Jaccard:0.12957
 |
Component:395; Jaccard:0.1198
 |
Component:395; Jaccard:0.098484
 |
Correlation:0.068894
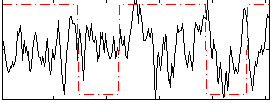 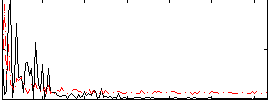 |
Correlation:0.44702
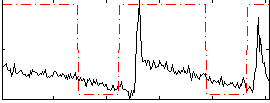 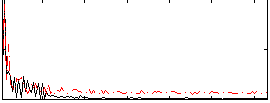 |
Correlation:0.44702
  |
Correlation:0.44702
  |
Correlation:0.44702
  |
Correlation:0.44702
  |
Correlation:0.44702
  |
Component:327; Jaccard:0.094918
 |
Component:327; Jaccard:0.10891
 |
Component:327; Jaccard:0.11714
 |
Component:327; Jaccard:0.11619
 |
Component:070; Jaccard:0.10628
 |
Component:070; Jaccard:0.096882
 |
Component:070; Jaccard:0.082908
 |
Correlation:0.32641
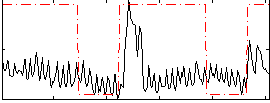 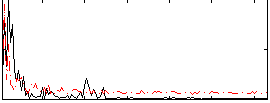 |
Correlation:0.32641
  |
Correlation:0.32641
  |
Correlation:0.32641
  |
Correlation:0.04516
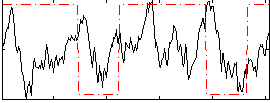 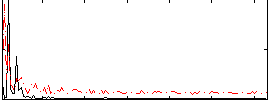 |
Correlation:0.04516
  |
Correlation:0.04516
  |
Component:395; Jaccard:0.093676
 |
Component:290; Jaccard:0.10575
 |
Component:290; Jaccard:0.10984
 |
Component:290; Jaccard:0.10952
 |
Component:327; Jaccard:0.10441
 |
Component:290; Jaccard:0.087076
 |
Component:290; Jaccard:0.071334
 |
Correlation:0.44702
  |
Correlation:0.068894
  |
Correlation:0.068894
  |
Correlation:0.068894
  |
Correlation:0.32641
  |
Correlation:0.068894
  |
Correlation:0.068894
  |
Component:070; Jaccard:0.089756
 |
Component:070; Jaccard:0.097908
 |
Component:070; Jaccard:0.10538
 |
Component:070; Jaccard:0.10767
 |
Component:290; Jaccard:0.10181
 |
Component:389; Jaccard:0.085524
 |
Component:389; Jaccard:0.069606
 |
Correlation:0.04516
  |
Correlation:0.04516
  |
Correlation:0.04516
  |
Correlation:0.04516
  |
Correlation:0.068894
  |
Correlation:0.15901
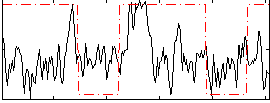 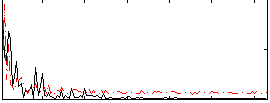 |
Correlation:0.15901
  |
Component:389; Jaccard:0.089581
 |
Component:389; Jaccard:0.096722
 |
Component:389; Jaccard:0.10124
 |
Component:389; Jaccard:0.10026
 |
Component:389; Jaccard:0.096746
 |
Component:327; Jaccard:0.082018
 |
Component:327; Jaccard:0.058311
 |
Correlation:0.15901
  |
Correlation:0.15901
  |
Correlation:0.15901
  |
Correlation:0.15901
  |
Correlation:0.15901
  |
Correlation:0.32641
  |
Correlation:0.32641
  |
Component:096; Jaccard:0.087518
 |
Component:331; Jaccard:0.088982
 |
Component:331; Jaccard:0.093112
 |
Component:331; Jaccard:0.089423
 |
Component:331; Jaccard:0.079752
 |
Component:094; Jaccard:0.06743
 |
Component:094; Jaccard:0.057253
 |
Correlation:0.21091
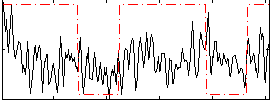 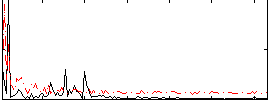 |
Correlation:0.043369
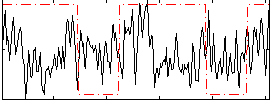 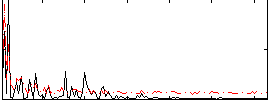 |
Correlation:0.043369
  |
Correlation:0.043369
  |
Correlation:0.043369
  |
Correlation:0.29411
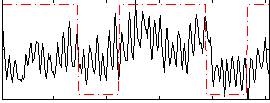 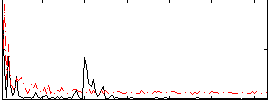 |
Correlation:0.29411
  |
Component:104; Jaccard:0.085087
 |
Component:096; Jaccard:0.087213
 |
Component:163; Jaccard:0.086685
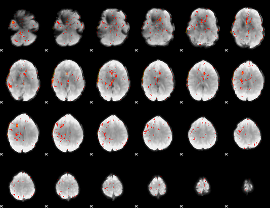 |
Component:163; Jaccard:0.081608
 |
Component:094; Jaccard:0.074231
 |
Component:331; Jaccard:0.06515
 |
Component:331; Jaccard:0.054754
 |
Correlation:0.27938
  |
Correlation:0.21091
  |
Correlation:0.23222
  |
Correlation:0.23222
  |
Correlation:0.29411
  |
Correlation:0.043369
  |
Correlation:0.043369
  |
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
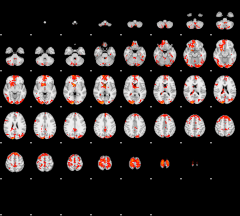 |
GLM binary result thresholded by 1
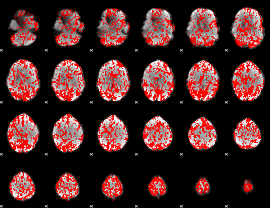 |
GLM binary result thresholded by 1.5
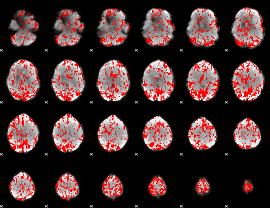 |
GLM binary result thresholded by 2
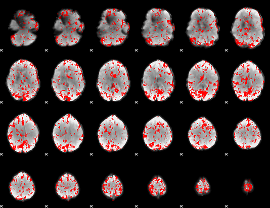 |
GLM binary result thresholded by 2.5
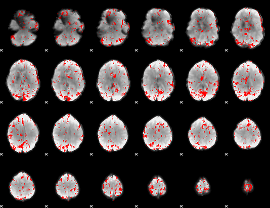 |
GLM binary result thresholded by 3
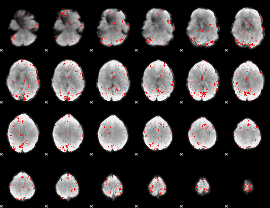 |
GLM binary result thresholded by 3.5
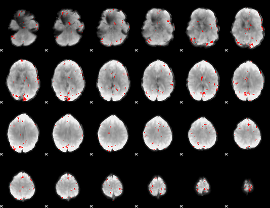 |
GLM binary result thresholded by 4
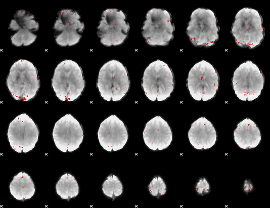 |
GLM Z-score result thresholded by 1
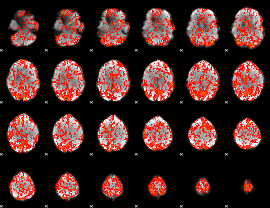 |
GLM Z-score result thresholded by 1.5
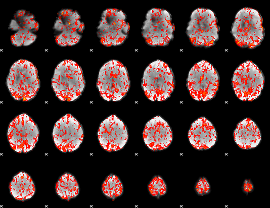 |
GLM Z-score result thresholded by 2
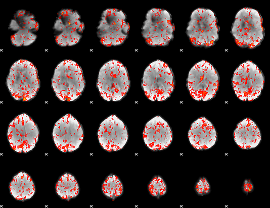 |
GLM Z-score result thresholded by 2.5
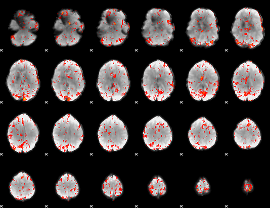 |
GLM Z-score result thresholded by 3
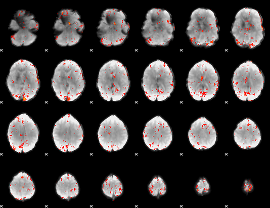 |
GLM Z-score result thresholded by 3.5
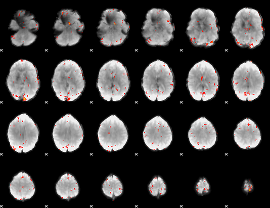 |
GLM Z-score result thresholded by 4
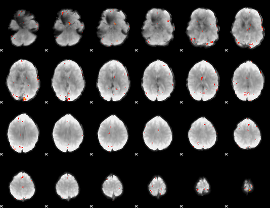 |
Component:236; Jaccard:0.10343
 |
Component:236; Jaccard:0.12019
 |
Component:236; Jaccard:0.13176
 |
Component:236; Jaccard:0.12781
 |
Component:010; Jaccard:0.11331
 |
Component:010; Jaccard:0.084587
 |
Component:337; Jaccard:0.050902
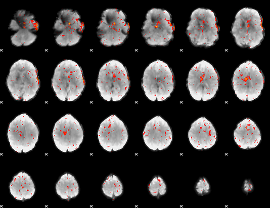 |
Correlation:0.12819
  |
Correlation:0.12819
  |
Correlation:0.12819
  |
Correlation:0.12819
  |
Correlation:0.13766
  |
Correlation:0.13766
  |
Correlation:0.39596
  |
Component:282; Jaccard:0.095494
 |
Component:010; Jaccard:0.10552
 |
Component:010; Jaccard:0.11957
 |
Component:010; Jaccard:0.12505
 |
Component:236; Jaccard:0.102
 |
Component:337; Jaccard:0.075297
 |
Component:010; Jaccard:0.050597
 |
Correlation:0.11584
  |
Correlation:0.13766
  |
Correlation:0.13766
  |
Correlation:0.13766
  |
Correlation:0.12819
  |
Correlation:0.39596
  |
Correlation:0.13766
  |
Component:332; Jaccard:0.089569
 |
Component:282; Jaccard:0.10261
 |
Component:332; Jaccard:0.10848
 |
Component:332; Jaccard:0.10552
 |
Component:337; Jaccard:0.094437
 |
Component:223; Jaccard:0.064656
 |
Component:223; Jaccard:0.040159
 |
Correlation:0.17528
  |
Correlation:0.11584
  |
Correlation:0.17528
  |
Correlation:0.17528
  |
Correlation:0.39596
  |
Correlation:0.21498
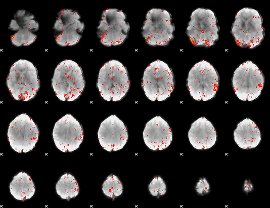
  |
Correlation:0.21498
  |
Component:010; Jaccard:0.088126
 |
Component:332; Jaccard:0.10197
 |
Component:282; Jaccard:0.10526
 |
Component:337; Jaccard:0.10033
 |
Component:223; Jaccard:0.090394
 |
Component:236; Jaccard:0.061756
 |
Component:127; Jaccard:0.035706
 |
Correlation:0.13766
  |
Correlation:0.17528
  |
Correlation:0.11584
  |
Correlation:0.39596
  |
Correlation:0.21498
  |
Correlation:0.12819
  |
Correlation:0.1317
  |
Component:121; Jaccard:0.087085
 |
Component:121; Jaccard:0.092488
 |
Component:223; Jaccard:0.094661
 |
Component:223; Jaccard:0.097986
 |
Component:332; Jaccard:0.087048
 |
Component:127; Jaccard:0.057878
 |
Component:400; Jaccard:0.029613
 |
Correlation:0.18998
  |
Correlation:0.18998
  |
Correlation:0.21498
  |
Correlation:0.21498
  |
Correlation:0.17528
  |
Correlation:0.1317
  |
Correlation:0.37846
  |
Component:056; Jaccard:0.079753
 |
Component:103; Jaccard:0.085707
 |
Component:342; Jaccard:0.090762
 |
Component:282; Jaccard:0.094607
 |
Component:127; Jaccard:0.077854
 |
Component:332; Jaccard:0.056283
 |
Component:236; Jaccard:0.029434
 |
Correlation:-0.022259
  |
Correlation:0.12052
  |
Correlation:0.052162
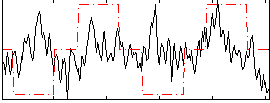 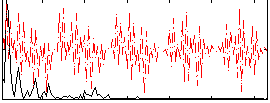 |
Correlation:0.11584
  |
Correlation:0.1317
  |
Correlation:0.17528
  |
Correlation:0.12819
  |
Component:103; Jaccard:0.07958
 |
Component:342; Jaccard:0.085481
 |
Component:337; Jaccard:0.089888
 |
Component:342; Jaccard:0.08787
 |
Component:012; Jaccard:0.073197
 |
Component:342; Jaccard:0.049209
 |
Component:044; Jaccard:0.029104
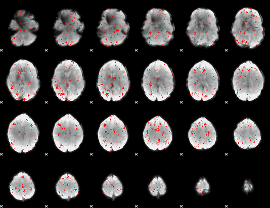 |
Correlation:0.12052
  |
Correlation:0.052162
  |
Correlation:0.39596
  |
Correlation:0.052162
  |
Correlation:-0.029402
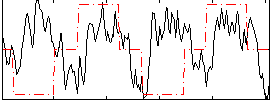  |
Correlation:0.052162
  |
Correlation:0.35056
  |
Component:248; Jaccard:0.079327
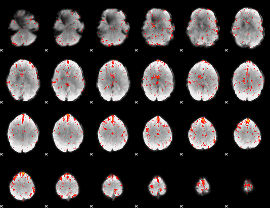 |
Component:223; Jaccard:0.083724
 |
Component:121; Jaccard:0.088807
 |
Component:180; Jaccard:0.084966
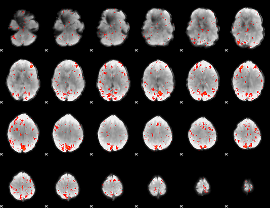 |
Component:342; Jaccard:0.072027
 |
Component:012; Jaccard:0.048109
 |
Component:332; Jaccard:0.027609
 |
Correlation:0.093925
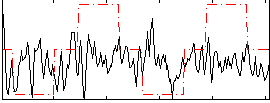 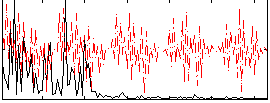 |
Correlation:0.21498
  |
Correlation:0.18998
  |
Correlation:0.34242
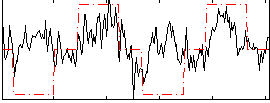  |
Correlation:0.052162
  |
Correlation:-0.029402
  |
Correlation:0.17528
  |
Component:176; Jaccard:0.077506
 |
Component:056; Jaccard:0.083237
 |
Component:180; Jaccard:0.087653
 |
Component:012; Jaccard:0.084626
 |
Component:180; Jaccard:0.070056
 |
Component:400; Jaccard:0.04778
 |
Component:342; Jaccard:0.026108
 |
Correlation:0.10774
  |
Correlation:-0.022259
  |
Correlation:0.34242
  |
Correlation:-0.029402
  |
Correlation:0.34242
  |
Correlation:0.37846
  |
Correlation:0.052162
  |
Component:342; Jaccard:0.076665
 |
Component:248; Jaccard:0.083222
 |
Component:012; Jaccard:0.08632
 |
Component:127; Jaccard:0.084019
 |
Component:282; Jaccard:0.0697
 |
Component:180; Jaccard:0.046012
 |
Component:180; Jaccard:0.025973
 |
Correlation:0.052162
  |
Correlation:0.093925
  |
Correlation:-0.029402
  |
Correlation:0.1317
  |
Correlation:0.11584
  |
Correlation:0.34242
  |
Correlation:0.34242
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































