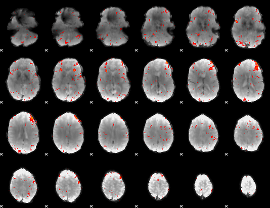
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
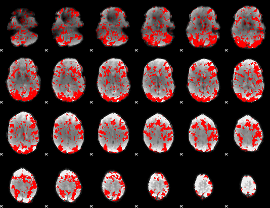 |
GLM binary result thresholded by 1.5
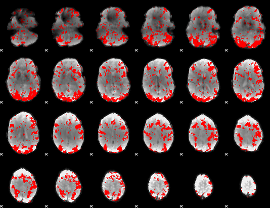 |
GLM binary result thresholded by 2
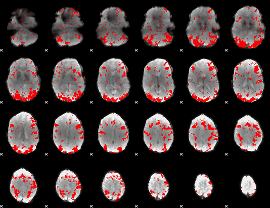 |
GLM binary result thresholded by 2.5
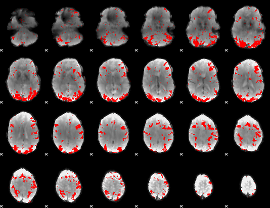 |
GLM binary result thresholded by 3
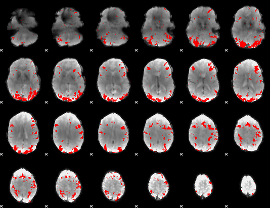 |
GLM binary result thresholded by 3.5
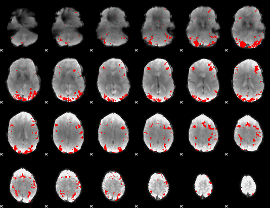 |
GLM binary result thresholded by 4
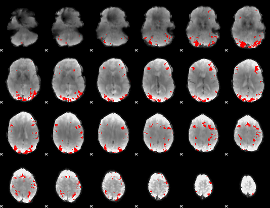 |
GLM Z-score result thresholded by 1
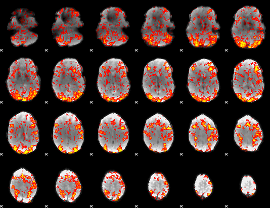 |
GLM Z-score result thresholded by 1.5
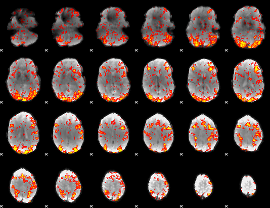 |
GLM Z-score result thresholded by 2
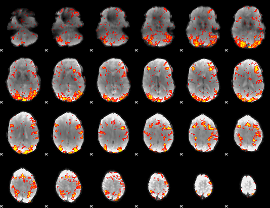 |
GLM Z-score result thresholded by 2.5
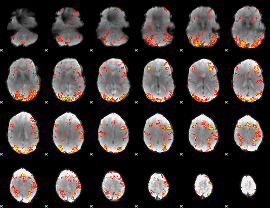 |
GLM Z-score result thresholded by 3
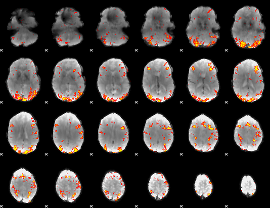 |
GLM Z-score result thresholded by 3.5
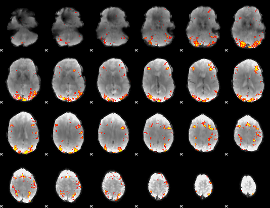 |
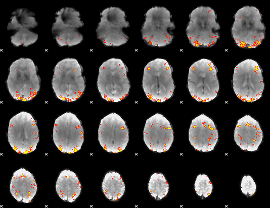
GLM Z-score result thresholded by 4
 |
Component:194; Jaccard:0.13274
 |
Component:096; Jaccard:0.16841
 |
Component:096; Jaccard:0.2215
 |
Component:096; Jaccard:0.28085
 |
Component:096; Jaccard:0.33361
 |
Component:096; Jaccard:0.3692
 |
Component:096; Jaccard:0.38324
 |
Correlation:0.059632
  |
Correlation:0.26452
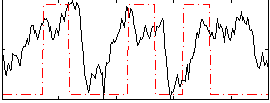 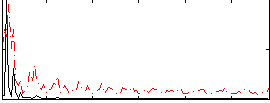 |
Correlation:0.26452
  |
Correlation:0.26452
  |
Correlation:0.26452
  |
Correlation:0.26452
  |
Correlation:0.26452
  |
Component:096; Jaccard:0.1249
 |
Component:194; Jaccard:0.16602
 |
Component:194; Jaccard:0.2002
 |
Component:194; Jaccard:0.22703
 |
Component:194; Jaccard:0.24253
 |
Component:194; Jaccard:0.24293
 |
Component:194; Jaccard:0.22752
 |
Correlation:0.26452
  |
Correlation:0.059632
  |
Correlation:0.059632
  |
Correlation:0.059632
  |
Correlation:0.059632
  |
Correlation:0.059632
  |
Correlation:0.059632
  |
Component:322; Jaccard:0.12088
 |
Component:322; Jaccard:0.14523
 |
Component:322; Jaccard:0.16565
 |
Component:061; Jaccard:0.17933
 |
Component:061; Jaccard:0.19298
 |
Component:061; Jaccard:0.19536
 |
Component:061; Jaccard:0.18701
 |
Correlation:0.33352
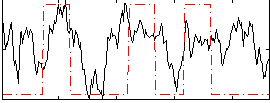 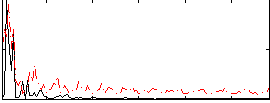 |
Correlation:0.33352
  |
Correlation:0.33352
  |
Correlation:0.013635
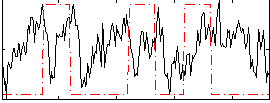 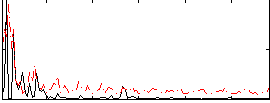 |
Correlation:0.013635
  |
Correlation:0.013635
  |
Correlation:0.013635
  |
Component:309; Jaccard:0.11421
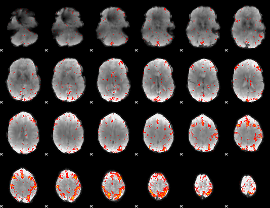 |
Component:061; Jaccard:0.13453
 |
Component:061; Jaccard:0.15983
 |
Component:322; Jaccard:0.17561
 |
Component:322; Jaccard:0.17874
 |
Component:048; Jaccard:0.17861
 |
Component:048; Jaccard:0.17195
 |
Correlation:0.039627
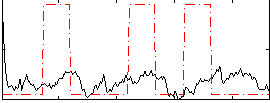 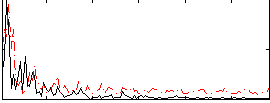 |
Correlation:0.013635
  |
Correlation:0.013635
  |
Correlation:0.33352
  |
Correlation:0.33352
  |
Correlation:0.15442
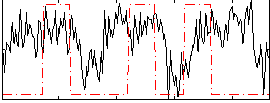 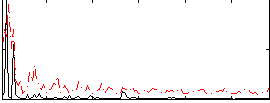 |
Correlation:0.15442
  |
Component:128; Jaccard:0.11417
 |
Component:338; Jaccard:0.13324
 |
Component:338; Jaccard:0.15307
 |
Component:338; Jaccard:0.16654
 |
Component:048; Jaccard:0.17417
 |
Component:338; Jaccard:0.17106
 |
Component:338; Jaccard:0.15924
 |
Correlation:0.2844
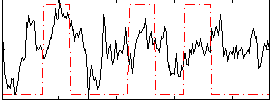 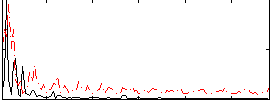 |
Correlation:0.037416
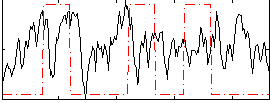  |
Correlation:0.037416
  |
Correlation:0.037416
  |
Correlation:0.15442
  |
Correlation:0.037416
  |
Correlation:0.037416
  |
Component:338; Jaccard:0.11165
 |
Component:128; Jaccard:0.12833
 |
Component:048; Jaccard:0.14082
 |
Component:048; Jaccard:0.16136
 |
Component:338; Jaccard:0.17177
 |
Component:322; Jaccard:0.17029
 |
Component:311; Jaccard:0.15539
 |
Correlation:0.037416
  |
Correlation:0.2844
  |
Correlation:0.15442
  |
Correlation:0.15442
  |
Correlation:0.037416
  |
Correlation:0.33352
  |
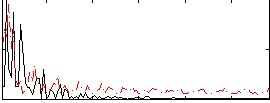
Correlation:0.14906
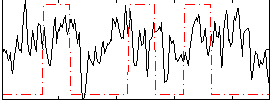  |
Component:061; Jaccard:0.10819
 |
Component:309; Jaccard:0.12388
 |
Component:311; Jaccard:0.14029
 |
Component:311; Jaccard:0.15582
 |
Component:311; Jaccard:0.1637
 |
Component:311; Jaccard:0.16451
 |
Component:322; Jaccard:0.15494
 |
Correlation:0.013635
  |
Correlation:0.039627
  |
Correlation:0.14906
  |
Correlation:0.14906
  |
Correlation:0.14906
  |
Correlation:0.14906
  |
Correlation:0.33352
  |
Component:341; Jaccard:0.10039
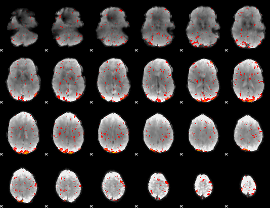 |
Component:311; Jaccard:0.12016
 |
Component:128; Jaccard:0.13626
 |
Component:107; Jaccard:0.13981
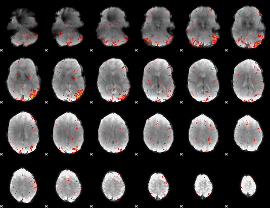 |
Component:107; Jaccard:0.1491
 |
Component:107; Jaccard:0.14769
 |
Component:107; Jaccard:0.1403
 |
Correlation:-0.10521
 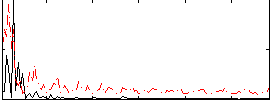 |
Correlation:0.14906
  |
Correlation:0.2844
  |
Correlation:0.15761
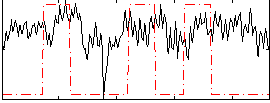 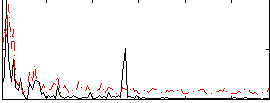 |
Correlation:0.15761
  |
Correlation:0.15761
  |
Correlation:0.15761
  |
Component:311; Jaccard:0.098441
 |
Component:048; Jaccard:0.11853
 |
Component:309; Jaccard:0.12805
 |
Component:128; Jaccard:0.13924
 |
Component:105; Jaccard:0.13411
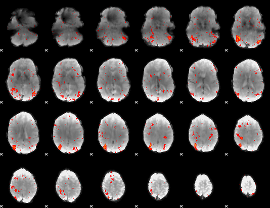 |
Component:105; Jaccard:0.12974
 |
Component:105; Jaccard:0.11986
 |
Correlation:0.14906
  |
Correlation:0.15442
  |
Correlation:0.039627
  |
Correlation:0.2844
  |
Correlation:0.12358
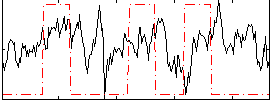 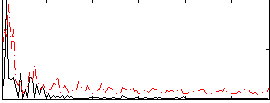 |
Correlation:0.12358
  |
Correlation:0.12358
  |
Component:250; Jaccard:0.097834
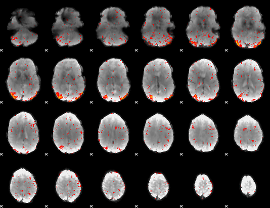 |
Component:250; Jaccard:0.11305
 |
Component:250; Jaccard:0.12685
 |
Component:250; Jaccard:0.13386
 |
Component:128; Jaccard:0.13221
 |
Component:341; Jaccard:0.12284
 |
Component:341; Jaccard:0.11155
 |
Correlation:0.063464
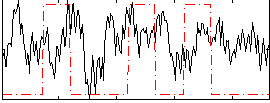 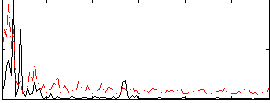 |
Correlation:0.063464
  |
Correlation:0.063464
  |
Correlation:0.063464
  |
Correlation:0.2844
  |
Correlation:-0.10521
  |
Correlation:-0.10521
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
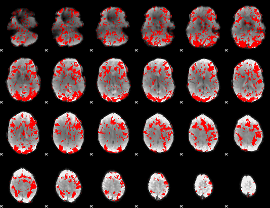 |
GLM binary result thresholded by 1.5
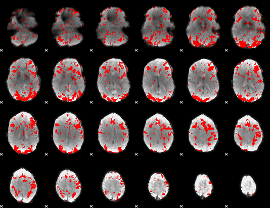 |
GLM binary result thresholded by 2
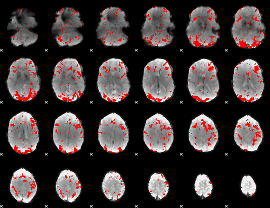 |
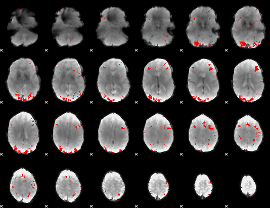
GLM binary result thresholded by 2.5
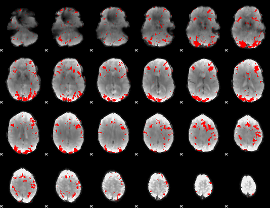 |
GLM binary result thresholded by 3
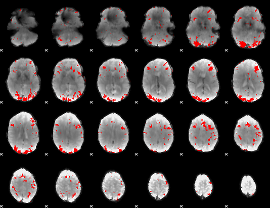 |
GLM binary result thresholded by 3.5
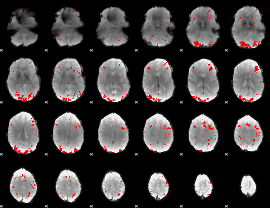 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
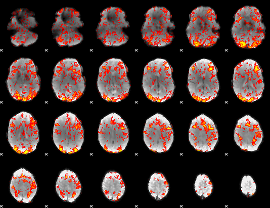 |
GLM Z-score result thresholded by 1.5
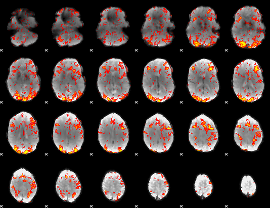 |
GLM Z-score result thresholded by 2
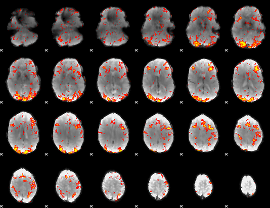 |
GLM Z-score result thresholded by 2.5
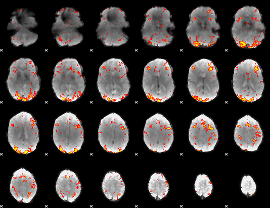 |
GLM Z-score result thresholded by 3
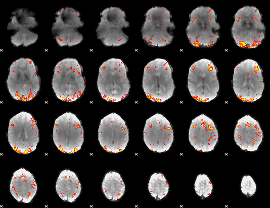 |
GLM Z-score result thresholded by 3.5
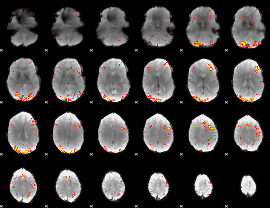 |
GLM Z-score result thresholded by 4
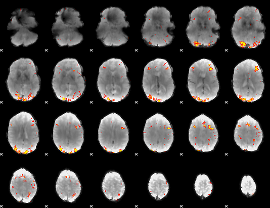 |
Component:096; Jaccard:0.13105
 |
Component:096; Jaccard:0.17241
 |
Component:096; Jaccard:0.22229
 |
Component:096; Jaccard:0.26881
 |
Component:096; Jaccard:0.30008
 |
Component:096; Jaccard:0.31011
 |
Component:096; Jaccard:0.29577
 |
Correlation:0.023038
 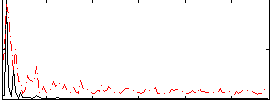 |
Correlation:0.023038
  |
Correlation:0.023038
  |
Correlation:0.023038
  |
Correlation:0.023038
  |
Correlation:0.023038
  |
Correlation:0.023038
  |
Component:194; Jaccard:0.12371
 |
Component:338; Jaccard:0.14679
 |
Component:338; Jaccard:0.16844
 |
Component:194; Jaccard:0.17988
 |
Component:338; Jaccard:0.18135
 |
Component:338; Jaccard:0.17692
 |
Component:061; Jaccard:0.16161
 |
Correlation:0.19681
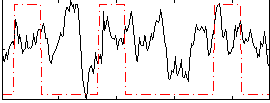 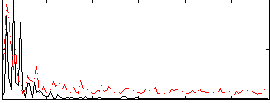 |
Correlation:0.11544
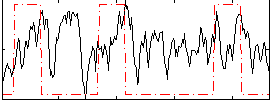  |
Correlation:0.11544
  |
Correlation:0.19681
  |
Correlation:0.11544
  |
Correlation:0.11544
  |
Correlation:0.29319
  |
Component:338; Jaccard:0.12226
 |
Component:194; Jaccard:0.14605
 |
Component:194; Jaccard:0.16769
 |
Component:338; Jaccard:0.17768
 |
Component:194; Jaccard:0.17794
 |
Component:061; Jaccard:0.17462
 |
Component:338; Jaccard:0.16076
 |
Correlation:0.11544
  |
Correlation:0.19681
  |
Correlation:0.19681
  |
Correlation:0.11544
  |
Correlation:0.19681
  |
Correlation:0.29319
  |
Correlation:0.11544
  |
Component:311; Jaccard:0.10922
 |
Component:311; Jaccard:0.13233
 |
Component:311; Jaccard:0.15439
 |
Component:061; Jaccard:0.16998
 |
Component:061; Jaccard:0.17682
 |
Component:194; Jaccard:0.16902
 |
Component:311; Jaccard:0.15098
 |
Correlation:0.26684
 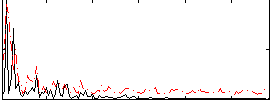 |
Correlation:0.26684
  |
Correlation:0.26684
  |
Correlation:0.29319
  |
Correlation:0.29319
  |
Correlation:0.19681
  |
Correlation:0.26684
  |
Component:061; Jaccard:0.10889
 |
Component:061; Jaccard:0.13205
 |
Component:061; Jaccard:0.1543
 |
Component:048; Jaccard:0.16839
 |
Component:048; Jaccard:0.17254
 |
Component:311; Jaccard:0.16514
 |
Component:194; Jaccard:0.14952
 |
Correlation:0.29319
  |
Correlation:0.29319
  |
Correlation:0.29319
  |
Correlation:0.25446
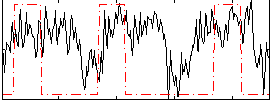 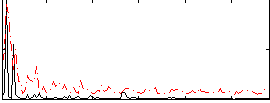 |
Correlation:0.25446
  |
Correlation:0.26684
  |
Correlation:0.19681
  |
Component:322; Jaccard:0.10852
 |
Component:048; Jaccard:0.13023
 |
Component:048; Jaccard:0.15338
 |
Component:311; Jaccard:0.16758
 |
Component:311; Jaccard:0.17176
 |
Component:048; Jaccard:0.16115
 |
Component:048; Jaccard:0.14036
 |
Correlation:-0.038017
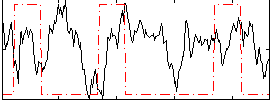 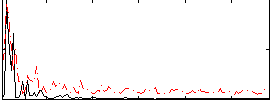 |
Correlation:0.25446
  |
Correlation:0.25446
  |
Correlation:0.26684
  |
Correlation:0.26684
  |
Correlation:0.25446
  |
Correlation:0.25446
  |
Component:048; Jaccard:0.10433
 |
Component:322; Jaccard:0.12733
 |
Component:322; Jaccard:0.14309
 |
Component:322; Jaccard:0.14849
 |
Component:171; Jaccard:0.14407
 |
Component:171; Jaccard:0.14487
 |
Component:171; Jaccard:0.13733
 |
Correlation:0.25446
  |
Correlation:-0.038017
  |
Correlation:-0.038017
  |
Correlation:-0.038017
  |
Correlation:0.3683
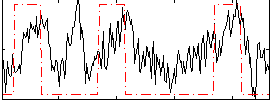 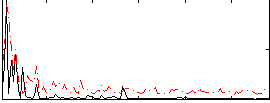 |
Correlation:0.3683
  |
Correlation:0.3683
  |
Component:366; Jaccard:0.097944
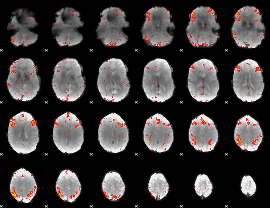 |
Component:080; Jaccard:0.10557
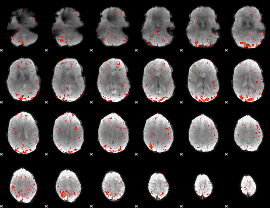 |
Component:171; Jaccard:0.12157
 |
Component:171; Jaccard:0.1364
 |
Component:322; Jaccard:0.14275
 |
Component:322; Jaccard:0.12688
 |
Component:322; Jaccard:0.10753
 |
Correlation:-0.16727
  |
Correlation:0.23677
  |
Correlation:0.3683
  |
Correlation:0.3683
  |
Correlation:-0.038017
  |
Correlation:-0.038017
  |
Correlation:-0.038017
  |
Component:080; Jaccard:0.09472
 |
Component:171; Jaccard:0.10535
 |
Component:080; Jaccard:0.11442
 |
Component:257; Jaccard:0.12038
 |
Component:080; Jaccard:0.11504
 |
Component:080; Jaccard:0.1067
 |
Component:080; Jaccard:0.097113
 |
Correlation:0.23677
  |
Correlation:0.3683
  |
Correlation:0.23677
  |
Correlation:-0.0002906
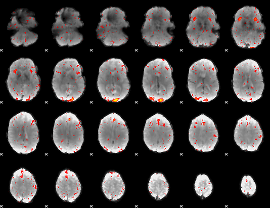
  |
Correlation:0.23677
  |
Correlation:0.23677
  |
Correlation:0.23677
  |
Component:341; Jaccard:0.094704
 |
Component:366; Jaccard:0.10487
 |
Component:257; Jaccard:0.11324
 |
Component:080; Jaccard:0.1182
 |
Component:257; Jaccard:0.11495
 |
Component:257; Jaccard:0.10252
 |
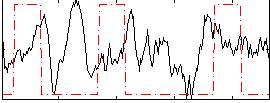
Component:137; Jaccard:0.091659
 |
Correlation:0.035387
 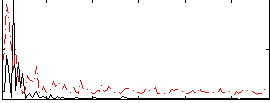 |
Correlation:-0.16727
  |
Correlation:-0.0002906
  |
Correlation:0.23677
  |
Correlation:-0.0002906
  |
Correlation:-0.0002906
  |
Correlation:0.22513
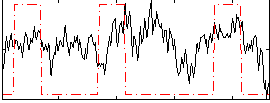 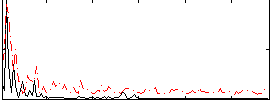 |
Cope: 03:MATCH-REL BACK
|
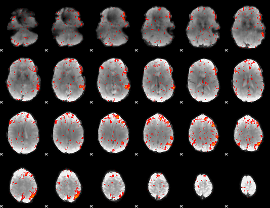
GLM result group-wise
 |
GLM binary result thresholded by 1
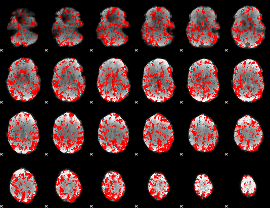 |
GLM binary result thresholded by 1.5
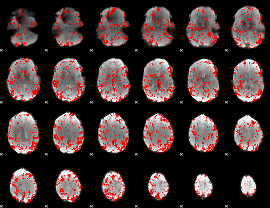 |
GLM binary result thresholded by 2
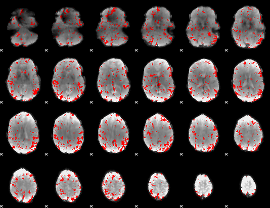 |
GLM binary result thresholded by 2.5
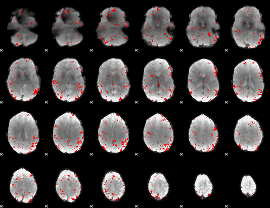 |
GLM binary result thresholded by 3
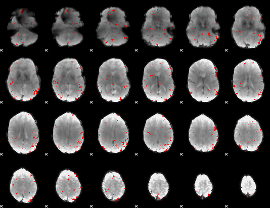 |
GLM binary result thresholded by 3.5
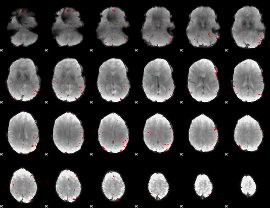 |
GLM binary result thresholded by 4
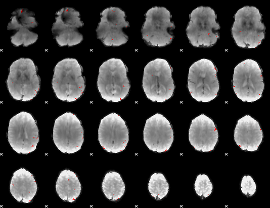 |
GLM Z-score result thresholded by 1
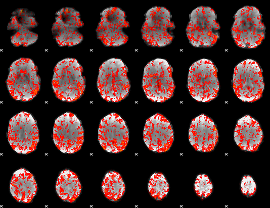 |
GLM Z-score result thresholded by 1.5
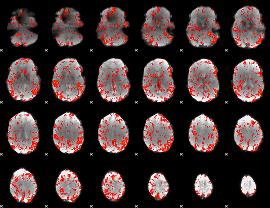 |
GLM Z-score result thresholded by 2
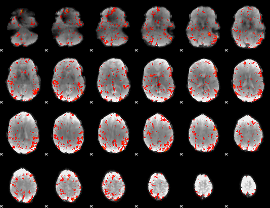 |
GLM Z-score result thresholded by 2.5
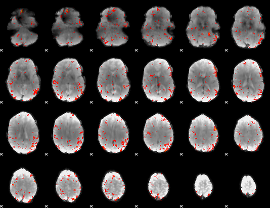 |
GLM Z-score result thresholded by 3
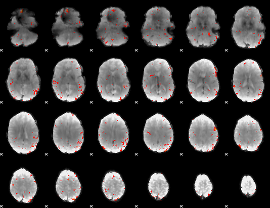 |
GLM Z-score result thresholded by 3.5
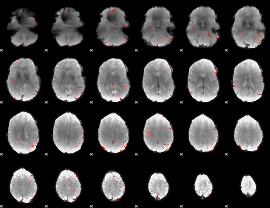 |
GLM Z-score result thresholded by 4
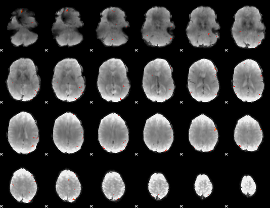 |
Component:210; Jaccard:0.09595
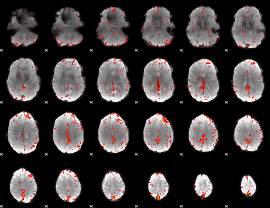 |
Component:128; Jaccard:0.10109
 |
Component:128; Jaccard:0.10663
 |
Component:128; Jaccard:0.098628
 |
Component:128; Jaccard:0.072112
 |
Component:128; Jaccard:0.042915
 |
Component:387; Jaccard:0.021102
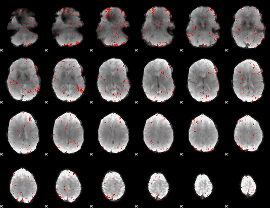 |
Correlation:0.063102
  |
Correlation:0.16966
 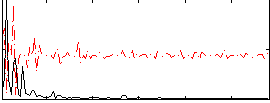 |
Correlation:0.16966
  |
Correlation:0.16966
  |
Correlation:0.16966
  |
Correlation:0.16966
  |
Correlation:0.33829
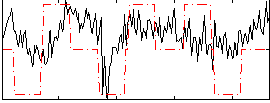 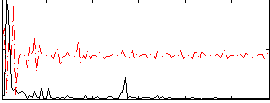 |
Component:123; Jaccard:0.091877
 |
Component:210; Jaccard:0.099407
 |
Component:224; Jaccard:0.09727
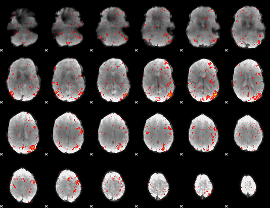 |
Component:224; Jaccard:0.088225
 |
Component:224; Jaccard:0.064492
 |
Component:387; Jaccard:0.04007
 |
Component:128; Jaccard:0.020728
 |
Correlation:0.19062
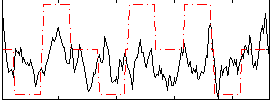 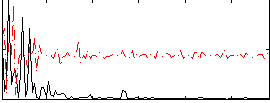 |
Correlation:0.063102
  |
Correlation:-0.0046029
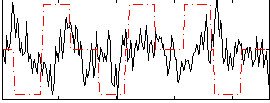  |
Correlation:-0.0046029
  |
Correlation:-0.0046029
  |
Correlation:0.33829
  |
Correlation:0.16966
  |
Component:220; Jaccard:0.08875
 |
Component:220; Jaccard:0.095679
 |
Component:105; Jaccard:0.096443
 |
Component:105; Jaccard:0.084855
 |
Component:365; Jaccard:0.063546
 |
Component:224; Jaccard:0.037468
 |
Component:224; Jaccard:0.01921
 |
Correlation:-0.019802
 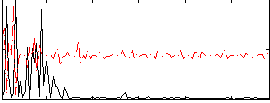 |
Correlation:-0.019802
  |
Correlation:0.099861
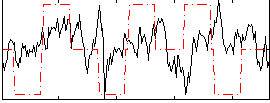 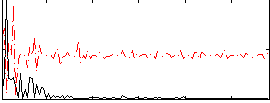 |
Correlation:0.099861
  |
Correlation:0.29415
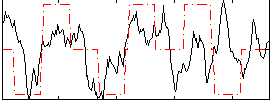 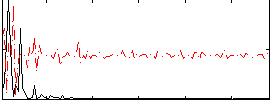 |
Correlation:-0.0046029
  |
Correlation:-0.0046029
  |
Component:128; Jaccard:0.086874
 |
Component:123; Jaccard:0.095495
 |
Component:210; Jaccard:0.094441
 |
Component:365; Jaccard:0.083687
 |
Component:387; Jaccard:0.061064
 |
Component:365; Jaccard:0.037383
 |
Component:365; Jaccard:0.019138
 |
Correlation:0.16966
  |
Correlation:0.19062
  |
Correlation:0.063102
  |
Correlation:0.29415
  |
Correlation:0.33829
  |
Correlation:0.29415
  |
Correlation:0.29415
  |
Component:399; Jaccard:0.085776
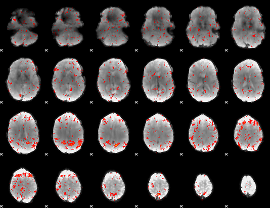 |
Component:399; Jaccard:0.092923
 |
Component:220; Jaccard:0.093067
 |
Component:387; Jaccard:0.079119
 |
Component:105; Jaccard:0.060397
 |
Component:322; Jaccard:0.034127
 |
Component:105; Jaccard:0.018337
 |
Correlation:0.011701
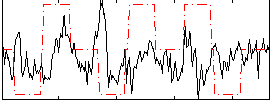 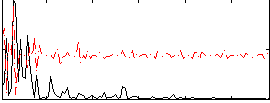 |
Correlation:0.011701
  |
Correlation:-0.019802
  |
Correlation:0.33829
  |
Correlation:0.099861
  |
Correlation:0.22021
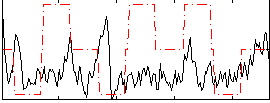
 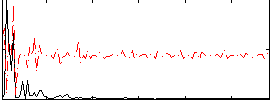 |
Correlation:0.099861
  |
Component:392; Jaccard:0.083325
 |
Component:224; Jaccard:0.092506
 |
Component:123; Jaccard:0.090367
 |
Component:210; Jaccard:0.074762
 |
Component:322; Jaccard:0.050004
 |
Component:105; Jaccard:0.033291
 |
Component:322; Jaccard:0.016678
 |
Correlation:-0.014937
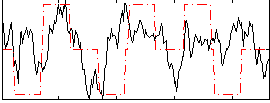
 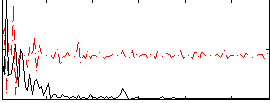 |
Correlation:-0.0046029
  |
Correlation:0.19062
  |
Correlation:0.063102
  |
Correlation:0.22021
  |
Correlation:0.099861
  |
Correlation:0.22021
  |
Component:396; Jaccard:0.080935
 |
Component:396; Jaccard:0.088015
 |
Component:399; Jaccard:0.089729
 |
Component:220; Jaccard:0.07367
 |
Component:210; Jaccard:0.049378
 |
Component:107; Jaccard:0.029608
 |
Component:107; Jaccard:0.015542
 |
Correlation:0.094207
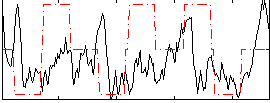 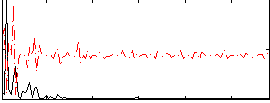 |
Correlation:0.094207
  |
Correlation:0.011701
  |
Correlation:-0.019802
  |
Correlation:0.063102
  |
Correlation:0.14711
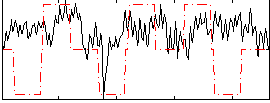 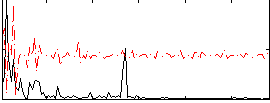 |
Correlation:0.14711
  |
Component:224; Jaccard:0.079801
 |
Component:105; Jaccard:0.087118
 |
Component:396; Jaccard:0.087239
 |
Component:399; Jaccard:0.072021
 |
Component:349; Jaccard:0.048655
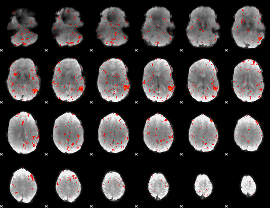 |
Component:210; Jaccard:0.028036
 |
Component:210; Jaccard:0.013442
 |
Correlation:-0.0046029
  |
Correlation:0.099861
  |
Correlation:0.094207
  |
Correlation:0.011701
  |
Correlation:0.18314
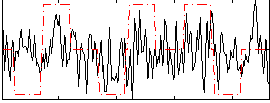 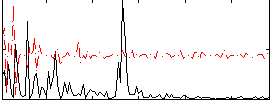 |
Correlation:0.063102
  |
Correlation:0.063102
  |
Component:339; Jaccard:0.079168
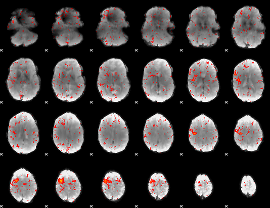 |
Component:390; Jaccard:0.084748
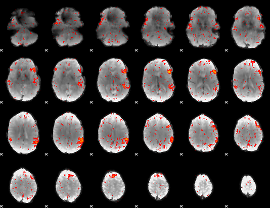 |
Component:365; Jaccard:0.086388
 |
Component:396; Jaccard:0.071986
 |
Component:220; Jaccard:0.048116
 |
Component:349; Jaccard:0.02703
 |
Component:041; Jaccard:0.013406
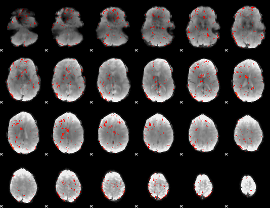 |
Correlation:0.11411
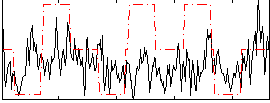 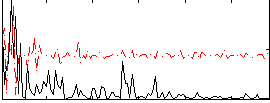 |
Correlation:0.085628
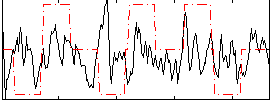 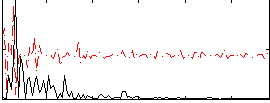 |
Correlation:0.29415
  |
Correlation:0.094207
  |
Correlation:-0.019802
  |
Correlation:0.18314
  |
Correlation:0.2057
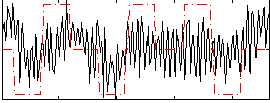 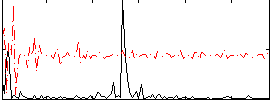 |
Component:309; Jaccard:0.078691
 |
Component:339; Jaccard:0.084288
 |
Component:387; Jaccard:0.083224
 |
Component:123; Jaccard:0.071602
 |
Component:107; Jaccard:0.046397
 |
Component:399; Jaccard:0.025324
 |
Component:349; Jaccard:0.01189
 |
Correlation:0.01166
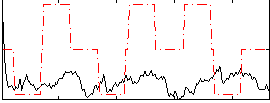 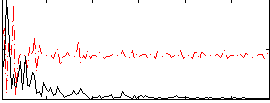 |
Correlation:0.11411
  |
Correlation:0.33829
  |
Correlation:0.19062
  |
Correlation:0.14711
  |
Correlation:0.011701
  |
Correlation:0.18314
  |
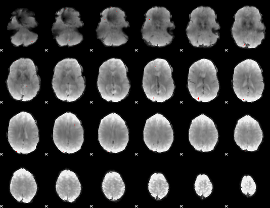
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
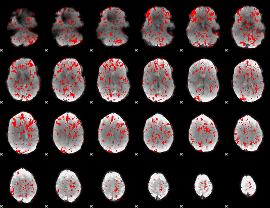 |
GLM binary result thresholded by 1.5
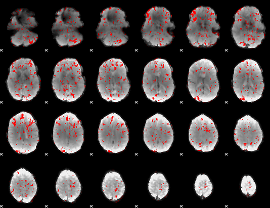 |
GLM binary result thresholded by 2
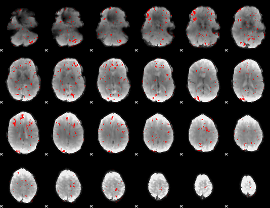 |
GLM binary result thresholded by 2.5
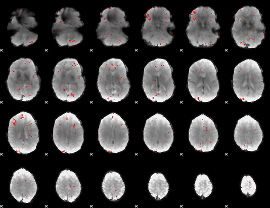 |
GLM binary result thresholded by 3
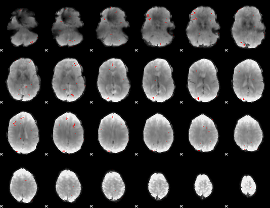 |
GLM binary result thresholded by 3.5
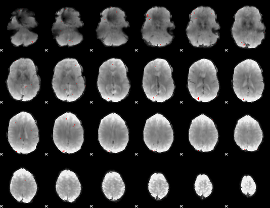 |
GLM binary result thresholded by 4
 |
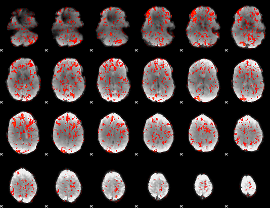
GLM Z-score result thresholded by 1
 |
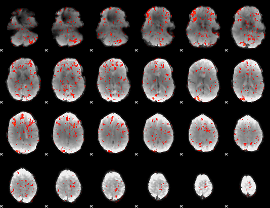
GLM Z-score result thresholded by 1.5
 |
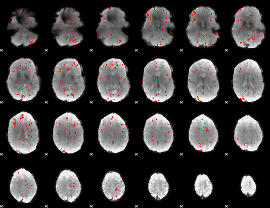
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
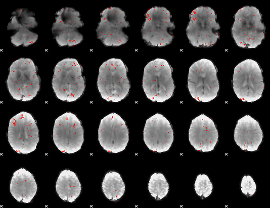 |
GLM Z-score result thresholded by 3
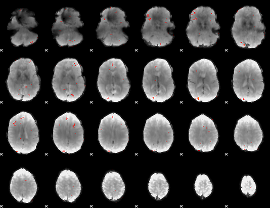 |
GLM Z-score result thresholded by 3.5
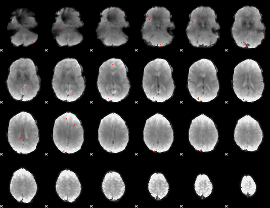 |
GLM Z-score result thresholded by 4
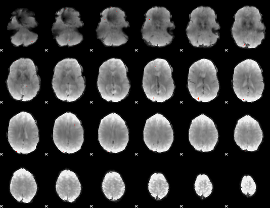 |
Component:231; Jaccard:0.11098
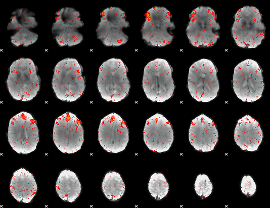 |
Component:231; Jaccard:0.11356
 |
Component:231; Jaccard:0.092148
 |
Component:171; Jaccard:0.072467
 |
Component:171; Jaccard:0.046217
 |
Component:171; Jaccard:0.025702
 |
Component:171; Jaccard:0.012215
 |
Correlation:0.0044379
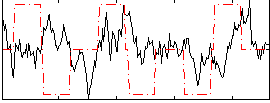 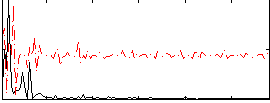 |
Correlation:0.0044379
  |
Correlation:0.0044379
  |
Correlation:0.28078
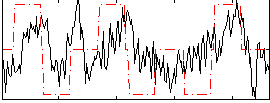  |
Correlation:0.28078
  |
Correlation:0.28078
  |
Correlation:0.28078
  |
Component:159; Jaccard:0.098961
 |
Component:159; Jaccard:0.10246
 |
Component:171; Jaccard:0.09015
 |
Component:231; Jaccard:0.059895
 |
Component:231; Jaccard:0.03014
 |
Component:231; Jaccard:0.012705
 |
Component:338; Jaccard:0.005038
 |
Correlation:0.12799
  |
Correlation:0.12799
  |
Correlation:0.28078
  |
Correlation:0.0044379
  |
Correlation:0.0044379
  |
Correlation:0.0044379
  |
Correlation:0.046248
 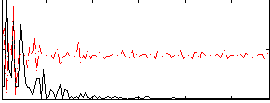 |
Component:273; Jaccard:0.080292
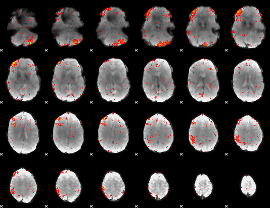 |
Component:171; Jaccard:0.089112
 |
Component:159; Jaccard:0.08752
 |
Component:159; Jaccard:0.055155
 |
Component:159; Jaccard:0.02876
 |
Component:007; Jaccard:0.010502
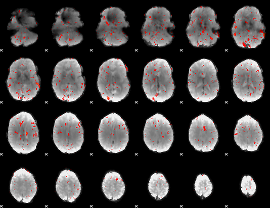 |
Component:007; Jaccard:0.004718
 |
Correlation:-0.18666
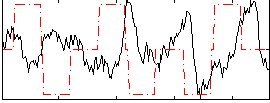 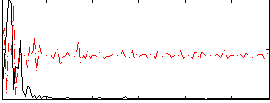 |
Correlation:0.28078
  |
Correlation:0.12799
  |
Correlation:0.12799
  |
Correlation:0.12799
  |
Correlation:0.094012
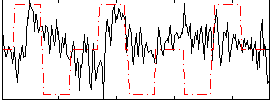 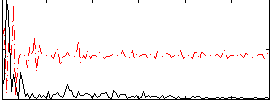 |
Correlation:0.094012
  |
Component:171; Jaccard:0.077054
 |
Component:273; Jaccard:0.077294
 |
Component:273; Jaccard:0.061503
 |
Component:260; Jaccard:0.042455
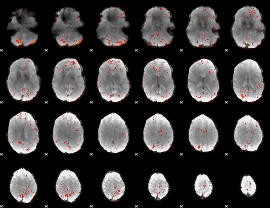 |
Component:007; Jaccard:0.023249
 |
Component:159; Jaccard:0.01047
 |
Component:231; Jaccard:0.004705
 |
Correlation:0.28078
  |
Correlation:-0.18666
  |
Correlation:-0.18666
  |
Correlation:0.20411
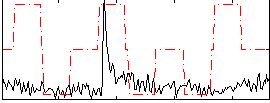 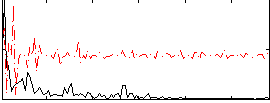 |
Correlation:0.094012
  |
Correlation:0.12799
  |
Correlation:0.0044379
  |
Component:246; Jaccard:0.072229
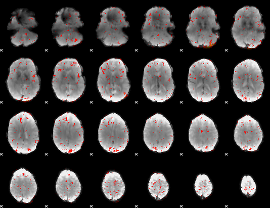 |
Component:260; Jaccard:0.07118
 |
Component:260; Jaccard:0.061318
 |
Component:007; Jaccard:0.041319
 |
Component:246; Jaccard:0.02126
 |
Component:246; Jaccard:0.010199
 |
Component:311; Jaccard:0.004669
 |
Correlation:0.27299
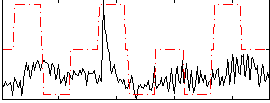 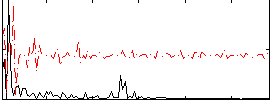 |
Correlation:0.20411
  |
Correlation:0.20411
  |
Correlation:0.094012
  |
Correlation:0.27299
  |
Correlation:0.27299
  |
Correlation:0.069812
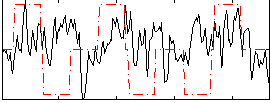 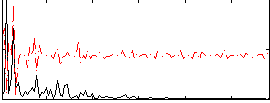 |
Component:248; Jaccard:0.072092
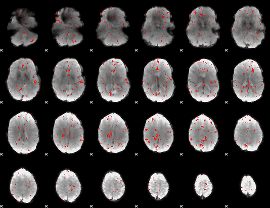 |
Component:246; Jaccard:0.070229
 |
Component:007; Jaccard:0.061239
 |
Component:246; Jaccard:0.040846
 |
Component:260; Jaccard:0.020428
 |
Component:260; Jaccard:0.009137
 |
Component:257; Jaccard:0.004433
 |
Correlation:0.15552
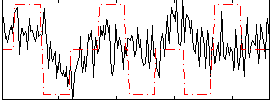 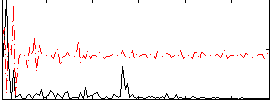 |
Correlation:0.27299
  |
Correlation:0.094012
  |
Correlation:0.27299
  |
Correlation:0.20411
  |
Correlation:0.20411
  |
Correlation:-0.1071
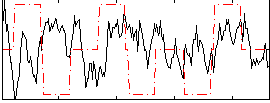 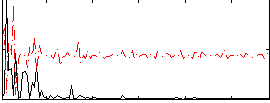 |
Component:260; Jaccard:0.070133
 |
Component:248; Jaccard:0.070019
 |
Component:246; Jaccard:0.060402
 |
Component:273; Jaccard:0.038565
 |
Component:021; Jaccard:0.018163
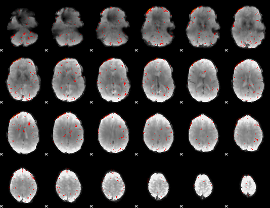 |
Component:338; Jaccard:0.008374
 |
Component:246; Jaccard:0.003981
 |
Correlation:0.20411
  |
Correlation:0.15552
  |
Correlation:0.27299
  |
Correlation:-0.18666
  |
Correlation:0.11719
  |
Correlation:0.046248
  |
Correlation:0.27299
  |
Component:274; Jaccard:0.068885
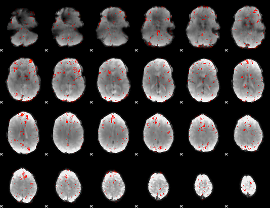 |
Component:007; Jaccard:0.066298
 |
Component:021; Jaccard:0.056829
 |
Component:021; Jaccard:0.036827
 |
Component:273; Jaccard:0.018029
 |
Component:342; Jaccard:0.007543
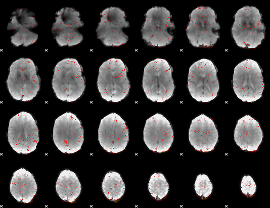 |
Component:159; Jaccard:0.003776
 |
Correlation:0.25359
  |
Correlation:0.094012
  |
Correlation:0.11719
  |
Correlation:0.11719
  |
Correlation:-0.18666
  |
Correlation:0.17316
  |
Correlation:0.12799
  |
Component:013; Jaccard:0.068863
 |
Component:021; Jaccard:0.065143
 |
Component:248; Jaccard:0.05682
 |
Component:248; Jaccard:0.035481
 |
Component:366; Jaccard:0.016726
 |
Component:366; Jaccard:0.007433
 |
Component:260; Jaccard:0.003084
 |
Correlation:-0.09303
  |
Correlation:0.11719
  |
Correlation:0.15552
  |
Correlation:0.15552
  |
Correlation:-0.18849
  |
Correlation:-0.18849
  |
Correlation:0.20411
  |
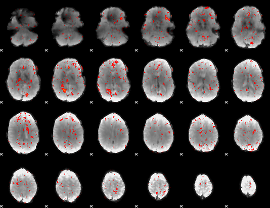
Component:025; Jaccard:0.067672
 |
Component:013; Jaccard:0.064023
 |
Component:274; Jaccard:0.048574
 |
Component:366; Jaccard:0.031063
 |
Component:248; Jaccard:0.016338
 |
Component:257; Jaccard:0.007289
 |
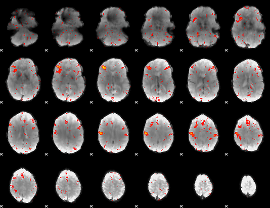
Component:293; Jaccard:0.002731
 |
Correlation:0.29807
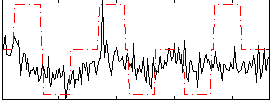 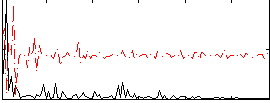 |
Correlation:-0.09303
  |
Correlation:0.25359
  |
Correlation:-0.18849
  |
Correlation:0.15552
  |
Correlation:-0.1071
  |
Correlation:0.031148
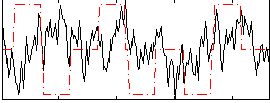 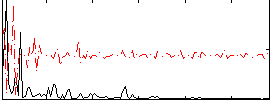 |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
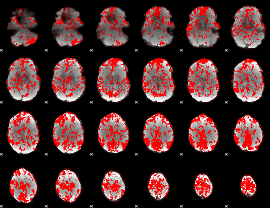 |
GLM binary result thresholded by 1.5
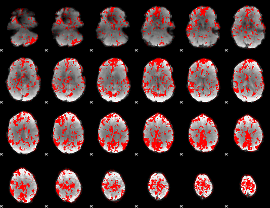 |
GLM binary result thresholded by 2
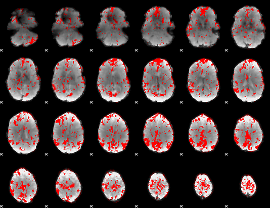 |
GLM binary result thresholded by 2.5
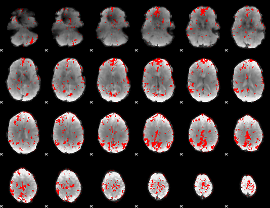 |
GLM binary result thresholded by 3
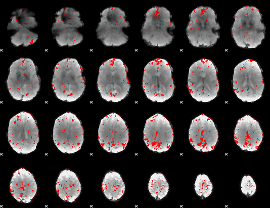 |
GLM binary result thresholded by 3.5
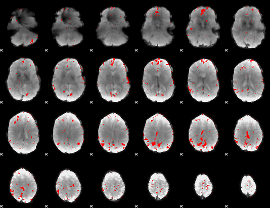 |
GLM binary result thresholded by 4
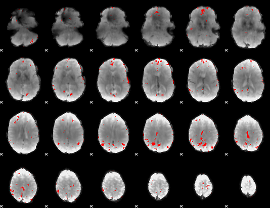 |
GLM Z-score result thresholded by 1
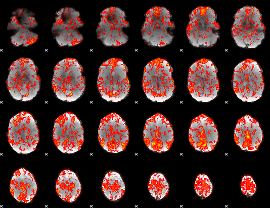 |
GLM Z-score result thresholded by 1.5
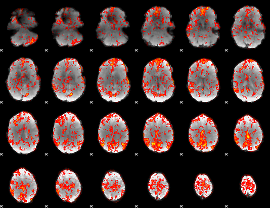 |
GLM Z-score result thresholded by 2
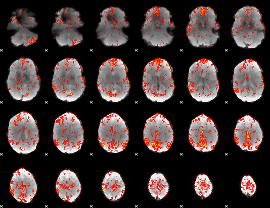 |
GLM Z-score result thresholded by 2.5
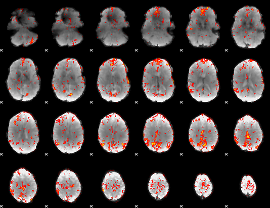 |
GLM Z-score result thresholded by 3
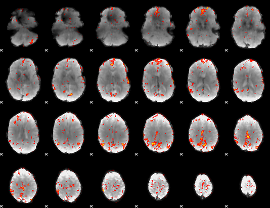 |
GLM Z-score result thresholded by 3.5
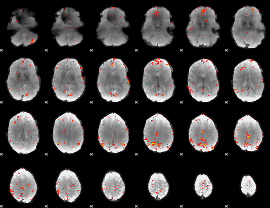 |
GLM Z-score result thresholded by 4
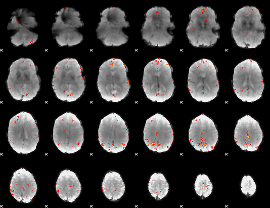 |
Component:268; Jaccard:0.10559
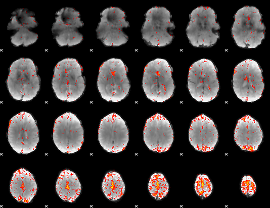 |
Component:102; Jaccard:0.11817
 |
Component:102; Jaccard:0.13919
 |
Component:289; Jaccard:0.15716
 |
Component:289; Jaccard:0.16864
 |
Component:289; Jaccard:0.15557
 |
Component:289; Jaccard:0.12549
 |
Correlation:0.1946
 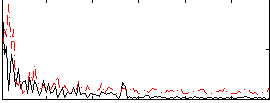 |
Correlation:0.2096
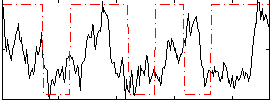  |
Correlation:0.2096
  |
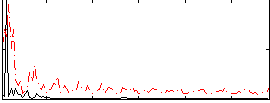
Correlation:0.2971
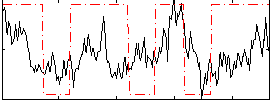  |
Correlation:0.2971
  |
Correlation:0.2971
  |
Correlation:0.2971
  |
Component:392; Jaccard:0.10003
 |
Component:396; Jaccard:0.11589
 |
Component:289; Jaccard:0.13612
 |
Component:102; Jaccard:0.15422
 |
Component:102; Jaccard:0.1542
 |
Component:102; Jaccard:0.13529
 |
Component:102; Jaccard:0.10466
 |
Correlation:0.045039
  |
Correlation:0.046132
 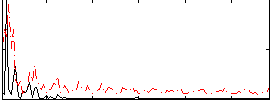 |
Correlation:0.2971
  |
Correlation:0.2096
  |
Correlation:0.2096
  |
Correlation:0.2096
  |
Correlation:0.2096
  |
Component:396; Jaccard:0.099816
 |
Component:289; Jaccard:0.11147
 |
Component:396; Jaccard:0.13372
 |
Component:396; Jaccard:0.14521
 |
Component:396; Jaccard:0.14507
 |
Component:396; Jaccard:0.12889
 |
Component:396; Jaccard:0.10357
 |
Correlation:0.046132
  |
Correlation:0.2971
  |
Correlation:0.046132
  |
Correlation:0.046132
  |
Correlation:0.046132
  |
Correlation:0.046132
  |
Correlation:0.046132
  |
Component:102; Jaccard:0.096828
 |
Component:280; Jaccard:0.10741
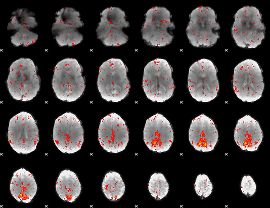 |
Component:280; Jaccard:0.12012
 |
Component:280; Jaccard:0.12484
 |
Component:280; Jaccard:0.1176
 |
Component:280; Jaccard:0.096781
 |
Component:280; Jaccard:0.072607
 |
Correlation:0.2096
  |
Correlation:0.20936
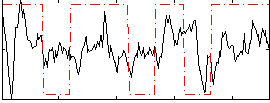 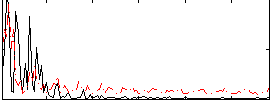 |
Correlation:0.20936
  |
Correlation:0.20936
  |
Correlation:0.20936
  |
Correlation:0.20936
  |
Correlation:0.20936
  |
Component:280; Jaccard:0.094476
 |
Component:268; Jaccard:0.10603
 |
Component:055; Jaccard:0.1054
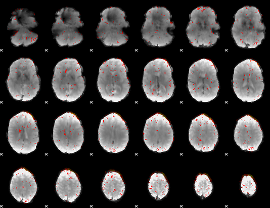 |
Component:055; Jaccard:0.10749
 |
Component:143; Jaccard:0.10144
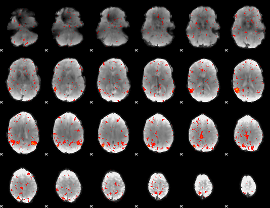 |
Component:164; Jaccard:0.090402
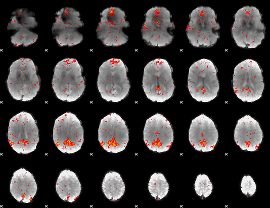 |
Component:164; Jaccard:0.070994
 |
Correlation:0.20936
  |
Correlation:0.1946
  |
Correlation:0.15318
 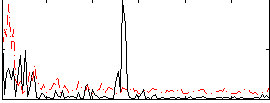 |
Correlation:0.15318
  |
Correlation:0.14005
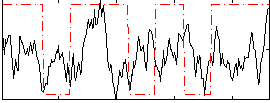 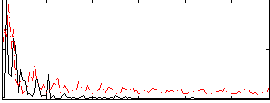 |
Correlation:-0.014112
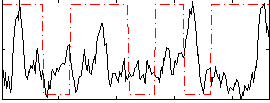  |
Correlation:-0.014112
  |
Component:263; Jaccard:0.093987
 |
Component:392; Jaccard:0.10346
 |
Component:392; Jaccard:0.1036
 |
Component:143; Jaccard:0.10606
 |
Component:164; Jaccard:0.10065
 |
Component:143; Jaccard:0.088956
 |
Component:143; Jaccard:0.066919
 |
Correlation:0.22816
  |
Correlation:0.045039
  |
Correlation:0.045039
  |
Correlation:0.14005
  |
Correlation:-0.014112
  |
Correlation:0.14005
  |
Correlation:0.14005
  |
Component:332; Jaccard:0.090919
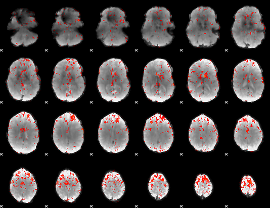 |
Component:263; Jaccard:0.096436
 |
Component:143; Jaccard:0.10113
 |
Component:164; Jaccard:0.10325
 |
Component:395; Jaccard:0.090582
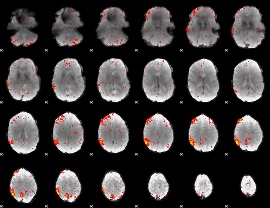 |
Component:395; Jaccard:0.07199
 |
Component:395; Jaccard:0.054317
 |
Correlation:0.34339
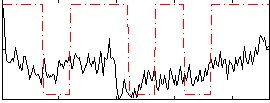 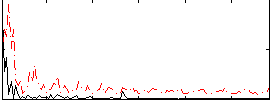 |
Correlation:0.22816
  |
Correlation:0.14005
  |
Correlation:-0.014112
  |
Correlation:-0.031181
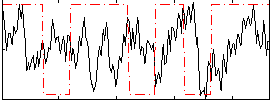 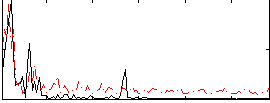 |
Correlation:-0.031181
  |
Correlation:-0.031181
  |
Component:289; Jaccard:0.090407
 |
Component:159; Jaccard:0.093818
 |
Component:159; Jaccard:0.10047
 |
Component:159; Jaccard:0.098907
 |
Component:159; Jaccard:0.088644
 |
Component:159; Jaccard:0.070434
 |
Component:159; Jaccard:0.054011
 |
Correlation:0.2971
  |
Correlation:0.23627
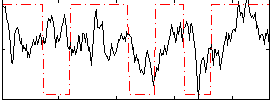 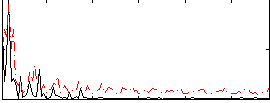 |
Correlation:0.23627
  |
Correlation:0.23627
  |
Correlation:0.23627
  |
Correlation:0.23627
  |
Correlation:0.23627
  |
Component:153; Jaccard:0.089776
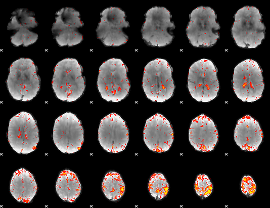 |
Component:055; Jaccard:0.092509
 |
Component:395; Jaccard:0.098231
 |
Component:395; Jaccard:0.098707
 |
Component:055; Jaccard:0.088134
 |
Component:027; Jaccard:0.068973
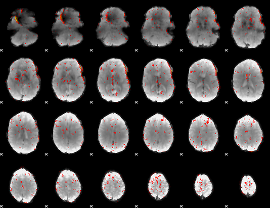 |
Component:027; Jaccard:0.049459
 |
Correlation:0.16213
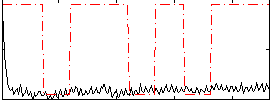 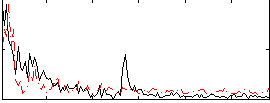 |
Correlation:0.15318
  |
Correlation:-0.031181
  |
Correlation:-0.031181
  |
Correlation:0.15318
  |
Correlation:0.16597
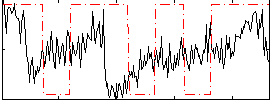 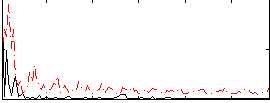 |
Correlation:0.16597
  |
Component:159; Jaccard:0.083751
 |
Component:395; Jaccard:0.09153
 |
Component:268; Jaccard:0.098183
 |
Component:027; Jaccard:0.095792
 |
Component:027; Jaccard:0.083878
 |
Component:114; Jaccard:0.063713
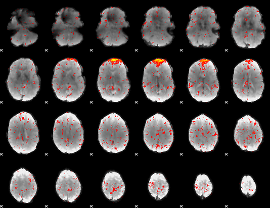 |
Component:062; Jaccard:0.043225
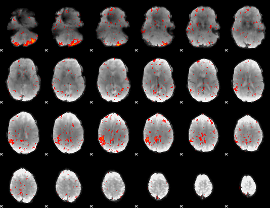 |
Correlation:0.23627
  |
Correlation:-0.031181
  |
Correlation:0.1946
  |
Correlation:0.16597
  |
Correlation:0.16597
  |
Correlation:0.18065
  |
Correlation:-0.050219
  |
Cope: 06:neg_REL BACK
|
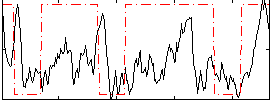
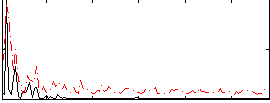
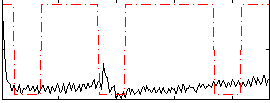
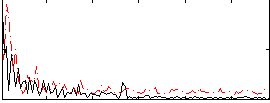
GLM result group-wise
 |
GLM binary result thresholded by 1
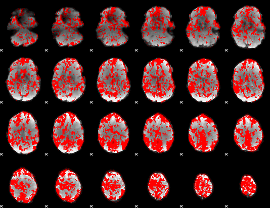 |
GLM binary result thresholded by 1.5
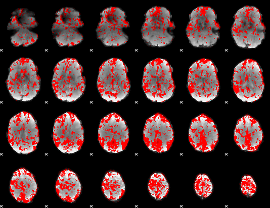 |
GLM binary result thresholded by 2
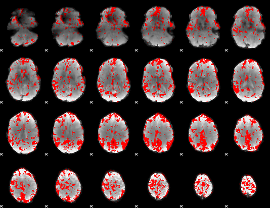 |
GLM binary result thresholded by 2.5
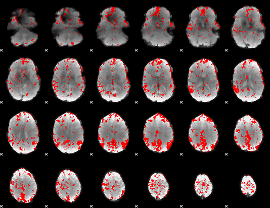 |
GLM binary result thresholded by 3
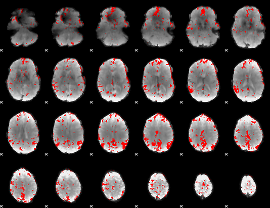 |
GLM binary result thresholded by 3.5
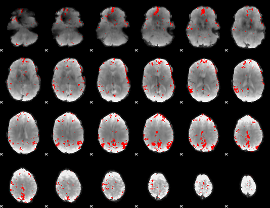 |
GLM binary result thresholded by 4
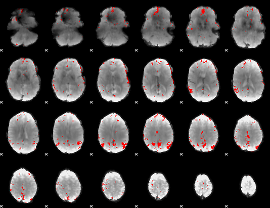 |
GLM Z-score result thresholded by 1
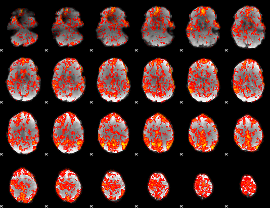 |
GLM Z-score result thresholded by 1.5
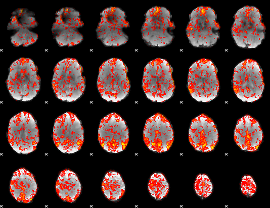 |
GLM Z-score result thresholded by 2
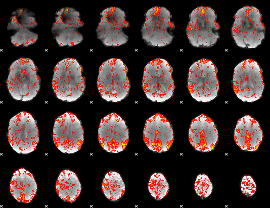 |
GLM Z-score result thresholded by 2.5
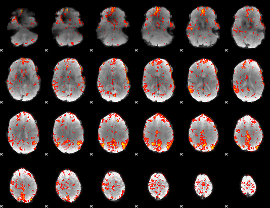 |
GLM Z-score result thresholded by 3
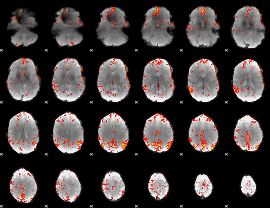 |
GLM Z-score result thresholded by 3.5
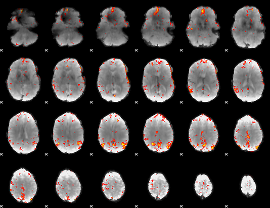 |
GLM Z-score result thresholded by 4
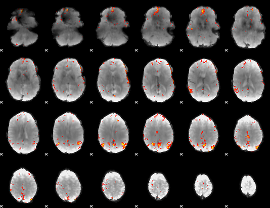 |
Component:392; Jaccard:0.10977
 |
Component:396; Jaccard:0.13034
 |
Component:396; Jaccard:0.16304
 |
Component:396; Jaccard:0.19778
 |
Component:396; Jaccard:0.22456
 |
Component:396; Jaccard:0.22625
 |
Component:396; Jaccard:0.2003
 |
Correlation:0.019837
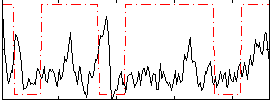 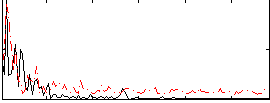 |
Correlation:0.20508
  |
Correlation:0.20508
  |
Correlation:0.20508
  |
Correlation:0.20508
  |
Correlation:0.20508
  |
Correlation:0.20508
  |
Component:268; Jaccard:0.10735
 |
Component:392; Jaccard:0.12351
 |
Component:392; Jaccard:0.13328
 |
Component:102; Jaccard:0.15142
 |
Component:102; Jaccard:0.17093
 |
Component:102; Jaccard:0.16671
 |
Component:102; Jaccard:0.14459
 |
Correlation:0.031414
  |
Correlation:0.019837
  |
Correlation:0.019837
  |
Correlation:0.033597
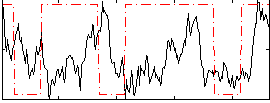 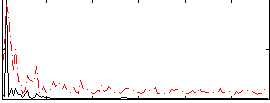 |
Correlation:0.033597
  |
Correlation:0.033597
  |
Correlation:0.033597
  |
Component:396; Jaccard:0.10385
 |
Component:268; Jaccard:0.11539
 |
Component:102; Jaccard:0.1282
 |
Component:392; Jaccard:0.13838
 |
Component:164; Jaccard:0.13893
 |
Component:164; Jaccard:0.13868
 |
Component:164; Jaccard:0.12201
 |
Correlation:0.20508
  |
Correlation:0.031414
  |
Correlation:0.033597
  |
Correlation:0.019837
  |
Correlation:-0.0060583
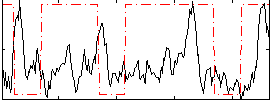 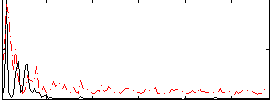 |
Correlation:-0.0060583
  |
Correlation:-0.0060583
  |
Component:263; Jaccard:0.095551
 |
Component:102; Jaccard:0.10627
 |
Component:027; Jaccard:0.11423
 |
Component:164; Jaccard:0.12679
 |
Component:392; Jaccard:0.12845
 |
Component:143; Jaccard:0.12064
 |
Component:143; Jaccard:0.10668
 |
Correlation:0.0094392
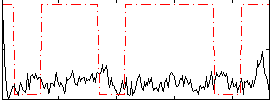 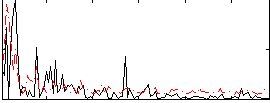 |
Correlation:0.033597
  |
Correlation:0.20011
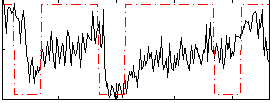 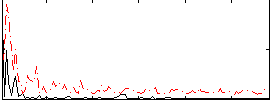 |
Correlation:-0.0060583
  |
Correlation:0.019837
  |
Correlation:0.030074
  |
Correlation:0.030074
  |
Component:332; Jaccard:0.093858
 |
Component:263; Jaccard:0.10281
 |
Component:268; Jaccard:0.1104
 |
Component:027; Jaccard:0.12541
 |
Component:143; Jaccard:0.12805
 |
Component:289; Jaccard:0.11716
 |
Component:289; Jaccard:0.099398
 |
Correlation:-0.06973
  |
Correlation:0.0094392
  |
Correlation:0.031414
  |
Correlation:0.20011
  |
Correlation:0.030074
  |
Correlation:0.13602
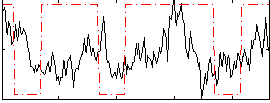 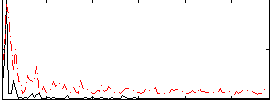 |
Correlation:0.13602
  |
Component:153; Jaccard:0.091996
 |
Component:332; Jaccard:0.09836
 |
Component:164; Jaccard:0.10913
 |
Component:289; Jaccard:0.12087
 |
Component:289; Jaccard:0.12648
 |
Component:027; Jaccard:0.11361
 |
Component:027; Jaccard:0.087597
 |
Correlation:0.086872
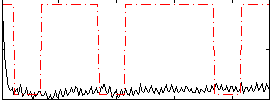 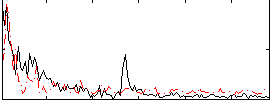 |
Correlation:-0.06973
  |
Correlation:-0.0060583
  |
Correlation:0.13602
  |
Correlation:0.13602
  |
Correlation:0.20011
  |
Correlation:0.20011
  |
Component:214; Jaccard:0.089445
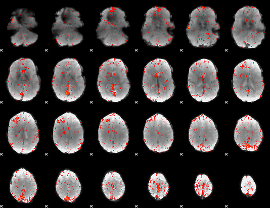 |
Component:027; Jaccard:0.09773
 |
Component:289; Jaccard:0.10701
 |
Component:143; Jaccard:0.12072
 |
Component:027; Jaccard:0.12509
 |
Component:392; Jaccard:0.10713
 |
Component:211; Jaccard:0.079571
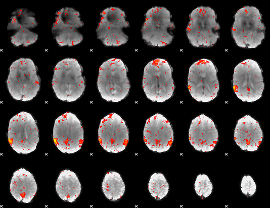 |
Correlation:0.1652
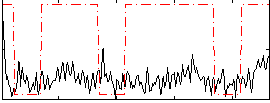 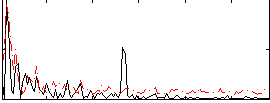 |
Correlation:0.20011
  |
Correlation:0.13602
  |
Correlation:0.030074
  |
Correlation:0.20011
  |
Correlation:0.019837
  |
Correlation:-0.069451
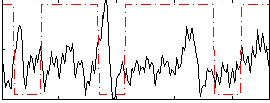 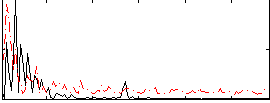 |
Component:102; Jaccard:0.087258
 |
Component:214; Jaccard:0.097125
 |
Component:211; Jaccard:0.10646
 |
Component:211; Jaccard:0.11606
 |
Component:211; Jaccard:0.11606
 |
Component:211; Jaccard:0.10357
 |
Component:392; Jaccard:0.079425
 |
Correlation:0.033597
  |
Correlation:0.1652
  |
Correlation:-0.069451
  |
Correlation:-0.069451
  |
Correlation:-0.069451
  |
Correlation:-0.069451
  |
Correlation:0.019837
  |
Component:220; Jaccard:0.083276
 |
Component:153; Jaccard:0.094084
 |
Component:143; Jaccard:0.10637
 |
Component:214; Jaccard:0.099771
 |
Component:214; Jaccard:0.092882
 |
Component:280; Jaccard:0.083659
 |
Component:280; Jaccard:0.068137
 |
Correlation:0.083606
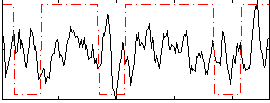 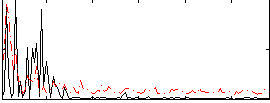 |
Correlation:0.086872
  |
Correlation:0.030074
  |
Correlation:0.1652
  |
Correlation:0.1652
  |
Correlation:-0.1557
  |
Correlation:-0.1557
  |
Component:211; Jaccard:0.081952
 |
Component:211; Jaccard:0.093997
 |
Component:263; Jaccard:0.10294
 |
Component:220; Jaccard:0.097624
 |
Component:280; Jaccard:0.091927
 |
Component:214; Jaccard:0.078223
 |
Component:210; Jaccard:0.057543
 |
Correlation:-0.069451
  |
Correlation:-0.069451
  |
Correlation:0.0094392
  |
Correlation:0.083606
  |
Correlation:-0.1557
  |
Correlation:0.1652
  |
Correlation:0.13482
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































