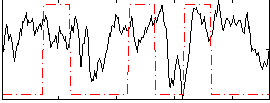
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:163; Jaccard:0.10639
 |
Component:163; Jaccard:0.13299
 |
Component:163; Jaccard:0.16722
 |
Component:163; Jaccard:0.20585
 |
Component:063; Jaccard:0.25088
 |
Component:063; Jaccard:0.29631
 |
Component:063; Jaccard:0.33077
 |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.30804
  |
Correlation:0.30804
  |
Correlation:0.30804
  |
Component:291; Jaccard:0.10513
 |
Component:291; Jaccard:0.12435
 |
Component:063; Jaccard:0.15936
 |
Component:063; Jaccard:0.20346
 |
Component:163; Jaccard:0.24611
 |
Component:163; Jaccard:0.27922
 |
Component:163; Jaccard:0.29882
 |
Correlation:-0.025536
  |
Correlation:-0.025536
  |
Correlation:0.30804
  |
Correlation:0.30804
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Correlation:0.13895
  |
Component:176; Jaccard:0.10084
 |
Component:063; Jaccard:0.12373
 |
Component:291; Jaccard:0.1463
 |
Component:356; Jaccard:0.16985
 |
Component:356; Jaccard:0.19888
 |
Component:356; Jaccard:0.22314
 |
Component:388; Jaccard:0.24186
 |
Correlation:0.13418
  |
Correlation:0.30804
  |
Correlation:-0.025536
  |
Correlation:0.13299
  |
Correlation:0.13299
  |
Correlation:0.13299
  |
Correlation:0.19574
  |
Component:063; Jaccard:0.097329
 |
Component:176; Jaccard:0.11637
 |
Component:356; Jaccard:0.13969
 |
Component:291; Jaccard:0.16426
 |
Component:250; Jaccard:0.18891
 |
Component:388; Jaccard:0.21863
 |
Component:356; Jaccard:0.23706
 |
Correlation:0.30804
  |
Correlation:0.13418
  |
Correlation:0.13299
  |
Correlation:-0.025536
  |
Correlation:0.29845
  |
Correlation:0.19574
  |
Correlation:0.13299
  |
Component:263; Jaccard:0.095909
 |
Component:263; Jaccard:0.11422
 |
Component:263; Jaccard:0.13618
 |
Component:250; Jaccard:0.1599
 |
Component:388; Jaccard:0.18792
 |
Component:250; Jaccard:0.21494
 |
Component:250; Jaccard:0.23201
 |
Correlation:0.1276
  |
Correlation:0.1276
  |
Correlation:0.1276
  |
Correlation:0.29845
  |
Correlation:0.19574
  |
Correlation:0.29845
  |
Correlation:0.29845
  |
Component:280; Jaccard:0.094494
 |
Component:356; Jaccard:0.11382
 |
Component:176; Jaccard:0.13314
 |
Component:263; Jaccard:0.15785
 |
Component:263; Jaccard:0.17674
 |
Component:263; Jaccard:0.19133
 |
Component:263; Jaccard:0.19853
 |
Correlation:-0.032076
  |
Correlation:0.13299
  |
Correlation:0.13418
  |
Correlation:0.1276
  |
Correlation:0.1276
  |
Correlation:0.1276
  |
Correlation:0.1276
  |
Component:356; Jaccard:0.092485
 |
Component:280; Jaccard:0.11128
 |
Component:250; Jaccard:0.13161
 |
Component:388; Jaccard:0.15557
 |
Component:291; Jaccard:0.17663
 |
Component:291; Jaccard:0.18029
 |
Component:370; Jaccard:0.19031
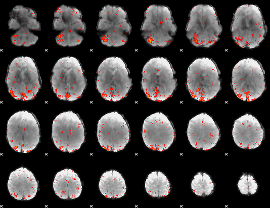 |
Correlation:0.13299
  |
Correlation:-0.032076
  |
Correlation:0.29845
  |
Correlation:0.19574
  |
Correlation:-0.025536
  |
Correlation:-0.025536
  |
Correlation:0.19113
  |
Component:242; Jaccard:0.091003
 |
Component:250; Jaccard:0.10706
 |
Component:280; Jaccard:0.1301
 |
Component:280; Jaccard:0.149
 |
Component:280; Jaccard:0.16522
 |
Component:370; Jaccard:0.1763
 |
Component:291; Jaccard:0.17662
 |
Correlation:0.18727
  |
Correlation:0.29845
  |
Correlation:-0.032076
  |
Correlation:-0.032076
  |
Correlation:-0.032076
  |
Correlation:0.19113
  |
Correlation:-0.025536
  |
Component:185; Jaccard:0.088215
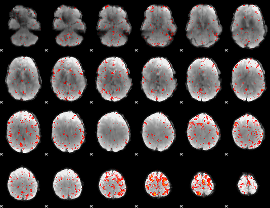 |
Component:242; Jaccard:0.10602
 |
Component:388; Jaccard:0.12587
 |
Component:176; Jaccard:0.14745
 |
Component:176; Jaccard:0.15771
 |
Component:280; Jaccard:0.17203
 |
Component:280; Jaccard:0.17292
 |
Correlation:-0.010096
  |
Correlation:0.18727
  |
Correlation:0.19574
  |
Correlation:0.13418
  |
Correlation:0.13418
  |
Correlation:-0.032076
  |
Correlation:-0.032076
  |
Component:250; Jaccard:0.086857
 |
Component:059; Jaccard:0.099937
 |
Component:242; Jaccard:0.12298
 |
Component:242; Jaccard:0.13882
 |
Component:370; Jaccard:0.15742
 |
Component:242; Jaccard:0.16191
 |
Component:242; Jaccard:0.16396
 |
Correlation:0.29845
  |
Correlation:-0.050742
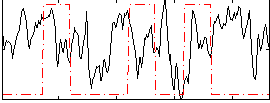 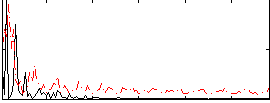 |
Correlation:0.18727
  |
Correlation:0.18727
  |
Correlation:0.19113
  |
Correlation:0.18727
  |
Correlation:0.18727
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
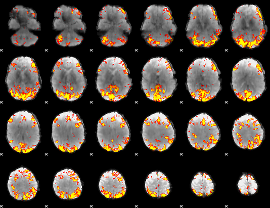 |
GLM Z-score result thresholded by 4
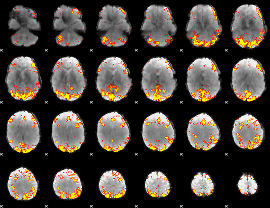 |
Component:291; Jaccard:0.12333
 |
Component:291; Jaccard:0.1468
 |
Component:291; Jaccard:0.16881
 |
Component:163; Jaccard:0.19009
 |
Component:163; Jaccard:0.21224
 |
Component:163; Jaccard:0.23279
 |
Component:163; Jaccard:0.24792
 |
Correlation:0.31442
  |
Correlation:0.31442
  |
Correlation:0.31442
  |
Correlation:0.25759
 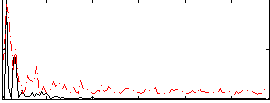 |
Correlation:0.25759
  |
Correlation:0.25759
  |
Correlation:0.25759
  |
Component:163; Jaccard:0.1173
 |
Component:163; Jaccard:0.14086
 |
Component:163; Jaccard:0.16541
 |
Component:291; Jaccard:0.18828
 |
Component:063; Jaccard:0.20246
 |
Component:063; Jaccard:0.22329
 |
Component:063; Jaccard:0.24353
 |
Correlation:0.25759
  |
Correlation:0.25759
  |
Correlation:0.25759
  |
Correlation:0.31442
  |
Correlation:0.19687
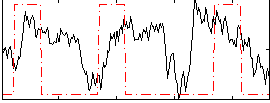  |
Correlation:0.19687
  |
Correlation:0.19687
  |
Component:242; Jaccard:0.11188
 |
Component:242; Jaccard:0.13334
 |
Component:242; Jaccard:0.1545
 |
Component:356; Jaccard:0.17854
 |
Component:291; Jaccard:0.20215
 |
Component:356; Jaccard:0.22153
 |
Component:356; Jaccard:0.23653
 |
Correlation:0.30167
  |
Correlation:0.30167
  |
Correlation:0.30167
  |
Correlation:0.35449
  |
Correlation:0.31442
  |
Correlation:0.35449
  |
Correlation:0.35449
  |
Component:280; Jaccard:0.10902
 |
Component:356; Jaccard:0.12964
 |
Component:356; Jaccard:0.15427
 |
Component:063; Jaccard:0.17777
 |
Component:356; Jaccard:0.20125
 |
Component:291; Jaccard:0.21244
 |
Component:388; Jaccard:0.22081
 |
Correlation:0.33943
  |
Correlation:0.35449
  |
Correlation:0.35449
  |
Correlation:0.19687
  |
Correlation:0.35449
  |
Correlation:0.31442
  |
Correlation:0.21874
  |
Component:356; Jaccard:0.1069
 |
Component:063; Jaccard:0.12856
 |
Component:063; Jaccard:0.153
 |
Component:242; Jaccard:0.1745
 |
Component:242; Jaccard:0.19241
 |
Component:242; Jaccard:0.20639
 |
Component:291; Jaccard:0.21844
 |
Correlation:0.35449
  |
Correlation:0.19687
  |
Correlation:0.19687
  |
Correlation:0.30167
  |
Correlation:0.30167
  |
Correlation:0.30167
  |
Correlation:0.31442
  |
Component:063; Jaccard:0.10613
 |
Component:280; Jaccard:0.12834
 |
Component:280; Jaccard:0.14638
 |
Component:280; Jaccard:0.16264
 |
Component:294; Jaccard:0.1791
 |
Component:388; Jaccard:0.19944
 |
Component:242; Jaccard:0.21687
 |
Correlation:0.19687
  |
Correlation:0.33943
  |
Correlation:0.33943
  |
Correlation:0.33943
  |
Correlation:0.30846
  |
Correlation:0.21874
  |
Correlation:0.30167
  |
Component:263; Jaccard:0.10541
 |
Component:263; Jaccard:0.12269
 |
Component:294; Jaccard:0.14066
 |
Component:294; Jaccard:0.16076
 |
Component:388; Jaccard:0.17801
 |
Component:294; Jaccard:0.19385
 |
Component:294; Jaccard:0.20656
 |
Correlation:0.10431
  |
Correlation:0.10431
  |
Correlation:0.30846
  |
Correlation:0.30846
  |
Correlation:0.21874
  |
Correlation:0.30846
  |
Correlation:0.30846
  |
Component:176; Jaccard:0.10342
 |
Component:059; Jaccard:0.12112
 |
Component:059; Jaccard:0.13926
 |
Component:059; Jaccard:0.15722
 |
Component:280; Jaccard:0.17533
 |
Component:250; Jaccard:0.18867
 |
Component:250; Jaccard:0.20383
 |
Correlation:0.12232
  |
Correlation:0.30342
  |
Correlation:0.30342
  |
Correlation:0.30342
  |
Correlation:0.33943
  |
Correlation:0.26661
  |
Correlation:0.26661
  |
Component:059; Jaccard:0.10269
 |
Component:294; Jaccard:0.1195
 |
Component:263; Jaccard:0.13834
 |
Component:388; Jaccard:0.15496
 |
Component:250; Jaccard:0.17179
 |
Component:280; Jaccard:0.18587
 |
Component:280; Jaccard:0.19297
 |
Correlation:0.30342
  |
Correlation:0.30846
  |
Correlation:0.10431
  |
Correlation:0.21874
  |
Correlation:0.26661
  |
Correlation:0.33943
  |
Correlation:0.33943
  |
Component:011; Jaccard:0.10258
 |
Component:250; Jaccard:0.11585
 |
Component:250; Jaccard:0.13521
 |
Component:250; Jaccard:0.15389
 |
Component:059; Jaccard:0.17148
 |
Component:059; Jaccard:0.18172
 |
Component:059; Jaccard:0.18621
 |
Correlation:0.15522
  |
Correlation:0.26661
  |
Correlation:0.26661
  |
Correlation:0.26661
  |
Correlation:0.30342
  |
Correlation:0.30342
  |
Correlation:0.30342
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
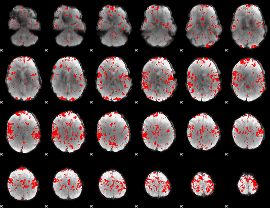 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
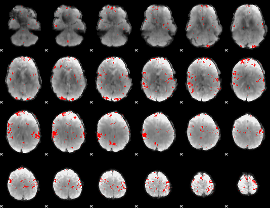 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
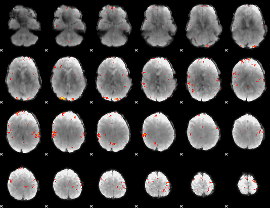 |
GLM Z-score result thresholded by 4
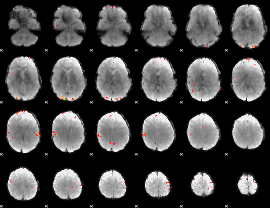 |
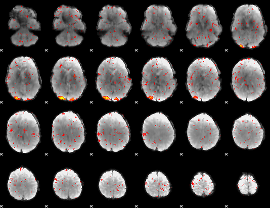
Component:129; Jaccard:0.10985
 |
Component:129; Jaccard:0.12949
 |
Component:129; Jaccard:0.14089
 |
Component:129; Jaccard:0.13445
 |
Component:151; Jaccard:0.11472
 |
Component:151; Jaccard:0.09165
 |
Component:151; Jaccard:0.066862
 |
Correlation:0.020665
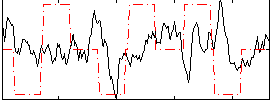  |
Correlation:0.020665
  |
Correlation:0.020665
  |
Correlation:0.020665
  |
Correlation:0.38698
  |
Correlation:0.38698
  |
Correlation:0.38698
  |
Component:131; Jaccard:0.10146
 |
Component:131; Jaccard:0.11899
 |
Component:131; Jaccard:0.13059
 |
Component:131; Jaccard:0.12296
 |
Component:129; Jaccard:0.1077
 |
Component:129; Jaccard:0.070529
 |
Component:129; Jaccard:0.041419
 |
Correlation:0.10479
 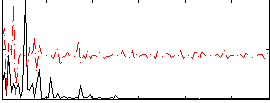 |
Correlation:0.10479
  |
Correlation:0.10479
  |
Correlation:0.10479
  |
Correlation:0.020665
  |
Correlation:0.020665
  |
Correlation:0.020665
  |
Component:109; Jaccard:0.08312
 |
Component:109; Jaccard:0.094943
 |
Component:151; Jaccard:0.11014
 |
Component:151; Jaccard:0.12061
 |
Component:131; Jaccard:0.091748
 |
Component:131; Jaccard:0.052901
 |
Component:131; Jaccard:0.028177
 |
Correlation:0.27296
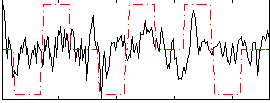 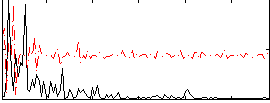 |
Correlation:0.27296
  |
Correlation:0.38698
  |
Correlation:0.38698
  |
Correlation:0.10479
  |
Correlation:0.10479
  |
Correlation:0.10479
  |
Component:030; Jaccard:0.082132
 |
Component:151; Jaccard:0.091293
 |
Component:109; Jaccard:0.10025
 |
Component:109; Jaccard:0.093519
 |
Component:109; Jaccard:0.072378
 |
Component:123; Jaccard:0.044957
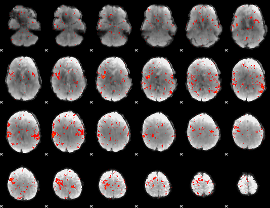 |
Component:218; Jaccard:0.025183
 |
Correlation:0.3363
  |
Correlation:0.38698
  |
Correlation:0.27296
  |
Correlation:0.27296
  |
Correlation:0.27296
  |
Correlation:0.18789
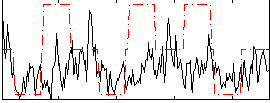 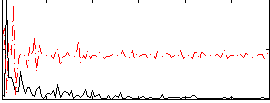 |
Correlation:0.20387
  |
Component:245; Jaccard:0.079711
 |
Component:030; Jaccard:0.090125
 |
Component:030; Jaccard:0.092773
 |
Component:123; Jaccard:0.086106
 |
Component:123; Jaccard:0.070063
 |
Component:109; Jaccard:0.043336
 |
Component:123; Jaccard:0.024251
 |
Correlation:-0.088246
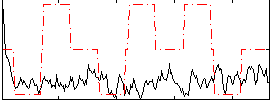  |
Correlation:0.3363
  |
Correlation:0.3363
  |
Correlation:0.18789
  |
Correlation:0.18789
  |
Correlation:0.27296
  |
Correlation:0.18789
  |
Component:302; Jaccard:0.079554
 |
Component:302; Jaccard:0.088793
 |
Component:302; Jaccard:0.09257
 |
Component:302; Jaccard:0.084347
 |
Component:302; Jaccard:0.065314
 |
Component:076; Jaccard:0.042145
 |
Component:254; Jaccard:0.023104
 |
Correlation:0.10589
  |
Correlation:0.10589
  |
Correlation:0.10589
  |
Correlation:0.10589
  |
Correlation:0.10589
  |
Correlation:0.026176
 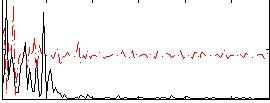 |
Correlation:0.12608
 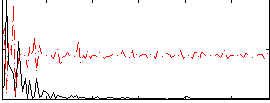 |
Component:309; Jaccard:0.079149
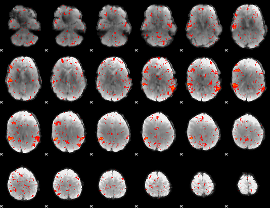 |
Component:123; Jaccard:0.08815
 |
Component:123; Jaccard:0.092171
 |
Component:030; Jaccard:0.081899
 |
Component:076; Jaccard:0.063323
 |
Component:302; Jaccard:0.04041
 |
Component:223; Jaccard:0.023068
 |
Correlation:0.021806
  |
Correlation:0.18789
  |
Correlation:0.18789
  |
Correlation:0.3363
  |
Correlation:0.026176
  |
Correlation:0.10589
  |
Correlation:0.16005
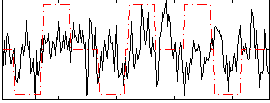  |
Component:123; Jaccard:0.078685
 |
Component:076; Jaccard:0.082415
 |
Component:076; Jaccard:0.086486
 |
Component:076; Jaccard:0.079951
 |
Component:030; Jaccard:0.061792
 |
Component:218; Jaccard:0.038536
 |
Component:385; Jaccard:0.022944
 |
Correlation:0.18789
  |
Correlation:0.026176
  |
Correlation:0.026176
  |
Correlation:0.026176
  |
Correlation:0.3363
  |
Correlation:0.20387
  |
Correlation:0.38693
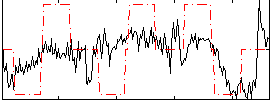 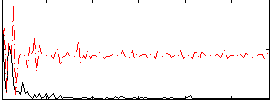 |
Component:076; Jaccard:0.074308
 |
Component:309; Jaccard:0.081569
 |
Component:204; Jaccard:0.080833
 |
Component:204; Jaccard:0.072432
 |
Component:385; Jaccard:0.05713
 |
Component:385; Jaccard:0.037779
 |
Component:076; Jaccard:0.022749
 |
Correlation:0.026176
  |
Correlation:0.021806
  |
Correlation:0.33373
  |
Correlation:0.33373
  |
Correlation:0.38693
  |
Correlation:0.38693
  |
Correlation:0.026176
  |
Component:151; Jaccard:0.073453
 |
Component:245; Jaccard:0.080142
 |
Component:254; Jaccard:0.078007
 |
Component:254; Jaccard:0.069582
 |
Component:204; Jaccard:0.057011
 |
Component:223; Jaccard:0.037472
 |
Component:109; Jaccard:0.022041
 |
Correlation:0.38698
  |
Correlation:-0.088246
  |
Correlation:0.12608
  |
Correlation:0.12608
  |
Correlation:0.33373
  |
Correlation:0.16005
  |
Correlation:0.27296
  |
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
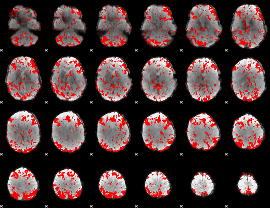 |
GLM binary result thresholded by 1.5
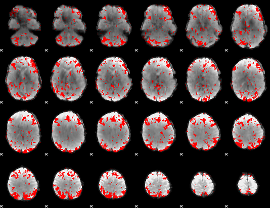 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
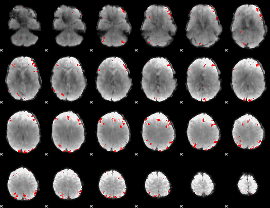 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
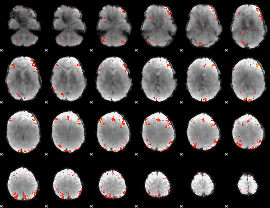 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
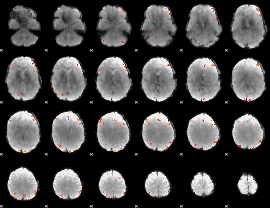 |
Component:242; Jaccard:0.13437
 |
Component:242; Jaccard:0.1552
 |
Component:307; Jaccard:0.16642
 |
Component:307; Jaccard:0.1748
 |
Component:307; Jaccard:0.16416
 |
Component:307; Jaccard:0.1391
 |
Component:307; Jaccard:0.1048
 |
Correlation:0.067802
  |
Correlation:0.067802
  |
Correlation:0.20872
  |
Correlation:0.20872
  |
Correlation:0.20872
  |
Correlation:0.20872
  |
Correlation:0.20872
  |
Component:294; Jaccard:0.1314
 |
Component:294; Jaccard:0.15408
 |
Component:294; Jaccard:0.16632
 |
Component:294; Jaccard:0.16012
 |
Component:294; Jaccard:0.13715
 |
Component:294; Jaccard:0.10692
 |
Component:294; Jaccard:0.073593
 |
Correlation:0.20962
  |
Correlation:0.20962
  |
Correlation:0.20962
  |
Correlation:0.20962
  |
Correlation:0.20962
  |
Correlation:0.20962
  |
Correlation:0.20962
  |
Component:100; Jaccard:0.12689
 |
Component:307; Jaccard:0.14475
 |
Component:242; Jaccard:0.165
 |
Component:242; Jaccard:0.1547
 |
Component:242; Jaccard:0.12898
 |
Component:242; Jaccard:0.097593
 |
Component:242; Jaccard:0.067405
 |
Correlation:0.4081
  |
Correlation:0.20872
  |
Correlation:0.067802
  |
Correlation:0.067802
  |
Correlation:0.067802
  |
Correlation:0.067802
  |
Correlation:0.067802
  |
Component:011; Jaccard:0.12437
 |
Component:100; Jaccard:0.14304
 |
Component:100; Jaccard:0.15078
 |
Component:100; Jaccard:0.13784
 |
Component:100; Jaccard:0.11257
 |
Component:253; Jaccard:0.086338
 |
Component:253; Jaccard:0.067081
 |
Correlation:0.10213
  |
Correlation:0.4081
  |
Correlation:0.4081
  |
Correlation:0.4081
  |
Correlation:0.4081
  |
Correlation:0.3165
  |
Correlation:0.3165
  |
Component:307; Jaccard:0.1175
 |
Component:011; Jaccard:0.13744
 |
Component:011; Jaccard:0.14002
 |
Component:011; Jaccard:0.12678
 |
Component:253; Jaccard:0.10442
 |
Component:100; Jaccard:0.080018
 |
Component:146; Jaccard:0.054102
 |
Correlation:0.20872
  |
Correlation:0.10213
  |
Correlation:0.10213
  |
Correlation:0.10213
  |
Correlation:0.3165
  |
Correlation:0.4081
  |
Correlation:0.37904
  |
Component:291; Jaccard:0.11639
 |
Component:291; Jaccard:0.12711
 |
Component:291; Jaccard:0.1277
 |
Component:291; Jaccard:0.11482
 |
Component:011; Jaccard:0.10216
 |
Component:146; Jaccard:0.079642
 |
Component:011; Jaccard:0.050295
 |
Correlation:0.20149
  |
Correlation:0.20149
  |
Correlation:0.20149
  |
Correlation:0.20149
  |
Correlation:0.10213
  |
Correlation:0.37904
  |
Correlation:0.10213
  |
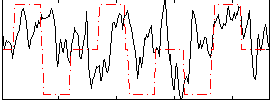
Component:059; Jaccard:0.11157
 |
Component:059; Jaccard:0.12258
 |
Component:059; Jaccard:0.12492
 |
Component:154; Jaccard:0.11402
 |
Component:146; Jaccard:0.099719
 |
Component:011; Jaccard:0.073907
 |
Component:100; Jaccard:0.049988
 |
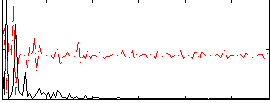
Correlation:0.20991
  |
Correlation:0.20991
  |
Correlation:0.20991
  |
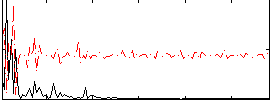
Correlation:0.12209
  |
Correlation:0.37904
  |
Correlation:0.10213
  |
Correlation:0.4081
  |
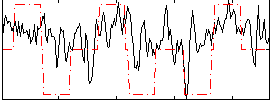
Component:366; Jaccard:0.10897
 |
Component:154; Jaccard:0.11969
 |
Component:154; Jaccard:0.12347
 |
Component:059; Jaccard:0.11393
 |
Component:291; Jaccard:0.093813
 |
Component:154; Jaccard:0.070845
 |
Component:154; Jaccard:0.049573
 |
Correlation:0.40138
  |
Correlation:0.12209
  |
Correlation:0.12209
  |
Correlation:0.20991
  |
Correlation:0.20149
  |
Correlation:0.12209
  |
Correlation:0.12209
  |
Component:399; Jaccard:0.10833
 |
Component:399; Jaccard:0.11579
 |
Component:399; Jaccard:0.11144
 |
Component:146; Jaccard:0.11205
 |
Component:154; Jaccard:0.092369
 |
Component:291; Jaccard:0.070233
 |
Component:291; Jaccard:0.045688
 |
Correlation:0.4085
  |
Correlation:0.4085
  |
Correlation:0.4085
  |
Correlation:0.37904
  |
Correlation:0.12209
  |
Correlation:0.20149
  |
Correlation:0.20149
  |
Component:165; Jaccard:0.10572
 |
Component:366; Jaccard:0.11529
 |
Component:165; Jaccard:0.11094
 |
Component:253; Jaccard:0.11159
 |
Component:059; Jaccard:0.089921
 |
Component:059; Jaccard:0.064291
 |
Component:280; Jaccard:0.043294
 |
Correlation:0.2591
  |
Correlation:0.40138
  |
Correlation:0.2591
  |
Correlation:0.3165
  |
Correlation:0.20991
  |
Correlation:0.20991
  |
Correlation:0.22019
  |
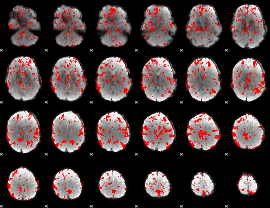
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
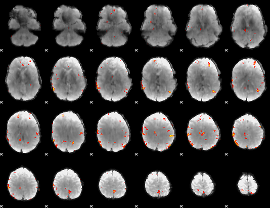 |
GLM Z-score result thresholded by 4
 |
Component:026; Jaccard:0.15993
 |
Component:026; Jaccard:0.21063
 |
Component:026; Jaccard:0.25461
 |
Component:026; Jaccard:0.27396
 |
Component:026; Jaccard:0.25838
 |
Component:026; Jaccard:0.21626
 |
Component:026; Jaccard:0.16703
 |
Correlation:0.1889
  |
Correlation:0.1889
  |
Correlation:0.1889
  |
Correlation:0.1889
  |
Correlation:0.1889
  |
Correlation:0.1889
  |
Correlation:0.1889
  |
Component:359; Jaccard:0.11629
 |
Component:359; Jaccard:0.13131
 |
Component:359; Jaccard:0.13747
 |
Component:359; Jaccard:0.12958
 |
Component:021; Jaccard:0.11595
 |
Component:021; Jaccard:0.098645
 |
Component:021; Jaccard:0.076992
 |
Correlation:0.18478
  |
Correlation:0.18478
  |
Correlation:0.18478
  |
Correlation:0.18478
  |
Correlation:0.24951
  |
Correlation:0.24951
  |
Correlation:0.24951
  |
Component:021; Jaccard:0.10514
 |
Component:021; Jaccard:0.11926
 |
Component:021; Jaccard:0.13001
 |
Component:257; Jaccard:0.12805
 |
Component:359; Jaccard:0.11578
 |
Component:359; Jaccard:0.094504
 |
Component:359; Jaccard:0.068976
 |
Correlation:0.24951
  |
Correlation:0.24951
  |
Correlation:0.24951
  |
Correlation:0.29308
  |
Correlation:0.18478
  |
Correlation:0.18478
  |
Correlation:0.18478
  |
Component:046; Jaccard:0.10444
 |
Component:257; Jaccard:0.11923
 |
Component:257; Jaccard:0.12942
 |
Component:021; Jaccard:0.12616
 |
Component:257; Jaccard:0.10886
 |
Component:257; Jaccard:0.087038
 |
Component:257; Jaccard:0.063457
 |
Correlation:0.27449
  |
Correlation:0.29308
  |
Correlation:0.29308
  |
Correlation:0.24951
  |
Correlation:0.29308
  |
Correlation:0.29308
  |
Correlation:0.29308
  |
Component:257; Jaccard:0.10219
 |
Component:046; Jaccard:0.10776
 |
Component:046; Jaccard:0.10419
 |
Component:046; Jaccard:0.089685
 |
Component:264; Jaccard:0.076551
 |
Component:264; Jaccard:0.063814
 |
Component:183; Jaccard:0.049263
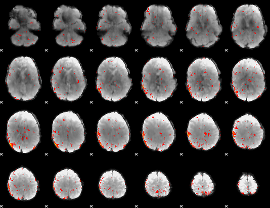 |
Correlation:0.29308
  |
Correlation:0.27449
  |
Correlation:0.27449
  |
Correlation:0.27449
  |
Correlation:-0.12331
 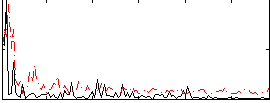 |
Correlation:-0.12331
  |
Correlation:0.03824
  |
Component:302; Jaccard:0.08901
 |
Component:302; Jaccard:0.090029
 |
Component:264; Jaccard:0.090171
 |
Component:264; Jaccard:0.086905
 |
Component:046; Jaccard:0.07251
 |
Component:183; Jaccard:0.059654
 |
Component:264; Jaccard:0.048246
 |
Correlation:0.12383
  |
Correlation:0.12383
  |
Correlation:-0.12331
  |
Correlation:-0.12331
  |
Correlation:0.27449
  |
Correlation:0.03824
  |
Correlation:-0.12331
  |
Component:001; Jaccard:0.081454
 |
Component:264; Jaccard:0.085105
 |
Component:302; Jaccard:0.087667
 |
Component:302; Jaccard:0.079268
 |
Component:183; Jaccard:0.068883
 |
Component:046; Jaccard:0.053813
 |
Component:058; Jaccard:0.042098
 |
Correlation:0.19971
 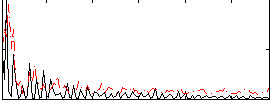 |
Correlation:-0.12331
  |
Correlation:0.12383
  |
Correlation:0.12383
  |
Correlation:0.03824
  |
Correlation:0.27449
  |
Correlation:-0.034785
  |
Component:058; Jaccard:0.080011
 |
Component:058; Jaccard:0.081912
 |
Component:058; Jaccard:0.081289
 |
Component:058; Jaccard:0.075793
 |
Component:302; Jaccard:0.066492
 |
Component:058; Jaccard:0.053122
 |
Component:024; Jaccard:0.040622
 |
Correlation:-0.034785
  |
Correlation:-0.034785
  |
Correlation:-0.034785
  |
Correlation:-0.034785
  |
Correlation:0.12383
  |
Correlation:-0.034785
  |
Correlation:0.096159
  |
Component:272; Jaccard:0.078954
 |
Component:272; Jaccard:0.081425
 |
Component:272; Jaccard:0.078826
 |
Component:183; Jaccard:0.074571
 |
Component:058; Jaccard:0.065114
 |
Component:024; Jaccard:0.052459
 |
Component:302; Jaccard:0.038952
 |
Correlation:0.24738
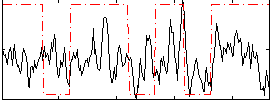  |
Correlation:0.24738
  |
Correlation:0.24738
  |
Correlation:0.03824
  |
Correlation:-0.034785
  |
Correlation:0.096159
  |
Correlation:0.12383
  |
Component:110; Jaccard:0.076726
 |
Component:001; Jaccard:0.076601
 |
Component:183; Jaccard:0.074548
 |
Component:272; Jaccard:0.070639
 |
Component:024; Jaccard:0.062634
 |
Component:302; Jaccard:0.052241
 |
Component:272; Jaccard:0.037947
 |
Correlation:0.33585
  |
Correlation:0.19971
  |
Correlation:0.03824
  |
Correlation:0.24738
  |
Correlation:0.096159
  |
Correlation:0.12383
  |
Correlation:0.24738
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:026; Jaccard:0.12731
 |
Component:026; Jaccard:0.17189
 |
Component:026; Jaccard:0.22207
 |
Component:026; Jaccard:0.26935
 |
Component:026; Jaccard:0.30263
 |
Component:026; Jaccard:0.30555
 |
Component:026; Jaccard:0.28491
 |
Correlation:0.22338
  |
Correlation:0.22338
  |
Correlation:0.22338
  |
Correlation:0.22338
  |
Correlation:0.22338
  |
Correlation:0.22338
  |
Correlation:0.22338
  |
Component:359; Jaccard:0.1195
 |
Component:359; Jaccard:0.14155
 |
Component:359; Jaccard:0.16076
 |
Component:359; Jaccard:0.17267
 |
Component:359; Jaccard:0.17174
 |
Component:359; Jaccard:0.15934
 |
Component:359; Jaccard:0.14027
 |
Correlation:0.25328
  |
Correlation:0.25328
  |
Correlation:0.25328
  |
Correlation:0.25328
  |
Correlation:0.25328
  |
Correlation:0.25328
  |
Correlation:0.25328
  |
Component:129; Jaccard:0.1169
 |
Component:046; Jaccard:0.12843
 |
Component:046; Jaccard:0.14016
 |
Component:046; Jaccard:0.14116
 |
Component:046; Jaccard:0.13187
 |
Component:021; Jaccard:0.11737
 |
Component:021; Jaccard:0.11057
 |
Correlation:0.16647
  |
Correlation:0.25672
  |
Correlation:0.25672
  |
Correlation:0.25672
  |
Correlation:0.25672
  |
Correlation:0.075765
  |
Correlation:0.075765
  |
Component:302; Jaccard:0.11483
 |
Component:302; Jaccard:0.12787
 |
Component:302; Jaccard:0.13444
 |
Component:302; Jaccard:0.13284
 |
Component:302; Jaccard:0.12661
 |
Component:046; Jaccard:0.11586
 |
Component:302; Jaccard:0.098717
 |
Correlation:0.30248
  |
Correlation:0.30248
  |
Correlation:0.30248
  |
Correlation:0.30248
  |
Correlation:0.30248
  |
Correlation:0.25672
  |
Correlation:0.30248
  |
Component:046; Jaccard:0.11258
 |
Component:129; Jaccard:0.12685
 |
Component:129; Jaccard:0.13027
 |
Component:129; Jaccard:0.12364
 |
Component:021; Jaccard:0.11879
 |
Component:302; Jaccard:0.11364
 |
Component:046; Jaccard:0.097842
 |
Correlation:0.25672
  |
Correlation:0.16647
  |
Correlation:0.16647
  |
Correlation:0.16647
  |
Correlation:0.075765
  |
Correlation:0.30248
  |
Correlation:0.25672
  |
Component:021; Jaccard:0.089774
 |
Component:021; Jaccard:0.10199
 |
Component:021; Jaccard:0.11309
 |
Component:021; Jaccard:0.119
 |
Component:129; Jaccard:0.11459
 |
Component:129; Jaccard:0.098571
 |
Component:129; Jaccard:0.080552
 |
Correlation:0.075765
  |
Correlation:0.075765
  |
Correlation:0.075765
  |
Correlation:0.075765
  |
Correlation:0.16647
  |
Correlation:0.16647
  |
Correlation:0.16647
  |
Component:058; Jaccard:0.088893
 |
Component:058; Jaccard:0.096081
 |
Component:251; Jaccard:0.098646
 |
Component:058; Jaccard:0.097641
 |
Component:058; Jaccard:0.092569
 |
Component:058; Jaccard:0.082312
 |
Component:058; Jaccard:0.072999
 |
Correlation:0.2444
  |
Correlation:0.2444
  |
Correlation:0.092256
  |
Correlation:0.2444
  |
Correlation:0.2444
  |
Correlation:0.2444
  |
Correlation:0.2444
  |
Component:251; Jaccard:0.087102
 |
Component:251; Jaccard:0.095352
 |
Component:058; Jaccard:0.098411
 |
Component:251; Jaccard:0.095985
 |
Component:251; Jaccard:0.090071
 |
Component:251; Jaccard:0.079335
 |
Component:076; Jaccard:0.065727
 |
Correlation:0.092256
  |
Correlation:0.092256
  |
Correlation:0.2444
  |
Correlation:0.092256
  |
Correlation:0.092256
  |
Correlation:0.092256
  |
Correlation:0.2335
  |
Component:390; Jaccard:0.085766
 |
Component:076; Jaccard:0.091205
 |
Component:076; Jaccard:0.095005
 |
Component:076; Jaccard:0.093702
 |
Component:076; Jaccard:0.086928
 |
Component:076; Jaccard:0.078693
 |
Component:251; Jaccard:0.064569
 |
Correlation:0.040642
  |
Correlation:0.2335
  |
Correlation:0.2335
  |
Correlation:0.2335
  |
Correlation:0.2335
  |
Correlation:0.2335
  |
Correlation:0.092256
  |
Component:076; Jaccard:0.083997
 |
Component:390; Jaccard:0.088129
 |
Component:390; Jaccard:0.086577
 |
Component:264; Jaccard:0.084944
 |
Component:264; Jaccard:0.082013
 |
Component:264; Jaccard:0.073074
 |
Component:264; Jaccard:0.061096
 |
Correlation:0.2335
  |
Correlation:0.040642
  |
Correlation:0.040642
  |
Correlation:0.27904
  |
Correlation:0.27904
  |
Correlation:0.27904
  |
Correlation:0.27904
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































