Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
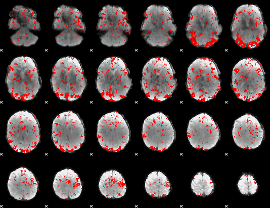 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
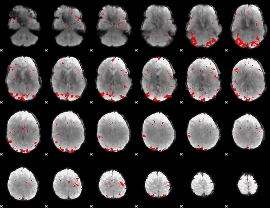 |
GLM binary result thresholded by 3.5
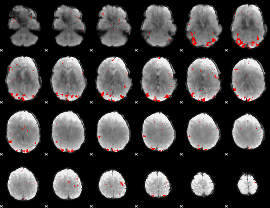 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
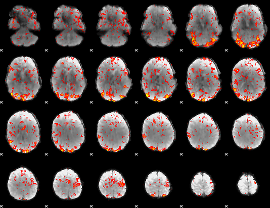 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
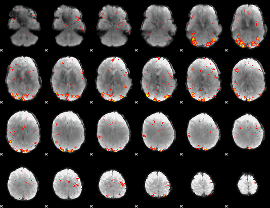 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:369; Jaccard:0.11226
 |
Component:224; Jaccard:0.15158
 |
Component:224; Jaccard:0.20273
 |
Component:224; Jaccard:0.24865
 |
Component:224; Jaccard:0.27345
 |
Component:224; Jaccard:0.25171
 |
Component:224; Jaccard:0.20964
 |
Correlation:0.26588
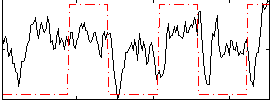  |
Correlation:0.61023
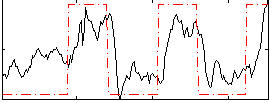 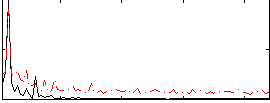 |
Correlation:0.61023
  |
Correlation:0.61023
  |
Correlation:0.61023
  |
Correlation:0.61023
  |
Correlation:0.61023
  |
Component:224; Jaccard:0.11193
 |
Component:369; Jaccard:0.13747
 |
Component:369; Jaccard:0.15974
 |
Component:369; Jaccard:0.16895
 |
Component:103; Jaccard:0.16397
 |
Component:103; Jaccard:0.15267
 |
Component:103; Jaccard:0.1338
 |
Correlation:0.61023
  |
Correlation:0.26588
  |
Correlation:0.26588
  |
Correlation:0.26588
  |
Correlation:-0.11385
  |
Correlation:-0.11385
  |
Correlation:-0.11385
  |
Component:009; Jaccard:0.10541
 |
Component:009; Jaccard:0.11951
 |
Component:103; Jaccard:0.13608
 |
Component:103; Jaccard:0.15831
 |
Component:369; Jaccard:0.15927
 |
Component:369; Jaccard:0.13501
 |
Component:369; Jaccard:0.10639
 |
Correlation:0.095543
  |
Correlation:0.095543
  |
Correlation:-0.11385
  |
Correlation:-0.11385
  |
Correlation:0.26588
  |
Correlation:0.26588
  |
Correlation:0.26588
  |
Component:321; Jaccard:0.097653
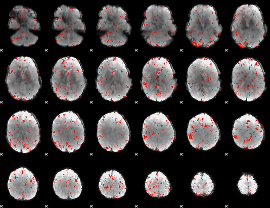 |
Component:186; Jaccard:0.11238
 |
Component:186; Jaccard:0.12973
 |
Component:186; Jaccard:0.13434
 |
Component:186; Jaccard:0.12819
 |
Component:186; Jaccard:0.11137
 |
Component:239; Jaccard:0.09761
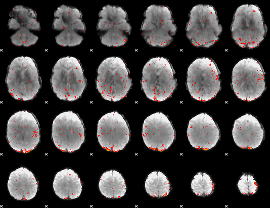 |
Correlation:0.10842
  |
Correlation:0.54992
  |
Correlation:0.54992
  |
Correlation:0.54992
  |
Correlation:0.54992
  |
Correlation:0.54992
  |
Correlation:0.25089
  |
Component:186; Jaccard:0.093318
 |
Component:103; Jaccard:0.10865
 |
Component:009; Jaccard:0.12853
 |
Component:009; Jaccard:0.12355
 |
Component:239; Jaccard:0.11397
 |
Component:239; Jaccard:0.11085
 |
Component:186; Jaccard:0.089374
 |
Correlation:0.54992
  |
Correlation:-0.11385
  |
Correlation:0.095543
  |
Correlation:0.095543
  |
Correlation:0.25089
  |
Correlation:0.25089
  |
Correlation:0.54992
  |
Component:197; Jaccard:0.090427
 |
Component:321; Jaccard:0.1042
 |
Component:197; Jaccard:0.10937
 |
Component:197; Jaccard:0.11159
 |
Component:197; Jaccard:0.10562
 |
Component:197; Jaccard:0.091965
 |
Component:197; Jaccard:0.075367
 |
Correlation:0.16432
  |
Correlation:0.10842
  |
Correlation:0.16432
  |
Correlation:0.16432
  |
Correlation:0.16432
  |
Correlation:0.16432
  |
Correlation:0.16432
  |
Component:103; Jaccard:0.086085
 |
Component:197; Jaccard:0.10205
 |
Component:321; Jaccard:0.10348
 |
Component:239; Jaccard:0.1037
 |
Component:009; Jaccard:0.10474
 |
Component:009; Jaccard:0.081896
 |
Component:009; Jaccard:0.06205
 |
Correlation:-0.11385
  |
Correlation:0.16432
  |
Correlation:0.10842
  |
Correlation:0.25089
  |
Correlation:0.095543
  |
Correlation:0.095543
  |
Correlation:0.095543
  |
Component:252; Jaccard:0.084592
 |
Component:252; Jaccard:0.089457
 |
Component:239; Jaccard:0.089548
 |
Component:321; Jaccard:0.090757
 |
Component:384; Jaccard:0.071423
 |
Component:384; Jaccard:0.060865
 |
Component:384; Jaccard:0.049071
 |
Correlation:-0.023376
  |
Correlation:-0.023376
  |
Correlation:0.25089
  |
Correlation:0.10842
  |
Correlation:0.28139
  |
Correlation:0.28139
  |
Correlation:0.28139
  |
Component:344; Jaccard:0.080241
 |
Component:344; Jaccard:0.084216
 |
Component:252; Jaccard:0.084788
 |
Component:334; Jaccard:0.077761
 |
Component:321; Jaccard:0.071345
 |
Component:075; Jaccard:0.060298
 |
Component:075; Jaccard:0.048689
 |
Correlation:0.24422
  |
Correlation:0.24422
  |
Correlation:-0.023376
  |
Correlation:0.18027
 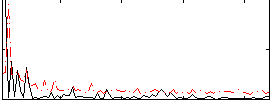 |
Correlation:0.10842
  |
Correlation:-0.06512
  |
Correlation:-0.06512
  |
Component:297; Jaccard:0.075645
 |
Component:334; Jaccard:0.079851
 |
Component:344; Jaccard:0.083354
 |
Component:384; Jaccard:0.077405
 |
Component:075; Jaccard:0.067794
 |
Component:334; Jaccard:0.056621
 |
Component:334; Jaccard:0.043248
 |
Correlation:-0.023718
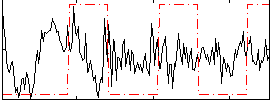 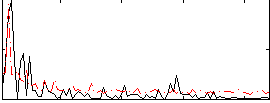 |
Correlation:0.18027
  |
Correlation:0.24422
  |
Correlation:0.28139
  |
Correlation:-0.06512
  |
Correlation:0.18027
  |
Correlation:0.18027
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
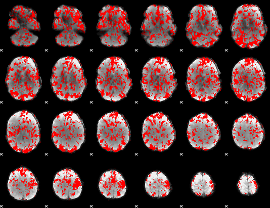 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
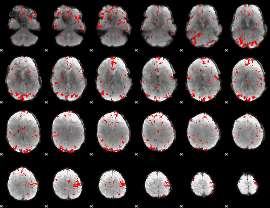 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
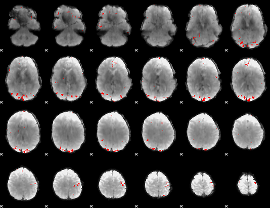 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
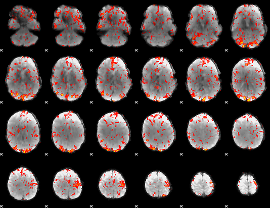 |
GLM Z-score result thresholded by 2.5
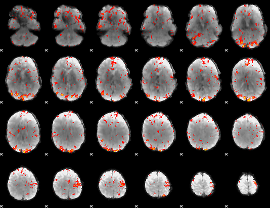 |
GLM Z-score result thresholded by 3
 |
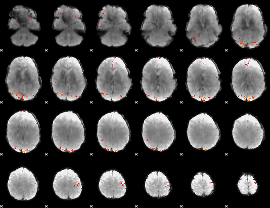
GLM Z-score result thresholded by 3.5
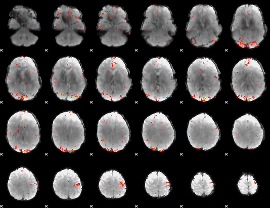 |
GLM Z-score result thresholded by 4
 |
Component:103; Jaccard:0.10295
 |
Component:103; Jaccard:0.13999
 |
Component:103; Jaccard:0.18693
 |
Component:103; Jaccard:0.22874
 |
Component:103; Jaccard:0.24325
 |
Component:103; Jaccard:0.21994
 |
Component:103; Jaccard:0.1705
 |
Correlation:0.34939
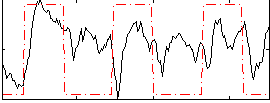  |
Correlation:0.34939
  |
Correlation:0.34939
  |
Correlation:0.34939
  |
Correlation:0.34939
  |
Correlation:0.34939
  |
Correlation:0.34939
  |
Component:076; Jaccard:0.10004
 |
Component:323; Jaccard:0.11912
 |
Component:323; Jaccard:0.12881
 |
Component:323; Jaccard:0.12284
 |
Component:323; Jaccard:0.098866
 |
Component:239; Jaccard:0.088799
 |
Component:239; Jaccard:0.071237
 |
Correlation:0.1262
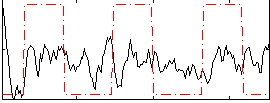  |
Correlation:0.19562
  |
Correlation:0.19562
  |
Correlation:0.19562
  |
Correlation:0.19562
  |
Correlation:-0.017143
  |
Correlation:-0.017143
  |
Component:323; Jaccard:0.099801
 |
Component:009; Jaccard:0.10848
 |
Component:009; Jaccard:0.11618
 |
Component:009; Jaccard:0.11212
 |
Component:239; Jaccard:0.097105
 |
Component:369; Jaccard:0.076687
 |
Component:369; Jaccard:0.056836
 |
Correlation:0.19562
  |
Correlation:0.15943
  |
Correlation:0.15943
  |
Correlation:0.15943
  |
Correlation:-0.017143
  |
Correlation:-0.15436
  |
Correlation:-0.15436
  |
Component:009; Jaccard:0.0972
 |
Component:076; Jaccard:0.10784
 |
Component:076; Jaccard:0.10782
 |
Component:369; Jaccard:0.10457
 |
Component:009; Jaccard:0.09565
 |
Component:009; Jaccard:0.073253
 |
Component:009; Jaccard:0.047332
 |
Correlation:0.15943
  |
Correlation:0.1262
  |
Correlation:0.1262
  |
Correlation:-0.15436
  |
Correlation:0.15943
  |
Correlation:0.15943
  |
Correlation:0.15943
  |
Component:297; Jaccard:0.092089
 |
Component:297; Jaccard:0.099599
 |
Component:369; Jaccard:0.10534
 |
Component:239; Jaccard:0.097159
 |
Component:369; Jaccard:0.091856
 |
Component:323; Jaccard:0.061692
 |
Component:224; Jaccard:0.045983
 |
Correlation:0.038544
  |
Correlation:0.038544
  |
Correlation:-0.15436
  |
Correlation:-0.017143
  |
Correlation:-0.15436
  |
Correlation:0.19562
  |
Correlation:-0.49789
  |
Component:108; Jaccard:0.089855
 |
Component:369; Jaccard:0.097863
 |
Component:026; Jaccard:0.098312
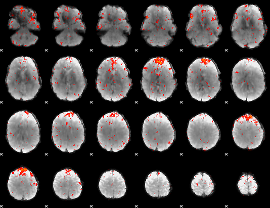 |
Component:076; Jaccard:0.093952
 |
Component:075; Jaccard:0.077106
 |
Component:224; Jaccard:0.060318
 |
Component:075; Jaccard:0.041938
 |
Correlation:0.059388
  |
Correlation:-0.15436
  |
Correlation:0.191
  |
Correlation:0.1262
  |
Correlation:0.14192
  |
Correlation:-0.49789
  |
Correlation:0.14192
  |
Component:369; Jaccard:0.087288
 |
Component:026; Jaccard:0.09747
 |
Component:297; Jaccard:0.098146
 |
Component:026; Jaccard:0.086624
 |
Component:224; Jaccard:0.069883
 |
Component:075; Jaccard:0.05801
 |
Component:323; Jaccard:0.032125
 |
Correlation:-0.15436
  |
Correlation:0.191
  |
Correlation:0.038544
  |
Correlation:0.191
  |
Correlation:-0.49789
  |
Correlation:0.14192
  |
Correlation:0.19562
  |
Component:026; Jaccard:0.087076
 |
Component:108; Jaccard:0.09431
 |
Component:108; Jaccard:0.090332
 |
Component:297; Jaccard:0.085578
 |
Component:076; Jaccard:0.069679
 |
Component:151; Jaccard:0.046092
 |
Component:384; Jaccard:0.030421
 |
Correlation:0.191
  |
Correlation:0.059388
  |
Correlation:0.059388
  |
Correlation:0.038544
  |
Correlation:0.1262
  |
Correlation:0.49332
  |
Correlation:-0.18201
  |
Component:283; Jaccard:0.07983
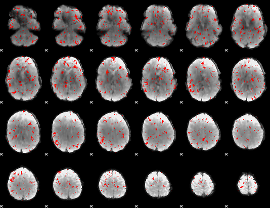 |
Component:283; Jaccard:0.081389
 |
Component:239; Jaccard:0.085605
 |
Component:075; Jaccard:0.084612
 |
Component:151; Jaccard:0.065028
 |
Component:076; Jaccard:0.045726
 |
Component:334; Jaccard:0.02903
 |
Correlation:0.006307
  |
Correlation:0.006307
  |
Correlation:-0.017143
  |
Correlation:0.14192
  |
Correlation:0.49332
  |
Correlation:0.1262
  |
Correlation:0.025376
  |
Component:105; Jaccard:0.076171
 |
Component:030; Jaccard:0.078575
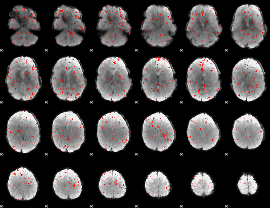 |
Component:075; Jaccard:0.082426
 |
Component:030; Jaccard:0.076754
 |
Component:297; Jaccard:0.064274
 |
Component:137; Jaccard:0.044219
 |
Component:186; Jaccard:0.028024
 |
Correlation:0.22115
 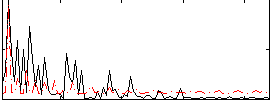 |
Correlation:0.14351
  |
Correlation:0.14192
  |
Correlation:0.14351
  |
Correlation:0.038544
  |
Correlation:0.10903
  |
Correlation:-0.25572
 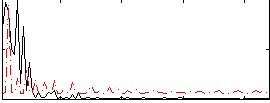 |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
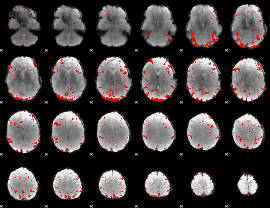 |
GLM binary result thresholded by 3.5
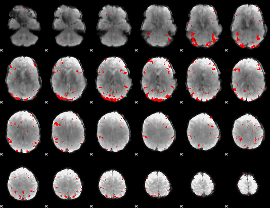 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
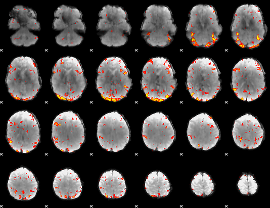 |
GLM Z-score result thresholded by 3.5
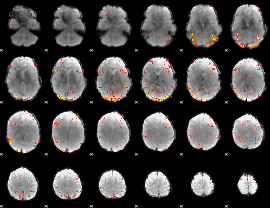 |
GLM Z-score result thresholded by 4
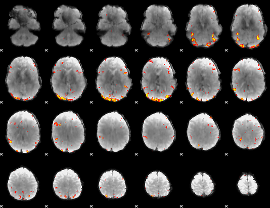 |
Component:155; Jaccard:0.12672
 |
Component:224; Jaccard:0.15775
 |
Component:224; Jaccard:0.21319
 |
Component:224; Jaccard:0.26915
 |
Component:224; Jaccard:0.30243
 |
Component:224; Jaccard:0.30465
 |
Component:224; Jaccard:0.28238
 |
Correlation:0.35162
  |
Correlation:0.60106
  |
Correlation:0.60106
  |
Correlation:0.60106
  |
Correlation:0.60106
  |
Correlation:0.60106
  |
Correlation:0.60106
  |
Component:224; Jaccard:0.11407
 |
Component:155; Jaccard:0.1567
 |
Component:155; Jaccard:0.18234
 |
Component:155; Jaccard:0.19187
 |
Component:129; Jaccard:0.18884
 |
Component:129; Jaccard:0.17921
 |
Component:129; Jaccard:0.15804
 |
Correlation:0.60106
  |
Correlation:0.35162
  |
Correlation:0.35162
  |
Correlation:0.35162
  |
Correlation:0.49232
  |
Correlation:0.49232
  |
Correlation:0.49232
  |
Component:129; Jaccard:0.11052
 |
Component:129; Jaccard:0.13718
 |
Component:129; Jaccard:0.16453
 |
Component:129; Jaccard:0.18506
 |
Component:155; Jaccard:0.17638
 |
Component:155; Jaccard:0.14637
 |
Component:155; Jaccard:0.11106
 |
Correlation:0.49232
  |
Correlation:0.49232
  |
Correlation:0.49232
  |
Correlation:0.49232
  |
Correlation:0.35162
  |
Correlation:0.35162
  |
Correlation:0.35162
  |
Component:197; Jaccard:0.097529
 |
Component:186; Jaccard:0.11333
 |
Component:186; Jaccard:0.13053
 |
Component:186; Jaccard:0.13785
 |
Component:186; Jaccard:0.13482
 |
Component:186; Jaccard:0.12199
 |
Component:186; Jaccard:0.10314
 |
Correlation:0.15732
  |
Correlation:0.43699
 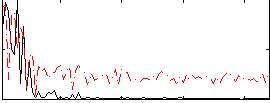 |
Correlation:0.43699
  |
Correlation:0.43699
  |
Correlation:0.43699
  |
Correlation:0.43699
  |
Correlation:0.43699
  |
Component:186; Jaccard:0.094216
 |
Component:257; Jaccard:0.11075
 |
Component:257; Jaccard:0.12103
 |
Component:257; Jaccard:0.12112
 |
Component:197; Jaccard:0.10921
 |
Component:197; Jaccard:0.092559
 |
Component:284; Jaccard:0.082724
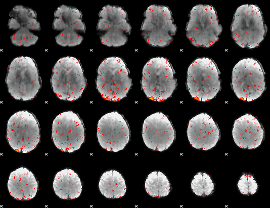 |
Correlation:0.43699
  |
Correlation:0.37292
 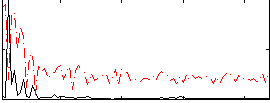 |
Correlation:0.37292
  |
Correlation:0.37292
  |
Correlation:0.15732
  |
Correlation:0.15732
  |
Correlation:0.35888
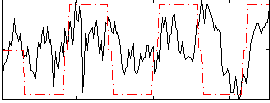  |
Component:257; Jaccard:0.094147
 |
Component:197; Jaccard:0.1087
 |
Component:197; Jaccard:0.11619
 |
Component:197; Jaccard:0.1181
 |
Component:257; Jaccard:0.10776
 |
Component:284; Jaccard:0.092233
 |
Component:197; Jaccard:0.076742
 |
Correlation:0.37292
  |
Correlation:0.15732
  |
Correlation:0.15732
  |
Correlation:0.15732
  |
Correlation:0.37292
  |
Correlation:0.35888
  |
Correlation:0.15732
  |
Component:329; Jaccard:0.09236
 |
Component:209; Jaccard:0.099166
 |
Component:209; Jaccard:0.105
 |
Component:284; Jaccard:0.10452
 |
Component:284; Jaccard:0.10339
 |
Component:257; Jaccard:0.086835
 |
Component:257; Jaccard:0.064254
 |
Correlation:0.25923
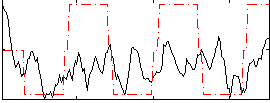  |
Correlation:0.42622
  |
Correlation:0.42622
  |
Correlation:0.35888
  |
Correlation:0.35888
  |
Correlation:0.37292
  |
Correlation:0.37292
  |
Component:209; Jaccard:0.088799
 |
Component:329; Jaccard:0.098813
 |
Component:329; Jaccard:0.10146
 |
Component:209; Jaccard:0.10373
 |
Component:209; Jaccard:0.094661
 |
Component:209; Jaccard:0.076606
 |
Component:209; Jaccard:0.060076
 |
Correlation:0.42622
  |
Correlation:0.25923
  |
Correlation:0.25923
  |
Correlation:0.42622
  |
Correlation:0.42622
  |
Correlation:0.42622
  |
Correlation:0.42622
  |
Component:210; Jaccard:0.084286
 |
Component:210; Jaccard:0.093354
 |
Component:284; Jaccard:0.098273
 |
Component:329; Jaccard:0.093905
 |
Component:303; Jaccard:0.083655
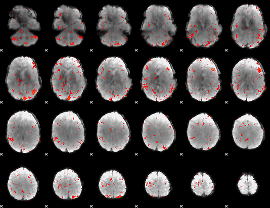 |
Component:303; Jaccard:0.068979
 |
Component:369; Jaccard:0.057013
 |
Correlation:0.23121
  |
Correlation:0.23121
  |
Correlation:0.35888
  |
Correlation:0.25923
  |
Correlation:0.25081
  |
Correlation:0.25081
  |
Correlation:0.22795
 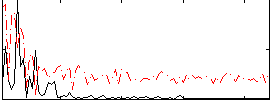 |
Component:203; Jaccard:0.082698
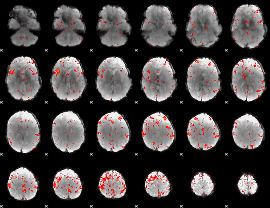 |
Component:303; Jaccard:0.091815
 |
Component:210; Jaccard:0.09672
 |
Component:303; Jaccard:0.093297
 |
Component:369; Jaccard:0.081117
 |
Component:369; Jaccard:0.068925
 |
Component:303; Jaccard:0.056417
 |
Correlation:0.32121
  |
Correlation:0.25081
  |
Correlation:0.23121
  |
Correlation:0.25081
  |
Correlation:0.22795
  |
Correlation:0.22795
  |
Correlation:0.25081
  |
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
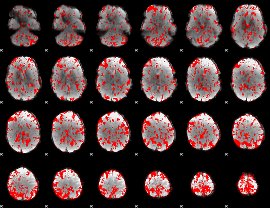 |
GLM binary result thresholded by 1.5
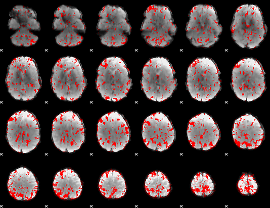 |
GLM binary result thresholded by 2
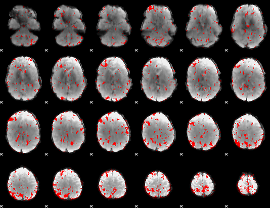 |
GLM binary result thresholded by 2.5
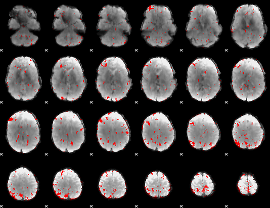 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
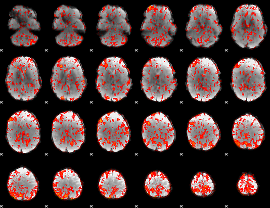 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
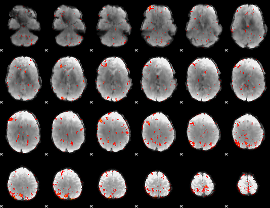 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:351; Jaccard:0.10494
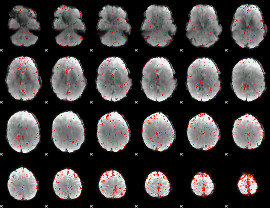 |
Component:351; Jaccard:0.11457
 |
Component:062; Jaccard:0.12038
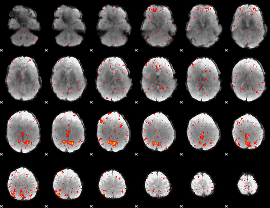 |
Component:268; Jaccard:0.12004
 |
Component:268; Jaccard:0.099063
 |
Component:268; Jaccard:0.066043
 |
Component:287; Jaccard:0.038105
 |
Correlation:0.096041
  |
Correlation:0.096041
  |
Correlation:0.13021
  |
Correlation:0.038034
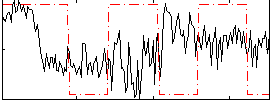  |
Correlation:0.038034
  |
Correlation:0.038034
  |
Correlation:0.16852
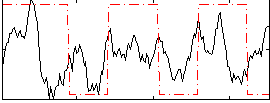  |
Component:267; Jaccard:0.10286
 |
Component:267; Jaccard:0.11201
 |
Component:268; Jaccard:0.11949
 |
Component:062; Jaccard:0.11717
 |
Component:062; Jaccard:0.094254
 |
Component:062; Jaccard:0.06226
 |
Component:268; Jaccard:0.036542
 |
Correlation:-0.13695
  |
Correlation:-0.13695
  |
Correlation:0.038034
  |
Correlation:0.13021
  |
Correlation:0.13021
  |
Correlation:0.13021
  |
Correlation:0.038034
  |
Component:287; Jaccard:0.094592
 |
Component:062; Jaccard:0.10994
 |
Component:287; Jaccard:0.11242
 |
Component:287; Jaccard:0.10909
 |
Component:287; Jaccard:0.087996
 |
Component:287; Jaccard:0.062022
 |
Component:062; Jaccard:0.031219
 |
Correlation:0.16852
  |
Correlation:0.13021
  |
Correlation:0.16852
  |
Correlation:0.16852
  |
Correlation:0.16852
  |
Correlation:0.16852
  |
Correlation:0.13021
  |
Component:062; Jaccard:0.094168
 |
Component:287; Jaccard:0.10597
 |
Component:351; Jaccard:0.11015
 |
Component:312; Jaccard:0.094287
 |
Component:378; Jaccard:0.069105
 |
Component:378; Jaccard:0.044406
 |
Component:378; Jaccard:0.023619
 |
Correlation:0.13021
  |
Correlation:0.16852
  |
Correlation:0.096041
  |
Correlation:0.22116
  |
Correlation:-0.13226
  |
Correlation:-0.13226
  |
Correlation:-0.13226
  |
Component:329; Jaccard:0.086695
 |
Component:268; Jaccard:0.10352
 |
Component:267; Jaccard:0.10915
 |
Component:351; Jaccard:0.091334
 |
Component:312; Jaccard:0.067486
 |
Component:267; Jaccard:0.038637
 |
Component:399; Jaccard:0.021928
 |
Correlation:-0.14764
  |
Correlation:0.038034
  |
Correlation:-0.13695
  |
Correlation:0.096041
  |
Correlation:0.22116
  |
Correlation:-0.13695
  |
Correlation:-0.012663
  |
Component:312; Jaccard:0.085704
 |
Component:312; Jaccard:0.097992
 |
Component:312; Jaccard:0.10333
 |
Component:267; Jaccard:0.089329
 |
Component:267; Jaccard:0.063464
 |
Component:399; Jaccard:0.036553
 |
Component:267; Jaccard:0.018847
 |
Correlation:0.22116
  |
Correlation:0.22116
  |
Correlation:0.22116
  |
Correlation:-0.13695
  |
Correlation:-0.13695
  |
Correlation:-0.012663
  |
Correlation:-0.13695
  |
Component:268; Jaccard:0.085075
 |
Component:279; Jaccard:0.0907
 |
Component:279; Jaccard:0.089465
 |
Component:378; Jaccard:0.081709
 |
Component:279; Jaccard:0.059491
 |
Component:312; Jaccard:0.035802
 |
Component:249; Jaccard:0.017425
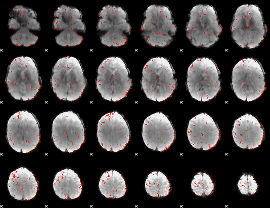 |
Correlation:0.038034
  |
Correlation:0.13753
  |
Correlation:0.13753
  |
Correlation:-0.13226
  |
Correlation:0.13753
  |
Correlation:0.22116
  |
Correlation:0.2627
  |
Component:279; Jaccard:0.084961
 |
Component:249; Jaccard:0.08782
 |
Component:249; Jaccard:0.086153
 |
Component:279; Jaccard:0.078305
 |
Component:351; Jaccard:0.057509
 |
Component:279; Jaccard:0.035588
 |
Component:312; Jaccard:0.017197
 |
Correlation:0.13753
  |
Correlation:0.2627
  |
Correlation:0.2627
  |
Correlation:0.13753
  |
Correlation:0.096041
  |
Correlation:0.13753
  |
Correlation:0.22116
  |
Component:247; Jaccard:0.081822
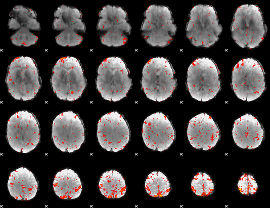 |
Component:329; Jaccard:0.086756
 |
Component:378; Jaccard:0.084062
 |
Component:249; Jaccard:0.074401
 |
Component:399; Jaccard:0.056134
 |
Component:051; Jaccard:0.031808
 |
Component:279; Jaccard:0.016548
 |
Correlation:0.061903
  |
Correlation:-0.14764
  |
Correlation:-0.13226
  |
Correlation:0.2627
  |
Correlation:-0.012663
  |
Correlation:0.11241
  |
Correlation:0.13753
  |
Component:363; Jaccard:0.081445
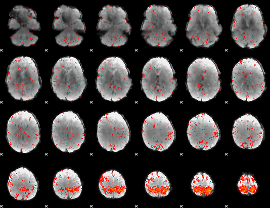 |
Component:247; Jaccard:0.083376
 |
Component:395; Jaccard:0.083045
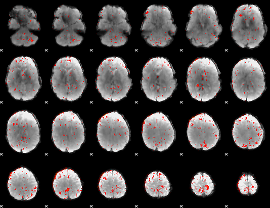 |
Component:395; Jaccard:0.073215
 |
Component:249; Jaccard:0.053023
 |
Component:249; Jaccard:0.030723
 |
Component:051; Jaccard:0.016443
 |
Correlation:0.1223
  |
Correlation:0.061903
  |
Correlation:-0.019335
 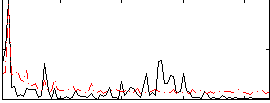 |
Correlation:-0.019335
  |
Correlation:0.2627
  |
Correlation:0.2627
  |
Correlation:0.11241
  |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:329; Jaccard:0.11951
 |
Component:329; Jaccard:0.13364
 |
Component:329; Jaccard:0.1387
 |
Component:268; Jaccard:0.14158
 |
Component:268; Jaccard:0.1426
 |
Component:378; Jaccard:0.13069
 |
Component:378; Jaccard:0.10638
 |
Correlation:0.33028
  |
Correlation:0.33028
  |
Correlation:0.33028
  |
Correlation:0.16643
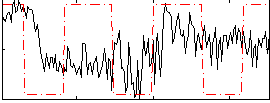  |
Correlation:0.16643
  |
Correlation:0.16414
  |
Correlation:0.16414
  |
Component:267; Jaccard:0.10866
 |
Component:267; Jaccard:0.12223
 |
Component:399; Jaccard:0.12946
 |
Component:378; Jaccard:0.13624
 |
Component:378; Jaccard:0.1393
 |
Component:268; Jaccard:0.12631
 |
Component:268; Jaccard:0.099129
 |
Correlation:0.32145
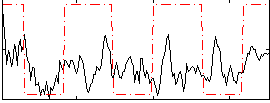  |
Correlation:0.32145
  |
Correlation:0.33804
  |
Correlation:0.16414
  |
Correlation:0.16414
  |
Correlation:0.16643
  |
Correlation:0.16643
  |
Component:287; Jaccard:0.10462
 |
Component:287; Jaccard:0.11867
 |
Component:267; Jaccard:0.12793
 |
Component:329; Jaccard:0.1345
 |
Component:287; Jaccard:0.12165
 |
Component:129; Jaccard:0.10919
 |
Component:129; Jaccard:0.089296
 |
Correlation:0.0016926
  |
Correlation:0.0016926
  |
Correlation:0.32145
  |
Correlation:0.33028
  |
Correlation:0.0016926
  |
Correlation:0.49638
  |
Correlation:0.49638
  |
Component:210; Jaccard:0.10369
 |
Component:210; Jaccard:0.11828
 |
Component:210; Jaccard:0.12749
 |
Component:399; Jaccard:0.12984
 |
Component:129; Jaccard:0.12077
 |
Component:287; Jaccard:0.1069
 |
Component:287; Jaccard:0.085331
 |
Correlation:0.24955
  |
Correlation:0.24955
  |
Correlation:0.24955
  |
Correlation:0.33804
  |
Correlation:0.49638
  |
Correlation:0.0016926
  |
Correlation:0.0016926
  |
Component:247; Jaccard:0.10194
 |
Component:399; Jaccard:0.11722
 |
Component:268; Jaccard:0.12731
 |
Component:287; Jaccard:0.12944
 |
Component:248; Jaccard:0.12021
 |
Component:399; Jaccard:0.10062
 |
Component:399; Jaccard:0.074424
 |
Correlation:0.10613
  |
Correlation:0.33804
  |
Correlation:0.16643
  |
Correlation:0.0016926
  |
Correlation:0.41811
  |
Correlation:0.33804
  |
Correlation:0.33804
  |
Component:399; Jaccard:0.10016
 |
Component:129; Jaccard:0.11202
 |
Component:287; Jaccard:0.12727
 |
Component:248; Jaccard:0.12568
 |
Component:399; Jaccard:0.1199
 |
Component:248; Jaccard:0.09781
 |
Component:248; Jaccard:0.068551
 |
Correlation:0.33804
  |
Correlation:0.49638
  |
Correlation:0.0016926
  |
Correlation:0.41811
  |
Correlation:0.33804
  |
Correlation:0.41811
  |
Correlation:0.41811
  |
Component:129; Jaccard:0.097087
 |
Component:247; Jaccard:0.111
 |
Component:378; Jaccard:0.12523
 |
Component:129; Jaccard:0.12446
 |
Component:329; Jaccard:0.1181
 |
Component:210; Jaccard:0.093838
 |
Component:210; Jaccard:0.065895
 |
Correlation:0.49638
  |
Correlation:0.10613
  |
Correlation:0.16414
  |
Correlation:0.49638
  |
Correlation:0.33028
  |
Correlation:0.24955
  |
Correlation:0.24955
  |
Component:351; Jaccard:0.096332
 |
Component:268; Jaccard:0.1092
 |
Component:129; Jaccard:0.12389
 |
Component:267; Jaccard:0.12436
 |
Component:210; Jaccard:0.11182
 |
Component:329; Jaccard:0.091031
 |
Component:329; Jaccard:0.065077
 |
Correlation:0.1927
  |
Correlation:0.16643
  |
Correlation:0.49638
  |
Correlation:0.32145
  |
Correlation:0.24955
  |
Correlation:0.33028
  |
Correlation:0.33028
  |
Component:279; Jaccard:0.091573
 |
Component:378; Jaccard:0.1081
 |
Component:248; Jaccard:0.11611
 |
Component:210; Jaccard:0.12391
 |
Component:267; Jaccard:0.108
 |
Component:062; Jaccard:0.087072
 |
Component:062; Jaccard:0.06459
 |
Correlation:0.068751
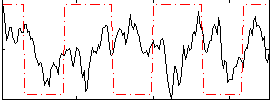 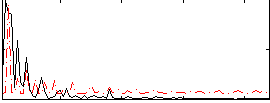 |
Correlation:0.16414
  |
Correlation:0.41811
  |
Correlation:0.24955
  |
Correlation:0.32145
  |
Correlation:0.056328
  |
Correlation:0.056328
  |
Component:378; Jaccard:0.089307
 |
Component:351; Jaccard:0.10428
 |
Component:247; Jaccard:0.11578
 |
Component:247; Jaccard:0.11258
 |
Component:062; Jaccard:0.099323
 |
Component:267; Jaccard:0.086831
 |
Component:267; Jaccard:0.059708
 |
Correlation:0.16414
  |
Correlation:0.1927
  |
Correlation:0.10613
  |
Correlation:0.10613
  |
Correlation:0.056328
  |
Correlation:0.32145
  |
Correlation:0.32145
  |
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
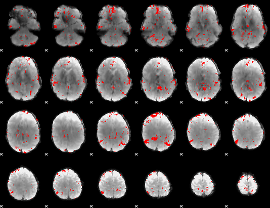 |
GLM binary result thresholded by 3
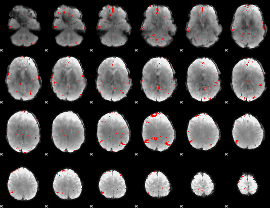 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
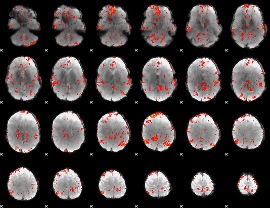 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
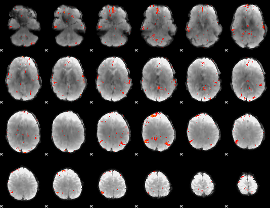 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:323; Jaccard:0.10636
 |
Component:323; Jaccard:0.12855
 |
Component:323; Jaccard:0.14401
 |
Component:323; Jaccard:0.14227
 |
Component:323; Jaccard:0.11643
 |
Component:323; Jaccard:0.081534
 |
Component:323; Jaccard:0.049758
 |
Correlation:0.26972
  |
Correlation:0.26972
  |
Correlation:0.26972
  |
Correlation:0.26972
  |
Correlation:0.26972
  |
Correlation:0.26972
  |
Correlation:0.26972
  |
Component:165; Jaccard:0.10023
 |
Component:059; Jaccard:0.11991
 |
Component:059; Jaccard:0.13264
 |
Component:059; Jaccard:0.12869
 |
Component:059; Jaccard:0.10071
 |
Component:059; Jaccard:0.062617
 |
Component:151; Jaccard:0.038559
 |
Correlation:0.29174
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.2949
  |
Correlation:0.50492
  |
Component:333; Jaccard:0.10021
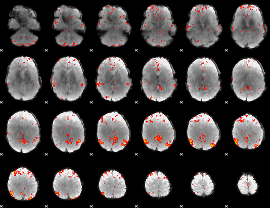 |
Component:333; Jaccard:0.11401
 |
Component:333; Jaccard:0.11659
 |
Component:333; Jaccard:0.1077
 |
Component:151; Jaccard:0.081183
 |
Component:151; Jaccard:0.060461
 |
Component:059; Jaccard:0.03539
 |
Correlation:0.28221
  |
Correlation:0.28221
  |
Correlation:0.28221
  |
Correlation:0.28221
  |
Correlation:0.50492
  |
Correlation:0.50492
  |
Correlation:0.2949
  |
Component:059; Jaccard:0.09951
 |
Component:165; Jaccard:0.11069
 |
Component:165; Jaccard:0.11387
 |
Component:165; Jaccard:0.10252
 |
Component:333; Jaccard:0.080896
 |
Component:333; Jaccard:0.051665
 |
Component:126; Jaccard:0.030086
 |
Correlation:0.2949
  |
Correlation:0.29174
  |
Correlation:0.29174
  |
Correlation:0.29174
  |
Correlation:0.28221
  |
Correlation:0.28221
  |
Correlation:0.33968
  |
Component:126; Jaccard:0.097108
 |
Component:126; Jaccard:0.10904
 |
Component:126; Jaccard:0.11173
 |
Component:126; Jaccard:0.10132
 |
Component:126; Jaccard:0.078279
 |
Component:126; Jaccard:0.050692
 |
Component:333; Jaccard:0.029589
 |
Correlation:0.33968
  |
Correlation:0.33968
  |
Correlation:0.33968
  |
Correlation:0.33968
  |
Correlation:0.33968
  |
Correlation:0.33968
  |
Correlation:0.28221
  |
Component:105; Jaccard:0.084597
 |
Component:105; Jaccard:0.088648
 |
Component:151; Jaccard:0.089552
 |
Component:151; Jaccard:0.090363
 |
Component:165; Jaccard:0.075863
 |
Component:165; Jaccard:0.047052
 |
Component:165; Jaccard:0.026027
 |
Correlation:0.2109
  |
Correlation:0.2109
  |
Correlation:0.50492
  |
Correlation:0.50492
  |
Correlation:0.29174
  |
Correlation:0.29174
  |
Correlation:0.29174
  |
Component:206; Jaccard:0.081724
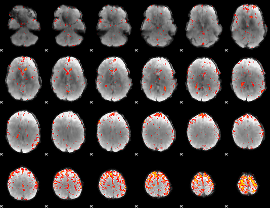 |
Component:311; Jaccard:0.081308
 |
Component:105; Jaccard:0.085391
 |
Component:311; Jaccard:0.0721
 |
Component:137; Jaccard:0.0553
 |
Component:137; Jaccard:0.037295
 |
Component:137; Jaccard:0.023184
 |
Correlation:0.015029
  |
Correlation:0.23355
  |
Correlation:0.2109
  |
Correlation:0.23355
  |
Correlation:0.13174
  |
Correlation:0.13174
  |
Correlation:0.13174
  |
Component:363; Jaccard:0.080289
 |
Component:151; Jaccard:0.08035
 |
Component:311; Jaccard:0.08118
 |
Component:105; Jaccard:0.069885
 |
Component:082; Jaccard:0.052329
 |
Component:082; Jaccard:0.034675
 |
Component:082; Jaccard:0.020342
 |
Correlation:0.073592
  |
Correlation:0.50492
  |
Correlation:0.23355
  |
Correlation:0.2109
  |
Correlation:0.29488
  |
Correlation:0.29488
  |
Correlation:0.29488
  |
Component:311; Jaccard:0.076479
 |
Component:082; Jaccard:0.076425
 |
Component:082; Jaccard:0.075732
 |
Component:082; Jaccard:0.068751
 |
Component:311; Jaccard:0.051201
 |
Component:311; Jaccard:0.031542
 |
Component:311; Jaccard:0.017906
 |
Correlation:0.23355
  |
Correlation:0.29488
  |
Correlation:0.29488
  |
Correlation:0.29488
  |
Correlation:0.23355
  |
Correlation:0.23355
  |
Correlation:0.23355
  |
Component:306; Jaccard:0.074272
 |
Component:306; Jaccard:0.075759
 |
Component:306; Jaccard:0.070995
 |
Component:137; Jaccard:0.066529
 |
Component:105; Jaccard:0.046769
 |
Component:105; Jaccard:0.025874
 |
Component:380; Jaccard:0.016351
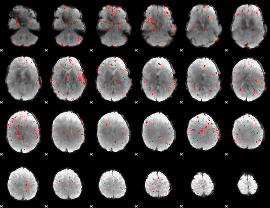 |
Correlation:0.0090353
  |
Correlation:0.0090353
  |
Correlation:0.0090353
  |
Correlation:0.13174
  |
Correlation:0.2109
  |
Correlation:0.2109
  |
Correlation:-0.05837
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































