Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
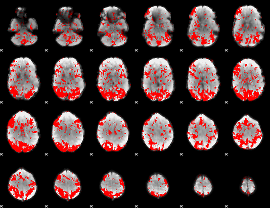
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
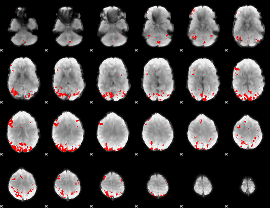
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
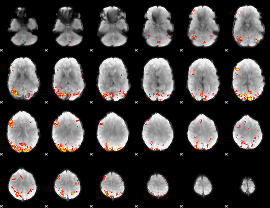
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:393; Jaccard:0.19302
 |
Component:393; Jaccard:0.2554
 |
Component:393; Jaccard:0.31536
 |
Component:393; Jaccard:0.35577
 |
Component:393; Jaccard:0.36693
 |
Component:393; Jaccard:0.34968
 |
Component:393; Jaccard:0.307
 |
Correlation:0.080905
  |
Correlation:0.080905
  |
Correlation:0.080905
  |
Correlation:0.080905
  |
Correlation:0.080905
  |
Correlation:0.080905
  |
Correlation:0.080905
  |
Component:262; Jaccard:0.15171
 |
Component:262; Jaccard:0.18261
 |
Component:262; Jaccard:0.20289
 |
Component:337; Jaccard:0.21804
 |
Component:337; Jaccard:0.21862
 |
Component:337; Jaccard:0.2023
 |
Component:337; Jaccard:0.17558
 |
Correlation:0.076761
  |
Correlation:0.076761
  |
Correlation:0.076761
  |
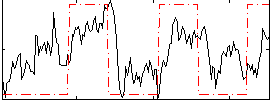
Correlation:0.43709
 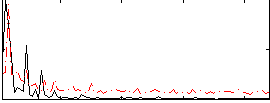 |
Correlation:0.43709
  |
Correlation:0.43709
  |
Correlation:0.43709
  |
Component:166; Jaccard:0.14536
 |
Component:166; Jaccard:0.1742
 |
Component:337; Jaccard:0.19949
 |
Component:262; Jaccard:0.20945
 |
Component:262; Jaccard:0.1972
 |
Component:262; Jaccard:0.17526
 |
Component:262; Jaccard:0.14779
 |
Correlation:0.11245
  |
Correlation:0.11245
  |
Correlation:0.43709
  |
Correlation:0.076761
  |
Correlation:0.076761
  |
Correlation:0.076761
  |
Correlation:0.076761
  |
Component:200; Jaccard:0.14042
 |
Component:337; Jaccard:0.16801
 |
Component:166; Jaccard:0.19092
 |
Component:166; Jaccard:0.19301
 |
Component:166; Jaccard:0.18258
 |
Component:166; Jaccard:0.16163
 |
Component:213; Jaccard:0.1341
 |
Correlation:0.068145
  |
Correlation:0.43709
  |
Correlation:0.11245
  |
Correlation:0.11245
  |
Correlation:0.11245
  |
Correlation:0.11245
  |
Correlation:0.39368
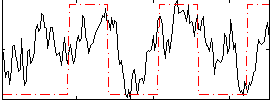  |
Component:337; Jaccard:0.1346
 |
Component:200; Jaccard:0.16126
 |
Component:200; Jaccard:0.17423
 |
Component:213; Jaccard:0.17452
 |
Component:213; Jaccard:0.17085
 |
Component:213; Jaccard:0.15673
 |
Component:166; Jaccard:0.13332
 |
Correlation:0.43709
  |
Correlation:0.068145
  |
Correlation:0.068145
  |
Correlation:0.39368
  |
Correlation:0.39368
  |
Correlation:0.39368
  |
Correlation:0.11245
  |
Component:391; Jaccard:0.11782
 |
Component:213; Jaccard:0.1422
 |
Component:213; Jaccard:0.16369
 |
Component:200; Jaccard:0.17432
 |
Component:200; Jaccard:0.16218
 |
Component:200; Jaccard:0.14403
 |
Component:200; Jaccard:0.11834
 |
Correlation:0.16918
 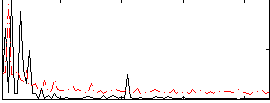 |
Correlation:0.39368
  |
Correlation:0.39368
  |
Correlation:0.068145
  |
Correlation:0.068145
  |
Correlation:0.068145
  |
Correlation:0.068145
  |
Component:213; Jaccard:0.11554
 |
Component:391; Jaccard:0.13116
 |
Component:391; Jaccard:0.13661
 |
Component:391; Jaccard:0.13097
 |
Component:043; Jaccard:0.12441
 |
Component:043; Jaccard:0.11598
 |
Component:043; Jaccard:0.10307
 |
Correlation:0.39368
  |
Correlation:0.16918
  |
Correlation:0.16918
  |
Correlation:0.16918
  |
Correlation:0.32421
  |
Correlation:0.32421
  |
Correlation:0.32421
  |
Component:380; Jaccard:0.10997
 |
Component:110; Jaccard:0.11953
 |
Component:110; Jaccard:0.12333
 |
Component:043; Jaccard:0.12542
 |
Component:391; Jaccard:0.11937
 |
Component:391; Jaccard:0.098271
 |
Component:391; Jaccard:0.081842
 |
Correlation:0.19798
 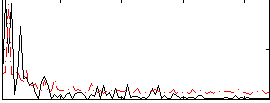 |
Correlation:0.1039
  |
Correlation:0.1039
  |
Correlation:0.32421
  |
Correlation:0.16918
  |
Correlation:0.16918
  |
Correlation:0.16918
  |
Component:110; Jaccard:0.10741
 |
Component:380; Jaccard:0.11798
 |
Component:380; Jaccard:0.12189
 |
Component:380; Jaccard:0.11656
 |
Component:380; Jaccard:0.10763
 |
Component:380; Jaccard:0.093482
 |
Component:380; Jaccard:0.076077
 |
Correlation:0.1039
  |
Correlation:0.19798
  |
Correlation:0.19798
  |
Correlation:0.19798
  |
Correlation:0.19798
  |
Correlation:0.19798
  |
Correlation:0.19798
  |
Component:052; Jaccard:0.096065
 |
Component:043; Jaccard:0.10779
 |
Component:043; Jaccard:0.12013
 |
Component:110; Jaccard:0.11597
 |
Component:052; Jaccard:0.097135
 |
Component:052; Jaccard:0.086228
 |
Component:052; Jaccard:0.071126
 |
Correlation:0.053192
  |
Correlation:0.32421
  |
Correlation:0.32421
  |
Correlation:0.1039
  |
Correlation:0.053192
  |
Correlation:0.053192
  |
Correlation:0.053192
  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
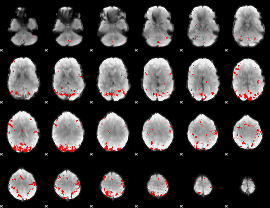
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
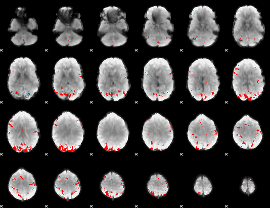
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
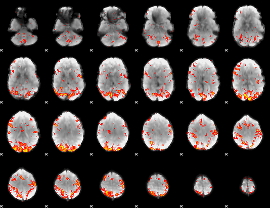 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
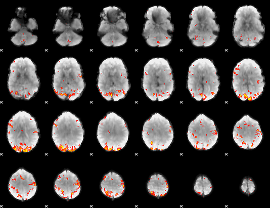 |
GLM Z-score result thresholded by 4
 |
Component:393; Jaccard:0.15456
 |
Component:393; Jaccard:0.20674
 |
Component:393; Jaccard:0.26678
 |
Component:393; Jaccard:0.31234
 |
Component:393; Jaccard:0.33082
 |
Component:393; Jaccard:0.31417
 |
Component:393; Jaccard:0.26846
 |
Correlation:0.09911
  |
Correlation:0.09911
  |
Correlation:0.09911
  |
Correlation:0.09911
  |
Correlation:0.09911
  |
Correlation:0.09911
  |
Correlation:0.09911
  |
Component:262; Jaccard:0.13934
 |
Component:262; Jaccard:0.17961
 |
Component:262; Jaccard:0.22072
 |
Component:262; Jaccard:0.24718
 |
Component:262; Jaccard:0.25225
 |
Component:262; Jaccard:0.22428
 |
Component:262; Jaccard:0.18222
 |
Correlation:0.085114
  |
Correlation:0.085114
  |
Correlation:0.085114
  |
Correlation:0.085114
  |
Correlation:0.085114
  |
Correlation:0.085114
  |
Correlation:0.085114
  |
Component:166; Jaccard:0.12216
 |
Component:166; Jaccard:0.14868
 |
Component:166; Jaccard:0.17241
 |
Component:166; Jaccard:0.1835
 |
Component:166; Jaccard:0.17759
 |
Component:166; Jaccard:0.16098
 |
Component:166; Jaccard:0.13096
 |
Correlation:0.096542
  |
Correlation:0.096542
  |
Correlation:0.096542
  |
Correlation:0.096542
  |
Correlation:0.096542
  |
Correlation:0.096542
  |
Correlation:0.096542
  |
Component:270; Jaccard:0.12144
 |
Component:270; Jaccard:0.14466
 |
Component:270; Jaccard:0.16479
 |
Component:270; Jaccard:0.17458
 |
Component:270; Jaccard:0.16606
 |
Component:270; Jaccard:0.14802
 |
Component:270; Jaccard:0.12091
 |
Correlation:0.34102
  |
Correlation:0.34102
  |
Correlation:0.34102
  |
Correlation:0.34102
  |
Correlation:0.34102
  |
Correlation:0.34102
  |
Correlation:0.34102
  |
Component:273; Jaccard:0.10932
 |
Component:273; Jaccard:0.12064
 |
Component:273; Jaccard:0.12638
 |
Component:273; Jaccard:0.12544
 |
Component:273; Jaccard:0.1139
 |
Component:380; Jaccard:0.097912
 |
Component:380; Jaccard:0.079161
 |
Correlation:0.27688
  |
Correlation:0.27688
  |
Correlation:0.27688
  |
Correlation:0.27688
  |
Correlation:0.27688
  |
Correlation:-0.043042
  |
Correlation:-0.043042
  |
Component:332; Jaccard:0.10559
 |
Component:332; Jaccard:0.11309
 |
Component:380; Jaccard:0.11622
 |
Component:380; Jaccard:0.11753
 |
Component:380; Jaccard:0.11245
 |
Component:273; Jaccard:0.094789
 |
Component:273; Jaccard:0.072406
 |
Correlation:0.17194
  |
Correlation:0.17194
  |
Correlation:-0.043042
  |
Correlation:-0.043042
  |
Correlation:-0.043042
  |
Correlation:0.27688
  |
Correlation:0.27688
  |
Component:380; Jaccard:0.098391
 |
Component:380; Jaccard:0.10978
 |
Component:332; Jaccard:0.11492
 |
Component:391; Jaccard:0.11093
 |
Component:391; Jaccard:0.10086
 |
Component:337; Jaccard:0.084199
 |
Component:337; Jaccard:0.067025
 |
Correlation:-0.043042
  |
Correlation:-0.043042
  |
Correlation:0.17194
  |
Correlation:0.055291
  |
Correlation:0.055291
  |
Correlation:-0.26309
  |
Correlation:-0.26309
  |
Component:200; Jaccard:0.095941
 |
Component:391; Jaccard:0.10616
 |
Component:200; Jaccard:0.11207
 |
Component:200; Jaccard:0.10862
 |
Component:200; Jaccard:0.10037
 |
Component:391; Jaccard:0.084007
 |
Component:200; Jaccard:0.065514
 |
Correlation:-0.093139
  |
Correlation:0.055291
  |
Correlation:-0.093139
  |
Correlation:-0.093139
  |
Correlation:-0.093139
  |
Correlation:0.055291
  |
Correlation:-0.093139
  |
Component:205; Jaccard:0.095759
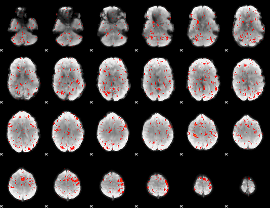 |
Component:200; Jaccard:0.10589
 |
Component:391; Jaccard:0.11203
 |
Component:005; Jaccard:0.1079
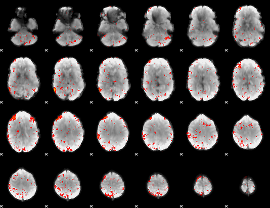 |
Component:005; Jaccard:0.098203
 |
Component:200; Jaccard:0.083959
 |
Component:378; Jaccard:0.065116
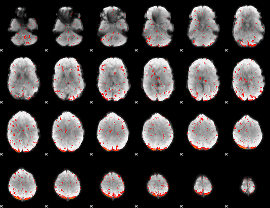 |
Correlation:0.32512
  |
Correlation:-0.093139
  |
Correlation:0.055291
  |
Correlation:0.098812
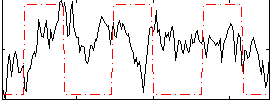  |
Correlation:0.098812
  |
Correlation:-0.093139
  |
Correlation:0.34774
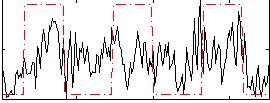  |
Component:391; Jaccard:0.094839
 |
Component:042; Jaccard:0.10479
 |
Component:042; Jaccard:0.11029
 |
Component:332; Jaccard:0.10719
 |
Component:042; Jaccard:0.096416
 |
Component:005; Jaccard:0.081884
 |
Component:391; Jaccard:0.064437
 |
Correlation:0.055291
  |
Correlation:0.2796
  |
Correlation:0.2796
  |
Correlation:0.17194
  |
Correlation:0.2796
  |
Correlation:0.098812
  |
Correlation:0.055291
  |
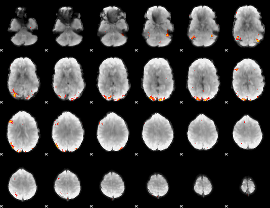
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
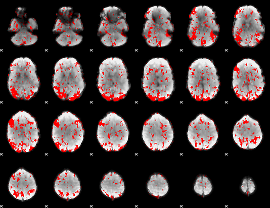 |
GLM binary result thresholded by 1.5
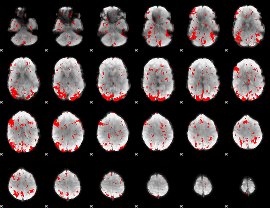 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
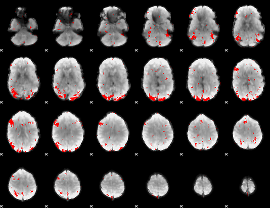 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
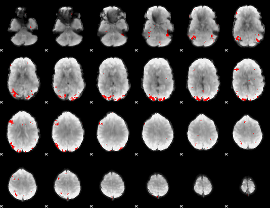 |
GLM binary result thresholded by 4
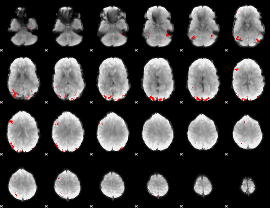 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
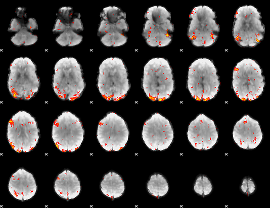 |
GLM Z-score result thresholded by 3
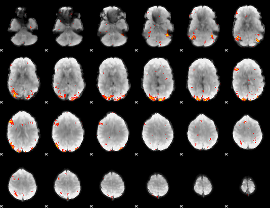 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:337; Jaccard:0.1721
 |
Component:043; Jaccard:0.21981
 |
Component:043; Jaccard:0.25735
 |
Component:043; Jaccard:0.26677
 |
Component:043; Jaccard:0.25393
 |
Component:043; Jaccard:0.21849
 |
Component:043; Jaccard:0.17689
 |
Correlation:0.37979
 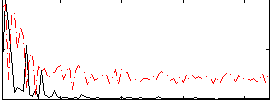 |
Correlation:0.28371
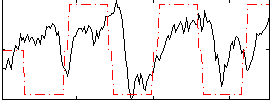 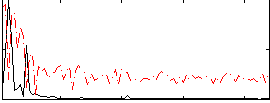 |
Correlation:0.28371
  |
Correlation:0.28371
  |
Correlation:0.28371
  |
Correlation:0.28371
  |
Correlation:0.28371
  |
Component:043; Jaccard:0.17195
 |
Component:337; Jaccard:0.20859
 |
Component:337; Jaccard:0.23308
 |
Component:337; Jaccard:0.22599
 |
Component:337; Jaccard:0.19831
 |
Component:337; Jaccard:0.16517
 |
Component:337; Jaccard:0.12935
 |
Correlation:0.28371
  |
Correlation:0.37979
  |
Correlation:0.37979
  |
Correlation:0.37979
  |
Correlation:0.37979
  |
Correlation:0.37979
  |
Correlation:0.37979
  |
Component:200; Jaccard:0.13822
 |
Component:213; Jaccard:0.15892
 |
Component:213; Jaccard:0.17088
 |
Component:213; Jaccard:0.15983
 |
Component:213; Jaccard:0.13372
 |
Component:213; Jaccard:0.10454
 |
Component:213; Jaccard:0.081718
 |
Correlation:0.087482
  |
Correlation:0.36985
  |
Correlation:0.36985
  |
Correlation:0.36985
  |
Correlation:0.36985
  |
Correlation:0.36985
  |
Correlation:0.36985
  |
Component:213; Jaccard:0.13772
 |
Component:200; Jaccard:0.1508
 |
Component:200; Jaccard:0.15084
 |
Component:200; Jaccard:0.13196
 |
Component:358; Jaccard:0.10638
 |
Component:358; Jaccard:0.094509
 |
Component:358; Jaccard:0.081102
 |
Correlation:0.36985
  |
Correlation:0.087482
  |
Correlation:0.087482
  |
Correlation:0.087482
  |
Correlation:0.17527
  |
Correlation:0.17527
  |
Correlation:0.17527
  |
Component:393; Jaccard:0.10236
 |
Component:393; Jaccard:0.11084
 |
Component:393; Jaccard:0.1132
 |
Component:358; Jaccard:0.11498
 |
Component:200; Jaccard:0.10518
 |
Component:200; Jaccard:0.077211
 |
Component:200; Jaccard:0.058253
 |
Correlation:-0.009875
  |
Correlation:-0.009875
  |
Correlation:-0.009875
  |
Correlation:0.17527
  |
Correlation:0.087482
  |
Correlation:0.087482
  |
Correlation:0.087482
  |
Component:391; Jaccard:0.084247
 |
Component:358; Jaccard:0.10189
 |
Component:358; Jaccard:0.1128
 |
Component:393; Jaccard:0.10395
 |
Component:393; Jaccard:0.084249
 |
Component:393; Jaccard:0.063288
 |
Component:393; Jaccard:0.04738
 |
Correlation:0.061776
  |
Correlation:0.17527
  |
Correlation:0.17527
  |
Correlation:-0.009875
  |
Correlation:-0.009875
  |
Correlation:-0.009875
  |
Correlation:-0.009875
  |
Component:094; Jaccard:0.083578
 |
Component:094; Jaccard:0.086789
 |
Component:094; Jaccard:0.086175
 |
Component:094; Jaccard:0.074375
 |
Component:094; Jaccard:0.060215
 |
Component:034; Jaccard:0.04827
 |
Component:034; Jaccard:0.038964
 |
Correlation:0.21537
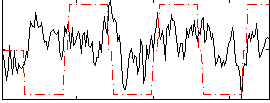 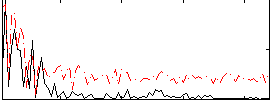 |
Correlation:0.21537
  |
Correlation:0.21537
  |
Correlation:0.21537
  |
Correlation:0.21537
  |
Correlation:0.24461
  |
Correlation:0.24461
  |
Component:358; Jaccard:0.083291
 |
Component:391; Jaccard:0.085149
 |
Component:391; Jaccard:0.080633
 |
Component:370; Jaccard:0.06878
 |
Component:370; Jaccard:0.058814
 |
Component:094; Jaccard:0.04726
 |
Component:094; Jaccard:0.037788
 |
Correlation:0.17527
  |
Correlation:0.061776
  |
Correlation:0.061776
  |
Correlation:-0.23657
 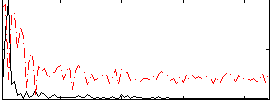 |
Correlation:-0.23657
  |
Correlation:0.21537
  |
Correlation:0.21537
  |
Component:370; Jaccard:0.077804
 |
Component:370; Jaccard:0.079262
 |
Component:370; Jaccard:0.075453
 |
Component:391; Jaccard:0.068266
 |
Component:034; Jaccard:0.058147
 |
Component:370; Jaccard:0.046218
 |
Component:370; Jaccard:0.035239
 |
Correlation:-0.23657
  |
Correlation:-0.23657
  |
Correlation:-0.23657
  |
Correlation:0.061776
  |
Correlation:0.24461
  |
Correlation:-0.23657
  |
Correlation:-0.23657
  |
Component:121; Jaccard:0.07427
 |
Component:121; Jaccard:0.073654
 |
Component:052; Jaccard:0.072077
 |
Component:052; Jaccard:0.066304
 |
Component:391; Jaccard:0.058104
 |
Component:391; Jaccard:0.045182
 |
Component:391; Jaccard:0.034314
 |
Correlation:0.16044
 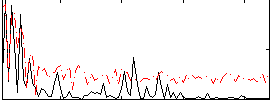 |
Correlation:0.16044
  |
Correlation:-0.015921
  |
Correlation:-0.015921
  |
Correlation:0.061776
  |
Correlation:0.061776
  |
Correlation:0.061776
  |
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:091; Jaccard:0.12087
 |
Component:091; Jaccard:0.15483
 |
Component:091; Jaccard:0.19464
 |
Component:091; Jaccard:0.23447
 |
Component:091; Jaccard:0.26006
 |
Component:091; Jaccard:0.25592
 |
Component:091; Jaccard:0.22522
 |
Correlation:0.36633
  |
Correlation:0.36633
  |
Correlation:0.36633
  |
Correlation:0.36633
  |
Correlation:0.36633
  |
Correlation:0.36633
  |
Correlation:0.36633
  |
Component:069; Jaccard:0.11238
 |
Component:366; Jaccard:0.13901
 |
Component:366; Jaccard:0.17204
 |
Component:366; Jaccard:0.20209
 |
Component:366; Jaccard:0.21722
 |
Component:366; Jaccard:0.20726
 |
Component:366; Jaccard:0.18217
 |
Correlation:0.35411
  |
Correlation:0.30137
  |
Correlation:0.30137
  |
Correlation:0.30137
  |
Correlation:0.30137
  |
Correlation:0.30137
  |
Correlation:0.30137
  |
Component:366; Jaccard:0.10954
 |
Component:069; Jaccard:0.13898
 |
Component:069; Jaccard:0.16528
 |
Component:069; Jaccard:0.18723
 |
Component:069; Jaccard:0.19461
 |
Component:069; Jaccard:0.17987
 |
Component:069; Jaccard:0.15117
 |
Correlation:0.30137
  |
Correlation:0.35411
  |
Correlation:0.35411
  |
Correlation:0.35411
  |
Correlation:0.35411
  |
Correlation:0.35411
  |
Correlation:0.35411
  |
Component:162; Jaccard:0.10182
 |
Component:219; Jaccard:0.12326
 |
Component:219; Jaccard:0.14687
 |
Component:219; Jaccard:0.16564
 |
Component:099; Jaccard:0.17305
 |
Component:099; Jaccard:0.16596
 |
Component:099; Jaccard:0.14619
 |
Correlation:0.46486
  |
Correlation:0.21171
  |
Correlation:0.21171
  |
Correlation:0.21171
  |
Correlation:0.38197
  |
Correlation:0.38197
  |
Correlation:0.38197
  |
Component:397; Jaccard:0.10046
 |
Component:099; Jaccard:0.12149
 |
Component:099; Jaccard:0.14604
 |
Component:099; Jaccard:0.16527
 |
Component:219; Jaccard:0.17116
 |
Component:219; Jaccard:0.16274
 |
Component:219; Jaccard:0.14292
 |
Correlation:0.30407
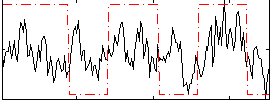 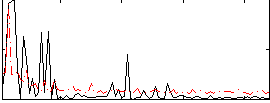 |
Correlation:0.38197
  |
Correlation:0.38197
  |
Correlation:0.38197
  |
Correlation:0.21171
  |
Correlation:0.21171
  |
Correlation:0.21171
  |
Component:219; Jaccard:0.10024
 |
Component:397; Jaccard:0.11808
 |
Component:397; Jaccard:0.1338
 |
Component:397; Jaccard:0.14743
 |
Component:397; Jaccard:0.14762
 |
Component:190; Jaccard:0.13635
 |
Component:190; Jaccard:0.1197
 |
Correlation:0.21171
  |
Correlation:0.30407
  |
Correlation:0.30407
  |
Correlation:0.30407
  |
Correlation:0.30407
  |
Correlation:0.30917
  |
Correlation:0.30917
  |
Component:099; Jaccard:0.098571
 |
Component:162; Jaccard:0.11347
 |
Component:190; Jaccard:0.12425
 |
Component:190; Jaccard:0.13985
 |
Component:190; Jaccard:0.14678
 |
Component:397; Jaccard:0.13278
 |
Component:397; Jaccard:0.11023
 |
Correlation:0.38197
  |
Correlation:0.46486
  |
Correlation:0.30917
  |
Correlation:0.30917
  |
Correlation:0.30917
  |
Correlation:0.30407
  |
Correlation:0.30407
  |
Component:293; Jaccard:0.094916
 |
Component:190; Jaccard:0.10687
 |
Component:162; Jaccard:0.12311
 |
Component:162; Jaccard:0.12584
 |
Component:162; Jaccard:0.1178
 |
Component:191; Jaccard:0.10223
 |
Component:191; Jaccard:0.083263
 |
Correlation:0.11283
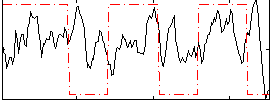 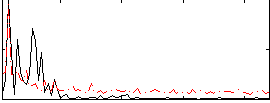 |
Correlation:0.30917
  |
Correlation:0.46486
  |
Correlation:0.46486
  |
Correlation:0.46486
  |
Correlation:0.46288
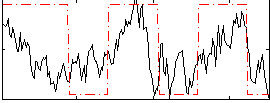  |
Correlation:0.46288
  |
Component:175; Jaccard:0.090059
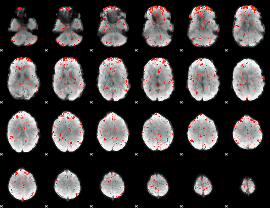 |
Component:293; Jaccard:0.10415
 |
Component:293; Jaccard:0.11105
 |
Component:193; Jaccard:0.11371
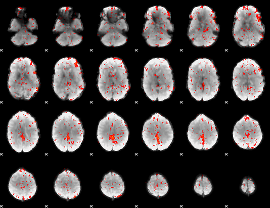 |
Component:191; Jaccard:0.111
 |
Component:162; Jaccard:0.10175
 |
Component:291; Jaccard:0.080746
 |
Correlation:0.4494
  |
Correlation:0.11283
  |
Correlation:0.11283
  |
Correlation:0.25993
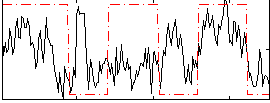  |
Correlation:0.46288
  |
Correlation:0.46486
  |
Correlation:0.034666
  |
Component:291; Jaccard:0.08975
 |
Component:175; Jaccard:0.099539
 |
Component:193; Jaccard:0.10975
 |
Component:147; Jaccard:0.11161
 |
Component:193; Jaccard:0.10876
 |
Component:147; Jaccard:0.09396
 |
Component:162; Jaccard:0.07973
 |
Correlation:0.034666
  |
Correlation:0.4494
  |
Correlation:0.25993
  |
Correlation:0.23478
  |
Correlation:0.25993
  |
Correlation:0.23478
  |
Correlation:0.46486
  |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
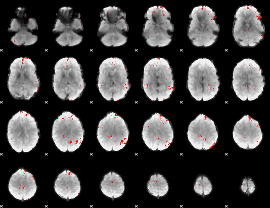 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
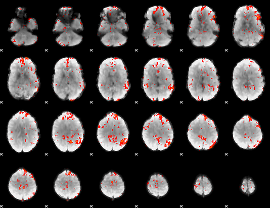 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
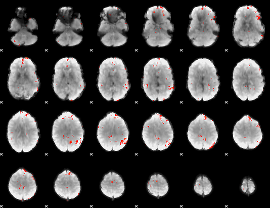 |
GLM Z-score result thresholded by 4
 |
Component:091; Jaccard:0.12683
 |
Component:091; Jaccard:0.15289
 |
Component:091; Jaccard:0.17256
 |
Component:091; Jaccard:0.16359
 |
Component:091; Jaccard:0.12676
 |
Component:091; Jaccard:0.080015
 |
Component:091; Jaccard:0.040144
 |
Correlation:-0.23515
 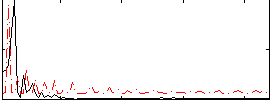 |
Correlation:-0.23515
  |
Correlation:-0.23515
  |
Correlation:-0.23515
  |
Correlation:-0.23515
  |
Correlation:-0.23515
  |
Correlation:-0.23515
  |
Component:366; Jaccard:0.11411
 |
Component:366; Jaccard:0.13737
 |
Component:366; Jaccard:0.1516
 |
Component:366; Jaccard:0.14388
 |
Component:366; Jaccard:0.1126
 |
Component:366; Jaccard:0.073625
 |
Component:366; Jaccard:0.038312
 |
Correlation:-0.22833
  |
Correlation:-0.22833
  |
Correlation:-0.22833
  |
Correlation:-0.22833
  |
Correlation:-0.22833
  |
Correlation:-0.22833
  |
Correlation:-0.22833
  |
Component:069; Jaccard:0.10252
 |
Component:069; Jaccard:0.11816
 |
Component:190; Jaccard:0.12895
 |
Component:190; Jaccard:0.12276
 |
Component:190; Jaccard:0.10111
 |
Component:190; Jaccard:0.066335
 |
Component:190; Jaccard:0.033423
 |
Correlation:-0.22372
  |
Correlation:-0.22372
  |
Correlation:-0.39722
  |
Correlation:-0.39722
  |
Correlation:-0.39722
  |
Correlation:-0.39722
  |
Correlation:-0.39722
  |
Component:190; Jaccard:0.10102
 |
Component:190; Jaccard:0.11713
 |
Component:069; Jaccard:0.12399
 |
Component:069; Jaccard:0.11251
 |
Component:291; Jaccard:0.085154
 |
Component:291; Jaccard:0.054387
 |
Component:291; Jaccard:0.028295
 |
Correlation:-0.39722
  |
Correlation:-0.39722
  |
Correlation:-0.22372
  |
Correlation:-0.22372
  |
Correlation:-0.1029
  |
Correlation:-0.1029
  |
Correlation:-0.1029
  |
Component:291; Jaccard:0.10009
 |
Component:291; Jaccard:0.10976
 |
Component:291; Jaccard:0.11273
 |
Component:291; Jaccard:0.10644
 |
Component:069; Jaccard:0.084405
 |
Component:219; Jaccard:0.050653
 |
Component:219; Jaccard:0.026176
 |
Correlation:-0.1029
  |
Correlation:-0.1029
  |
Correlation:-0.1029
  |
Correlation:-0.1029
  |
Correlation:-0.22372
  |
Correlation:-0.24012
  |
Correlation:-0.24012
  |
Component:219; Jaccard:0.092858
 |
Component:219; Jaccard:0.10464
 |
Component:219; Jaccard:0.11024
 |
Component:219; Jaccard:0.10022
 |
Component:219; Jaccard:0.076367
 |
Component:069; Jaccard:0.049092
 |
Component:182; Jaccard:0.025508
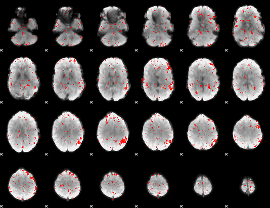 |
Correlation:-0.24012
  |
Correlation:-0.24012
  |
Correlation:-0.24012
  |
Correlation:-0.24012
  |
Correlation:-0.24012
  |
Correlation:-0.22372
  |
Correlation:-0.087843
  |
Component:193; Jaccard:0.09163
 |
Component:193; Jaccard:0.099139
 |
Component:133; Jaccard:0.10204
 |
Component:133; Jaccard:0.095106
 |
Component:099; Jaccard:0.074337
 |
Component:099; Jaccard:0.048108
 |
Component:397; Jaccard:0.02425
 |
Correlation:-0.16382
  |
Correlation:-0.16382
  |
Correlation:-0.016309
  |
Correlation:-0.016309
  |
Correlation:-0.30546
  |
Correlation:-0.30546
  |
Correlation:-0.31793
  |
Component:397; Jaccard:0.089211
 |
Component:397; Jaccard:0.098309
 |
Component:193; Jaccard:0.099927
 |
Component:099; Jaccard:0.092491
 |
Component:133; Jaccard:0.073463
 |
Component:133; Jaccard:0.047569
 |
Component:099; Jaccard:0.023979
 |
Correlation:-0.31793
  |
Correlation:-0.31793
  |
Correlation:-0.16382
  |
Correlation:-0.30546
  |
Correlation:-0.016309
  |
Correlation:-0.016309
  |
Correlation:-0.30546
  |
Component:147; Jaccard:0.088463
 |
Component:133; Jaccard:0.096158
 |
Component:099; Jaccard:0.099248
 |
Component:397; Jaccard:0.089341
 |
Component:235; Jaccard:0.070762
 |
Component:182; Jaccard:0.046436
 |
Component:133; Jaccard:0.023531
 |
Correlation:-0.040629
  |
Correlation:-0.016309
  |
Correlation:-0.30546
  |
Correlation:-0.31793
  |
Correlation:0.093175
  |
Correlation:-0.087843
  |
Correlation:-0.016309
  |
Component:133; Jaccard:0.085421
 |
Component:147; Jaccard:0.095539
 |
Component:397; Jaccard:0.098231
 |
Component:193; Jaccard:0.08919
 |
Component:397; Jaccard:0.069178
 |
Component:397; Jaccard:0.045512
 |
Component:235; Jaccard:0.022901
 |
Correlation:-0.016309
  |
Correlation:-0.040629
  |
Correlation:-0.31793
  |
Correlation:-0.16382
  |
Correlation:-0.31793
  |
Correlation:-0.31793
  |
Correlation:0.093175
  |
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
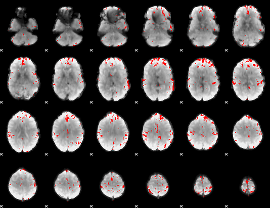 |
GLM Z-score result thresholded by 1
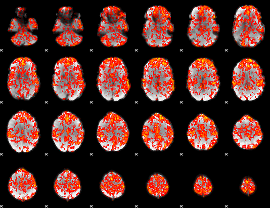 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
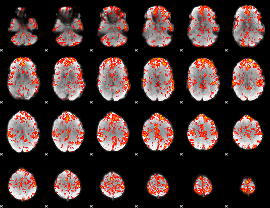 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
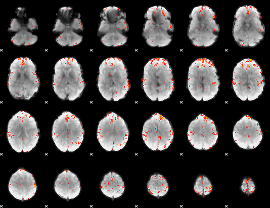 |
Component:162; Jaccard:0.10083
 |
Component:162; Jaccard:0.11526
 |
Component:091; Jaccard:0.13436
 |
Component:091; Jaccard:0.15584
 |
Component:091; Jaccard:0.16339
 |
Component:091; Jaccard:0.15751
 |
Component:099; Jaccard:0.13412
 |
Correlation:0.45547
  |
Correlation:0.45547
  |
Correlation:0.32626
  |
Correlation:0.32626
  |
Correlation:0.32626
  |
Correlation:0.32626
  |
Correlation:0.37287
  |
Component:293; Jaccard:0.097458
 |
Component:091; Jaccard:0.11416
 |
Component:189; Jaccard:0.13145
 |
Component:189; Jaccard:0.14665
 |
Component:189; Jaccard:0.1533
 |
Component:189; Jaccard:0.14942
 |
Component:091; Jaccard:0.13271
 |
Correlation:0.17058
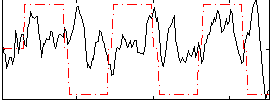  |
Correlation:0.32626
  |
Correlation:0.35368
  |
Correlation:0.35368
  |
Correlation:0.35368
  |
Correlation:0.35368
  |
Correlation:0.32626
  |
Component:189; Jaccard:0.096285
 |
Component:189; Jaccard:0.11331
 |
Component:069; Jaccard:0.13083
 |
Component:069; Jaccard:0.14588
 |
Component:099; Jaccard:0.15178
 |
Component:099; Jaccard:0.14936
 |
Component:189; Jaccard:0.13161
 |
Correlation:0.35368
  |
Correlation:0.35368
  |
Correlation:0.31343
 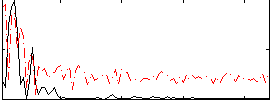 |
Correlation:0.31343
  |
Correlation:0.37287
  |
Correlation:0.37287
  |
Correlation:0.35368
  |
Component:091; Jaccard:0.095157
 |
Component:069; Jaccard:0.11203
 |
Component:162; Jaccard:0.1288
 |
Component:099; Jaccard:0.1405
 |
Component:069; Jaccard:0.15167
 |
Component:069; Jaccard:0.144
 |
Component:069; Jaccard:0.11946
 |
Correlation:0.32626
  |
Correlation:0.31343
  |
Correlation:0.45547
  |
Correlation:0.37287
  |
Correlation:0.31343
  |
Correlation:0.31343
  |
Correlation:0.31343
  |
Component:069; Jaccard:0.094067
 |
Component:293; Jaccard:0.11183
 |
Component:293; Jaccard:0.12627
 |
Component:293; Jaccard:0.13655
 |
Component:162; Jaccard:0.1382
 |
Component:293; Jaccard:0.12795
 |
Component:162; Jaccard:0.10572
 |
Correlation:0.31343
  |
Correlation:0.17058
  |
Correlation:0.17058
  |
Correlation:0.17058
  |
Correlation:0.45547
  |
Correlation:0.17058
  |
Correlation:0.45547
  |
Component:305; Jaccard:0.087258
 |
Component:099; Jaccard:0.10135
 |
Component:099; Jaccard:0.12184
 |
Component:162; Jaccard:0.13649
 |
Component:293; Jaccard:0.13798
 |
Component:162; Jaccard:0.12676
 |
Component:219; Jaccard:0.10541
 |
Correlation:0.14234
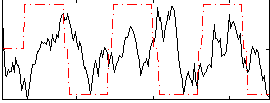 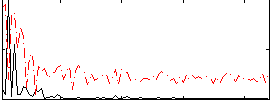 |
Correlation:0.37287
  |
Correlation:0.37287
  |
Correlation:0.45547
  |
Correlation:0.17058
  |
Correlation:0.45547
  |
Correlation:0.24507
  |
Component:126; Jaccard:0.084537
 |
Component:366; Jaccard:0.098804
 |
Component:366; Jaccard:0.11414
 |
Component:366; Jaccard:0.12776
 |
Component:366; Jaccard:0.13024
 |
Component:219; Jaccard:0.12408
 |
Component:293; Jaccard:0.10409
 |
Correlation:0.23574
  |
Correlation:0.28731
  |
Correlation:0.28731
  |
Correlation:0.28731
  |
Correlation:0.28731
  |
Correlation:0.24507
  |
Correlation:0.17058
  |
Component:366; Jaccard:0.08389
 |
Component:305; Jaccard:0.098241
 |
Component:219; Jaccard:0.11172
 |
Component:219; Jaccard:0.1259
 |
Component:219; Jaccard:0.12861
 |
Component:366; Jaccard:0.11949
 |
Component:366; Jaccard:0.099285
 |
Correlation:0.28731
  |
Correlation:0.14234
  |
Correlation:0.24507
  |
Correlation:0.24507
  |
Correlation:0.24507
  |
Correlation:0.28731
  |
Correlation:0.28731
  |
Component:099; Jaccard:0.083496
 |
Component:219; Jaccard:0.097158
 |
Component:305; Jaccard:0.10862
 |
Component:305; Jaccard:0.11681
 |
Component:305; Jaccard:0.11807
 |
Component:305; Jaccard:0.11134
 |
Component:305; Jaccard:0.091971
 |
Correlation:0.37287
  |
Correlation:0.24507
  |
Correlation:0.14234
  |
Correlation:0.14234
  |
Correlation:0.14234
  |
Correlation:0.14234
  |
Correlation:0.14234
  |
Component:397; Jaccard:0.082838
 |
Component:397; Jaccard:0.095199
 |
Component:397; Jaccard:0.10746
 |
Component:397; Jaccard:0.11565
 |
Component:397; Jaccard:0.11531
 |
Component:397; Jaccard:0.10383
 |
Component:397; Jaccard:0.083688
 |
Correlation:0.33738
  |
Correlation:0.33738
  |
Correlation:0.33738
  |
Correlation:0.33738
  |
Correlation:0.33738
  |
Correlation:0.33738
  |
Correlation:0.33738
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































